Abstract
Translational Science Search (TSS; http://tscience.nlm.nih.gov) is a web application for finding MEDLINE/PubMed journal articles that are regarded by their authors as novel, promising, or may have potential clinical application. A set of “translational” filters and related terms was created by reviewing journal articles published in clinical and translational science (TS) journals. Through E‐Utilities, a user’s query and TS filters are submitted to PubMed, and then, the retrieved PubMed citations are matched with a database of MeSH terms (for disease conditions) and RxNorm (for interventions) to locate the search term, translational filters found, and associated interventions in the title and abstract. An algorithm ranks the interventions and conditions, and then highlights them in the results page for quick reading and evaluation. Using previously searched terms and standard formulas, the precision and recall of TSS were 0.99 and 0.47, compared to 0.58 and 1.0 for PubMed Entrez, respectively. Clin Trans Sci 2011; Volume 4: 455–459
Keywords: translational science search, MEDLINE/PubMed, RxNorm, MeSH
Introduction
Despite billions of dollars spent on publicly and privately funded research, the translation of basic research to clinical trials and eventually to clinical applications has not yet resulted in the delivery of improved healthcare. 1 The fate of even highly promising basic science studies is uncertain. Obstacles abound—adequate sustained funding through the clinical translation process, the demanding process of clinical trials, and determining research direction under competing programs. 1 , 2
In a review of six leading basic science journals, Contopoulos‐Ioannidis et al. found that only 25% of promising technologies led to randomized controlled trials published in journals and less than 10% found their way to routine clinical application. 2 In an accompanying editorial, Crowley raised concerns about the thoroughness of the review but nevertheless agreed that the frequency of basic science to clinical translations is still very low. 3 Not only is the rate low but often the interval is long, usually lasting more than 10 years. 2
This study approaches the issue from a different perspective—with more than 20 million citations from about 5,800 journals currently archived in MEDLINE/PubMed, could the failure in discovery of relevant research be a causative factor as well? Might adding filters and limiters enhance the discovery of basic science or clinical research papers, especially research identified by authors as novel, enhance finding these types of journal articles?
Background
An article in Medscape Today 5 reported of an abstract that was presented at the 34th Critical Care Congress in 2005 entitled, “S‐100 Measurement in Patients With Minor Head Injury Can Reduce CT Use.” Using askMEDLINE, 6 , 7 a search was made using the question, “Is serum S‐100 useful in assessing head injury.” At that time, 23 citations were retrieved, one of which was the first report found on the use of S‐100 as a marker for head injury published in 1987. The translational delay was 18 years from the first report to its clinical application for a significant research finding with major implications on healthcare cost. This is not surprising and quite consistent with the lag cited previously. 2 Based on this experience and several others, it was decided to develop a translational science search (TSS) application to provide researchers an alternative tool for discovering relevant interventions of disease processes and conditions of interest.
The aim to this project is to develop a set of translational science (TS) filters that are not yet available in PubMed that would focus on prior studies that are deemed as novel by researchers and considered to lead to potential therapies or diagnostic modalities for disease processes and other medical conditions. The tool itself would make it convenient to identify these disease processes and interventions in the retrieved citations. A preliminary announcement of this resource was made in 2009. 4 Technical details and an evaluation on precision and recall are discussed in this paper.
Methods
Overview of TSS
The overall strategy was to develop a TSS tool by using terms that would likely find promising “translational” articles in PubMed. The search is initiated by using an intervention (drug, chemical, target molecule, gene locus, test, etc.) or a particular disease entity, process, or condition (Rett syndrome, hereditary persistence of fetal hemoglobin, autistic disorder, inflammatory bowel disease, etc). The query could be made more specific by using both intervention and condition, such as, postmenopausal osteoporosis AND lasofoxifene; sickle cell disease AND nitric oxide; HIV AND acyclovir, etc. The results obtained will list a maximum of 100 of the most recent citations on the first results page, depending on the publication date selected. More citations can be displayed if needed. The web application currently runs on a Windows enterprise server with Apache, MySQL, and PHP.
Creation of “Translational Science”(TS) filters
TS filters are a collection of words or phrases that point to the potential use of an intervention or condition. The initial set of filters were created by manually reviewing journal articles, where the authors identified their results as “novel,”“promising,”“unique,” or had “potential use” and “potential application,” These are papers that are within the category of what might be considered “translational”—basic science research with potential for clinical trials and systematic clinical research, and clinical research trials that might have clinical applications for patients. Related terms were found using a term matching program that expanded the initial set. A thesaurus was also used to find additional terms. The database originally had 7,497 unique term combinations (two or three words, e.g., “future therapy,”“potential therapeutic target,”“novel promising application”). Using E‐Utilities, these filters were used to search PubMed for citations containing these words and phrases in their titles and abstracts. TS filters were continuously modified to optimize the time needed to search and maximize the yield of translational publications retrieved. Each one of the original filters was sent to PubMed each time a query was made. Examples of filters and the corresponding number of PubMed citations are shown in Table 1 .
Table 1.
Examples of TS filters and the number of citations retrieved based on searches of PubMed on August 5, 2009 and earlier. Most efficient (highest yield) are listed in the left two columns, least efficient (lowest yield), excluding the filters retrieving no citations, in the right two columns.
| Most efficient (highest yield) | Least efficient (lowest yield) | ||
|---|---|---|---|
| TS filter terms | Number of citations | TS filter terms | Number of citations |
| Effective treatment | 27,414 | Eventual therapeutic outcome | 2 |
| Beneficial effect | 20,791 | Ultimate therapeutic target | 2 |
| Potential use | 10,849 | Ultimate remedy | 2 |
| Effective therapy | 7,933 | Vital therapeutic target | 2 |
| Potential target | 5,110 | Beneficial basis | 2 |
| Potential application | 4,732 | Potential clinical | 1 |
| Possible use | 4,179 | Potential candidate treatment | 1 |
The filters were ranked by efficiency from the most efficient (highest yield) to the least efficient, excluding the filters that retrieved no citations, by examining the number of returned PubMed citations for each query. Figure 1 shows the relation between ranking and the number of citations. The top 42 ranked filters retrieve more than 800 PubMed citations each. The slope becomes flat after the top 42 filters. From the results, only the top 124 filters retrieved more than 100 citations. The remaining filters produced fewer than 100 citations each. More than 7,000 terms did not retrieve any citations at all. Table 1 shows examples of high and low yielding search terms.
Figure 1.
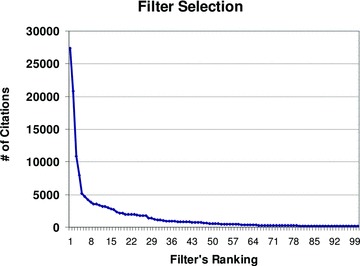
Top 100 ranked filters and the number of PubMed citations retrieved.
Since PubMed had to be queried using each of the 7,497 filters for each search term initially, the retrieval was slow (more than 30 seconds) despite multiple server optimization procedures. A decision was made to optimize the time to retrieve results and number of filters. The filters that retrieved relatively small numbers of citations (fewer than 100 citations of more than 19 million total abstracts in PubMed) were deemed less effective for developing a translation science search application and discarded. The filter set (term combinations) was pared down to 59 search limiters derived through repeated searching and ranking of terms. In some cases, word combinations filters were reduced to one or two terms, or eliminated entirely after discovering that they were not effective.
Another issue considered in lessening the size of the filter set is that in PubMed, only “meaningful” phrases are included in the searchable index. Not all word strings found repeatedly in the database is considered meaningful. In PubMed, new compound words (phrases) are generated twice a month by analyzing data using a parts of speech analyzer 8 to find noun phrases from the title and abstract fields. Noun phrases with frequency counts of fewer than three are removed. Other criteria include: a phrase must contain at least one alphabetic character and a phrase may have at most six words. New compound words are merged with the existing list of phrases. The number of compound words is now more than 24 million. This element may explain the lack of retrievals for many term combinations in the filter set.
Optimization of TSS is ongoing by finding possible filters to add to the current set. This is done by comparing filter retrievals with PubMed using search terms obtained from recent clinical and translational journals. When potential filter terms that might find translational articles are found, they are added to the current set. Suggestions from colleagues are also considered and added after testing and review.
Database configuration
Several possible sources were considered to identify and match the citations from PubMed with the search terms and related interventions and conditions and the following were selected: for the intervention database, the “Ingredient” and “Brand name” records from RxNorm, 9 a standardized nomenclature for clinical drugs and drug‐delivery devices from the National Library of Medicine (NLM) was chosen. After removing suppressed and duplicate terms in RxNorm, 14,119 terms remained in the table.
For the conditions database, MeSH, 10 a controlled vocabulary thesaurus from NLM was preferred. A computer program was written to process MeSH tree files and MeSH terms in the “Diseases” branch of MeSH tree were selected, then entries for MeSH terms were retrieved. The resulting table has 34,593 unique diseases or entries from the MeSH tree.
To enhance searching performance, database optimization was done on both databases including table simplification, column indexing, and storage engine selection.
Search processing
An online search interface ( Figure 2 ) allows researchers to enter a condition, an intervention, or both for a narrower, a more focused search. Users can also set the date of publication in the search interface. The query is then combined with more than 60 TS filters in the current set, and then sent to PubMed through “E‐Utilities.” 12
Figure 2.
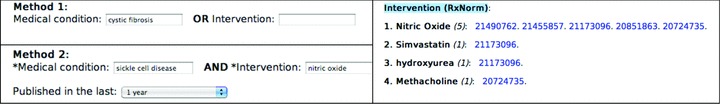
TSS search examples (left column)—Method 1, medical conditions only (top) and combined (below). Right side shows an example of results retrieved for Method 2. Numbers are PubMed IDs of journal articles.
A multistep parsing algorithm identifies the intervention or condition terms in citations and then “Stopwords” in each article are deleted. PubMed’s “Stopwords” list (examples: of, the, what, quite) contains 132 most common words that are considered uninformative for information retrieval. The parsing algorithm then checks the remainder of the content and then marks intervention or condition terms. To accelerate TSS tool performance, other words in the content that are not found in the intervention and disease database are held in a temporary “uninformative” list. In succeeding citations, words in this temporary list will not be searched again in the intervention or condition databases for this particular search. Figure 3 illustrates the dataflow in TSS processing.
Figure 3.
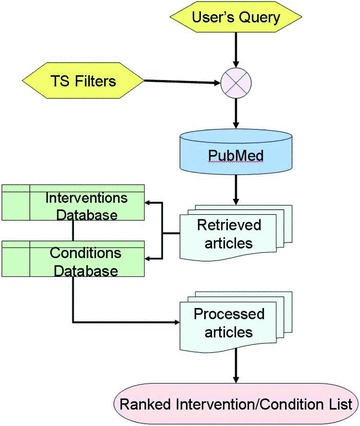
TSS processing flowchart.
The results from PubMed are a combination of citations retrieved from all the filter terms in the set and the user’s search term. From the retrieved PubMed citations, the most recent 100 articles are selected and ranked. Condition (MeSH) or intervention (RxNorm) terms with the highest count of citations are ranked higher and are listed on top. If the search term is a medical condition, related interventions are returned first, then related conditions after. The reverse is done if an Intervention is the search term. For combined searches (medical condition and intervention), interventions are returned first.
Words in the search string and phrases found in the intervention and condition’s databases are highlighted in the title and abstracts. The abstract can be shown in a new browser window by clicking on the PubMed Identifier (PMID). Each extracted medical condition term in the results is both a valid MeSH entry, which appears in the citation and its formal MeSH heading. The types of interventions are categorized in the abstract page. In the current version, only RxNorm’s “Ingredient” and “Brand Name” elements are included.
For queries that retrieve more than 100 articles, a link is provided to view the next 100 PMIDs, or more if there are any. Terms are rank ordered according to frequency count of the retrieved citations (PMIDs). For example, in Figure 2 , “Nitric oxide” has the highest citations count (19 citations) and is listed first. Within the listing itself, (Nitric oxide), the PMIDs are ordered according to chronology, so the newer articles are listed first. Citations that are not classified in either one of the two headings are listed separately at the bottom of the page. PMIDs may be listed (duplicated) in several “Condition” or “Intervention” categories. When this occurs, the web browser will indicate that the abstract had been viewed previously by a change in the link color.
Search terms, conditions, intervention, and translational terms, and TS filters in the title and abstract are highlighted when the abstract is displayed (see Figure 4 ). This feature lets the researcher focus on terms and phrases of interest in the article and facilitates review.
Figure 4.

Abstract with highlighted search terms, filter terms, conditions, and interventions.
Evaluation
Fifteen previous searches (five from each category: “Condition” and “Intervention,”“Condition” only, and “Intervention” only) were randomly selected. For each term, TSS and PubMed were searched for articles published between October 1, 2009 and September 30, 2010, and the retrievals from each were compared. For calculating precision and recall, all abstracts of citations retrieved by TSS were reviewed by one of the authors (PF) for “significance” or “relevance.” For this evaluation, the use of “significance” and “relevance” are subjective evaluations to denote the “translational” nature of the journal articles found. Abstracts are determined as “significant” or “relevant” if they are “novel,” and have data and information that could likely lead to a clinical application, clinical trial or clinical intervention, or become the basis of further research on the topic. It is a qualitative “Yes” or “No” assessment of abstracts. This definition is based on the concepts of effectiveness of databases reported by Stokes et al., 12 but in a qualitative sense only, not the quantitative manner measured and discussed in their article. The assessment on the translational nature of an article was quite tolerant—even a minimal indication of the translational nature of the citation was deemed to be a “Yes.” Errors on the determination of the relevance and significance then would tend to favor an increase in PubMed’s precision and lower TSS recall because of the reciprocal nature of the relationship between precision and recall.
All citations retrieved with TSS were reviewed. All abstracts retrieved using PubMed were also reviewed if they were less than 50. If the total number of articles from PubMed search was 50 or more than TSS, a set of 50 citations were randomly selected by a machine randomization algorithm for evaluation on their significance and relevance. The ratio of read and unread citations in PubMed search result set is taken into account when estimating the total number of “relevant” citations. A total of 1,234 abstracts were reviewed.
Precision and recall were determined using conventional methods according to the following formulas:
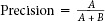 , where A is the number of documents retrieved and relevant and B is the number of citations retrieved and nonrelevant. A+B is the number of total citations retrieved by this method.
, where A is the number of documents retrieved and relevant and B is the number of citations retrieved and nonrelevant. A+B is the number of total citations retrieved by this method.
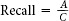 , where A is the number of citations retrieved and relevant and C is the number of total relevant citations in the database.
, where A is the number of citations retrieved and relevant and C is the number of total relevant citations in the database.
Results
All 364 citations retrieved by TSS were reviewed for relevance or significance, compared to 54% (870/1,608) for PubMed. For both TSS and PubMed, searching using two terms (more focused, narrower) retrieved fewer citations, 11% (19/172, 19/173) and 14–17% (114/804, 114/690) for TSS and PM, respectively ( Table 2 ). Using the same search terms shown in Table 2 and for the same year‐long review period, TSS retrieved on the average, 23% as many citations as PubMed.
Table 2.
Precision and recall (P&R) of TSS and PubMed (PM). Qualitative significance and relevance criteria were applied in determining P&R.
| Search term | TSS | PM | Precision (TSS) | Recall (TSS) | Precision (PM) | Recall (PM) |
|---|---|---|---|---|---|---|
| Condition and intervention | ||||||
| Systemic sclerosis AND Rituximab | 5 | 14 | 1.00 | 0.50 | 0.71 | 1 |
| Asthma AND Matrix Metalloproteinase 12 | 1 | 3 | 1.00 | 1.00 | 0.33 | 1 |
| Varicella AND Acyclovir | 2 | 48 | 1.00 | 0.33 | 0.13 | 1 |
| Sickle cell disease AND Nitric oxide | 7 | 31 | 1.00 | 0.54 | 0.42 | 1 |
| Brain trauma AND Serum S‐100 | 4 | 18 | 1.00 | 0.50 | 0.44 | 1 |
| Condition | ||||||
| Ochronosis | 2 | 17 | 1.00 | 0.50 | 0.24 | 1 |
| Medullary carcinoma thyroid | 41 | 182 | 1.00 | 0.43 | 0.66 | 1 |
| Schistosomiasis | 115 | 520 | 0.99 | 0.29 | 0.90 | 1 |
| Dupuytren’s contracture | 5 | 39 | 1.00 | 0.19 | 0.67 | 1 |
| Pseudoxanthoma Elasticum | 10 | 46 | 0.90 | 0.25 | 0.78 | 1 |
| Intervention | ||||||
| Rivaroxaban | 33 | 72 | 1.00 | 0.77 | 0.60 | 1 |
| Fenofibrate | 35 | 186 | 0.94 | 0.18 | 0.96 | 1 |
| Mineralocorticoid receptor | 57 | 274 | 1.00 | 0.21 | 0.99 | 1 |
| P2Y12 antagonists | 28 | 65 | 1.00 | 0.44 | 0.97 | 1 |
| Phosphodiesterase III | 19 | 93 | 1.00 | 0.25 | 0.84 | 1 |
| Totals (citations retrieved/citations reviewed) | 364/364 | 1,608/870 | ||||
| Average | 0.99 | 0.47 | 0.58 | 1.00 | ||
The retrievals from PubMed were used to calculate the recall rate since there is currently no “gold standard” to compare with TSS. Using the formula above, the recall rate for TSS is 0.47 while PubMed’s recall rate is 1.0 since PubMed contains “all” the relevant articles documents in the dataset. Precision rate for the TSS is 0.99 compared to PubMed’s 0.58. A “Related articles” link is available for each citation retrieved. A review of retrieved abstracts obtained when a search for “Related articles” for one intervention (Rivaroxaban) showed that 45% of related PMIDs were the same PMIDs discovered through PubMed search.
Discussion
The evaluation results ( Table 2 ) showing the comparison of precision and recall between TSS and PubMed illustrate the classic inverse relationship between precision and recall. The high precision of TSS (99%) is associated with a 47% recall rate. Conversely, PubMed’s high recall (100%) rate is associated with a 58% precision rate. Buckland and Gey suggested a two‐stage strategy to improve the search by using a high recall method first followed by a more precise method. 13 A different approach was adapted in TSS by starting off with a highly precise search, with links provided in the results page to search PubMed (high recall) if the researcher finds the TSS results unsatisfactory. This approach was decided on because the search algorithm uses a set of filters especially designed to find research articles identified by authors to be novel or promising. A quick review comparing TSS and PubMed results did not indicate that fewer results in TSS was detrimental to the discovery of possible basic science research that could be translated to clinical applications. A review of abstracts retrieved using the filter sets were also found to be highly translational in nature with high potential to lead to clinical trials, clinical applications, or as a starting point for further research. Moreover, a search for “Related articles” of the TSS search result citations retrieved 40% of abstracts found using PubMed. This could signify that many of the publication dealt with similar or related interventions. It is likely then that the low recall may be compensated by a secondary PubMed search step or a search for related items through links in individual abstracts. A two‐step approach, high precision (TSS) search followed by a high recall (PubMed) search may be done if there is a concern that important research publications are being missed.
TSS results of searches are continuously reviewed to determine if the search results are relevant. Filter terms are incorporated after testing and optimization. Due to the complex and multistep algorithm of TSS, latency is a limitation. The source of the latency has been narrowed down to the ranking and highlighting of interventions and conditions terms. Eliminating these two features was considered, but it was felt that these features were essential to the usefulness of TSS, so a decision was made to keep them. Although TSS is slower than a PubMed Entrez search, most queries take less than 30 seconds or even shorter if the publication date is limited and citations retrieved are fewer. Server optimization is also continuing. Citations obtained through TSS searches were also found to be almost all translational papers and to be “high quality.”
To facilitate review, TSS returns either intervention or conditions first, depending on the term searched. We are continuing to optimize the TSS to decrease search latency. Suggestions are welcome on optimization procedures.
TSS’ biggest advantage is its clinical focus and its capability to concentrate on a condition or intervention of interest, in finding filtered published research identified as promising, novel, and having potential clinical applications. Highlighting relevant terms is also a benefit because it draws the researcher to terms of interest in the abstract so scanning for potential references is quicker.
As anticipated, TSS locates publications that have potential for clinical applications. The more important assessment will be its usefulness to TS researchers. Its ultimate success will come if it leads to an increase in basic science to clinical applications research, especially if it shortens the time from basic research to clinical application.
Conclusion
TSS is a clinically focused tool for discovering possible novel research, therapies, and potential interventions. As intended, this tool retrieves publications that have potential for clinical applications. TSS extracts medical conditions and interventions from PubMed, sorts, and ranks, and then highlights them based on RxNorm and MeSH terms to make it convenient for researches to find prospective clinical interventions.
Disclaimer
The views and opinions of the author expressed herein do not necessarily state or reflect those of the National Library of Medicine, National Institutes of Health, or the US Department of Health and Human Services.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM) and Lister Hill National Center for Biomedical Communications (LHNCBC).
REFERENCES
- 1. Sung NS, Crowley WF, Jr ., Genel M, Salber P, Sandy L, Sherwood LM, Johnson SB, Catanese V, Tilson H, Getz K, et al Central challenges facing the national clinical research enterprise. JAMA. 2003; 289: 1278–1287. [DOI] [PubMed] [Google Scholar]
- 2. Contopoulos‐Ioannidis DG, Ntzani E, Ioannidis JP. Translation of highly promising basic science research into clinical applications. Am J Med. 2003; 114(6): 477–484. [DOI] [PubMed] [Google Scholar]
- 3. Crowley WF Jr. Translation of basic research into useful treatments: how often does it occur? Am J Med. Apr 152003; 114(6): 503–505. [DOI] [PubMed] [Google Scholar]
- 4. Fontelo P, Liu F. A novel tool for translational research discovery. Clin Transl Sci. 2009. Dec; 2(6): 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. http://www.medscape.com/viewarticle/497751. Accessed June 19, 2011.
- 6. askMEDLINE: http://askmedline.nlm.nih.gov
- 7. Fontelo P, Liu F, Ackerman M, Schardt CM, Keitz SA. askMEDLINE: a report on a year‐long experience. AMIA Annu Symp Proc. 2006; 923. [PMC free article] [PubMed] [Google Scholar]
- 8. Smith L, Rindflesch T, Wilbur WJ. MedPost: a part‐of‐speech tagger for bioMedical text. Bioinformatics. Sep 222004; 20(14): 2320–2321. [DOI] [PubMed] [Google Scholar]
- 9. RxNorm:http://www.nlm.nih.gov/research/umls/rxnorm. Accessed June 19, 2011.
- 10. MeSH: http://www.nlm.nih.gov/mesh. Accessed June 19, 2011.
- 11. E‐Utilities: http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html. Accessed June 19, 2011.
- 12. Stokes P, Foster A, Urquhart C. Beyond relevance and recall: testing new user‐centred measures of database performance. Health Info Libr J. Sep2009; 26(3): 220–231. [DOI] [PubMed] [Google Scholar]
- 13. Buckland M, Gey F. The relationship between Recall and Precision. J. Am Soc Inf Sci. 45(1): 12–19. [Google Scholar]


