Abstract
In our previous study (Chen et al. J Biol Chem 2005, 280:12316–12329), we utilized an α-helical antimicrobial peptide V681 as the framework to study the effects of peptide hydrophobicity, amphipathicity, and helicity on biologic activities where we obtained several V681 analogs with dramatic improvement in peptide therapeutic indices against gram-negative and gram-positive bacteria. In the present study, the d-enantiomers of three peptides – V681, V13AD and V13KL were synthesized to compare biophysical and biologic properties with their enantiomeric isomers. Each d-enantiomer was shown by circular dichroism spectroscopy to be a mirror image of the corresponding l-isomer in benign conditions and in the presence of 50% trifluoroethanol. l- and d-enantiomers exhibited equivalent antimicrobial activities against a diverse group of Pseudomonas aeruginosa clinical isolates, various gram-negative and gram-positive bacteria and a fungus. In addition, l- and d-enantiomeric peptides were equally active in their ability to lyse human red blood cells. The similar activity of l- and d-enantiomeric peptides on prokaryotic or eukaryotic cell membranes suggests that there are no chiral receptors and the cell membrane is the sole target for these peptides. Peptide d-V13KD showed significant improvements in the therapeutic indices compared with the parent peptide V681 by 53-fold against P. aeruginosa strains, 80-fold against gram-negative bacteria, 69-fold against gram-positive bacteria, and 33-fold against Candida albicans. The excellent stability of d-enantiomers to trypsin digestion (no proteolysis by trypsin) compared with the rapid breakdown of the l-enantiomers highlights the advantage of the d-enantiomers and their potential as clinical therapeutics.
Keywords: α-helical peptides, antimicrobial activity, antimicrobial peptides, enantiomers, hemolytic activity, mechanism
The widespread use of traditional antibiotics has resulted in the emergence of many antibiotic-resistant strains, prompting an urgent need for a new class of antibiotics (1,2). Cationic antimicrobial peptides have become important candidates as potential therapeutic agents (3–5) and have been shown to be active in in vivo animal studies (6). Although the exact mode of action of antimicrobial peptides has not been established (7–9), it has been proposed that the cytoplasmic membrane is the main target for many of these peptides, whereby peptide accumulation in the membrane causes increased permeability and loss of barrier function (10,11). The development of resistance to membrane active peptides whose sole target is the cytoplasmic membrane is not expected because this would require substantial changes in the lipid composition of cell membranes of micro-organisms. However, the major barrier to the use of antimicrobial peptides as antibiotics is their toxicity or ability to lyse eukaryotic cells, normally expressed as hemolytic activity (toxicity to human red blood cells), which is a main reason preventing their applications as injectable therapeutics.
Enantiomeric forms of antimicrobial peptides with all-d-amino acids were used to study the membrane-binding mechanism (12–14), as it was previously thought that cell membrane chirality would require a specific peptide chirality for it to be active. However, many studies have shown that all-d-amino acid peptides have equal activities to their all-l-enantiomers (12–19), suggesting that the antimicrobial mechanism of these peptides does not involve a stereoselective interaction with a chiral enzyme or lipid or protein receptor. In addition, all-d-peptides are resistant to proteolytic enzyme degradation, which enhances their potential as clinical therapeutics.
In our previous study (20), we utilized a de novo design approach to alter the secondary structure, hydrophobicity, and amphipathicity of an α-helical amphipathic antimicrobial peptide V681 (21,22) and obtained lead compounds with high antimicrobial activities and extremely low hemolytic activity. In the present study, we compare the biophysical and biologic properties between peptide enantiomers with all-l- or all-d-amino acids of our lead compounds, including antimicrobial activities against various bacterial strains, especially a diverse group of Pseudomonas aeruginosa clinical isolates with a wide range of susceptibility to the antibiotic ciprofloxacin. It is well known that respiratory infections by P. aeruginosa are the major cause of morbidity and mortality in adult patients with cystic fibrosis (23–25). Pseudomonas aeruginosa infection is also a serious problem in patients hospitalized with cancer and burns, the case fatality in such patients being 50% (24,25). According to data collected from 1990 to 1996 by the US Centers for Disease Control and Prevention (CDC), P. aeruginosa was the second most common cause of nosocomial pneumonia (17% of isolates), the third most common cause of urinary tract infections (11%), the fourth most common cause of surgical site infections (8%), the seventh most common isolated pathogen from the bloodstream (3%), and the fifth most common isolate overall (9%). We believe that, this comparative de novo design study of l- and d-antimicrobial peptides is the critical step toward the development of new antimicrobial therapeutics and understanding the mechanism of action of α-helical antimicrobial peptides.
Materials and Methods
Peptide synthesis and purification
Syntheses of the peptides were carried out by solid-phase peptide synthesis using t-butyloxycarbonyl chemistry and 4-methylbenzhydrylamine (MBHA) resin (0.97 mmol/g), as described previously (26). The crude peptides were purified by preparative reversed-phase high-performance liquid chromatography (RP-HPLC) using a Zorbax 300 SB-C8 column (250 × 9.4 mm I.D.; 6.5 µm particle size, 300 Å pore size; Agilent Technologies, Little Falls, DE, USA) with a linear AB gradient (0.1% acetonitrile/min) at a flow rate of 2 mL/min, where eluent A was 0.2% aqueous trifluoroacetic acid (TFA), pH 2 and eluent B was 0.2% TFA in acetonitrile (27). The purity of peptides was verified by analytical RP-HPLC as described below. The peptides were further characterized by mass spectrometry and amino acid analysis.
Analytical RP-HPLC of peptides
Peptides were analyzed on an Agilent 1100 series liquid chromatograph (Little Falls, DE, USA). Runs were performed on a Zorbax 300 SB-C8 column (150 × 2.1 mm I.D.; 5 µm particle size, 300 Å pore size) from Agilent Technologies using a linear AB gradient (1% acetonitrile/min) and a flow rate of 0.25 mL/min, where eluent A was 0.2% aqueous TFA, pH 2 and eluent B was 0.2% TFA in acetonitrile. Temperature profiling analyses were performed in 3 °C increments, from 5 to 80 °C using a linear AB gradient of 0.5% acetonitrile/min.
Characterization of helical structure
The mean residue molar ellipticities of peptides were determined by circular dichroism (CD) spectroscopy, using a Jasco J-720 spectropolarimeter (Easton, MD, USA), at 5 °C under benign (non-denaturing) conditions (50 mm KH2PO4/K2HPO4/100 mm KCl, pH 7.4), hereafter referred to as KP buffer, as well as in the presence of an α-helix inducing solvent, 2,2,2-trifluoroethanol (TFE; 50 mm KH2PO4/K2HPO4/100 mm KC1, pH 7.4 buffer/50% TFE). A 10-fold dilution of an approximately 500 µm stock solution of the peptide analogs was loaded into a 0.02 cm quartz cell and its ellipticity scanned from 190 to 250 nm.
Measurement of antimicrobial activity (MIC)
Minimal inhibitory concentrations (MIC) were determined using a standard microtiter dilution method in a Mueller-Hinton (MH) medium. Briefly, cells were grown overnight at 37 °C in MH broth and diluted in the same medium. Serial dilutions of the peptides were added to the microtiter plates in a volume of 100 µL followed by 10 µL of bacteria to give a final inoculum of 1 × 105 colony-forming units (CFU)/mL. Plates were incubated at 37 °C for 24 h and MICs determined as the lowest peptide concentration that inhibited growth. However, for MIC determination of P. aeruginosa clinical isolates, brain heart infusion (BH1) medium was used instead of MH broth. In addition, the bacteria were diluted to a final inoculum of 1 × 106 CFU/mL.
Measurement of hemolytic activity (MHC)
Peptide samples were added to 1% human erythrocytes in phosphate-buffered saline (100 mm NaCl; 80 mm Na2HPO4; 20 mm NaH2PO4, pH 7.4) and reactions were incubated at 37 °C for 18 h in microtiter plates. Peptide samples were diluted twofold in order to determine the concentration that produced no hemolysis. This determination was made by withdrawing aliquots from the hemolysis assays, removing unlysed erythrocytes by centrifugation (800 × g) and determining which concentration of peptide failed to cause the release of hemoglobin. Hemoglobin release was determined spectrophotometrically at 570 nm. The hemolytic titre was the highest twofold dilution of the peptide that still caused release of hemoglobin from erythrocytes. The control for no release of hemoglobin was a sample of 1% erythrocytes without any peptide added. As erythrocytes were in an isotonic medium, no detectable release (<1% of that released upon complete hemolysis) of hemoglobin was observed from this control during the course of the assay. For the hemolysis time study, hemolytic activity of peptides at concentrations of 8, 16, 32, 64, 125, 250 and 500 µg/mL was measured at 0, 1, 2, 4 and 8 h at 37 °C.
Calculation of therapeutic index (MHC/MIC ratio)
It should be noted that both MHC and MIC values were determined by serial twofold dilutions. Thus, for individual bacteria and individual peptides the therapeutic index (MHC/MIC) could vary by as much as fourfold if the peptide is very active in both hemolytic and antimicrobial activities; of course, if a peptide has poor or no hemolytic activity, the major variation in the therapeutic index (MHC/MIC) comes from the variation in the MIC value (as much as twofold).
Proteolytic stability assay
Proteolytic stability of the peptides was carried out with trypsin in a molar ratio of 1:20 000 (trypsin:peptide = 0.1 µm:2 mm). The buffer used was 50 mm NH4HCO3 at pH 7.4 for both peptides and enzyme. The mixtures of peptide and trypsin were incubated at 37 °C. Samples were collected at time-points of 0, 5, 10, 20 and 30 min, 1, 2, 4 and 8 h. Equal volumes of 20% aqueous TFA were added to each sample to stop the reaction and peptide degradation was checked by RP-HPLC. Runs were performed on a Zorbax 300 SB-C8 column (150 × 2.1 mm I.D.; 5 µm particle size, 300 Å pore size) from Agilent Technologies at room temperature using a linear AB gradient (1% acetonitrile/min) and a flow rate of 0.25 mL/min, where eluent A was 0.2% aqueous TFA, pH 2 and eluent B was 0.2% TFA in acetonitrile. The change in integrated peak area of the peptides was used to monitor the degree of proteolysis during the time study.
Results
Peptide design
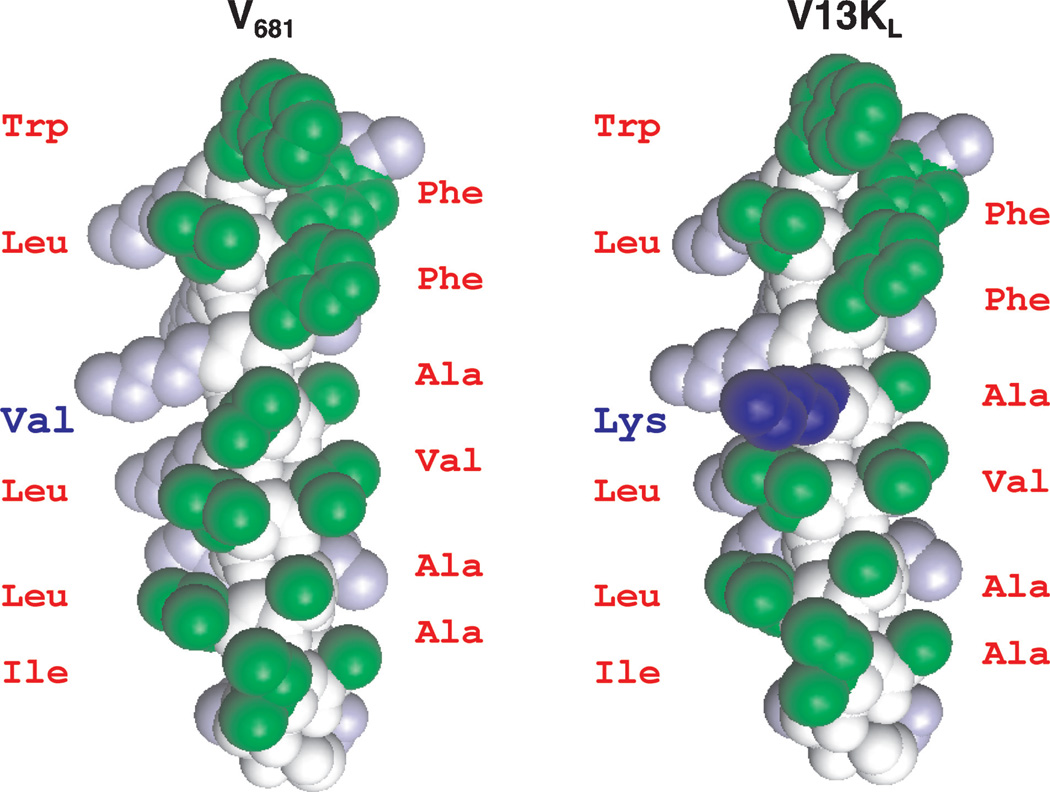
Peptide V681 is a 26-residue α-helical amphipathic antimicrobial peptide (Figure 1 and Table 1) with excellent antimicrobial activity but high toxicity to human cells. Peptides V13KL and V13AD are peptides with l-Lys and d-Ala substitutions at position 13 of V681 on the non-polar face of the helix (Table 1) and were chosen as lead peptide analogs for the present study because they demonstrated the best overall therapeutic indices against gram-negative and gram-positive bacteria, and the lowest toxicity against eukaryotic cells from a previous study (20). In the present study, we prepared the enantiomeric peptides of V681 and analogs V13KL and V13AD. Peptides V681 and V13KL contain all-l-amino acids and d-V681 and d-V13KD contain all-d-amino acids. In the case of V13AD and d-V13AL, position 13 is d-alanine and l-alanine, respectively (Table 1). Thus, d-V681, d-V13KD, and d-V13AL are opposite in stereochemistry to the corresponding l-peptides, V681, V13KL and V13AD, respectively. A control peptide C designed to exhibit negligible secondary structure, i.e. a random coil, was employed as a standard peptide for temperature profiling during RP-HPLC to monitor peptide association (Table 1) (20,28,29).
Figure 1. Space-filling model of peptides V681 and V13KL.
Hydrophobic amino acids on the non-polar face of the helix are green; hydrophilic amino acids on the polar face of the helix are gray; peptide backbone is colored white. The Lys substitution at position 13 (V13KL) on the non-polar face of the helix is blue. The models were created by the pymol v0.98. The peptide sequences are shown in Table 1.
Table 1.
Peptides used in this study
 |
d- denotes that all amino acids in the peptide are d-amino acids except position 13, which is denoted as l or d depending on the peptide.
Peptide sequences are shown using the one-letter code for amino acid residues; subscripts of l or d denote the l-amino acid and d-amino acid, respectively; Ac- denotes Nα-acetyl and -amide denotes C-terminal amide; in sequences, bold letters denote the amino acid substitutions at position 13 of the non-polar face of V681 or d-V681.
Peptide C is a random coil peptide used as a control in reversed-phase high-performance liquid chromatography (RP-HPLC) temperature profiling.
Secondary structure of peptides
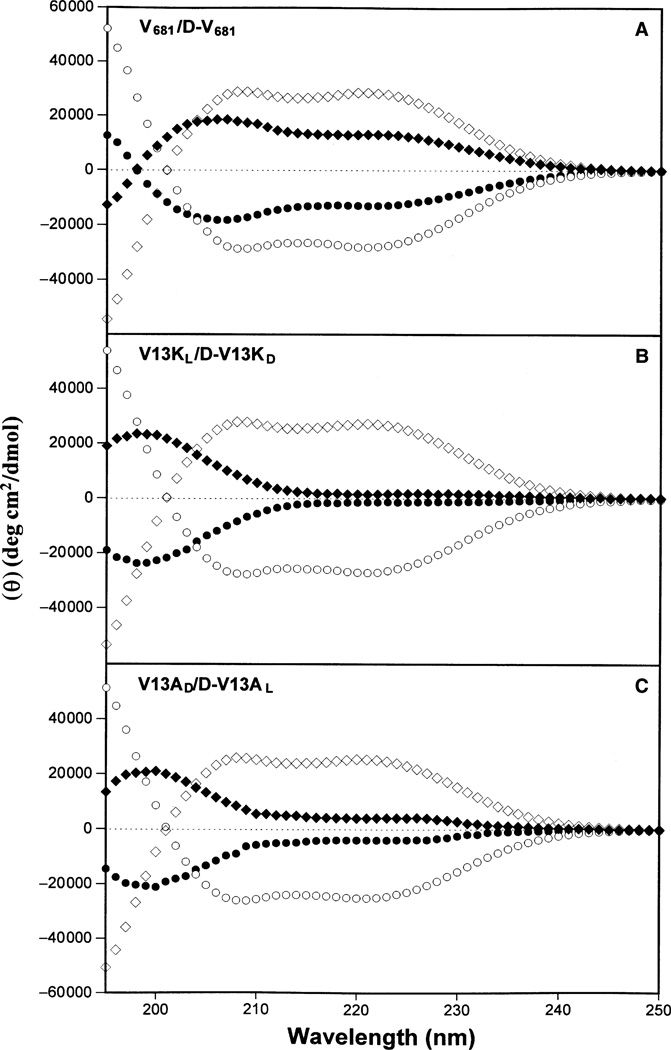
To determine the secondary structure of peptides in different environments, CD spectra of the peptide analogs were measured under benign conditions (100 mm KCl, 50 mm KH2PO4/K2HPO4, pH 7.4, referred to as KP buffer) and also in 50% TFE to mimic the hydrophobic environment of the membrane. As illustrated in Figure 2, the parent peptide, V681, was only partially helical in KP buffer; peptides V13KL and V13AD exhibited negligible secondary structure in KP buffer due to disruption of the non-polar face of the helix by introducing a hydrophilic l-lysine residue into peptide V13KL or a helix-disruptive d-alanine residue into peptide V13AD. However, in the presence of 50% TFE, all three l-peptides were fully folded α-helical structures with similar ellipticities and helicity (Table 2). As expected, the d-peptides showed spectra that were exact mirror images compared to their l-enantiomers, with ellipticities equivalent but of opposite sign both in benign KP buffer and in 50% TFE (Table 2).
Figure 2. Circular dichroism (CD) spectra of peptides V681 and d-V681 (panel A), V13KL and d-V13KD (panel B) and peptides V13AD and d-V13AL (panel C) at pH 7.4 and 5 °C, in 50 mm aq. potassium phosphate buffer (KP buffer) containing 100 mm KCl.
In panels (A–C), solid symbols represent the CD spectra of peptide analogs in KP buffer without trifluoroethanol (TFE), whilst open symbols represent CD spectra obtained in the presence of 50% TFE; the symbols used are: circles for l-peptides V681, V13KL, and V13AD and diamonds for d-peptides d-V681, d-V13KD, and d-V13AL.
Table 2.
Biophysical data of peptide analogs
| Hydrophobicityb | Benign | 50% TFE | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Peptidea | [θ]222c | % helixd | [θ]222c | % helixd | PAe | ||||
| V681 | 96.6 | 88.8 | −13 000 | 46 | −27 950 | 99 | 7.2 | ||
| d-V681 | 96.6 | 88.8 | 13 050 | 46 | 28 100 | 100 | 7.2 | ||
| V13AD | 81.2 | 73.2 | −4000 | 14 | −25 000 | 89 | 4.1 | ||
| d-V13AL | 81.2 | 73.2 | 4000 | 14 | 25 050 | 89 | 4.1 | ||
| V13KL | 74.9 | 64.7 | −1400 | 5 | −27 000 | 96 | 2.1 | ||
| d-V13KD | 74.9 | 64.7 | 1500 | 5 | 26 950 | 96 | 2.1 | ||
Peptide sequences are shown in Table 1.
Peptides are ordered by decreasing retention time (tR) in RP-HPLC at pH 2 at temperatures of 5 and 80 °C which is a measure of overall hydrophobicity.
The mean residue molar ellipticities [θ]222 (deg/cm2/dmol) at wavelength 222 nm were measured at 5 °C in benign conditions (100 mm KCl, 50 mm PO4, pH 7.4) or in benign buffer containing 50% TFE by circular dichroism spectroscopy. The negative values in molar ellipticity denote the left-handed helices and the positive values denote the right-handed helices.
The helical content (in percentage) of a peptide relative to the molar ellipticity value of the peptide d-V681 in the presence of 50% TFE.
PA denotes the dimerization parameter of each peptide during the RP-HPLC temperature profiling, which is the maximal retention time difference of [( for peptide analogs) − ( for control peptide C)] within the temperature range, and is the retention time difference of a peptide at a specific temperature (t) compared with that at 5 °C.
TFE, trifluoroethanol; RP-HPLC, reversed-phase high-performance liquid chromatography.
Peptide self-association
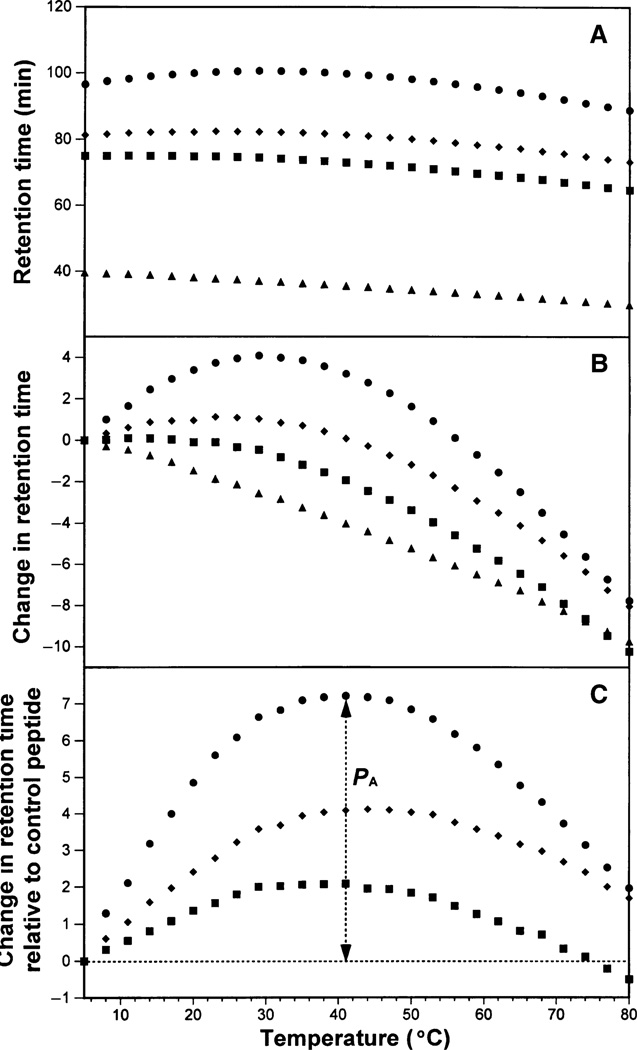
The ability of the peptides to self-associate was determined by RP-HPLC temperature profiling. Figure 3A shows the change in RP-HPLC retention times of the three pairs of enantiomers and the control peptide C over a temperature range of 5–80 °C. As expected, l- and d-peptide enantiomers were totally inseparable over this temperature range, because each pair of peptides is identical in sequence and must adopt identical conformations on interacting with the RP matrix, whether in an all-l- or all-d-conformation. RP-HPLC retention behavior has been frequently utilized to represent overall peptide hydrophobicity (20,26,30,31). In the present study, the hydrophobicity of the three peptide pairs is in the order V681/d-V681 > V13AD/d-V13AL > V13KL/d-V13KD (Table 2), which agrees with the change in hydrophobicity of the substitutions at position 13 in order of the most hydrophobic to the least hydrophobic amino acid residue (Val in V681 > Ala in V13A > Lys in V13K) (32). Figure 3B shows the retention behavior of the peptides after normalization of the retention times to the corresponding retention time at 5 °C in order to highlight differences in the elution behavior of peptides as the temperature is increased from 5 to 80 °C. For example, the retention times of peptides V681/d-V681 increase with increasing temperature (up to approximately 30 °C) followed by a retention time decrease with a further temperature increase. Such a temperature profile is characteristic of a peptide exhibiting self-association (20,28,29,33). As illustrated in Figure 3C, the peptide self-association parameter, PA, represents the maximum change in peptide retention time relative to the random coil peptide C. As peptide C is a monomeric random coil peptide in both aqueous and hydrophobic media, its retention behavior over the temperature ranging from 5 to 80 °C represents only general temperature effects on peptide retention behavior, i.e. a linear decrease in peptide retention time with increasing temperature because of greater solute diffusivity and enhanced mass transfer between the stationary and mobile phases at higher temperatures (34). Thus, after normalization to the retention times of peptide C, the retention behavior of the peptides represents only peptide self-association ability. Note that the higher the PA value, the greater the self-association ability. The order of peptide self-association ability of the three pairs of peptide enantiomers is identical to the order of peptide hydrophobicity, i.e. V681/d-V681 have the highest oligomerization ability (e.g. dimerization) in solution among the three pairs of peptide enantiomers (PA = 7.2, Table 2); in contrast, V13AD/d-V13AL showed a weaker ability to self-associate when compared with V681/d-V681 (PA = 4.1, Table 2); V13KL/d-V13KD exhibited the lowest self-association (PA = 2.1, Table 2). As shown in Table 2, it is also clear that the peptide retention times at 80 °C are dramatically lower than those at 5 °C. Apart from the decrease in retention time because of the general temperature effects noted above, unraveling of the α-helix will also occur with increasing temperature, resulting in the loss of the non-polar face of the amphipathic α-helical peptides and, hence, reduced retention times as the peptides become increasingly random coils.
Figure 3. Peptide retention behavior during reversed-phase high-performance liquid chromatography (RP-HPLC) with increasing temperature.
Column and conditions: RP-HPLC, SB-C8 column (150 × 2.1 mm I.D.; 5 µm particle size, 300 Å pore size), linear A–B gradient (0.5% acetonitrile/min) at a flow rate of 0.35 mL/min, where eluent A is 0.2% aqueous trifluoroacetic acid (TFA) and eluent B is 0.2% TFA in acetonitrile. The retention times of the peptides during the change of temperature are shown in panel (A). In panel (B), the retention time of peptides are normalized to 5 °C through the expression , where is the retention time at a specific temperature of an antimicrobial peptide or the random coil peptide, and is the retention time at 5 °C. In panel (C), the retention behavior of the peptides was normalized to that of the random coil peptide C through the expression ( for peptides) minus ( for C). In panels (A–C), the symbols used are: circles for V681 and d-V681, diamonds for V13AD and d-V13AL, squares for V13KL, d-V13KD and triangles for random coil peptide C.
Hemolytic activity
The hemolytic activity of the peptides against human erythrocytes was determined as the maximal peptide concentration that produces no hemolysis after 18 h of incubation at 37 °C. The parent peptide, V681, exhibited the strongest hemolytic activity with a value of 7.8 µg/mL, compared with peptides V13KL and V13AD (250.0 µg/mL and 31.3 µg/mL, respectively; Table 3). The activity of the d-enantiomers was quantitatively equivalent to that of l-enantiomers.
Table 3.
Hemolytic activities of peptides and MIC against Pseudomonas aeruginosa strains
| Hemolytic activity | Antimicrobial activity | Therapeutic index | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (µg/mL)c | ||||||||||||
| Peptide | MHC (µg/mL)a | Foldb | PAO 1 | WR 5 | PAK | PA 14 | M 2 | CP 204 | GMd | Folde | MHC/MICf | Foldg |
| V681 | 7.8 | 1 | 15.6 | 62.5 | 31.3 | 62.5 | 15.6 | 15.6 | 27.8 | 1.0 | 0.3 | 1.0 |
| d-V681 | 7.8 | 1 | 15.6 | 31.3 | 31.3 | 15.6 | 31.3 | 31.3 | 24.8 | 1.1 | 0.3 | 1.0 |
| V13AD | 31.3 | 4 | 15.6 | 62.5 | 62.5 | 31.3 | 7.8 | 7.8 | 22.1 | 1.3 | 1.4 | 4.7 |
| d-V13AL | 31.3 | 4 | 7.8 | 31.3 | 31.3 | 7.8 | 15.6 | 7.8 | 13.9 | 2.0 | 2.2 | 7.3 |
| V13KL | 250.0 | 32 | 31.3 | 250.0 | 125.0 | 62.5 | 31.3 | 7.8 | 49.6 | 0.6 | 5.0 | 16.7 |
| d-V13KD | 250.0h | 32 | 15.6 | 62.5 | 125.0 | 15.6 | 31.3 | 15.6 | 31.2 | 0.9 | 8.0 | 26.7 (53.4)h |
| Ciprofloxacini | 4 | 1 | 4 | 8 | 8 | 64 | ||||||
MHC denotes the maximal peptide concentration that produces no hemolysis after 18 h in the standard microtiter dilution method.
Denotes the fold decrease in hemolytic activity compared with the parent peptide V681.
MIC is minimal inhibitory concentration that inhibited growth of six P. aeruginosa strains. MIC is given based on three sets or more of determinations.
Denotes the geometric mean of a diverse group of P. aeruginosa clinical isolates.
Denotes the fold improvement in antimicrobial activity (geometric mean data) compared with the parent peptide V681.
Therapeutic index is the ratio of the MHC value (µg/mL) over the geometric mean MIC value (µg/mL). Large values indicate greater antimicrobial specificity.
Denotes the fold improvement in therapeutic index compared with the parent peptide V681.
In the hemolysis time study, d-V13KD showed no hemolysis at 500 µg/mL for 8 h, which would increase the fold improvement to 53.4-fold in the therapeutic index.
Relative susceptibility of these Pseudomonas strains to ciprofloxacin with the number 1 denoting the most susceptible strain.
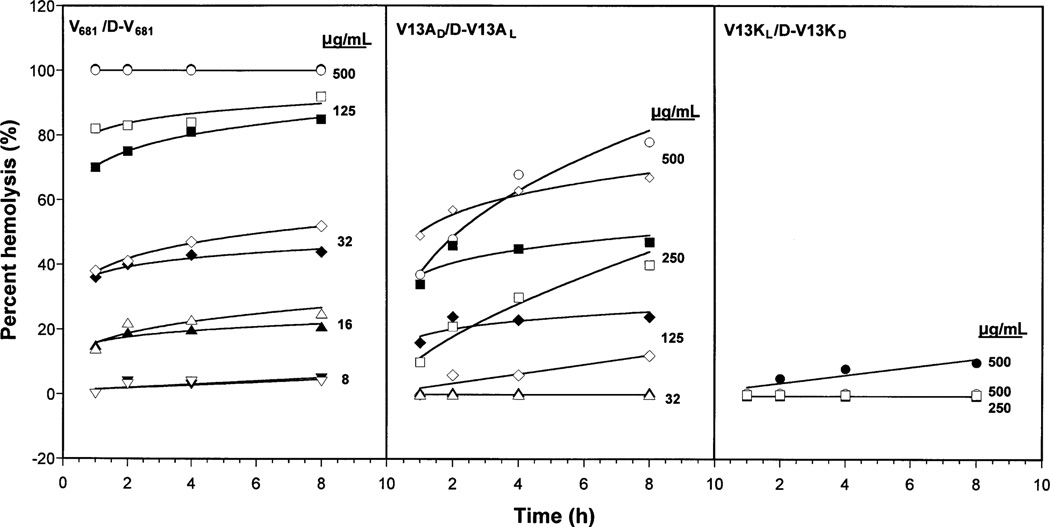
In our previous study (20), the hemolytic activities of peptides V681, V13AD and V13KL were determined as 15.6 µg/mL, 250.0 µg/mL and >250.0 µg/mL, respectively, following incubation for 12 h at 37 °C instead of the 18 h incubation of the present study. Hence, in order to explore the relationship between hemolysis and incubation time, a hemolysis time study was carried out to investigate the extent of hemolysis during different periods of incubation and different peptide concentrations. As illustrated in Figure 4, a hemolysis time study was carried out over 8 h at peptide concentrations of 8, 16, 32, 64, 125, 250 and 500 µg/mL. The percentage hemolysis of cells was determined spectrophotometrically by comparison with the complete hemolysis of cells in water. As illustrated in Figure 4, peptides V681, d-V681, V13AD, and d-V13AL exhibited an increase in erythrocyte hemolysis with increasing incubation time at most peptide concentrations. Significantly, peptides V13KL and d-V13KD showed negligible (<10%) or no hemolysis against human red blood cells after 8 h even at the extremely high peptide concentration of 500 µg/mL. As illustrated in Figure 4, it is clear that the enantiomeric peptide pair V681/d-V681 exhibit cell lysis to a significantly greater extent than the V13AD/d-V13AL peptide pair. Thus, 100% hemolysis was already apparent after just 1 h incubation for V681 and d-V681 at a peptide concentration of 500 µg/mL, whilst this value was approximately 40% for peptides V13AD and d-V13AL. In addition, while a small increase in hemolysis was apparent for V681 and d-V681 at a peptide concentration of just 8 µg/mL, negligible hemolysis was seen for V13AD and d-V13AL at a peptide concentration of 32 µg/mL. It is interesting to note that for our lead compounds, peptides V13AD/d-V13AL and V13KL/d-V13KD, d-enantiomers generally exhibited slightly weaker hemolytic activity compared with l-enantiomers at most peptide concentrations, the only exception being V13AD and d-V13AL at 500 µg/mL, where the activity of the l-peptide is greater at <4 h incubation but weaker from 4 h onwards.
Figure 4. Time study of peptide hemolysis of human red blood cells.
Closed symbols were used for l-peptides and open symbols were used for d-peptides. Different symbols were used to represent the different concentrations of peptides during the hemolysis study.
Peptide antimicrobial activity and therapeutic index against Pseudomonas aeruginosa strains
Pseudomonas aeruginosa strains used in this study are a diverse group of clinical isolates from different places in the world. Antibiotic susceptibility tests show that these P. aeruginosa strains share similar susceptibility to most antibiotics except that there is about a 64-fold difference for the range of ciprofloxacin susceptibility (Table 3). Antimicrobial activities of peptide enantiomers against P. aeruginosa strains are shown in Table 3. In general, the antimicrobial activity of l- and d-enantiomers against P. aeruginosa varied within fourfold. CP204, a P. aeruginosa strain from cystic fibrosis patients, is apparently the most susceptible strain to these peptides with MIC values from 7.8 to 31.3 µg/mL. The geometric mean MIC values from six P. aeruginosa strains were calculated to provide an overall evaluation of antimicrobial activity of these peptides against P. aeruginosa. Fold improvement was calculated by comparing the geometric mean MIC value of each peptide to that of the parent peptide V681. It is clear that, in most cases, all-d-peptides exhibited slightly better antimicrobial activity than their l-enantiomers.
The therapeutic index is a widely accepted parameter to represent the specificity of antimicrobial reagents. It is calculated by the ratio of MHC (hemolytic activity) and MIC (antimicrobial activity); thus, larger values of therapeutic index indicate greater antimicrobial specificity. The therapeutic indices of the peptides against P. aeruginosa are shown in Table 3. By replacing l-valine with d-alanine or l-lysine, we significantly increased the therapeutic index against P. aeruginosa strains by 4.7- (peptide V13AD) and 16.7-fold (peptide V13KL). In addition, by making enantiomeric d-peptides, we further improved the therapeutic index against P. aeruginosa by 7.3- (peptide d-V13AL) and 26.7-fold (peptide d-V13KD) as measured by the standard microtiter dilution method. Peptide d-V13KD showed no hemolysis at 500 µg/mL after 8 h in our time study (Figure 4) which is a more stringent test of hemolytic activity. The hemolytic activities of V681 were not different when determined by the standard microtiter dilution method at 18 h or by the time study at 8 h using a peptide concentration of 500 µg/mL (Table 3 and Figure 4). Thus, using a peptide concentration of 500 µg/mL, the MHC value of d-V13KD increased the fold improvement of the therapeutic index compared with V681 to 53-fold. Our overall conclusion is that the key differences in biologic activities lie in the much more dramatic variations in hemolytic activity (MHC values) because the differences in antimicrobial activity between the six peptides are relatively minor (Table 3). Thus, the dramatic increases in therapeutic indices arise from the difference in hemolytic activity.
Peptide antimicrobial activity and therapeutic index against various gram-negative bacteria
Table 4 shows peptide antimicrobial activity and therapeutic indices against six different gram-negative bacterium strains. It is clear to see that, in general, all-l-peptides have equivalent antimicrobial activity against gram-negative bacteria compared with their d-enantiomers, because MIC values of l- and d-enantiomers are all within a twofold difference. In Table 4, geometric mean MIC was calculated to provide an overall view of antimicrobial activity of the peptides against gram-negative bacteria and was also used to calculate the therapeutic index. Peptides V13KL and d-V13KD both show a 40-fold improvement in therapeutic index against gram-negative bacteria compared with peptide V681 as measured by the standard microtiter dilution method. For this improvement in the therapeutic index of peptides against gram-negative bacteria, peptide hemolytic activity is again the key factor. Using our most stringent test of hemolytic activity, the MHC value of 500 µg/mL for peptide d-V13KD showed no hemolysis after 8 h in our time study (Figure 4) and the fold improvement of the therapeutic index compared with V681 was 80-fold.
Table 4.
Antimicrobial (MIC) and hemolytic (MHC) activities of peptide analogs against Gram-negative bacteria and human red blood cells
| MHCa (µg/mL) | MICb (µg/mL) | Therapeutic indexf | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptides | hRBC |
E. coli C498 (wt)c |
E. coli C500 (abs)c |
St C587 (wt)c |
St C610 (abs)c |
Pa H187 (wt)c |
Pa H188 (abs)c |
GMd | Folde | MHC/MIC | Foldg |
| V681 | 7.8 | 6.3 | 3.1 | 12.5 | 3.1 | 6.3 | 3.1 | 5.0 | 1.0 | 1.6 | 1.0 |
| d-V681 | 7.8 | 6.3 | 3.1 | 12.5 | 6.3 | 6.3 | 6.3 | 6.3 | 0.8 | 1.2 | 0.8 |
| V13AD | 31.3 | 3.1 | 1.6 | 12.5 | 3.1 | 3.1 | 1.6 | 3.1 | 1.6 | 10.1 | 6.3 |
| d-V13AL | 31.3 | 3.1 | 3.1 | 12.5 | 3.1 | 3.1 | 3.1 | 3.9 | 1.3 | 8.0 | 5.0 |
| V13KL | 250.0 | 3.1 | 3.1 | 6.3 | 3.1 | 6.3 | 3.1 | 3.9 | 1.3 | 64.1 | 40.1 |
| d-V13Kc | 250.0h | 3.1 | 3.1 | 6.3 | 3.1 | 6.3 | 3.1 | 3.9 | 1.3 | 64.1 | 40.1 (80.2)h |
Hemolytic activity (maximal peptide concentration with no hemolysis after 18 h) was determined on human red blood cells (hRBC).
Antimicrobial activity (minimal inhibitory concentration) is from three sets of determinations.
E. coli, Escherichia coli; St, Salmonella typhimurium; Pa, Pseudomonas aeruginosa; wt, wild-type laboratory strain; abs, antibiotic-sensitive strain.
GM denotes the geometric mean of MIC values from all six microbial strains in this table.
Denotes fold improvement in the geometric mean of MIC compared with parent peptide V681.
Therapeutic index = MHC (µg/mL)/geometric mean of MIC (µg/mL). Larger values indicate greater antimicrobial specificity.
Denotes fold improvement in the therapeutic index compared with parent peptide V681.
In the hemolysis time study, d-V13KD showed no hemolysis at 500 µg/mL for 8 h which would increase the fold improvement in the therapeutic index to 80.2-fold.
Peptide antimicrobial activity and therapeutic index against various gram-positive bacteria and a fungus
The antimicrobial activity against gram-positive bacteria and a fungus of l- and d-enantiomeric peptides is compared in Table 5. It is clear that our peptides were effective in killing the gram-positive bacteria tested. The least sensitive gram-positive bacterium to the enantiomeric peptides was Enterococcus faecalis. The effectiveness of the peptides to lyse cells of the fungus Candida albicans was similar to E. faecalis. For gram-positive bacteria and the fungus, d-peptides again generally showed the same or better antimicrobial activity than l-enantiomers. Although the geometric mean MIC values of peptides V13KL/d-V13KD were among the two lowest, because of the high MHC values (poor or no hemolytic activity), the therapeutic indices of V13KL and d-V13KD were the highest against gram-positive bacteria and the fungus. Similar to that seen against gram-negative bacteria, the therapeutic index of peptide d-V13KD was increased 34.7- and 16.7-fold against gram-positive bacteria and C. albicans, respectively, because of the poor or no hemolytic activity as measured by the standard microtiter dilution method. Using our most stringent test of hemolytic activity, the MHC value of 500 µg/mL for peptide d-V13KD showed no hemolysis after 8 h in our time study (Figure 4) and the fold improvement of the therapeutic indices compared with V681 were 69- and 33-fold against gram-negative bacteria and fungus, respectively.
Table 5.
Antimicrobial (MIC) and hemolytic (MHC) activities of peptide analogs against Gram-positive bacteria, fungus and human red blood cells
| MHCa (µg/mL) | MICb (µg/mL) | Therapeutic indexe | MIC b (µg/mL) | Therapeutic indexf | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptides | hRBC |
S. aureus 622 (wt)c |
S. aureus 623 (methR)c |
S. epidermidis C621 (wt)c |
B. subtilis C971 (wt)c |
E. faecalis C625 (wt)c |
C. xerosis C875 (wt)c |
GMd | Foldd | MHC/MIC | Foldg | C. albicans | MHC/MIC | Foldg |
| V681 | 7.8 | 3.1 | 6.3 | 6.3 | 3.1 | 6.3 | 3.1 | 4.4 | 1.0 | 1.8 | 1.0 | 12.5 | 0.6 | 1.0 |
| d-V681 | 7.8 | 6.3 | 6.3 | 1.6 | 1.6 | 6.3 | 3.1 | 3.5 | 1.3 | 2.2 | 1.2 | 6.3 | 1.2 | 2.0 |
| V13AD | 31.3 | 1.6 | 3.1 | 1.6 | 3.1 | 12.5 | 1.6 | 2.8 | 1.6 | 11.2 | 6.2 | 12.5 | 2.5 | 4.2 |
| d-V13AL | 31.3 | 3.1 | 3.1 | 1.6 | 1.6 | 12.5 | 1.6 | 2.8 | 1.6 | 11.2 | 6.2 | 12.5 | 2.5 | 4.2 |
| V13KL | 250.0 | 12.5 | 12.5 | 6.3 | 3.1 | 50.0 | 1.6 | 7.9 | 0.6 | 31.6 | 17.6 | 50.0 | 5.0 | 8.3 |
| d-V13KD | 250.0h | 6.3 | 6.3 | 3.1 | 1.6 | 12.5 | 1.6 | 4.0 | 1.1 | 62.5 | 34.7 (69.4)h | 25.0 | 10.0 | 16.7 (33.4)h |
Hemolytic activity (maximal peptide concentration with no hemolysis after 18 h) was determined on human red blood cells (hRBC).
Antimicrobial and antifungal activities (minimal inhibitory concentration) are from three sets of determinations.
S. aureus, Staphylococcus aureus; S. epidermidis, Staphylococcus epidermidis; B. subtilis, Bacillus subtilis; E. faecalis, Enterococcus faecalis; C. xerosis, Corynebacterium xerosis; C. albicans, Candida albicans; wt, wild-type strain; methR, methicillin-resistant clinical strain.
GM denotes the geometric mean of MIC values from all six gram-positive microbial strains in this table. Fold denotes fold improvement in the geometric mean MIC compared with parent peptide V681.
Therapeutic index = MHC (µg/mL)/geometric mean MIC (µg/mL). Larger values indicate greater antimicrobial specificity.
Denotes the therapeutic index of the peptides against a fungus C. albicans.
Denotes fold improvement in the therapeutic index compared with parent peptide V681.
In the hemolysis time study, d-V13KD showed no hemolysis at 500 µg/mL for 8 h which would increase the fold improvement in the therapeutic index to 69.4- and 33.4-fold for gram-positive bacteria and C. albicans, respectively.
Proteolytic degradation study
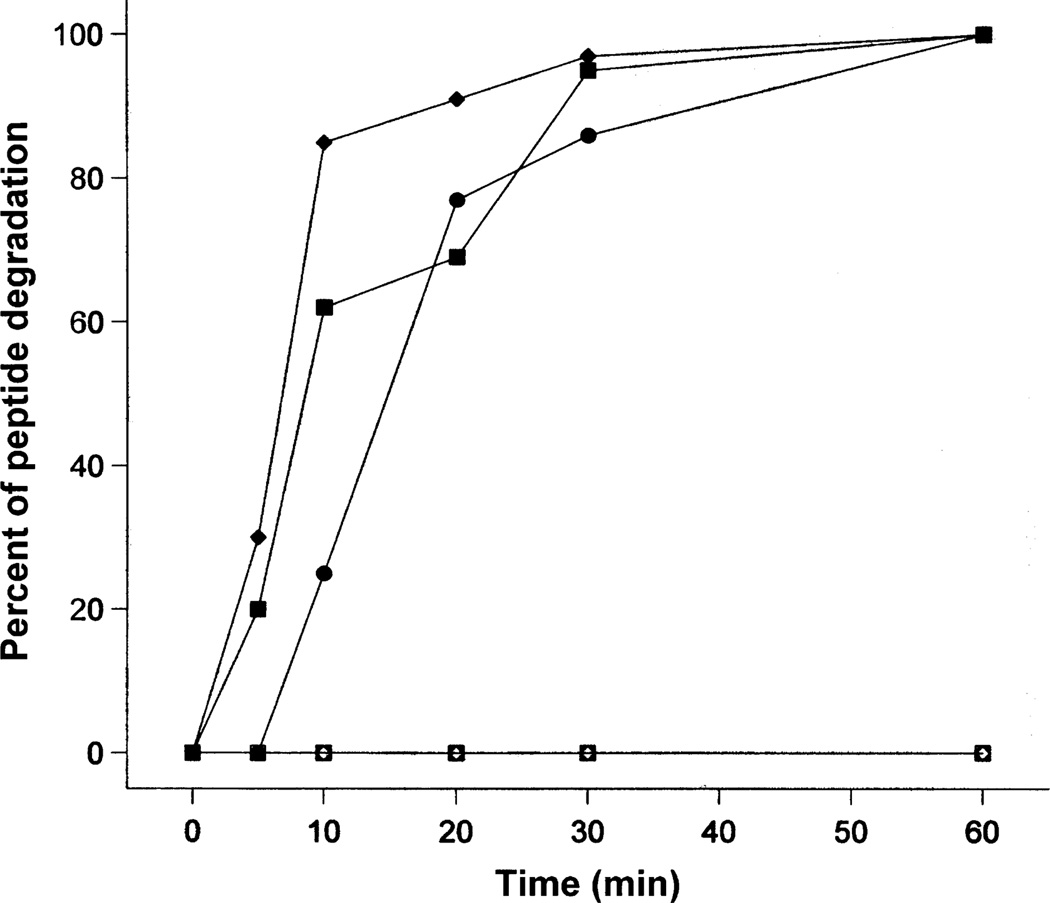
Figure 5 illustrates the stability of l- and d-enantiomeric peptides in the presence of trypsin at a molar ratio of 20 000:1 or 20 mm: 1 µm (peptide:trypsin) at 37 °C. The assay was carried out for 8 h; however, only the stability of the peptides in the first 60 min is illustrated in Figure 5, due to the reason that all three l-peptides were 100% digested by trypsin after 1 h of incubation at 37 °C. It is interesting to see that peptide V681 showed slightly more resistance against trypsin than V13KL and V13AD; in contrast, V13KL and V13AD generally showed a similar trend of degradation in the presence of trypsin. All three peptides were 86–97% digested after 30 min. In sharp contrast, d-peptides were completely stable (no degradation) to trypsin even at a 10-fold higher molar concentration ratio of trypsin versus peptide (2 mm peptide:1 µm trypsin) for 8 h (data not shown).
Figure 5. Peptide stability to proteolysis by trypsin.
Closed symbols were used for l-peptides and open symbols were used for d-peptides. Circles denote V681 and d-V681, squares denote V13KL and d-V13KD, and diamonds denote V13AD and d-V13AL.
Discussion
Enantiomers are non-superimposable mirror images of one another. When a peptide is composed of all-d-amino acids, it becomes the d-enantiomer of the all-l-peptide in the same sequence. In such cases, the original left-handed helix of the l-peptide will be present as a right-handed helix in its all-d-enantiomeric form, as demonstrated by the CD data illustrated in Figure 2. The l- and d-enantiomeric pairs of V681 exhibited <50% α-helical structure in benign buffer while the two analog pairs exhibited little or negligible α-helical structure in benign buffer; however, all three enantiomeric pairs were fully helical in the presence of 50% TFE. This characteristic ensures α-helical structure when the peptides interact with the biomembrane, as TFE has been frequently employed as a mimic of the hydrophobic environment characteristic of biomembranes (18,20). We believe having no α-helical structure in benign medium and inducible α-helical structure in the hydrophobic environment of the membrane is a critical aspect for antimicrobial activity.
In the present study, we have clearly shown that each enantiomeric peptide pair has the same activities against prokaryotic and eukaryotic cell membranes which suggest that the sole target for these antimicrobial peptides is the cell membrane. We previously proposed a model (20) to explain the peptide specificity between prokaryotic and eukaryotic membranes due to the different compositions of lipids between prokaryotic and eukaryotic membranes. Eukaryotic cell membranes, in contrast to prokaryotic membranes, are generally characterized by zwitterionic phospholipids, a relatively large amount of cholesterol and sphigomyelin, and the absence of the high, inside-negative transmembrane potential of prokaryotic membranes (35–38). This model proposes that hemolysis of eukaryotic cells requires the peptides to be inserted into the hydrophobic core of the membrane, perpendicular to the membrane surface, and interaction of the non-polar face of the amphipathic α-helix with the hydrophobic lipid core of the bilayer. The peptide may thus form transmembrane channels/pores and the hydrophilic surfaces point inward, producing an aqueous pore ('barrel-stave' mechanism) (39). In contrast, antimicrobial activity in prokaryotic cells, while maintaining specificity, requires the peptide to lie at the membrane interface parallel with the membrane surface and interaction of the non-polar face of the amphipathic α-helix with the hydrophobic component of the lipid and interaction of the positively charged residues with the negatively charged head groups of the phospholipid ('carpet' mechanism) (40,41). What dictates the two different modes of interaction is the difference in lipid composition of prokaryotic and eukaryotic membranes. We refer to this mode of interaction of antimicrobial peptides which combines the above two mechanisms as a 'membrane discrimination mechanism'.
Using this model, it is easy to understand why peptide V13KL and d-V13KD of the present study are non-hemolytic but at the same time possess excellent antimicrobial activity compared with the native sequence V681 (VL13) or d-V681 (VD13). Thus, the single substitution of Lys for Val at position 13 (V13KL and d-V13KD) in the center of the non-polar face disrupts the hydrophobic surface due to the presence of the positive charge, preventing the peptide from penetrating the bilayer as a transmembrane helix in eukaryotic cells. The peptide is then excluded from the bilayer and, hence, is non-hemolytic. In prokaryotic cells, the peptide is also excluded from penetrating the bilayer as a transmembrane helix but this is not required for excellent antimicrobial activity. Instead, the peptide can enter the interface region of the bilayer where disruption of the peptide hydrophobic surface by Lys can be tolerated and antimicrobial activity maintained. These are consistent with the results of previous studies of model membranes which demonstrated that pore formation mechanism ('barrel-stave' mechanism) was used by antimicrobial peptides in zwitterionic membranes whilst a detergent-like mechanism ('carpet' mechanism) for the peptides was shown when the peptides interact with negatively charged membranes (42,43). Thus, the position of the Lys residue in the center of the non-polar face becomes a critical factor in maintaining specificity between prokaryotic and eukaryotic membranes. This approach was previously used by our laboratory with cyclic β-sheet peptides (44–47).
The technique of RP-HPLC temperature profiling to monitor molecule self-association has been applied to several types of polypeptides, including cyclic β-sheet peptides (29), monomeric α-helices and α-helices that dimerize (20,28), including α-helices that dimerize to form coiled-coils (33). We believe that peptide self-association (i.e. the ability to dimerize) in aqueous solution is a very important parameter to understand the mechanism of action of antimicrobial peptides. α-Helical antimicrobial peptides are amphipathic; thus, if the self-association ability of a peptide (forming dimers by interaction of the two non-polar faces of two molecules) is too strong in aqueous media, it could decrease the ability of the peptide monomers to dissociate, pass through the cell wall of micro-organisms and penetrate into the biomembranes to kill target cells. In the present study, there is a direct correlation of the ability of peptides to dimerize and specificity, i.e. disruption of dimerization generates specificity between eukaryotic and prokaryotic cells. As shown in Table 2, the PA values of peptides derived from their temperature profiling data (Figure 3) reflect the ability of the amphipathic α-helices to associate/dimerize. Clearly, V681 and d-V681, because of their uniform non-polar faces, show the greatest ability to associate in aqueous solution and lowest specificity or the strongest ability to lyse human erythrocytes. This result supports the view that a peptide with a fully accessible non-polar face tends to form pores/ channels in the membranes of eukaryotic cells. In the case of V13AD and d-V13AL, the introduction of d-Ala and l-Ala into all-l- and all-d-amino acid peptides, respectively, disrupts α-helical structure and, thus, lowers dimerization ability relative to V681 and d-V681 and improves specificity. The introduction of Lys into nonpolar position 13 of V13KL and d-V13KD lowers this association ability even further and further improves specificity. Thus, the lack of ability of a peptide to oligomerize or dimerize, as exemplified by its PA value, is an excellent measure of the peptide's ability to be nonhemolytic concomitant with maintenance of sufficient hydrophobicity of the non-polar face to ensure antimicrobial activity. It is important to note that, in the present study, d-peptides exhibited the same self-association ability as their l-enantiomers; thus, similar biologic activities can be expected. This is shown by the fact that hemolytic activity and antimicrobial activity of d-peptides against human red blood cells and microbial cells, respectively, were quantitatively equivalent to that of the l-enantiomers, further demonstrating that there is no chiral selectivity by the membrane or other stereoselective interactions in the cytoplasm during hemolytic and antimicrobial activities.
Because of the dramatic different results on hemolytic activity of peptide V13AD in the previous (20) and the present study (250 µg/ mL after 12 h versus 31.3 µg/mL after 18 h, respectively), using the standard microtiter dilution method (see Materials and Methods), it became apparent that an investigation of the relationship between hemolysis and time was required. It is noteworthy that there is no universal protocol of determination of hemolytic activity, which makes it difficult to compare data from different sources. For example, some researchers use 4 h of incubation and take the minimal concentration of peptide to give 100% hemolysis as peptide hemolytic activity (21,48); in contrast, some use 12 h or longer (e.g. 18 h in this study) of incubation and take the maximal concentration of peptide to give no hemolysis as peptide hemolytic activity [(12,20) and the present study]. Hence, the hemolysis time study is important to understand the process of erythrocyte lysis. It is clear that the degree of cell lysis is correlated with time, which may be the main reason for the different values of hemolytic activity of V13AD in the two studies. Hence, we have established a stringent criterion for toxicity, which is no hemolysis at a peptide concentration of 500 µg/mL after 8 h. We believe that this time study at this very high peptide concentration gives a much more accurate evaluation of hemolytic activity and this method should be established as the gold standard test.
As mentioned before, P. aeruginosa is a family of notorious gram-negative bacterial strains which are resistant to most current antibiotics; thus, it is one of the most severe threats to human health (23–25). Only a few antibiotics are effective against Pseudomonas, including fluoroquinolones (49), gentamicin (50), and imipenem (51), and even these antibiotics are not effective against all strains. In this study, it is encouraging to see that our lead peptides V13AD and V13KL are effective against a diverse group of P. aeruginosa clinical isolates. Peptide d-V13AL exhibited the highest antimicrobial activity against P. aeruginosa strains; in contrast, dV13KD has the best overall therapeutic index due to its lack of hemolytic activity. It has to be pointed out here that, in this study, MIC values for P. aeruginosa and other gram-negative and gram-positive bacteria were determined in two different collaborating laboratories; in addition to different media used, the inoculum numbers of cells were also different (see details in the Materials and Methods), which may cause some variations of MIC values of P. aeruginosa strains when comparing the results reported in Tables 3 and 4.
It is important to note that, in general, there is no significant difference in peptide antimicrobial activities against P. aeruginosa strains, other gram-negative and gram-positive bacteria and a fungus between l- and d-enantiomeric peptides, or among peptides with different amino acid substitutions, i.e. V681, V13AD, and V13KL (Tables 3–5). This observation provides understanding of the mechanism of action of α-helical antimicrobial enantiomeric peptides as follows: there is a dramatic difference in peptide hydrophobicity at position 13 between Val and Lys. The Lys disrupts the continuous non-polar surface because of the positive charge and causes the peptide to locate in the interface region of the microbial membrane. This supports the view that the 'carpet' mechanism is essential for strong antimicrobial activity, i.e. for both l- and d-peptide enantiomers, the peptides kill bacteria by a detergent-like mechanism, without penetrating deeply into the hydrophobic core of membrane.
Based on the peptide degradation study, all-d-peptides were totally resistant to enzymatic digestion; hence, this may explain the slightly higher antimicrobial activity of d-peptides than that of their l-enantiomers against P. aeruginosa and gram-positive bacteria. The relatively high susceptibility of l-peptides to trypsin is no doubt due to the presence of multiple lysine residues in sequences, i.e. six lysines for V681 and V13AD, seven lysines for V13KL, resulting in the fast degradation of the l-peptides in 30 min even at a molar ratio of 20 000:1 (peptide:trypsin).
In conclusion, by comparing the biophysical and biologic properties of l- and d-enantiomeric peptides, we report here that l- and d-enantiomeric peptide pairs behaved the same in self-association ability in solution, had the same hemolytic activity against human red blood cells, and exhibited similar antimicrobial activity against P. aeruginosa strains, and other gram-negative and gram-positive bacteria and a fungus. No chiral selectivity was found in the antimicrobial and hemolytic activities of the peptides. In this study, our membrane discrimination mechanism has proved to be a good explanation of the mechanism of action for both l- and d-enantiomeric peptides. It is important to note that peptide d-V13KD has demonstrated dramatic improvements in therapeutic indices compared with the parent peptide V681 by 53-fold against P. aeruginosa strains, 80-fold against gram-negative bacteria, 69-fold against gram-positive bacteria, and 33-fold against C. albicans. The proteolytic stability of d-V13KD, its broad spectrum activity and lack of hemolytic activity demonstrate its clinical potential as a new therapeutic.
Acknowledgments
This study was supported by NIH grant RO1GM61855, RO1AI48717, the John Stewart Chair in Peptide Chemistry, and the University of Colorado Health Sciences Center (R.S.H.) and was in part supported by a grant from NIAID (AI15940) to M.L.V.
References
- 1.Travis J. Reviving the antibiotic miracle? Science. 1994;264:360–362. doi: 10.1126/science.8153615. [DOI] [PubMed] [Google Scholar]
- 2.Neu HC. The crisis in antibiotic resistance. Science. 1992;257:1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE. Peptide antibiotics. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 4.Andreu D, Rivas L. Animal antimicrobial peptides: an overview. Biopolymers. 1998;47:415–433. doi: 10.1002/(SICI)1097-0282(1998)47:6<415::AID-BIP2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 5.Sitaram N, Nagaraj R. Host-defense antimicrobial peptides: importance of structure for activity. Curr Pharm Des. 2002;8:727–742. doi: 10.2174/1381612023395358. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Parente J, Harris SM, Woods DE, Hancock RE, Falla TJ. Antimicrobial peptide therapeutics for cystic fibrosis. Antimicrob Agents Chemother. 2005;49:2921–2927. doi: 10.1128/AAC.49.7.2921-2927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy KV, Yedery RD, Aranha C. Antimicrobial peptides: premises and promises. Int J Antimicrob Agents. 2004;24:536–547. doi: 10.1016/j.ijantimicag.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 9.Toke O. Antimicrobial peptides: new candidates in the fight against bacterial infections. Biopolymers. 2005;80:717–735. doi: 10.1002/bip.20286. [DOI] [PubMed] [Google Scholar]
- 10.Hancock RE, Lehrer R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998;16:82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 11.Duclohier H, Molle G, Spach G. Antimicrobial peptide magainin I from xenopus skin forms anion-permeable channels in planar lipid bilayers. Biophys J. 1989;56:1017–1021. doi: 10.1016/S0006-3495(89)82746-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wade D, Boman A, Wahlin B, Drain CM, Andreu D, Boman HG, Merrifield RB. All-D amino acid-containing channel-forming antibiotic peptides. Proc Natl Acad Sci U S A. 1990;87:4761–4765. doi: 10.1073/pnas.87.12.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bland JM, De Lucca AJ, Jacks TJ, Vigo CB. All-D-cecropin B: synthesis, conformation, lipopolysaccharide binding, and antibacterial activity. Mol Cell Biochem. 2001;218:105–111. doi: 10.1023/a:1007293816634. [DOI] [PubMed] [Google Scholar]
- 14.De Lucca AJ, Bland JM, Vigo CB, Jacks TJ, Peter J, Walsh TJ. D-cecropin B: proteolytic resistance, lethality for pathogenic fungi and binding properties. Med Mycol. 2000;38:301–308. doi: 10.1080/mmy.38.4.301.308. [DOI] [PubMed] [Google Scholar]
- 15.Cribbs DH, Pike CJ, Weinstein SL, Velazquez P, Cotman CW. All-D-enantiomers of beta-amyloid exhibit similar biological properties to all-L-beta-a-myloids. J Biol Chem. 1997;272:7431–7436. doi: 10.1074/jbc.272.11.7431. [DOI] [PubMed] [Google Scholar]
- 16.Hamamoto K, Kida Y, Zhang Y, Shimizu T, Kuwano K. Antimicrobial activity and stability to proteolysis of small linear cationic peptides with D-amino acid substitutions. Microbiol Immunol. 2002;46:741–749. doi: 10.1111/j.1348-0421.2002.tb02759.x. [DOI] [PubMed] [Google Scholar]
- 17.Elmquist A, Langel U. In vitro uptake and stability study of pVEC and its all-D analog. Biol Chem. 2003;384:387–393. doi: 10.1515/BC.2003.044. [DOI] [PubMed] [Google Scholar]
- 18.Hong SY, Oh JE, Lee KH. Effect of D-amino acid substitution on the stability, the secondary structure, and the activity of membrane-active peptide. Biochem Pharmacol. 1999;58:1775–1780. doi: 10.1016/s0006-2952(99)00259-2. [DOI] [PubMed] [Google Scholar]
- 19.Wakabayashi H, Matsumoto H, Hashimoto K, Teraguchi S, Takase M, Hayasawa H. N-acylated and D enantiomer derivatives of a nonamer core peptide of lactoferricin B showing improved antimicrobial activity. Antimicrob Agents Chemother. 1999;43:1267–1269. doi: 10.1128/aac.43.5.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Mant CT, Farmer SW, Hancock RE, Vasil ML, Hodges RS. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J Biol Chem. 2005;280:12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Benz R, Hancock RE. Influence of proline residues on the antibacterial and synergistic activities of alpha-helical peptides. Biochemistry. 1999;38:8102–8111. doi: 10.1021/bi9904104. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Falla T, Wu M, Fidai S, Burian J, Kay W, Hancock RE. Determinants of recombinant production of antimicrobial cationic peptides and creation of peptide variants in bacteria. Biochem Biophys Res Commun. 1998;247:674–680. doi: 10.1006/bbrc.1998.8848. [DOI] [PubMed] [Google Scholar]
- 23.Hoiby N, Koch C. Cystic fibrosis: 1. Pseudomonas aeruginosa infection in cystic fibrosis and its management. Thorax. 1990;45:881–884. doi: 10.1136/thx.45.11.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elkin S, Geddes D. Pseudomonal infection in cystic fibrosis: the battle continues. Expert Rev Anti Infect Ther. 2003;1:609–618. doi: 10.1586/14787210.1.4.609. [DOI] [PubMed] [Google Scholar]
- 25.Pierce GE. Pseudomonas aeruginosa, Candida albicans, and device-related nosocomial infections: implications, trends, and potential approaches for control. J Ind Microbiol Biotechnol. 2005;32:309–318. doi: 10.1007/s10295-005-0225-2. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Mant CT, Hodges RS. Determination of stereochemistry stability coefficients of amino acid side-chains in an amphipathic alpha-helix. J Pept Res. 2002;59:18–33. doi: 10.1046/j.1397-002x.2001.10994.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Mehok AR, Mant CT, Hodges RS. Optimum concentration of trifluoroacetic acid for reversed-phase liquid chromatography of peptides revisited. J Chromatogr A. 2004;1043:9–18. doi: 10.1016/j.chroma.2004.03.070. [DOI] [PubMed] [Google Scholar]
- 28.Mant CT, Chen Y, Hodges RS. Temperature profiling of polypeptides in reversed-phase liquid chromatography: I. Monitoring of dimerization and unfolding of amphipathic alpha-helical peptides. J Chromatogr A. 2003;1009:29–43. doi: 10.1016/s0021-9673(03)00621-6. [DOI] [PubMed] [Google Scholar]
- 29.Lee DL, Mant CT, Hodges RS. A novel method to measure self-association of small amphipathic molecules: temperature profiling in reversed-phase chromatography. J Biol Chem. 2003;278:22918–22927. doi: 10.1074/jbc.M301777200. [DOI] [PubMed] [Google Scholar]
- 30.Monera OD, Sereda TJ, Zhou NE, Kay CM, Hodges RS. Relationship of side-chain hydrophobicity and α-helical propensity on the stability of the single-stranded amphipathic α-helix. J Pept Sci. 1995;1:319–329. doi: 10.1002/psc.310010507. [DOI] [PubMed] [Google Scholar]
- 31.Guo D, Mant CT, Taneja AK, Parker JMR, Hodges RS. Prediction of peptide retention times in reversed-phase high-performance liquid chromatography: I. Determination of retention coefficients of amino acid residues using model synthetic peptides. J Chromatogr. 1986;359:499–518. [Google Scholar]
- 32.Kovacs JM, Mant CT, Hodges RS. Determination of intrinsic hydrophilicity/hydrophobicity of amino acid side-chains in peptides in the absence of nearest-neighbor or conformational effects. Biopolymers (Pept Sci) 2006 doi: 10.1002/bip.20417. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mant CT, Tripet B, Hodges RS. Temperature profiling of polypeptides in reversed-phase liquid chromatography: II. Monitoring of folding and stability of two-stranded alpha-helical coiled-coils. J Chromatogr A. 2003;1009:45–59. doi: 10.1016/s0021-9673(03)00919-1. [DOI] [PubMed] [Google Scholar]
- 34.Dolan JW. Temperature selectivity in reversed-phase high performance liquid chromatography. J Chromatogr A. 2002;965:195–205. doi: 10.1016/s0021-9673(01)01321-8. [DOI] [PubMed] [Google Scholar]
- 35.Lugtenberg B, Van Alphen L. Molecular architecture and functioning of the outer membrane of Escherichia coli and other gram-negative bacteria. Biochim Biophys Acta. 1983;737:51–115. doi: 10.1016/0304-4157(83)90014-x. [DOI] [PubMed] [Google Scholar]
- 36.Zilberstein D, Schuldiner S, Padan E. Proton electrochemical gradient in Escherichia coli cells and its relation to active transport of lactose. Biochemistry. 1979;18:669–673. doi: 10.1021/bi00571a018. [DOI] [PubMed] [Google Scholar]
- 37.Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 38.Devaux PF, Seigneuret M. Specificity of lipid-protein interactions as determined by spectroscopic techniques. Biochim Biophys Acta. 1985;822:63–125. doi: 10.1016/0304-4157(85)90004-8. [DOI] [PubMed] [Google Scholar]
- 39.Ehrenstein G, Lecar H. Electrically gated ionic channels in lipid bilayers. Q Rev Biophys. 1977;10:1–34. doi: 10.1017/s0033583500000123. [DOI] [PubMed] [Google Scholar]
- 40.Pouny Y, Rapaport D, Mor A, Nicolas P, Shai Y. Interaction of antimicrobial dermaseptin and its fluorescently labeled analogues with phospholipid membranes. Biochemistry. 1992;31:12416–12423. doi: 10.1021/bi00164a017. [DOI] [PubMed] [Google Scholar]
- 41.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 42.Papo N, Shai Y. Exploring peptide membrane interaction using surface plasmon resonance: differentiation between pore formation versus membrane disruption by lytic peptides. Biochemistry. 2003;42:458–466. doi: 10.1021/bi0267846. [DOI] [PubMed] [Google Scholar]
- 43.Ladokhin AS, White SH. 'Detergent-like' permeabilization of anionic lipid vesicles by melittin. Biochim Biophys Acta. 2001;1514:253–260. doi: 10.1016/s0005-2736(01)00382-0. [DOI] [PubMed] [Google Scholar]
- 44.McInnes C, Kondejewski LH, Hodges RS, Sykes BD. Development of the structural basis for antimicrobial and hemolytic activities of peptides based on gramicidin S and design of novel analogs using NMR spectroscopy. J Biol Chem. 2000;275:14287–14294. doi: 10.1074/jbc.275.19.14287. [DOI] [PubMed] [Google Scholar]
- 45.Kondejewski LH, Jelokhani-Niaraki M, Farmer SW, Lix B, Kay CM, Sykes BD, Hancock RE, Hodges RS. Dissociation of antimicrobial and hemolytic activities in cyclic peptide diastereomers by systematic alterations in amphipathicity. J Biol Chem. 1999;274:13181–13192. doi: 10.1074/jbc.274.19.13181. [DOI] [PubMed] [Google Scholar]
- 46.Lee DL, Hodges RS. Structure-activity relationships of de novo designed cyclic antimicrobial peptides based on gramicidin S. Biopolymers (Pept Sci) 2003;71:28–48. doi: 10.1002/bip.10374. [DOI] [PubMed] [Google Scholar]
- 47.Lee DL, Powers JP, Pflegerl K, Vasil ML, Hancock RE, Hodges RS. Effects of single D-amino acid substitutions on disruption of beta-sheet structure and hydrophobicity in cyclic 14-residue antimicrobial peptide analogs related to gramicidin S. J Pept Res. 2004;63:69–84. doi: 10.1046/j.1399-3011.2003.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powers JP, Rozek A, Hancock RE. Structure-activity relationships for the beta-hairpin cationic antimicrobial peptide polyphemusin I. Biochim Biophys Acta. 2004;1698:239–250. doi: 10.1016/j.bbapap.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 49.Obritsch MD, Fish DN, Maclaren R, Jung R. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy. 2005;25:1353–1364. doi: 10.1592/phco.2005.25.10.1353. [DOI] [PubMed] [Google Scholar]
- 50.Al-Bakri AG, Gilbert P, Allison DG. Influence of gentamicin and tobramycin on binary biofilm formation by co-cultures of Burkholderia cepacia and Pseudomonas aeruginosa. J Basic Microbiol. 2005;45:392–396. doi: 10.1002/jobm.200510011. [DOI] [PubMed] [Google Scholar]
- 51.Bodmann KF. Current guidelines for the treatment of severe pneumonia and sepsis. Chemotherapy. 2005;51:227–233. doi: 10.1159/000087452. [DOI] [PubMed] [Google Scholar]