Abstract
The LOSS OF APOMEIOSIS (LOA) locus is one of two dominant loci known to control apomixis in the eudicot Hieracium praealtum. LOA stimulates the differentiation of somatic aposporous initial cells after the initiation of meiosis in ovules. Aposporous initial cells undergo nuclear proliferation close to sexual megaspores, forming unreduced aposporous embryo sacs, and the sexual program ceases. LOA-linked genetic markers were used to isolate 1.2 Mb of LOA-associated DNAs from H. praealtum. Physical mapping defined the genomic region essential for LOA function between two markers, flanking 400 kb of identified sequence and central unknown sequences. Cytogenetic and sequence analyses revealed that the LOA locus is located on a single chromosome near the tip of the long arm and surrounded by extensive, abundant complex repeat and transposon sequences. Chromosomal features and LOA-linked markers are conserved in aposporous Hieracium caespitosum and Hieracium piloselloides but absent in sexual Hieracium pilosella. Their absence in apomictic Hieracium aurantiacum suggests that meiotic avoidance may have evolved independently in aposporous subgenus Pilosella species. The structure of the hemizygous chromosomal region containing the LOA locus in the three Hieracium subgenus Pilosella species resembles that of the hemizygous apospory-specific genomic regions in monocot Pennisetum squamulatum and Cenchrus ciliaris. Analyses of partial DNA sequences at these loci show no obvious conservation, indicating that they are unlikely to share a common ancestral origin. This suggests convergent evolution of repeat-rich hemizygous chromosomal regions containing apospory loci in these monocot and eudicot species, which may be required for the function and maintenance of the trait.
Asexual seed formation, or apomixis, occurs in both monocot and eudicot plants and has evolved in more than 40 plant genera. Approximately 75% of apomicts exist in three families, the Poaceae, Asteraceae, and Rosaceae. Apomixis bypasses meiosis during female gametophyte formation. This contrasts with sexual female gametophyte development, which requires the meiosis of a megaspore mother cell (MMC) in the ovule (megasporogenesis) followed by nuclear proliferation of typically one of the meiotic products (megagametogenesis). Female gametophytes formed by the apomictic route of meiotic avoidance, or apomeiosis, are also termed unreduced. Seed development in sexual species requires one sperm cell to fuse with the egg cell in the female gametophyte to initiate embryogenesis and another sperm cell to fuse with the central cell nucleus for endosperm formation. Egg cells that differentiate in unreduced gametophytes of apomicts do not require fertilization to develop into an embryo, and endosperm formation may or may not require fertilization. Therefore, seedling progeny derived from apomictic reproduction retain a maternal genotype (Bicknell and Koltunow, 2004; Tucker and Koltunow, 2009). Apomixis is controlled by only a few dominant genetic loci in the currently studied monocot and eudicot species, but genes controlling these events have not been isolated (Ozias-Akins and van Dijk, 2007).
Apomictic plants of different evolutionary history have developed similar mechanisms of meiotic avoidance in order to form unreduced gametophytes. The two common modes observed are termed diplospory and apospory. Monocots in Poaceae such as Tripsacum and eudicot members of the Asteraceae including Taraxacum and Erigeron undergo diplospory, where the MMC or a cell that has aborted meiosis undergoes nuclear proliferation to form an unreduced embryo sac (Tucker and Koltunow, 2009). Apomicts in eudicot Hieracium subgenus Pilosella species (Asteraceae) and monocot Pennisetum (Poaceae) undergo apospory, whereby a somatic (sporophytic) cell termed an aposporous initial (AI) cell initiates embryo sac formation close to sexually programmed cells. Embryo and endosperm formation are both fertilization independent (autonomous) in apomictic Hieracium subgenus Pilosella, but in Pennisetum, fertilization is required for endosperm formation. A single dominant locus termed the Apospory-Specific Genomic Region (ASGR) is required for functional apomixis in Pennisetum. The ASGR has been cytogenetically and genetically analyzed and partially sequenced, and it stretches over 50 Mb on a hemizygous chromosomal region surrounded by transposons and repetitive sequences (Akiyama et al., 2004; Conner et al., 2008).
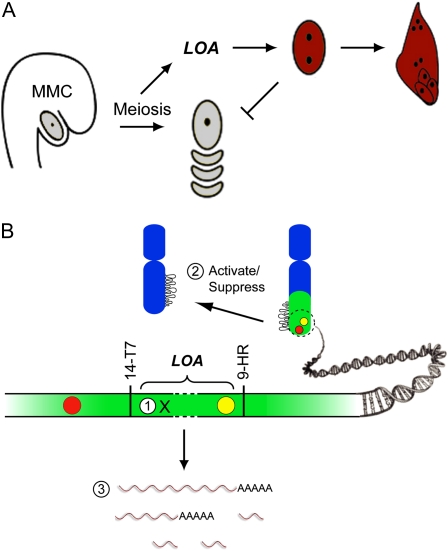
By contrast, two independent dominant loci control apomixis in Hieracium praealtum. Genetic markers linked to these loci have been determined using a deletion mapping approach (Catanach et al., 2006). The roles of these two loci and their interactions with the sexual pathway have been determined from the analyses of apomixis mutants, transgenic ablation of the sexual pathway, and spatial and temporal expression of developmental markers in sexual and apomictic species (Tucker et al., 2003; Koltunow et al., 2011b). The events of meiosis leading to megaspore tetrad formation appear necessary to activate the function of the LOSS OF APOMEIOSIS (LOA) locus, which stimulates the differentiation of somatic AI cells near sexually programmed cells (Koltunow et al., 2011b). As AI cells undergo nuclear proliferation, the sexual pathway terminates and the expanding aposporous embryo sac occupies the position vacated by degenerating sexual cells. Gametophytic function of the independent dominant locus LOSS OF PARTHENOGENESIS (LOP) in aposporous embryo sacs enables fertilization-independent embryo and endosperm development. Deletion of either locus leads to partial reversion to sexual reproduction, and loss of function in both loci results in complete reversion to sexual development. This indicates that sexual reproduction is the default reproductive mode upon which apomixis is superimposed in H. praealtum (Koltunow et al., 2011b). Thus, LOA and LOP loci are unlikely to encode factors essential for sexual reproduction. Both sexual and aposporous gametophytes show similar expression patterns of reproductive marker genes during the mitotic events of gametogenesis and early seed initiation (Tucker et al., 2003). Thus, LOA and LOP may function to heterochronically recruit the sexual machinery to enable apomixis, which resembles a truncated sexual pathway (Tucker et al., 2003; Koltunow et al., 2011b). The chromosomal locations of LOA and LOP have not been determined, and the genomic regions associated with the LOA and LOP loci have not been isolated.
Here, we identified a partial DNA contig across four markers linked to the central region of the LOA locus. Physical mapping and plant phenotyping were used to establish the genomic region critical for LOA function, which lies between two new genomic markers in H. praealtum. The chromosomal location of the LOA locus was determined by fluorescent in situ hybridization (FISH). LOA is located on a single chromosome near the distal tip of the long arm and is surrounded by repetitive sequences in H. praealtum and in two other Hieracium subgenus Pilosella species. Structural features of the hemizygous chromosomal region containing the LOA locus in these eudicot Hieracium species resemble those found in two other aposporous monocot species, suggesting that chromosomal structure might be functionally relevant for the induction and/or maintenance of apospory in these plants.
RESULTS
Identification of LOA-Associated Genomic Sequences and Specific Markers
Four sequence-characterized amplified region (SCAR) markers, LOA 300, LOA 267, LOA 275, and LOA 219, were previously found to be located in the central region of the LOA locus in H. praealtum R35 (Catanach et al., 2006; Koltunow et al., 2011b). These SCAR markers are also present in apomictic Hieracium caespitosum (C36) and Hieracium piloselloides (D36), but they are absent in sexual Hieracium pilosella (P36) and also in two other apomictic Hieracium aurantiacum accessions (A35 and A36; Table I; Koltunow et al., 2011b). All of the characterized H. praealtum deletion mutants that have lost LOA function lack these four SCAR markers except mutant 134, which is thought to contain a small deletion or translocation as a result of γ-irradiation (Supplemental Fig. S1; Koltunow et al., 2011b).
Table I.
Presence (+) or absence (−) of LOA-linked SCAR markers in Hieracium subgenus Pilosella accessions
| Marker |
Pilosella 2a |
Pilosella 1 |
||||||
| P36(sex) | P36(CR)(sex) | C36b(apo) | D36b(apo) | D18c(apo) | R35b(apo) | A35d(apo) | A36d(apo) | |
| LOA 219 | − | − | + | + | + | + | − | − |
| 21-T7e | − | − | + | + | + | + | − | − |
| 9-HRe | − | − | + | + | + | + | − | − |
| LOA 275e | − | − | + | + | + | + | − | − |
| 9-T7 | − | − | + | + | + | + | − | − |
| 12-T7 | − | − | − | + | + | + | − | − |
| 13-HR | − | − | + | + | + | + | − | − |
| LOA 267e | − | − | + | + | + | + | − | − |
| 14-T7 | − | − | + | + | + | + | − | − |
| 4-T7 | − | − | + | + | + | + | − | − |
| 5-HR | − | − | + | + | + | + | − | − |
| 18-T7 | − | − | + | + | + | + | − | − |
| LOA 300e | − | − | + | + | + | + | − | − |
| 5-T7 | − | − | − | + | + | + | − | − |
Subgenus Pilosella species belong to two divergent chloroplast haplotype networks, Pilosella 1 and Pilosella 2. The location of each accession in a particular network has been described previously by Koltunow et al. (2011b).
Apomicts with conserved modes of aposporous embryo sac formation. Modes for each species are explained in the text.
Polyhaploid plant with 18 chromosomes experimentally derived from a segregating D36 population (Bicknell et al., 2003).
Apomicts with conserved mode of aposporous embryo sac formation.
Sequences of amplified SCAR markers from C36, D36, and D18 have been determined and confirmed to be more than 98% identical to that of R35.
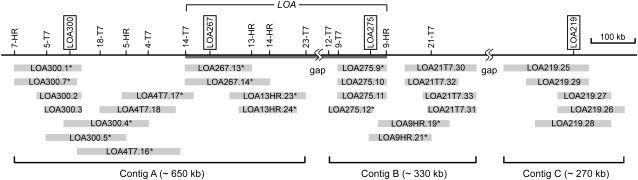
These four SCAR markers were used to screen an H. praealtum (R35) bacterial artificial chromosome (BAC) library in order to isolate genomic sequences associated with the LOA locus. Individual BACs containing the SCAR markers were extended by chromosome walking with the aim of obtaining the entire genomic sequence linking the four markers in the LOA locus. In this study, 28 BACs covering 1.2 Mb of sequence were identified, and they were assembled into three independent DNA contigs, A, B, and C, that provide partial coverage of sequences spanning the four SCAR markers (Fig. 1). LOA 300 and LOA 267 are physically linked in the largest contig, A, which comprises approximately 650 kb, while contigs B and C cover approximately 330 and 270 kb of genomic sequence, respectively (Fig. 1).
Figure 1.
A physical map of the region containing the LOA locus in H. praealtum. The SCAR markers linked to the LOA locus are indicated in order along the solid line representing the chromosome. Boxed markers indicate those initially used for the identification of BACs from the H. praealtum BAC library. Unboxed markers are SCARs developed in this study. The BAC clones identified and assembled into three contigs are shown in gray boxes. The term “gap” indicates sequences yet to be identified to complete the BAC contig. The genomic region essential for LOA function between 14-T7 and 9-HR required for AI cell formation, aposporous embryo sac formation, and sexual suppression is indicated. BAC clones pooled from contig A and contig B for 454 pyrosequencing are indicated with asterisks.
Thirteen new SCAR markers linked to the LOA locus were developed from BAC end sequences, bringing the total number of LOA-linked SCAR markers to 17 (Fig. 1). These LOA SCAR markers are based largely on repetitive sequences (Supplemental Fig. S2A). They detect sequences in H. praealtum (R35) and in the deletion mutant 134 but are absent in the remaining deletion mutants defective in LOA function (Koltunow et al., 2011b). Therefore, the majority of characterized apospory mutants contain physically large deletions that lack the three identified contig sequences (Supplemental Fig. S1).
The Three Identified DNA Contigs Associated with the LOA Locus Are Present on a Single, Elongated Chromosome in H. praealtum
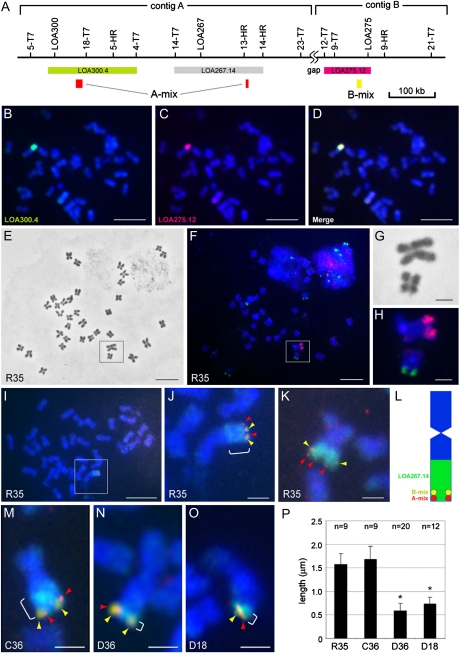
The physical linkage of the three isolated DNA contigs in the H. praealtum genome was investigated by BAC FISH analyses on metaphase chromosomes. Hieracium subgenus Pilosella species have a base chromosome number of nine, and apomicts are often tetraploid with 36 chromosomes. H. praealtum (R35) is an aneuploid apomict (3x + 8) containing 35 chromosomes, with an individual chromosome in the karyotype showing significant elongation (Koltunow et al., 2011b). Two BACs from contig A (LOA300.4 and LOA267.14) and one from contig B (LOA275.12; Figs. 1 and 2A) were fluorescently labeled and hybridized to metaphase spreads in different combinations in dual-color FISH analyses. These three probes from contigs A and B colocalized with intense hybridization to the same region of the single elongated chromosome (Fig. 2, B–D). Furthermore, these probes painted nearly half of the long arm of the elongated chromosome, even though the DNA inserts in the three BACs ranged from 110 to 185 kb (Fig. 2, B–H). This suggested that sequences present within these BACs are repeated over a very large region on the long arm of the chromosome. We estimated that the size of the region hybridized by the LOA267.14 BAC probe (185 kb) was approximately 228 ± 11 Mb (seven chromosomes), assuming a genome size of 6,800 Mb in H. praealtum R35 (Suda et al., 2007; http://genome.arizona.edu/orders/).
Figure 2.
Physical location of the LOA locus on Hieracium chromosomes. A, BAC clones and specific DNA probes (A-mix and B-mix) used for FISH analysis and their locations in contigs A and B. B, Hybridization of the LOA300.4 BAC probe (green) to a single chromosome in H. praealtum R35. C, Hybridization of the LOA275.12 BAC probe (red) to the same metaphase chromosome spread as in B. D, Merge of images in B and C. E, Phase-contrast image of metaphase chromosomes of H. praealtum R35. The elongated chromosome and an adjacent chromosome magnified in subsequent images is boxed. F, Chromosome spread in E, showing hybridization of the LOA267.14 probe (red) and 18S-5.8S-26S rDNA probe (green). G, Magnified phase-contrast image of the boxed region in E. H, Magnified fluorescent image of the chromosomes in F. I, Hybridization of three probes to the metaphase chromosomes of H. praealtum R35: LOA267.14 BAC (green) and specific DNA probes A-mix (red) and B-mix (yellow). J, Magnification of the long chromosome in I showing the location of contig A (red arrowheads) and contig B (yellow arrowheads) and surrounding repeats (green and bracket). K, Another chromosome showing the subtelomeric location of contigs A and B in R35. L, Schematic representation of the positions of contigs A and B on the elongated chromosome in R35. M to O, FISH analyses using LOA267.14 BAC (green and brackets) and A-mix (red arrowheads) and B-mix (yellow arrowheads) specific DNA probes in three other apomictic Hieracium accessions: H. caespitosum C36 (M), H. piloselloides D36 (N), and H. piloselloides D18 (O). P, Size of the region hybridized by the LOA267.14 BAC in H. praealtum R35, H. caespitosum C36, and H. piloselloides D36 and D18. The repetitive region hybridized by the LOA267.14 BAC is significantly smaller in D36 and D18 (asterisk; t test; P < 0.01). Bars = 10 μm (B–F and I) and 2 μm (G, H, J, K, and M–O).
Transposons and Repetitive Sequences Are Abundant in LOA Locus-Associated DNA Sequences
In order to examine the nature of the sequences present in contigs A and B that might be responsible for the extensive BAC probe hybridization on the long arm of the elongated chromosome, 454 pyrosequencing was used to determine the sequence of a pool of 10 BACs from contig A and another pool of four BACs from contig B (Fig. 1). The Hieracium genome has not been sequenced, and the absence of a reference genome coupled with the repetitive sequence nature of the BACs hampered the assembly of DNA sequences. A total of 379 nonredundant contigs were assembled from the contig A BAC pool and 241 nonredundant contigs from the contig B BAC pool. This resulted in a total of 620 nonredundant contigs and 760,082 bp of sequence coverage from the LOA locus (Table II).
Table II.
Repeat and transposon sequences in the Hieracium LOA locus and in Pennisetum and Cenchrus ASGR
| Parameter | LOA Hieracium | ASGR Pennisetum | ASGR Cenchrus |
| Repeat size (Mb) | 228a | 50b | >11c |
| BACs sequencedd | 14 | 20 | 13 |
| Nonredundant contigs | 620 | 760 | 581 |
| Sequence (bp) | 760,082 | 542,361 | 505,493 |
| Average length (bp) | 1,226 | 714 | 870 |
| Repeate | |||
| Simple repeat (%) | 1.8 | 0.4 | 0.4 |
| Low complex (%) | 1.3 | 0.5 | 0.7 |
| Complex repeat (%) | 31.1 | 16.8 | 28.0 |
| TransposonPSIf | |||
| Class I (retrotransposons) | |||
| Ty1-copia | 29 | 64 | 38 |
| Ty3-gypsy | 78 | 16 | 23 |
| LINE | 7 | 9 | 12 |
| Class II (DNA transposons) | |||
| hAT | 1 | 1 | 2 |
| ISa | 3 | 0 | 0 |
| MuDR_A_B | 1 | 2 | 1 |
| helitronORF | 1 | 3 | 5 |
| Total number of TE | 120 | 95 | 81 |
| Total TE (%)g | 14.9 | 9.7 | 8.8 |
Repeat region size on the chromosome containing the LOA locus measured in H. praealtum R35.
ASGR features two large blocks of repeat regions flanking a low-copy genomic region in P. squamulatum (Akiyama et al., 2004).
Distance between two nonrecombining BACs in C. ciliaris (Akiyama et al., 2005).
Pennisetum and Cenchrus ASGR sequences from Conner et al. (2008) and Hieracium LOA sequences from this study.
Simple repeat and low-complex DNA sequences were predicted by RepeatMasker, and complex repeat sequences were identified by DNA clustering (see “Materials and Methods”). Numbers indicate percentage of the masked repeat sequences against total sequenced length.
Number of transposon sequences predicted in the contig sequences by TransposonPSI.
Percentage of total sequence length predicted as transposable elements (TE) against the total sequence length of contigs.
Analyses using comparative genomics of open reading frames (ORFs) and ab initio gene prediction programs indicated that the sequenced region contained few ORFs and a large number of partially conserved ORFs, particularly transposon-related proteins. The sequenced genomic region of the LOA locus has a low GC content (38%), is AT rich, and contains simple repeat and low-complexity DNAs. The transposon prediction programs RepeatMasker, Censor, and TransposonPSI identified various transposon-related sequences, revealing that class I retrotransposons Ty1-copia and Ty3-gypsy (Kumar and Bennetzen, 1999) were particularly abundant in the LOA locus (Table II). In total, 120 transposon sequences were annotated, covering 14.9% of the 760-kb sequenced region. DNA clustering analysis identified 899 additional complex repeat fragments for which homologous sequences are present at least twice in the LOA locus that are not simple or low-complex DNAs or transposons. These complex repeat sequences cover 31.1% of the sequenced region (Table II). There does not appear to be a single sequence continuously repeated in the analyzed BAC pool sequences. The complex repeats, transposons, and simple/low-complex sequences constitute 49.1% of the sequence coverage at the LOA locus, and we conclude that a combination of these sequences accounts for the observed extensive BAC probe hybridization on the chromosome containing the LOA locus.
LOA Contigs A and B Are Located at the Distal Tip of the Long Chromosome in H. praealtum R35
To establish the location of contigs A and B within the repetitive region and concurrently gain an insight into the size of the intervening sequences (gap) between contigs A and B, we developed specific FISH probes for each contig. Potentially unique LOA-linked sequences derived from the 454 sequence contigs were selected and assessed for their suitability as FISH probes by genomic DNA-blot analysis (Supplemental Fig. S2B). A mixture of these probes derived from two regions in contig A (A-mix) and a single region in contig B (B-mix) was used for FISH (Fig. 2A). Multicolor FISH hybridization revealed that A-mix and B-mix probes hybridized as distinct but closely localized spots at the distal end of the long chromosome within the repetitive region labeled by the LOA267.14 BAC probe (Fig. 2, I–L; Supplemental Fig. S2C). Analysis of multiple chromosome samples indicated that the genomic region detected by the contig A-mix probes was located toward the distal tip of the chromosome, while the B-mix probes detected adjacent genomic sequences oriented toward the centromere (Fig. 2L).
Collectively, these results indicate that the identified contigs A and B associated with the LOA locus are closely located at the distal tip of a single long chromosome in H. praealtum. They are surrounded by a large array of complex repeats and transposons. The localization of the LOA locus linked to a single chromosome is also consistent with the predicted hemizygous genetic nature of the LOA locus (Catanach et al., 2006).
Conservation of SCAR Markers and Chromosomal Features Associated with the LOA Locus in Three Apomictic Hieracium Subgenus Pilosella Species
Next, we investigated whether the LOA locus-associated repetitive sequences, SCAR markers, and the contig A- and B-specific sequences in H. praealtum were present in other characterized sexual and aposporous Hieracium subgenus Pilosella species. These included two tetraploid accessions of sexual H. pilosella [P36 and P36(CR)] and three apomictic species, tetraploid H. caespitosum (C36) and H. piloselloides (D36), a diploid form of H. piloselloides (D18), and two accessions of H. aurantiacum, one tetraploid (A36) and the other with 35 chromosomes (A35). These plants fall into the two known Hieracium subgenus Pilosella chloroplast haplotype networks, Pilosella 1 and Pilosella 2, shown in Table I, based on their chloroplast trnT-trnL sequences (Koltunow et al., 2011b). Analyses using the 14 SCAR markers summarized in Table I revealed remarkable marker conservation in the two apomictic species H. caespitosum (C36) and H. piloselloides (D36 and D18). Sequencing of the PCR products obtained from five of the examined markers indicated in Table I confirmed the sequences to be greater than 98% identical to those in R35, supporting the conservation of markers. These SCAR markers were absent in two accessions of sexual H. pilosella [P36 and P36(CR)] and in two accessions of apomictic H. aurantiacum (A35 and A36; Table I).
FISH analyses using fluorescently labeled BAC LOA267.14 and the contig A- and B-specific probes (A- and B-mix; Fig. 2A) did not show significant hybridization to P36, A35, and A36 chromosomes (Supplemental Fig. S3; Table III). Therefore, the apomictic accessions of H. aurantiacum (A35 and A36) and sexual H. pilosella (P36) do not contain the same large block of repetitive sequences associated with the LOA locus in their genomes. Furthermore, they do not contain sequences that can be reproducibly detected by the A and B contig-specific probes in R35 (Supplemental Fig. S3).
Table III.
Summary of characteristics of the LOA locus and ribosomal DNA in sexual and apomictic Hieracium subgenus Pilosella
| Accession | Ploidy | Reprod Typea | Long Chromb | LOA SCARc | LOA Repeatd | LOA A-Mixe | LOA B-Mixe | 18S rDNAf | 5S rDNAg |
| H. praealtum R35 | 3x + 8 = 35 | Apo | + | + | + | + | + | 6 | 4 |
| H. caespitosum C36 | 4x = 36 | Apo | + | + | + | + | + | 8 | 4 |
| H. piloselloides D36 | 4x = 36 | Apo | − | + | + | + | + | 7 | 4 |
| H. piloselloides D18 | 2x = 18 | Apo | − | + | + | + | + | 3 | 2 |
| H. aurantiacum A35 | 3x + 8 = 35 | Apo | − | − | − | − | − | 8 | 4 |
| H. aurantiacum A36 | 4x = 36 | Apo | − | − | − | − | − | 8 | 4 |
| H. pilosella P36 | 4x = 36 | Sex | − | − | − | − | − | 10 | 4 |
Reproductive type, apomictic (Apo) or sexual (Sex).
Presence (+) or absence (−) of the long chromosome.
For details of the conservation of LOA SCAR markers, see Table I.
Presence of LOA-linked repeat sequences determined by LOA267.14 BAC FISH hybridization.
Specific hybridization of A and B-mix probes.
Number of 18S-5.8S-26S rDNA FISH signals in the karyotype.
Number of 5S rDNA FISH signals in the karyotype.
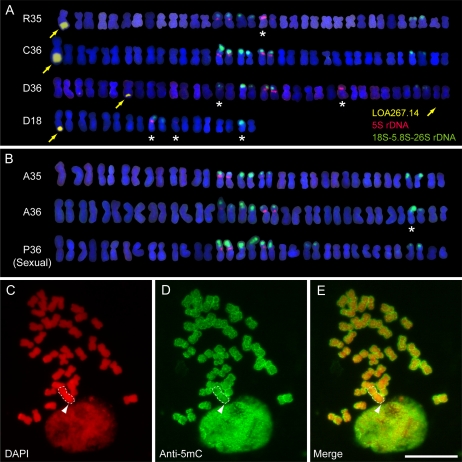
By contrast, the LOA-linked repetitive sequences were also observed on the long arm of a single chromosome in H. caespitosum (C36) and in the two H. piloselloides accessions (D36 and D18) when BAC LOA267.14 was used as a FISH probe (Fig. 3A). The LOA-associated repetitive sequences were found on an elongated long chromosome in C36 like that observed in R35, but the chromosomes containing repetitive sequences in D36 and D18 were not significantly elongated relative to the others (Fig. 3A). Furthermore, the contig A- and B-specific FISH probes hybridized at the distal end of the chromosomes containing the repetitive sequences in both H. caespitosum (C36) and H. piloselloides (D36 and D18; Fig. 2, M–O; Supplemental Fig. S4). It is also interesting that the size of the LOA-associated repeat region on the chromosomes of these plants varies. The repetitive region in D18 and D36, painted by the LOA267.14 BAC, is approximately one-third the size of that in R35 (Fig. 2, J, K, and M–P).
Figure 3.
FISH karyotypes of different Hieracium accessions hybridized with probes to detect LOA-associated repeat sequences and rDNA loci, and DNA methylation status in H. caespitosum C36. A, H. praealtum R35, H. caespitosum C36, and H. piloselloides D36 and D18 were hybridized with LOA267.14 BAC (yellow), 5S rDNA (red), and 18S-5.8S-26S rDNA (green) probes. Yellow arrows indicate hybridization of the LOA267.14 BAC probe. White asterisks indicate chromosomes containing rDNA that do not have a corresponding hybridization pattern match to another chromosome. B, H. aurantiacum A35 and A36 and sexual H. pilosella P36 chromosomes were hybridized with rDNA probes as in A. The LOA267.14 probe does not hybridize to chromosomes in these species (Supplemental Fig. S3). It was not used as a probe in this image to reduce background noise. C to E, Analysis of the methylation status of H. caespitosum chromosomes using anti-5-methylcytosine (Anti-5mC) antibody. Shown are 4′,6-diamidino-2-phenylindole (DAPI) staining in red (C), DNA methylation in green (D), and the merge of DAPI and DNA methylation images (E). The long chromosome is indicated by the white dotted lines. The arrowheads indicate the position of the LOA locus. Bar = 10 μm.
Collectively, these data show that features of the LOA locus, including the presence of LOA contig A and B-specific sequences near the tip of the long arm of a single chromosome surrounded by repetitive sequences that vary in length, are conserved in three apomictic Hieracium subgenus Pilosella species.
Ribosomal DNA Loci Are Hemizygous and Heterozygous in Some Apomictic Hieracium Species
The karyotypes of the different Hieracium accessions utilized in this study were determined by both chromosome constitutions and FISH hybridization patterns of 5S and 18S-5.8S-26S ribosomal DNA (rDNA) probes (Fig. 3, A and B). In the sexual plant H. pilosella (P36), all of the FISH rDNA hybridization signals were paired and similarly sized (Fig. 3B). This was not always the case for the apomictic plants (Fig. 3, A and B, white asterisks). H. praealtum (R35) and H. aurantiacum (A36) showed significant differences in the size of rDNA loci, indicating a heterozygous rDNA constitution (Fig. 3, A and B; Table III). Chromosomes of H. piloselloides D36 and D18 have rDNA regions that do not have corresponding partnering chromosomes with the same rDNA pattern, indicating hemizygous rDNA constitutions (Fig. 3A, white asterisks). Therefore, hemizygous chromosome constitutions extend beyond that observed for the LOA locus. The nonreductional mitotic nature of apomictic reproduction is likely to maintain both aneuploidy and hemizygotic chromosome constitutions in subgenus Pilosella.
The Chromosome Containing LOA and Surrounding Repeats Is Not Enriched for Heterochromatin Marks
Chromosomes containing extensive regions of repetitive DNA and retrotransposon-like sequences that were observed around the LOA locus (Fig. 2; Table II) are often highly heterochromatic and exhibit high levels of DNA methylation (Chan, 2008). Such chromosomes can also show reduced levels of euchromatic histone H3 methylation marks or be enriched in heterochromatic histone marks (Houben et al., 2003; Jasencakova et al., 2003). In order to examine if there were correlations between the transposon and repetitive regions around the LOA locus and chromatin structure, DNA and histone methylation status were investigated at a global chromosome level using antibody stains in apomictic H. caespitosum C36. Like R35, the C36 accession contains the LOA locus on a long chromosome, making it easier to track in chromosome squash preparations (Table III). Several antibodies specific for histone H3 methylation associated with euchromatin and transcriptional activation (H3K4me2 and H3K4me3) or heterochromatin formation and gene silencing (H3K9me2, H3K27me2, and H3K27me3) were used. No significant differences were observed in global histone H3 methylation patterning on the chromosome containing LOA-associated repeats relative to the other chromosomes (Supplemental Fig. S5). We also investigated DNA methylation using anti-5-methylcytosine antibody. Figure 3, C to E, shows that the distal half of the long arm of the long chromosome, where the LOA locus is located, did not exhibit increased DNA methylation relative to the other chromosomes. Taken together, the highly repetitive region around the LOA locus is not associated with a significantly higher degree of DNA methylation or unique posttranslational histone H3 modification relative to other chromosomes. Thus, the long arm of this chromosome predominantly containing repetitive sequences and transposons is unlikely to be completely transcriptionally silent. This analysis does not rule out the possibility that more localized DNA and histone modifications are playing a role in regulating the expression of genes at the LOA locus.
Delineation of the Genomic Region Essential for LOA Function
Next, we examined which parts of the genomic region defined by the three BAC contigs (Fig. 1) were critical for LOA function. As the genomic region covered by the identified BAC contigs is missing in all the loaLOP deletion mutants except mutant 134, which contains all 17 of the LOA-specific SCAR markers identified (Supplemental Fig. S1), we used a physical mapping approach to define the region critical for LOA function. H. praealtum R35 is an aneuploid, and the success of this approach is contingent upon recombination occurring at the LOA locus. Catanach et al. (2006) had previously reported the segregation of the LOA 300 marker from the apomixis initiation phenotype in two out of 37 plants within a small mapping population derived from a cross between sexual H. pilosella (P36) and apomictic H. praealtum (R35) as the pollen donor. We generated a population of 833 F1 progeny from this cross to investigate recombination at the LOA locus and the cosegregation of the markers with the capacity to form AI cells and aposporous embryo sacs in ovules.
The LOA-linked SCAR markers are not present in sexual H. pilosella P36 (Table I), so F1 plants carrying these markers inherit them from the apomictic pollen parent. We screened the 833 F1 progeny plants with the four SCAR markers LOA 300, LOA 267, LOA 275, and LOA 219, which span the LOA locus sequences (Fig. 1). A total of 125 plants containing one or more of these markers were identified, indicating low transmission efficiency (15%) of LOA-linked sequences. This supports the previous observations of segregation distortion for transmission of the LOA locus (Catanach et al., 2006). The 125 F1 progeny containing LOA-linked SCAR markers were further investigated for the presence of nine additional SCAR markers at the LOA locus, and they were divided into 11 classes based on the combinations of markers they contained (Table IV). Sixty-seven plants representative of these classes were examined cytologically for the presence of AI cells and aposporous embryo sacs. Linkage analysis of the SCAR markers in the F1 progeny confirmed the order of markers as shown in Figure 1, which was supported by an odds ratio of 109 over any other order of markers.
Table IV.
Analysis of the segregation of LOA-linked markers and AI cell formation in F1 progeny derived from a cross between sexual P36 (female) and apomict R35 (male)
| Classa | Contig A |
Contig B |
Contig C |
No. in F1c | No. Testedd | AI+e | AI−f | %g | ||||||||||
| 5-T7b | LOA 300 | 18-T7 | 5-HR | 4-T7 | 14-T7 | LOA 267 | 13-HR | 12-T7 | LOA 275 | 9-HR | 21-T7 | LOA 219 | ||||||
| R35 | + | + | + | + | + | + | + | + | + | + | + | + | + | NAh | 2 | 2 | 0 | 100 |
| 1 | + | + | + | + | + | + | + | + | + | + | + | + | + | 50 | 17 | 15 | 2i | 88 |
| 2 | + | + | + | + | + | + | + | + | + | + | + | + | − | 3 | 1 | 1 | 0 | 100 |
| 3 | + | + | + | + | + | + | − | + | + | + | + | + | + | 6 | 3 | 2 | 1 | 67 |
| 4 | + | + | + | − | + | + | + | + | − | + | + | + | + | 4 | 2 | 2 | 0 | 100 |
| 5 | − | − | − | − | − | − | + | + | + | + | + | + | − | 4 | 3 | 3 | 0 | 100 |
| 6 | + | − | + | − | − | − | − | + | − | − | − | − | − | 12 | 7 | 0 | 7 | 0 |
| 7 | − | − | − | − | − | − | − | − | − | + | − | + | + | 7 | 1 | 0 | 1 | 0 |
| 8 | − | − | − | − | − | − | − | − | − | − | − | + | + | 30 | 14 | 1i | 13 | 7 |
| 9 | − | − | − | − | − | − | − | − | − | − | − | − | + | 4 | 1 | 0 | 1 | 0 |
| 10 | − | − | − | − | − | − | − | − | − | − | − | + | − | 5 | 3 | 0 | 3 | 0 |
| 11 | − | − | − | − | − | − | − | − | − | − | − | − | − | 683 | 15 | 1i | 14 | 7 |
| P36 | − | − | − | − | − | − | − | − | − | − | − | − | − | NAh | 2 | 0 | 2 | 0 |
Sexual P36 (female) and apomictic R35 were crossed to generate F1 progeny that were divided into 11 classes according to the presence of LOA markers. Note that class 2 plants have all the markers except LOA 219. Class 3 plants contain LOA 219 and might lack any one of the other markers. Class 4 plants lack any two markers within the locus. Class 6 plants contain one to three markers in a random combination.
The LOA-linked SCAR markers used for recombination mapping. The presence (+) or absence (−) of SCAR markers in individual F1 plants and the parent plants is indicated.
Number of F1 plants in each class. Class 11 includes all the plants that did not have any of the four LOA SCAR markers used in the initial mapping.
Number of F1 plants whose AI cell formation was cytologically examined.
Number of F1 plants forming AI cells in the ovule.
Number of F1 plants that did not form AI cells in the ovule.
Percentage of plants with AI cell formation relative to the total number of plants examined by microscopy.
NA, not available.
These plants failed to show cosegregation of the LOA-linked markers with AI cell formation. This may relate to the low density of markers at the locus, particularly the inability to identify the presence or absence of DNA in the current gaps in the contigs that may be required for AI cell formation.
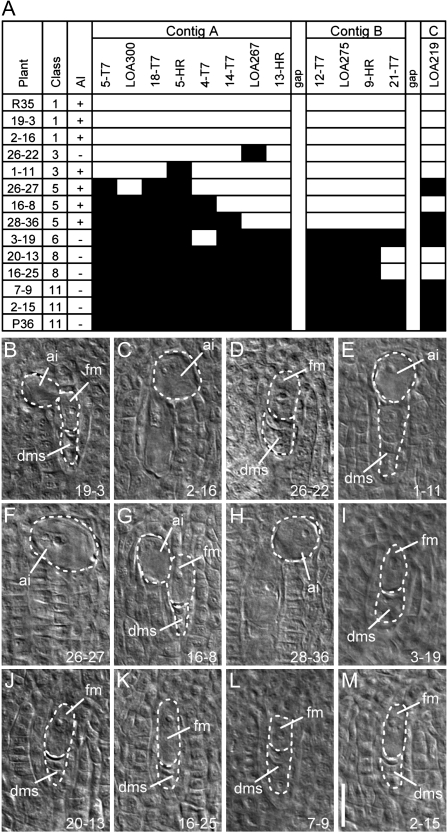
F1 plants containing all or most of the markers (classes 1–4) were able to form AI cells (Fig. 4, A–C and E). Plants containing only a few markers (class 6), or no markers (class 11) were unable to form AI cells (Fig. 4, I, L, and M), confirming a cosegregation of LOA-linked SCAR markers with the phenotype of AI cell formation. Overall consideration of the results in Table IV and Figure 4 suggested that the genomic region between marker 14-T7 on contig A and marker 21-T7 on contig B appears to be essential for LOA function (Fig. 4, A and F–H). One F1 progeny plant, 26-22, has all of the markers across the locus except for LOA 267 and is unable to form AI (Fig. 4, A and D), suggesting a close association of the LOA 267 marker with AI cell formation. However, we cannot exclude the possibility that recombination and loss of critical LOA sequences has also occurred in the genomic region that is yet to be identified.
Figure 4.
Analysis of the segregation of LOA-linked markers and AI cell formation in F1 progeny derived from a cross between sexual P36 (female) and apomict R35 (male). A, Presence (white) or absence (black) of SCAR markers in individual F1 plants and the parent plants used in the cross. Plants fall into classes containing markers that cross-reference with those shown in Table IV. The presence (+) or absence (−) of AI cells in the ovule is indicated. B to M, Microscopy images of cleared ovules of the plants in A. The plant identifier number is shown at the bottom right of each panel. ai, AI cell; dms, degenerating megaspores; fm, functional meiotic megaspore. Bar = 20 μm.
We considered that the aneuploid nature of R35 may have contributed in part to the low transmission of the LOA locus and associated markers. Therefore, we examined 671 F1 progeny derived from a cross between sexual H. pilosella P36 and apomictic H. caespitosum C36, where both parents are tetraploid. We also found low transmission efficiency (40%) of the LOA-linked markers in this F1 progeny. Surprisingly, the transmission of markers between 14-T7 and 21-T7 linked with the AI cell formation phenotype did not occur in this cross as frequently as in the previous P36 × R35 cross (Supplemental Table S1). Consistent with this, the progeny plants rarely developed AI cells and aposporous embryo sacs. Analyses of the progeny of the P36 × C36 cross support conclusions from the P36 × R35 cross that the genomic region between markers 9-HR and LOA 219 is not essential for the initiation of apomixis (classes 1–7; Supplemental Table S1). Given the poor transmission of markers in the P36 × C36 cross, it is not possible to confirm if the order of SCAR markers in H. caespitosum is the same as that in H. praealtum R35.
In summary, analyses of the mapping populations indicate that the genomic region between marker 14-T7 on contig A and marker 9-HR on contig B, containing 400 kb of isolated genomic sequences flanking a region yet to be identified, appears to be sufficient for the initiation of apomixis in H. praealtum (Fig. 1).
Comparison of Partial Genomic Sequences from Eudicot LOA and Monocot ASGR Loci
The ASGR in both aposporous Pennisetum squamulatum and Cenchrus ciliaris is located on a single chromosome that also contains transposons and repeated sequences (Akiyama et al., 2004, 2005). Genomic sequences associated with the Hieracium LOA locus and ASGR of both Pennisetum and Cenchrus were compared using BLASTN in order to investigate sequence conservation, and no significant similarity was found except for some simple repeat sequences. Next, we compared the LOA and ASGR-associated sequences using TBLASTX, which enabled a comparison of six-frame translated sequences, and matches to the putative genes currently identified at the ASGR were not found (Supplemental Table S2; Conner et al., 2008).
Transposons of the Ty3-gypsy and Ty1-copia type were found at both LOA and ASGR loci, with Ty3-gypsy-like retrotransposons being most abundant at the LOA locus and Ty1-copia-like sequences being most abundant in the ASGR (Table II). Ty1-copia-like retrotransposons are ubiquitous in plants. However, they are considerably diverse at the DNA sequence level, and multiple well-supported phylogenetic lineages have been identified in plants (Voytas et al., 1992; Kumar and Bennetzen, 1999). The retrotransposon sequences associated with the ASGR and the LOA locus did not show significant DNA conservation, indicating that they had originated from different ancestral sequences. Clustering analyses also identified 421 and 404 complex repeat fragments repeated at least twice in the Pennisetum and in the Cenchrus ASGR-associated contigs, covering 16.8% and 28% of the sequenced regions, respectively (Table II). There was no sequence conservation in the complex repeats found at the ASGR and the LOA locus.
Only partial DNA sequences have been obtained from the Hieracium LOA locus and the Pennisetum and Cenchrus ASGR, and as the current sequenced regions do not contain many genes, we are unable to make significant conclusions about candidate apomixis genes. However, we can conclude that the repetitive and transposon sequences present on the hemizygous chromosomal region containing these loci appear to have evolved independently. Therefore, this provides an example of convergent evolution of chromosomal structures on hemizygous chromosomal regions containing apospory loci in monocot and eudicot species.
DISCUSSION
The Repeat-Rich Chromosomal Structure Containing the LOA Locus Is Not Conserved in All Aposporous Hieracium Subgenus Pilosella Species
Hieracium subgenus Pilosella species exhibit tremendous variations in morphological form, and they are found in two divergent chloroplast haplotype network groups: Pilosella 1 and Pilosella 2 (Fehrer et al., 2007a, 2007b). Sexual and facultative apomictic species are self-incompatible and can interbreed, forming allopolyploid hybrids that enable the evolution of both sexual and apomictic species in this subgenus (Fehrer et al., 2007b; Koltunow et al., 2011a, 2011b). Here, we have identified genomic sequences and a suite of new SCAR markers associated with the LOA locus responsible for the initiation of apomixis in H. praealtum. Physical mapping using these markers has led to the delineation of sequences that are critical for LOA function being confined between two new markers with further intervening sequences yet to be identified. Cytogenetic analyses with BAC and LOA locus-specific probes indicate that the LOA locus is located near the tip of the long arm of a single chromosome in H. praealtum R35, surrounded by complex repetitive sequences. These features are not present in two accessions of sexual H. pilosella [P36 and P36(CR)].
Accessions from two other apomictic species, H. caespitosum (C36) and H. piloselloides (D36), share the LOA-specific SCAR markers and cytogenetic chromosomal repeat features found in H. praealtum (R35), suggesting conservation of the LOA locus in these three apomictic species. All three species have a similar growth habit, being tall with raceme inflorescences and predominantly yellow-colored florets. The events of meiosis leading to megaspore tetrad development are required for AI cell formation in H. piloselloides (D36). In accessions of the other two species containing the LOA locus and linked repeat sequences, AI cell formation is also most frequent during megaspore tetrad development, supporting the requirement for sexual reproduction to activate LOA (Fig. 5A; Koltunow et al., 2011b). These species are colocated in the same chloroplast haplotype network group, Pilosella 2. They also share similar modes of aposporous embryo sac formation with respect to the timing of AI cell nuclear division at megaspore degeneration and the frequent expansion of multiple aposporous embryo sac structures in the ovule (Koltunow et al., 1998, 2011a, 2011b). The observation that the size of the LOA-associated repeat varies in these species indicates that a constant repeat size is not essential for the progression of apospory.
Figure 5.
Functions of the LOA locus and models for LOA action to enable AI cell formation. A, The events of meiosis leading to megaspore tetrad formation in ovules are required to activate the function of the LOA locus. LOA stimulates the formation of somatic AI cells, and their nuclei undergo mitosis to form unreduced embryo sacs. During this process, the sexual reproductive pathway is suppressed. Factors that might activate LOA are discussed in the text. B, The LOA locus resides in a subtelomeric position on the long arm of a single hemizygous chromosomal region in three subgenus Pilosella species. In H. praealtum, it is flanked by the indicated markers. Three possible scenarios for LOA induction of AI cell formation are discussed in the text.
The presence of a long chromosome is not a perfect correlative marker for the presence of the LOA locus, because LOA locus-specific sequences were found on an elongated chromosome in apomictic H. praealtum and H. caespitosum but not in H. piloselloides, where the locus is not located on an elongated chromosome. It has been reported previously that some, but not all, aposporous subgenus Hieracium species have a long chromosome, and elongated chromosomes are also present in sexual species (Krahulcová and Krahulec, 1999; Krahulec et al., 2008; A. Krahulcová, personal communication).
LOA-linked repetitive sequences and SCAR markers were absent in two apomictic accessions of the orange-flowered H. aurantiacum (A35 and A36), which do not possess an elongated chromosome. In contrast to the other apomictic species studied here, both H. aurantiacum accessions are located in the Pilosella 1 chloroplast haplotype network (Koltunow et al., 2011b). These H. aurantiacum accessions differ slightly in their mode of aposporous embryo sac formation compared with those containing the LOA-linked markers and repeat sequences. In H. aurantiacum, AI cells are often observed before meiotic division of the MMC. The frequency of AI cell differentiation increases during meiosis in adjacent sexually programmed cells, and the AI cells undergo nuclear proliferation around meiosis to form aposporous embryo sacs. Interestingly, in both accessions of H. aurantiacum, multiple embryo sacs amalgamate during their expansion toward the sexually programmed cells. As a consequence, the sexual pathway ceases, and more than one aposporous embryo sac is rarely observed in mature H. aurantiacum ovules (Koltunow et al., 1998, 2000, 2011b).
Crosses between apomictic H. aurantiacum A35 and a H. praealtum (R35) mutant defective in LOA function (134, as female) restored apospory in the resulting hybrid progeny. The mode of apospory observed in these progeny resembled the H. aurantiacum type in some but not all cases (Koltunow et al., 2011b; S. Johnson, unpublished data). Therefore, apospory-specific genetic information is clearly transferable between the two species, even though H. aurantiacum A35 does not possess LOA-linked markers and the repeat-rich chromosome found in H. praealtum R35. We have only obtained partial sequence of the LOA region of H. praealtum R35, which is relatively gene poor, and genomic sequences critical for LOA function lying between the markers 14-T7 and 9-HR need to be identified. It is not possible to determine from metaphase chromosome spreads the exact size of the intervening genomic sequence between contig A and contig B probes, and FISH on pachytene chromosomes will provide a more reliable indication. It remains to be determined whether A35 and R35 contain the same types of apospory-inducing genes or different genes that act in the same or independent biological pathways.
Nevertheless, the lack of LOA-linked repetitive sequences and locus-specific markers, combined with the different chloroplast haplotype network location and altered developmental features of apospory in H. aurantiacum, suggest that there may have been independent routes for the evolution of apospory in Hieracium subgenus Pilosella. Analysis of a greater range of subgenus Pilosella species with characterized modes of reproduction using the H. praealtum LOA-specific markers and chromosome probes should determine the extent of conservation of the identified LOA locus and its repeat-associated chromosomal structure in Hieracium subgenus Pilosella species, providing further insight concerning the evolution of apospory in the subgenus.
Convergent Evolution of Chromosomes Containing Apospory Loci in Eudicot and Monocot Apomicts
Relationships between chromosome structure and the genetic control of apomixis have been proposed in other studies. For example, a satellite chromosome is present in diplosporous Taraxacum, and the diplospory locus is thought to be located on this satellite chromosome (van Dijk and Bakx-Schotman, 2004). Karyotype analyses in diplosporous Boechera species have revealed large-scale chromosome substitutions and the presence of a highly heterochromatic chromosome that has been postulated to have a role in the genetic control of apomixis (Kantama et al., 2007). However, the association of genetic loci controlling diplosporous apomixis with these chromosomal features in Boechera and Taraxacum has not been confirmed.
The LOA locus in Hieracium shares a number of cytogenetic features with the ASGR conferring apomixis in P. squamulatum (Ozias-Akins et al., 1998; Akiyama et al., 2004). Both LOA and ASGR loci are located at the distal end of a hemizygous chromosomal region containing repetitive sequences. The ASGR in aposporous C. ciliaris, a close relative of Pennisetum, is also surrounded by repetitive sequences, although the locus has a pericentromeric location (Akiyama et al., 2005). Differential chromosome location of ASGR in C. ciliaris and P. squamulatum may reflect a rearrangement of the locus, possibly through a translocation to the new location in P. squamulatum (Gualtieri et al., 2006). In Paspalum notatum, the apospory locus also has a hemizygous chromosomal location; however, current analyses have not revealed an association with repeats or heterochromatin (Calderini et al., 2006). Pennisetum, Cenchrus, and Paspalum belong to the same tribe, Paniceae, in the Poaceae family, and differences in the structures of chromosomes containing these loci may suggest polyphyletic evolution of the apospory locus within the grass family or a localized disruption of synteny caused by rearrangements (Calderini et al., 2006). Similarly, in the aposporous Hieracium subgenus Pilosella species analyzed in this study, LOA-specific sequences and associated chromosomal structures are not conserved.
Monocot Pennisetum and Cenchrus species are phylogenetically distant from eudicot Hieracium subgenus Pilosella. Monocots diverged from eudicots 140 to 150 million years ago (Chaw et al., 2004), and Asteraceae appeared 42 to 47 million years ago (Kim et al., 2005; Barreda et al., 2010). However, the loci for apospory in phylogenetically distant species are located on a single chromosome surrounded by abundant retrotransposon and complex repeat sequences. The ancestral origin of the repetitive sequences is clearly different, as there is no obvious conservation of repetitive or other DNA sequence currently identified at the LOA locus and ASGR, providing an example of convergent chromosome evolution. A similar example of convergent evolution of the chicken Z and human X sex chromosomes has recently been reported. These chromosomes evolved independently from different portions of an ancestral genome. However, they share genomic structural features, such as extensive repetition of a few genes at the distal end of the chromosome and an abundance of interspersed repeats and LINE transposons (Bellott et al., 2010).
Roles of the Repeat Sequences at Apospory Loci and Models for LOA Function
Akiyama et al. (2004) have suggested that the accumulation of repeats around the Pennisetum ASGR locus might be a consequence of sequence divergence due to the reproductive isolation of the apomictic genome coupled with the susceptibility of the hemizygous region to somatic invasion by retrotransposons. However, the complex repetitive sequences abundant in hemizygous chromosomal regions of the ASGR and LOA locus might also be functionally relevant for the induction and maintenance of the trait.
Recombination appears to be suppressed at the ASGR, making it difficult to currently define the critical region for apomixis below 50 Mb using map-based strategies (Akiyama et al., 2004; Ozias-Akins and van Dijk, 2007). This might relate to the genetic determinants for both aposporous embryo sac formation and autonomous embryo development residing at the ASGR as well as the unique structure of that hemizygous chromosomal region inhibiting their recombination (Akiyama et al., 2004). Recombination is not suppressed at the hemizygous chromosomal region containing LOA, as we have been able to delineate the region critical for LOA function by physical mapping. Thus, although another chromosome containing the extensive repetitive elements present at the LOA locus is not evident elsewhere in the genome, other genomic elements present at the LOA region must be participating in recombination during meiosis. Using FISH to track the LOA locus through the events of microsporogenesis may provide further insight into the behavior of the hemizygous chromosomal region during meiosis.
Although genomic regions containing loci with significant transposons and repeats are thought to be highly heterochromatic, transcription from these chromosomal regions does occur (Bellott et al., 2010; Stimpson and Sullivan, 2010). Recent transcriptome analyses of the ASGR-carrier chromosome of Pennisetum show that one transcript, which is tightly linked to the heterochromatic ASGR and has similarity to the Transposase_24 domain, is expressed in vegetative and reproductive tissues. A MADS domain-containing gene and a Lon protease present on the ASGR-carrier chromosome were also expressed only in reproductive tissues, but their cosegregation with the ASGR was not confirmed (Zeng et al., 2011). Our cytogenetic analysis of H. caespitosum C36 chromosomes indicated that the chromosomal region containing the LOA locus and associated repetitive sequences is not enriched in heterochromatic marks relative to other chromosomes, suggesting that it may also be transcriptionally active.
Models concerning the function of the LOA locus in apomixis initiation in Hieracium subgenus Pilosella need to take into consideration that while aposporous embryo sac formation requires information at the LOA locus for AI cell formation, the prior events of meiosis leading to megaspore tetrad development are required to activate LOA locus function (Fig. 5A; Koltunow et al., 2011b). Furthermore, factors at the LOA locus stimulating AI cell formation and potentially sexual suppression are not essential for sexual reproduction, because deletion of LOA results in the successful completion of sexual female gametophyte development (Koltunow et al., 2011b).
What factors associated with sexual reproduction might activate LOA function (Fig. 5A)? In Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and maize (Zea mays), the activity of genetic and epigenetic pathways is required to regulate the number of cells that can functionally undergo gametophytic development (Nonomura et al., 2003; Zhao et al., 2008; Tucker and Koltunow, 2009; Garcia-Aguilar et al., 2010; Olmedo-Monfil et al., 2010; Singh et al., 2011). Mutations in ARGONAUTE genes and associated genes involved in small RNA-processing pathways and the down-regulation of DNA methylation machinery lead to the formation of aposporous and diplosporous-like embryo sacs in Arabidopsis and maize (Garcia-Aguilar et al., 2010; Olmedo-Monfil et al., 2010; Singh et al., 2011). The diplosporous-like embryo sacs generated in the dominant argonaute104 mutant are functional and give rise to seedlings of increased ploidy after fertilization (Singh et al., 2011). If similar epigenetic pathways are involved in the regulation of early sexual reproductive development in Hieracium subgenus Pilosella species, the accumulated repetitive sequences and transposons associated with the LOA locus and/or genes present at the LOA locus may be targets of such epigenetic regulation. Similarly, other signaling pathways involving receptor-like kinases, which are required to limit multiple megaspore-like cells forming in rice, may be involved (Nonomura et al., 2003; Zhao et al., 2008).
Figure 5B summarizes three possible scenarios for the likely function of sequences at the LOA locus to enable AI cell formation. It is unlikely that a gene for AI cell formation at the LOA locus is a functional knockout and is not expressed (Fig. 5B, scenario 1). This hypothesis does not easily fit with the dominant nature of the LOA locus, nor can it explain the reversion to sexual reproduction in deletion mutants lacking LOA sequences (Koltunow et al., 2011b). The chromosomal structure of the LOA locus and associated sequences may itself influence gene expression at other loci in cis and trans, similar to paramutation in maize and the Ph1 locus regulating homeologous recombination in wheat (Triticum aestivum; Griffiths et al., 2006; Arteaga-Vazquez and Chandler, 2010; Fig. 5B, scenario 2). Deletions at the LOA locus would potentially modify the optimal chromatin structure and thereby prevent AI cell formation. Alternatively, transcripts produced from the LOA locus may directly enable the initiation of AI cell formation (Fig. 5B, scenario 3). These transcripts might be intact, truncated, or mutated gene products, alternative splicing products, noncoding RNA or small RNAs, and might be derived from a single gene or multiple, closely linked regions within the locus. Candidates include mutated genes involved in small RNA and DNA methylation pathways that give rise to apomixis-like embryo sac formation (Garcia-Aguilar et al., 2010; Olmedo-Monfil et al., 2010; Singh et al., 2011). Deletions in the LOA locus would remove such transcripts, disabling the capacity for AI cell formation. The involvement of the surrounding repetitive elements and transposons in regulating gene expression from the LOA locus cannot be excluded. They may directly or indirectly induce gene silencing, influence epigenetic marks and chromatin structure, or alter transcript levels (Grewal and Moazed, 2003; Martienssen et al., 2004; Arteaga-Vazquez and Chandler, 2010).
Isolation of the DNA sequences between the LOA-linked markers 14-T7 and 9-HR in the LOA locus and the identification of genes responsible for apomixis initiation in H. praealtum are our current priorities. The deletion mutants defective in LOA function provide a means to test candidate genes to examine if they restore apospory.
MATERIALS AND METHODS
Plant Materials
Eight Hieracium subgenus Pilosella accessions were used in this study. These included tetraploid (4x = 2n = 36) apomict Hieracium piloselloides D36, diploid (2n = 18) apomict H. piloselloides D18, aneuploid (3x + 8 = 35) apomict Hieracium praealtum R35, tetraploid (4x = 2n = 36) apomict Hieracium caespitosum C36, aneuploid (3x + 8 = 35) apomict Hieracium aurantiacum A35, tetraploid (4x = 2n = 36) apomict H. aurantiacum A36, and tetraploid (4x = 2n = 36) sexual Hieracium pilosella P36(CR) and P36 (Koltunow et al., 2011b). The γ-deletion mutants of H. praealtum described by Catanach et al. (2006) were also used. Plant growth conditions and reproductive features of these plants are described by Koltunow et al. (1998, 2011b).
Identification of Genomic Sequences Associated with the LOA Locus
The identification of BACs containing DNA associated with the LOA locus was initiated using four SCAR markers (LOA 300, LOA 267, LOA 275, and LOA 219) that were central to the LOA locus from prior analyses (Catanach et al., 2006; Koltunow et al., 2011b). An H. praealtum (R35) BAC library was screened using a combination of PCR and BAC filter hybridization as follows. First, superpools of BAC DNAs were screened by PCR with SCAR markers, and individual BAC DNA pools were identified. Then, BAC filters containing the identified BAC pools were hybridized with DNA probes to identify candidate BAC clones. DNA probes were generated from the SCAR marker sequences and labeled with [α-32P]dCTP. Individual BAC clone candidates were obtained from the Arizona Genomics Institute and tested with SCAR markers. Positive PCR fragments (150–400 bp) obtained by SCAR-PCR were sequenced to confirm that the sequences were identical to the original SCAR markers. An overlap of BAC contigs was confirmed by BAC fingerprint patterns using restriction enzyme digestion to classify the association with a BAC contig group. BAC end sequences were determined, and Southern-blot analysis was performed to assess the copy number and specificity of BAC end DNA sequences as described previously (Okada et al., 2000). New SCAR markers were developed from the BAC end sequences for further BAC library screening and physical mapping analyses. The development of SCAR markers from BAC end sequences is described in Supplemental Materials and Methods S1.
Sequencing of BAC Pools and Bioinformatic Analyses
Partial DNA sequences of BACs associated with the LOA locus were determined by 454 pyrosequencing at the Australian Genome Research Facility. Two pools of BACs were sequenced comprising the following BACs from contig A (LOA300.1, LOA300.7, LOA300.4, LOA300.5, LOA4T7.16, LOA4T7.17, LOA267.13, LOA267.14, LOA13HR.23, and LOA13HR.24) and the following BACs from contig B (LOA275.9, LOA275.12, LOA9HR.19, and LOA9HR.21; Fig. 1). Sequence data have been deposited in the National Center for Biotechnology Information Sequence Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/) with the accession number SRA035358.2. After contig assembly, 379 contigs of nonredundant sequence (532,951 bp in total) were obtained from the contig A sequencing pool and 241 contigs (227,131 bp) were obtained from the contig B sequencing pool. The strategy of pooling BACs for sequencing made it impossible to assign all the assembled contig sequences to specific BACs. DNA sequences from the ASGR sequences of Pennisetum and Cenchrus compiled as 1,341 contigs were obtained from http://asgr.uga.edu (Conner et al., 2008). For the purposes of bioinformatic comparison of DNA sequences between Pennisetum squamulatum, Cenchrus ciliaris, and H. praealtum, a series of random genomic DNA sequences generated using a sequence generator consisting of 134 sequences, average length of 7,164 bp and total length of 959,985 bp, served as a negative control.
The identification of repeats and transposons in Hieracium LOA contigs and Pennisetum and Cenchrus ASGR contigs was performed using TransposonPSI (http://transposonpsi.sourceforge.net), Censor version 4.2.22 (Jurka et al., 1996), and RepeatMasker version 3.2.9 (http://www.repeatmasker.org). Resulting output files obtained from the programs were processed using GALAXY (Goecks et al., 2010). In addition, a DNA sequence clustering method was used to identify complex repeat sequences that were not identified by the above-mentioned programs. Briefly, contig sequences were compared with each other by BLASTN with a low-complexity filter and a cutoff value of 1e−10. Sequence regions with significant similarity to other contigs were identified as candidate repeat clusters. Repeat cluster sequences, primarily containing transposons and simple/low-complex repeat sequences annotated by TransposonPSI and RepeatMasker, were removed from further analysis. Then, repeat cluster sequences were used to mask the contig sequences, and masked regions were counted and their sequence lengths were measured using Excel (Microsoft). In total, 899 sequence regions containing 236,401 bp were masked in LOA-associated contigs, 421 regions (91,289 bp) in Pennisetum, and 404 regions (141,420 bp) in Cenchrus ASGR-associated contigs. These masked regions have homologous sequences among the contigs and thus represent repeated sequences that are designated as complex repeats in Table II.
Sequences associated with the LOA locus (620 contigs) and the ASGR (1,341 contigs) were compared by BLASTN with an e-value cutoff of 1e−10 and TBLASTX with a cutoff of 1e−5. Sequence regions with significant similarity between LOA and ASGR were annotated by BLASTX with an e-value cutoff of 1e−5 against Arabidopsis (Arabidopsis thaliana) peptide sequences (http://www.arabidopsis.org/) and rice (Oryza sativa) peptide sequences (http://rice.plantbiology.msu.edu/). The results are summarized in Supplemental Table S2.
FISH Analysis
Vigorously growing root tips were collected from Hieracium plants aseptically grown on 0.5× Murashige and Skoog liquid medium (Koltunow et al., 1998). To accumulate metaphase cells, the root tips were pretreated in ice water for 17 h. The root tips were then fixed in 3:1 ethanol:acetic acid. Chromosome preparations for FISH were performed as described previously (Mukai et al., 1990). The LOA locus-specific FISH probes were designed from the BAC end sequences and partial BAC sequences. The specificity of probes was examined by genomic Southern-blot analysis, and four DNA fragments (A-mix) and two fragments (B-mix) were chosen for FISH (Supplemental Fig. S2B). Fluorescent probes were made, and FISH was carried out essentially as described by Mukai et al. (1990). A detailed protocol is provided in Supplemental Materials and Methods S1.
Analysis of the Segregation of LOA-Linked Markers and AI Cell Formation in F1 Progeny
Two mapping populations were generated. Sexual H. pilosella P36 was pollinated with apomict H. praealtum R35 or H. caespitosum C36 to generate F1 progeny plants. Leaf samples were collected from the F1 progeny, and DNA was extracted for SCAR-PCR (Tucker et al., 2001; Koltunow et al., 2011b). The quality of extracted DNA was assessed by PCR with control HDMC1 primers (Okada et al., 2007). Plants were selected containing one or more of the four SCAR markers, and these were further investigated for the presence of nine further SCAR markers linked to the locus. MapMaker exp 3.0 was used for the linkage analysis of the markers (Lander et al., 1987). Plants with or without LOA markers were phenotyped for AI cell and aposporous embryo sac formation at stages 4 and 10 of capitulum development (Koltunow et al., 1998).
Analysis of DNA and Histone Methylation by Indirect Immunofluorescence
The status of chromatin modifications along the long arm of the elongated chromosome containing the LOA locus in H. caespitosum C36 was investigated by immunostaining using the following specific antibodies, anti-H3K4me2 (Upstate; catalog no. 07-030; diluted 1:300), anti-H3K4me3 (Upstate; catalog no. 07-473; diluted 1:200), anti-H3K9me2 (Upstate; catalog no. 07-441; diluted 1:300), anti-H3K27me2 (Upstate; catalog no. 07-452; diluted 1:50), and anti-H3K27me3 (Upstate; catalog no. 07-449; diluted 1:100), as described previously (Houben et al., 2003). 5-Methylcytosine (Eurogentec; catalog no. MMS-900P-A) immunodetection was performed as described previously (Ruffini-Castiglione et al., 2002). Details of indirect immunofluorescence are described in Supplemental Materials and Methods S1.
Sequence data from this article can be found in the NCBI Sequence Read Archive under accession number SRA035358.2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Updated map for the presence (white) or absence (gray) of markers at LOA and LOP loci in H. praealtum R35.
Supplemental Figure S2. Development of SCAR markers and specific FISH probes from LOA locus sequences.
Supplemental Figure S3. FISH analysis of apomictic H. aurantiacum (A35 and A36) and sexual H. pilosella P36 using LOA probes developed for H. praealtum (R35).
Supplemental Figure S4. FISH analysis of three other apomictic Hieracium accessions using LOA probes developed for H. praealtum (R35).
Supplemental Figure S5. Distribution of histone H3 methylation in H. caespitosum C36 chromosomes.
Supplemental Table S1. Analysis of the segregation of LOA-linked markers and AI cell formation in F1 progeny derived from a cross between sexual P36 (female) and apomict C36 (male).
Supplemental Table S2. Comparison between LOA and ASGR contig sequences by TBLASTX and annotation of homologous regions.
Supplemental Table S3. List of primers for SCAR-PCR and PCR conditions.
Supplemental Materials and Methods S1. Methods for SCAR marker development, FISH, and DNA and histone methylation analyses.
Acknowledgments
We thank Deborah Blackman and Diyana Kamal Azlan for technical support and Peggy Ozias-Akins and Joann Conner for providing ASGR contig sequences. We thank Matthew Tucker, Michael Groszmann, Chris Helliwell, and Paul Boss for critical reading and comments on the manuscript and Andrew Spriggs, Darren Cullerne, and Jennifer Taylor for bioinformatic support.
References
- Akiyama Y, Conner JA, Goel S, Morishige DT, Mullet JE, Hanna WW, Ozias-Akins P. (2004) High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol 134: 1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y, Hanna WW, Ozias-Akins P. (2005) High-resolution physical mapping reveals that the apospory-specific genomic region (ASGR) in Cenchrus ciliaris is located on a heterochromatic and hemizygous region of a single chromosome. Theor Appl Genet 111: 1042–1051 [DOI] [PubMed] [Google Scholar]
- Arteaga-Vazquez MA, Chandler VL. (2010) Paramutation in maize: RNA mediated trans-generational gene silencing. Curr Opin Genet Dev 20: 156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreda VD, Palazzesi L, Tellería MC, Katinas L, Crisci JV, Bremer K, Passalia MG, Corsolini R, Rodríguez Brizuela R, Bechis F. (2010) Eocene Patagonia fossils of the daisy family. Science 329: 1621. [DOI] [PubMed] [Google Scholar]
- Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, Brown LG, Rozen S, Warren WC, Wilson RK, et al. (2010) Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell RA, Lambie SC, Butler RC. (2003) Quantification of progeny classes in two facultatively apomictic accessions of Hieracium. Hereditas 138: 11–20 [DOI] [PubMed] [Google Scholar]
- Bicknell RA, Koltunow AM. (2004) Understanding apomixis: recent advances and remaining conundrums. Plant Cell (Suppl) 16: S228–S245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderini O, Chang SB, de Jong H, Busti A, Paolocci F, Arcioni S, de Vries SC, Abma-Henkens MHC, Lankhorst RM, Donnison IS, et al. (2006) Molecular cytogenetics and DNA sequence analysis of an apomixis-linked BAC in Paspalum simplex reveal a non pericentromere location and partial microcolinearity with rice. Theor Appl Genet 112: 1179–1191 [DOI] [PubMed] [Google Scholar]
- Catanach AS, Erasmuson SK, Podivinsky E, Jordan BR, Bicknell R. (2006) Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc Natl Acad Sci USA 103: 18650–18655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW. (2008) Inputs and outputs for chromatin-targeted RNAi. Trends Plant Sci 13: 383–389 [DOI] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH. (2004) Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58: 424–441 [DOI] [PubMed] [Google Scholar]
- Conner JA, Goel S, Gunawan G, Cordonnier-Pratt MM, Johnson VE, Liang C, Wang H, Pratt LH, Mullet JE, Debarry J, et al. (2008) Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol 147: 1396–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrer J, Gemeinholzer B, Chrtek J, Jr, Bräutigam S. (2007a) Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). Mol Phylogenet Evol 42: 347–361 [DOI] [PubMed] [Google Scholar]
- Fehrer J, Krahulcová A, Krahulec F, Chrtek J, Rosenbaumová R, Bräutigam S. (2007b) Evolutionary aspects in Hieracium subgenus Pilosella. Hörandl E, Grossniklaus U, van Dijk PJ, Sharbel T, , Apomixis: Evolution, Mechanisms and Perspectives. Koeltz, Konigstein, Germany, pp 359–390 [Google Scholar]
- Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D. (2010) Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22: 3249–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy Team (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Moazed D. (2003) Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G. (2006) Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752 [DOI] [PubMed] [Google Scholar]
- Gualtieri G, Conner JA, Morishige DT, Moore LD, Mullet JE, Ozias-Akins P. (2006) A segment of the apospory-specific genomic region is highly microsyntenic not only between the apomicts Pennisetum squamulatum and buffelgrass, but also with a rice chromosome 11 centromeric-proximal genomic region. Plant Physiol 140: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben A, Demidov D, Gernand D, Meister A, Leach CR, Schubert I. (2003) Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J 33: 967–973 [DOI] [PubMed] [Google Scholar]
- Jasencakova Z, Soppe WJJ, Meister A, Gernand D, Turner BM, Schubert I. (2003) Histone modifications in Arabidopsis: high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J 33: 471–480 [DOI] [PubMed] [Google Scholar]
- Jurka J, Klonowski P, Dagman V, Pelton P. (1996) CENSOR: a program for identification and elimination of repetitive elements from DNA sequences. Comput Chem 20: 119–121 [DOI] [PubMed] [Google Scholar]
- Kantama L, Sharbel TF, Schranz ME, Mitchell-Olds T, de Vries S, de Jong H. (2007) Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proc Natl Acad Sci USA 104: 14026–14031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KJ, Choi KS, Jansen RK. (2005) Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol Biol Evol 22: 1783–1792 [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Johnson SD, Bicknell RA. (1998) Sexual and apomictic development in Hieracium. Sex Plant Reprod 11: 213–230 [Google Scholar]
- Koltunow AM, Johnson SD, Bicknell RA. (2000) Apomixis is not developmentally conserved in related, genetically characterized Hieracium plants of varying ploidy. Sex Plant Reprod 12: 253–266 [Google Scholar]
- Koltunow AMG, Johnson SD, Okada T. (2011a) Apomixis in hawkweed: Mendel’s experimental nemesis. J Exp Bot 62: 1699–1707 [DOI] [PubMed] [Google Scholar]
- Koltunow AMG, Johnson SD, Rodrigues JCM, Okada T, Hu Y, Tsuchiya T, Wilson S, Fletcher P, Ito K, Suzuki G, et al. (2011b) Sexual reproduction is the default mode in apomictic Hieracium subgenus Pilosella, in which two dominant loci function to enable apomixis. Plant J 66: 890–902 [DOI] [PubMed] [Google Scholar]
- Krahulcová A, Krahulec F. (1999) Chromosome numbers and reproductive systems in selected representatives of Hieracium subgen. Pilosella in the Krkonose Mts (the Sudeten Mts). Preslia 71: 217–234 [Google Scholar]
- Krahulec F, Krahulcová A, Fehrer J, Bräutigam S, Schuhwerk F. (2008) The structure of the agamic complex of Hieracium subgen. Pilosella in the Šumava Mts and its comparison with other regions in Central Europe. Preslia 80: 1–26 [Google Scholar]
- Kumar A, Bennetzen JL. (1999) Plant retrotransposons. Annu Rev Genet 33: 479–532 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA. (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181; erratum Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (2009) Genomics 93: 398 [DOI] [PubMed] [Google Scholar]
- Martienssen R, Lippman Z, May B, Ronemus M, Vaughn M. (2004) Transposons, tandem repeats, and the silencing of imprinted genes. Cold Spring Harb Symp Quant Biol 69: 371–379 [DOI] [PubMed] [Google Scholar]
- Mukai Y, Endo TR, Gill BS. (1990) Physical mapping of the 5S rRNA multigene family in common wheat. J Hered 81: 290–295 [Google Scholar]
- Nonomura K, Miyoshi K, Eiguchi M, Suzuki T, Miyao A, Hirochika H, Kurata N. (2003) The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Catanach AS, Johnson SD, Bicknell RA, Koltunow AM. (2007) An Hieracium mutant, loss of apomeiosis 1 (loa1) is defective in the initiation of apomixis. Sex Plant Reprod 20: 199–211 [Google Scholar]
- Okada T, Sasaki Y, Ohta R, Onozuka N, Toriyama K. (2000) Expression of Bra r 1 gene in transgenic tobacco and Bra r 1 promoter activity in pollen of various plant species. Plant Cell Physiol 41: 757–766 [DOI] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP. (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozias-Akins P, Roche D, Hanna WW. (1998) Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc Natl Acad Sci USA 95: 5127–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozias-Akins P, van Dijk PJ. (2007) Mendelian genetics of apomixis in plants. Annu Rev Genet 41: 509–537 [DOI] [PubMed] [Google Scholar]
- Ruffini-Castiglione M, Cremonini R, Frediani M. (2002) DNA methylation patterns on plant chromosomes. Caryologia 55: 275–282 [Google Scholar]
- Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D. (2011) Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23: 443–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Sullivan BA. (2010) Epigenomics of centromere assembly and function. Curr Opin Cell Biol 22: 772–780 [DOI] [PubMed] [Google Scholar]
- Suda J, Krahulcová A, Trávnícek P, Rosenbaumová R, Peckert T, Krahulec F. (2007) Genome size variation and species relationships in Hieracium sub-genus Pilosella (Asteraceae) as inferred by flow cytometry. Ann Bot (Lond) 100: 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Araujo AC, Paech NA, Hecht V, Schmidt ED, Rossell JB, De Vries SC, Koltunow AM. (2003) Sexual and apomictic reproduction in Hieracium subgenus Pilosella are closely interrelated developmental pathways. Plant Cell 15: 1524–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MR, Koltunow AM. (2009) Sexual and asexual (apomictic) seed development in flowering plants: molecular, morphological and evolutionary relationships. Funct Plant Biol 36: 1–15 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Paech NA, Willemse MTM, Koltunow AMG. (2001) Dynamics of callose deposition and beta-1,3-glucanase expression during reproductive events in sexual and apomictic Hieracium. Planta 212: 487–498 [DOI] [PubMed] [Google Scholar]
- van Dijk PJ, Bakx-Schotman JM. (2004) Formation of unreduced megaspores (diplospory) in apomictic dandelions (Taraxacum officinale, s.l.) is controlled by a sex-specific dominant locus. Genetics 166: 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF, Cummings MP, Koniczny A, Ausubel FM, Rodermel SR. (1992) copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci USA 89: 7124–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, de Palma J, Oane R, Gamuyao R, Luo M, Chaudhury A, Hervé P, Xue Q, Bennett J. (2008) OsTDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J 54: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Conner JA, Ozias-Akins P. (2011) Identification of ovule transcripts from the Apospory-Specific Genomic Region (ASGR)-carrier chromosome. BMC Genomics 12: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]