Abstract
As development unfolds, DNA replication is not only coordinated with cell proliferation, but is regulated uniquely in specific cell types and organs. This differential regulation of DNA synthesis requires crosstalk between DNA replication and differentiation. This dynamic aspect of DNA replication is highlighted by the finding that the distribution of replication origins varies between differentiated cell types and changes with differentiation. Moreover, differential DNA replication in some cell types can lead to increases or decreases in gene copy number along chromosomes. This review highlights the recent advances and technologies that have provided us with new insights into the developmental regulation of DNA replication.
Keywords: Amplification, Drosophila, Endo cycle, Mammalian differentiation, ORC, Replication origins
Introduction
Every cell division cycle requires the faithful duplication of the entire genome. In multicellular organisms, the rate of cell division of cycling cells can vary greatly, ranging from minutes to hours, depending on the developmental state of a particular cell within the organism. Further compounding the challenges of genome duplication is the epigenetic state of the genome, which is constantly changing during development. Although these requirements are daunting, the DNA replication machinery is precisely regulated to ensure that genome duplication occurs within the proper time frame without compromising genome stability.
It has been known for decades that the properties of DNA replication are exquisitely sensitive to developmental states. For example, in early Drosophila and Xenopus embryogenesis, the duration of S phase is in the order of minutes, with replication origins spaced <10 kb apart (Blumenthal et al., 1974; Hyrien and Mechali, 1993). This is in stark contrast to fully differentiated cell types in which S phase can last longer than ten hours, with origins of replication >100 kb apart (Spradling and Orr-Weaver, 1987). The developmental signals and underlying molecular mechanisms that impact the parameters of DNA replication during development are poorly understood. Recent technological advances combined with genome-wide approaches have given a more precise view of the changes that development imparts on the DNA replication program, providing a foundation from which to unravel the regulatory signals.
Here, we briefly summarize the mechanism of initiation of DNA replication and detail recent discoveries showing that mutations in key replication initiation proteins cause developmental defects in humans. We then examine how differentiation affects DNA replication, both the sites of replication origins and the timing of when origins are activated during genomic DNA replication. Finally, we present recent studies on the mechanism by which DNA copy number is altered through differential DNA replication during development.
Assembly and activation of the pre-replicative complex
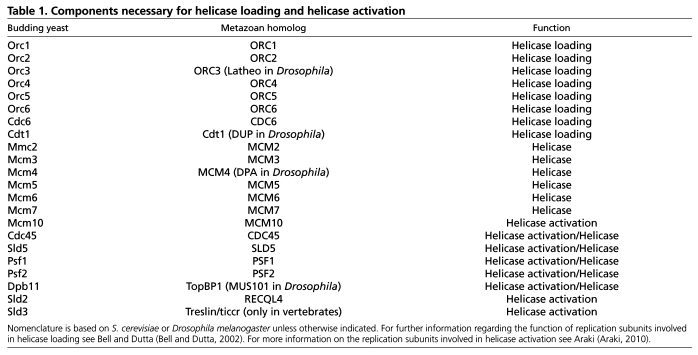
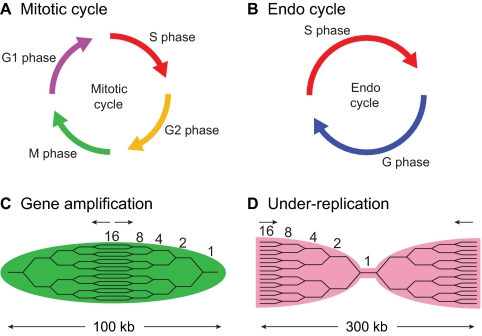
DNA replication initiates at cis-acting sites in the genome termed origins of replication (see Glossary, Box 1). For every round of DNA replication, thousands of replication origins are utilized in a coordinated manner. In metazoans, origins of replication are sequence-independent, highly influenced by chromatin structure and often change during the course of cell differentiation. The proteins involved in initiating DNA synthesis are conserved from yeast to humans (Table 1). The formation of replication complexes at origins of replication occurs in two phases: helicase loading or pre-replicative complex (pre-RC, see Glossary, Box 1) assembly; and helicase activation/replisome assembly. Helicase loading starts when the origin recognition complex (ORC, see Glossary, Box 1) binds to an origin of replication in late M and G1 phases of the cell cycle (Fig. 1A). ORC consists of six subunits (Orc1-Orc6), five of which are related to a large family of ATPases, the AAA+ ATPases (Bell and Dutta, 2002). Together with two additional replication factors, cell division cycle 6 (Cdc6) and cdc10-dependent transcript 1 (Cdt1), ORC loads a double hexamer of the minichromosome maintenance proteins 2-7 (Mcm2-Mcm7) onto DNA (Remus et al., 2009) (Fig. 1A).
Box 1. Glossary Amplicon. A defined region of increased gene copy number, either in a somatic cell or stably inherited in the germline.
Chorion. The eggshell surrounding an egg; composed of chorion proteins.
Endo cycle. A cell cycle in which cells undergo repeated S and G phases with no intervening mitoses.
Gene amplification. Increased gene copy number relative to overall ploidy.
Imaginal disc. A group of cells present in larvae that will form an adult organ such as eye, leg or wing; contains groups of determined diploid cells that divide during larval stages and differentiate into adult tissues during pupation.
Meirer-Grolin syndrome (MGS). A form of primordial dwarfism characterized by several developmental abnormalities, such as short stature, small ears and absent or underdeveloped patellae.
Origin of replication. Cis-acting DNA sequence where replication initiates. Origins of replication are sequence independent in metazoans.
Origin recognition complex (ORC). Six-subunit complex (Orc1-Orc6) that binds to origins of replication.
p19 cell line. A mouse pluripotent cell line derived from an embryonic carcinoma; can be induced to differentiate into multiple cell types.
Polyploidy. Having increased genomic content, i.e. more than two paired sets of chromosomes; occurs as a result of repeated S-G cycles with no intervening mitoses.
Polytene. Polyploid cells in which the sister chromatids are held in tight association.
Pre-replicative complex (pre-RC). Complex of Orc1-Orc6, Cdc6, Cdt1 and a double hexamer of the Mcm2-7 complex. The pre-RC refers to the loaded but inactive form of the replicative helicase.
Replication timing. The relative time during S phase when a specific genomic region is replicated.
Rothmund-Thomson syndrome (RTS). A rare autosomal recessive disorder characterized by several abnormalities including a facial rash, short stature, skeletal abnormalities, premature aging, chromosome fragility and a predisposition to tumor formation.
Under-replication. Reduced gene copy number relative to overall ploidy of a cell.
Table 1.
Components necessary for helicase loading and helicase activation
Fig. 1.
Helicase loading (pre-RC assembly) and activation. (A) Helicase loading (pre-RC assembly) at a potential origin of replication coinciding with a transcription start site (arrow). Although origins are sequence independent in metazoans, histone modifications are associated with transcriptional activation, and open/active chromatin correlates with ORC binding. Histone modifications associated with repressive chromatin are negatively associated with ORC binding. During helicase loading, binding of the ORC complex promotes recruitment of Cdc6, which in turn promotes the loading of a Cdt1–Mcm2-7 double hexamer. (B) Kinase activation. DDK promotes helicase activation by phosphorylating multiple Mcm subunits. S-CDK promotes helicase activation by phosphorylating Sld2 and Sld3, which in turn allow the recruitment of additional factors necessary for helicase activation. (C) Helicase activation is dependent on the recruitment of additional factors necessary to activate the replicative Mcm2-7 helicase, which is a complex of Mcm2-7, Cdc45 and GINS proteins (Sld5, Psf1, Psf2 and Psf3). Arrows indicate the direction of helicase movement.
Pre-RC complexes are ‘licensed’, or poised for replication, but are incapable of initiating DNA synthesis because the helicase is loaded in an inactive state. Activation of the helicase requires both S phase-specific cyclin-dependent and Dbf4-dependent kinases (S-CDK and DDK, respectively) (Fig. 1B). ORC binding and pre-RC formation occur in late M and G1 phases of the cell cycle when S-CDK activity is low. For helicase activation, S-CDK and DDK phosphorylation events must occur (Arias and Walter, 2007). In budding yeast, DDK phosphorylates multiple subunits of the Mcm2-Mcm7 complex (Randell et al., 2010; Sheu and Stillman, 2010). Also in budding yeast, two proteins required for activation of the helicase, Sld2 and Sld3, are the only S-CDK targets required for the initiation of DNA replication (Tanaka et al., 2007; Zegerman and Diffley, 2007). Once phosphorylated, Sld2 and Sld3 recruit the BRCT repeat-containing protein Dpb11 (TopBP1 in metazoans) to potential origins of replication. Homologs of both Sld2 (RECQL4) and Sld3 (Treslin, ticrr) have been identified in metazoans and recent work indicates that the S-CDK-dependent interaction between Sld3/Treslin and Dbp11/TopBP1 is conserved (Boos et al., 2011). Dpb11/TopBP1 is necessary for the recruitment of Mcm10, Cdc45 and the GINS complex [Sld5 (Gins4), Psf1 (Gins1), Psf2 (Gins2) and Psf3 (Gins3)], which result in CMG complex formation and activation of the helicase (Fig. 1C). Together, DDK and S-CDK activity regulate not only helicase loading, but also origin firing by the activation of the Mcm2-7/Cdc45/GINS complex, which is the active form of the replicative helicase (Ilves et al., 2010; Moyer et al., 2006).
Once replication has initiated, multiple mechanisms exist to ensure that DNA replication occurs once per cell cycle. As S phase progresses, S-CDK activity steadily increases and CDK-dependent phosphorylation of ORC subunits inhibits the further assembly and activation of pre-RCs (Arias and Walter, 2007; Chen and Bell, 2011). In metazoans, Cdt1 is degraded and the Geminin protein binds to and inhibits Cdt1, thereby preventing further recruitment of the Mcm complex to sites of DNA replication (Havens and Walter, 2009; McGarry and Kirschner, 1998; Wohlschlegel et al., 2000). Failure to inhibit re-replication can result in gene amplification and have deleterious effects on genome stability (Davidson et al., 2006; Green et al., 2010). Surprisingly, Geminin not only controls DNA replication through Cdt1 but also plays crucial developmental roles in Xenopus and mouse embryos by inhibiting lineage commitment or differentiation (Lim et al., 2011; Yang et al., 2011).
Mutations in pre-RC components cause developmental abnormalities
The differentiation of particular cell types and the development of organs are uniquely dependent on the proper regulation of DNA replication, and a number of studies have shown that mutations in pre-RC subunits can give rise to specific developmental phenotypes. In Drosophila, latheo was initially identified in a genetic screen for olfactory learning and memory mutants (Boynton and Tully, 1992). Surprisingly, the latheo gene was shown to encode ORC3 (Pinto et al., 1999). Subsequent analysis showed that latheo mutants exhibit disrupted cell proliferation within the imaginal discs (see Glossary, Box 1) and the central nervous system (CNS), with the strongest alleles affecting all imaginal discs and the late larval CNS, whereas hypomorphic mutants affect the adult brain. Remarkably, hypomorphic latheo mutants display a reduction in overall volume of a specific anatomical region of the adult brain known to be involved in olfactory learning, the mushroom body, potentially explaining how a subunit of the ORC complex could have such a specific function in learning and memory (Pinto et al., 1999). It is still unknown, however, why cells within distinct regions of the adult brain are susceptible to a reduction in the amount of ORC3. One possibility is that the rate of cell division is not uniform throughout the adult brain. Regions of the brain that require a faster cell division cycle would, therefore, place a higher demand on replication components and be more susceptible to reduced levels of ORC3. This is consistent with the observation that developmental control of S phase length is modulated by the number of origins used, rather than changes in the rate of replication fork progression (Blumenthal et al., 1974).
Recent studies on Meirer-Gorlin syndrome (MGS; see Glossary, Box 1) highlight further the importance of proper DNA replication during human development (Bicknell et al., 2011a; Bicknell et al., 2011b; Guernsey et al., 2011). MGS is a form of primordial dwarfism characterized by several developmental abnormalities (Gorlin, 1992). MGS follows an autosomal recessive mode of inheritance with differing degrees of severity and until recently no genes or loci had been found to be associated with this syndrome. A series of elegant studies, however, has now identified mutations in several pre-RC components as the cause of MGS.
Through single nucleotide polymorphism (SNP) mapping and direct sequencing of candidate genes in affected individuals, mutations in ORC1, ORC4, ORC6, CDT1 or CDC6 have been identified as the cause of MGS (Bicknell et al., 2011a; Bicknell et al., 2011b; Guernsey et al., 2011). The identification of mutations within multiple pre-RC subunits strongly implies that a defect in DNA replication, rather than DNA replication-independent functions of ORC (Hemerly et al., 2009; Sasaki and Gilbert, 2007), is responsible for MGS. Consistent with this hypothesis, cell lines derived from MGS patients with a mutation in ORC1 display a prolonged G1 phase and slower progression through S phase, probably owing to a failure in proper activation of replication origins (Bicknell et al., 2011b). Furthermore, using a zebrafish model system, injection of a morpholino oligonucleotide that targets orc1 resulted in a reduction in body size that correlated with the degree of orc1 transcript level depletion. Similar results were also obtained with a morpholino oligonucleotide that targets mcm5, supporting the notion that failure to activate replication origins properly can cause developmental abnormalities associated with body size (Bicknell et al., 2011b).
Mutations potentially affecting helicase activation can also give rise to developmental abnormalities in humans, emphasizing the importance of proper DNA replication for organismal development. Rothmund-Thomson syndrome (RTS; see Glossary, Box 1) is an autosomal recessive disorder characterized by several abnormalities including short stature, skeletal abnormalities, premature aging and chromosome fragility (Larizza et al., 2010). Mutations in RECQL4, the metazoan homolog of yeast Sld2, are responsible for ∼66% of RTS cases (Kitao et al., 1999; Larizza et al., 2010). RECQL4 functions in initiation of DNA replication and DNA repair. Therefore, it is not possible to assess which symptoms of RTS result from a failure to properly initiate DNA replication versus those that are due to a defect in DNA repair.
Understanding how DNA replication and cell proliferation are coordinated during development is a fascinating and challenging area of developmental biology. Many highly proliferative cell and tissue types are essential for proper organismal development, but mutations affecting origin licensing must only affect a subset of those cells and tissues in order to give rise to a specific set of phenotypes.
The effects of differentiation on DNA replication
Potential origins of replication are subject to developmental control
In metazoan cells, ORC binding is sequence independent, therefore it is a challenge to identify origins of replication. Given the plasticity of chromatin modifications and gene expression during differentiation, origins have to be flexible to accommodate the ever-changing chromatin landscape while faithfully replicating the genome within a precise developmental timeframe. One approach to determining the location and properties of metazoan replication origins has been to establish the location of all potential origins by identifying the genome-wide binding profile of ORC. In Drosophila, genome-wide ORC binding profiles (see Box 2) have revealed that promoters of active genes, sites of open and active chromatin, are enriched with ORC binding sites (Eaton et al., 2011; MacAlpine et al., 2010). Furthermore, the frequency of ORC binding sites is nearly three times greater in regions of the genome that replicate early in S phase compared with those that replicate late in S phase (MacAlpine et al., 2010). A more comprehensive analysis of ORC binding sites in multiple Drosophila cell lines has uncovered cell-type differences in ORC binding (Eaton et al., 2011). Additionally, ORC binding has now been analyzed in a primary tissue, the salivary gland of larvae (Sher et al., 2012). This analysis revealed that ∼30% of ORC binding sites are unique to the salivary gland. Taken together, these data indicate that potential origins of replication are subject to developmental regulation.
Box 2. Techniques used to study DNA replication
ORC binding profiles. Generated using chromatin immunoprecipitation (ChIP) of ORC subunits followed by hybridization of precipitated DNA to microarrays or high-throughput sequencing. ORC binding profiles can be used to identify all potential replication origins on a genome-wide scale.
Short nascent strand analysis. Utilizes the characteristics of newly synthesized DNA (i.e. size and presence of an RNA-DNA hybrid at the 5′ end because of the RNA primer) to isolate small leading strand fragments. Fragments can then be hybridized to a microarray or directly sequenced to identify origins of replication.
Bubble-trap analysis. Restriction enzyme-digested genomic DNA is mixed and slowly cooled in an agarose matrix, which selectively traps replication bubbles as a result of the polymerization of agarose through a replication bubble. Following electrophoresis, bubbles recovered from the gel can be hybridized to a microarray or directly sequenced to identify origins of replication.
Replication timing profiles. DNA is isolated at different times during S phase (e.g. early S phase versus late S phase) followed by copy number analysis to determine when a genomic region replicates as a function of S phase progression.
Comparative genomic hybridization (aCGH). Hybridization to microarray slides of genomic DNA derived from an experimental tissue or cell type together with control DNA. Allows for the analysis of copy number variation as well as genomic structural aberrations, such as insertions and deletions.
Single molecule analysis of replicated DNA (SMARD). Direct visualization of newly replicated single DNA molecules pulsed with thymidine analogs. In vivo pulse-labeled DNA molecules are extracted and stretched onto a glass slide for subsequent fluorescence in situ hybridization (FISH) analysis of specific DNA loci. This technique can be used to analyze both origin firing and replication fork progression at a single molecule level.
Defining origins of replication has been a rate-limiting factor in studying the properties of origin firing in mammalian cells. Recently, multiple techniques for mapping replication initiation sites have been developed and adapted for genome-wide identification (see Box 2); this is no small feat given that replication products are short lived and extremely low in abundance (Gilbert, 2010). Short nascent-strand analysis, for example, relies on the isolation and purification of small DNA fragments derived from leading strand replication products. Using this approach, profiles of initiation sites have been generated for 0.4% of the mouse genome and for 1% of the human genome from cell lines (Cadoret et al., 2008; Karnani et al., 2010; Sequeira-Mendes et al., 2009). Together, these studies have confirmed the positive relationship between origins, transcriptional activity, and markers of open chromatin. One caveat to this approach is that <14% of origins identified by small nascent-strand analysis in HeLa cells from two independent studies show overlap (Cadoret et al., 2008; Karnani et al., 2010). The reasons for such low concordance could be biological or technical, making future studies necessary to address these disparities.
More recently, two genome-scale studies have used nascent-strand analysis (see Box 2) to map replication origins in two human cancer cell lines, three mouse cell lines and a Drosophila cell line (Cayrou et al., 2011; Martin et al., 2011). In all lines, the majority of initiation events were associated with active transcription units. But, in contrast to ORC-binding profiles in Drosophila (MacAlpine et al., 2010), gene bodies rather than promoter regions of active genes were enriched with initiation events (Cayrou et al., 2011). Furthermore, both studies found that initiation events were under-represented at the actual transcription start sites (TSS) and that CpG-rich sequences were enriched with initiation events (Cayrou et al., 2011; Martin et al., 2011).
An independent method to identify initiation sites entails isolating early replication structures, or bubble structures, by the bubble-trap method (see Box 2) (Mesner et al., 2006). Genome-wide analysis of replication origins in mammalian cells by this method demonstrated that replication origins are clustered within zones and are associated with active and open chromatin (Mesner et al., 2011). Furthermore, analysis of replication origins by the bubble-trap method revealed cell-type specificity to origin usage. Perhaps not surprisingly, analysis of origin usage by this method also revealed that cell synchronization alters origin usage. Numerous replication origins were identified in synchronized cells that were not identified in unperturbed log-phase cells, demonstrating the need for limited cell manipulation for accurate assessment of replication properties (Mesner et al., 2011).
These studies in Drosophila and mammalian cell lines demonstrate that there is tissue- and cell type-specific regulation for which genomic sites serve as replication origins. The association between active transcription and replication origins suggests that the replication program is heavily influenced by the transcription program that accompanies differentiation.
The timing of replication is developmentally controlled
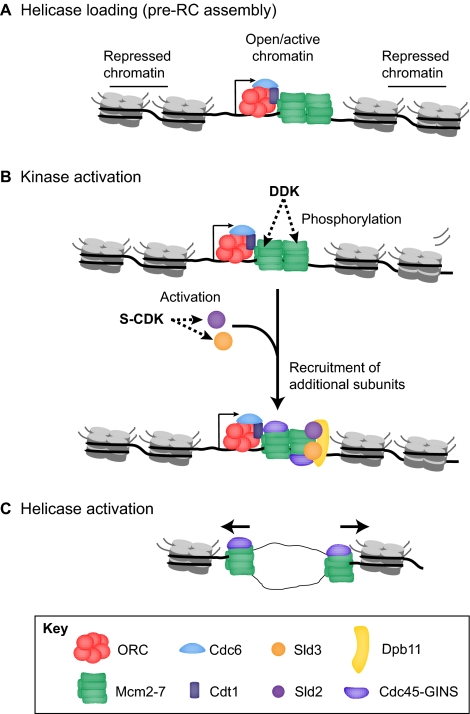
Replication timing (see Glossary, Box 1) is a fundamental property of genome duplication; some regions of the genome are replicated early in S phase, whereas other regions replicate later in S phase (Fig. 2). The significance of these timing differences remains obscure. For example, replication timing could passively reflect the chromatin state and, therefore, accessibility of replication factors to chromosomal regions or domains. Alternatively, replication of particular chromatin domains, such as heterochromatin, could require the coordination of chromatin-modifying enzymes with late-acting replication forks to propagate epigenetic information. The activity of replication fork-associated chromatin-modifying enzymes could be confined to the time in S phase when they are needed to establish and propagate particular epigenetic states. This might provide a mechanism to maintain the chromatin state characteristic of each genomic region.
Fig. 2.
Replication domains encompass numerous potential origins of replication. (A) Replication timing experiments monitor changes in gene copy number or new DNA synthesis during multiple intervals of S phase, defining zones of early or late replication. Typically, cells can be separated into early (green box) and late (pink box) replication fractions based on their DNA content. DNA isolated from each fraction can be labeled and hybridized to a microarray, or directly sequenced to monitor copy number changes during S phase. (B) Replication domains typically extend from 200 kb to 2 Mb in size, encompassing numerous individual origins of replication (diagram shows the pre-RC as illustrated in Fig. 1). Consequently, population-scale replication timing experiments lack the resolution to monitor changes in individual origin usage during differentiation.
Recent advances in microarray and DNA sequencing technologies have permitted rapid and cost effective identification of genome-wide replication timing profiles (see Box 2) for numerous cell types (Farkash-Amar et al., 2008; Hansen et al., 2010; Hiratani et al., 2008; Macalpine et al., 2004; Pope et al., 2010; Schübeler et al., 2002; Schwaiger et al., 2009). These studies rely on the isolation of DNA from defined times during S phase and the subsequent analysis of the twofold changes in copy number diagnostic of a region having been replicated. Additionally, a more recent method for monitoring the temporal order of replication during S phase has been developed that relies on monitoring newly synthesized DNA during S phase progression (Hansen et al., 2010). Replication timing experiments have revealed that large domains, typically 200-2000 kb in mammalian cells, are each replicated at distinct times in S phase (Desprat et al., 2009; Hansen et al., 2010; Hiratani et al., 2008; Pope et al., 2010). Generally, early-replicating regions of the genome are correlated with active transcription and chromatin marks associated with open forms of chromatin, whereas late-replicating regions are transcriptionally silent and enriched in chromatin marks associated with repressive chromatin. This is in agreement with previous correlations of replication timing and transcription from individual replication origins. These studies have been extensively reviewed, as has been the link between replication timing and imprinting, the process in which gene expression occurs from either the maternal or paternal allele (for details, see Gilbert, 2002; Göndör and Ohlsson, 2009). Consequently, we will focus on how differentiation affects not only replication timing, but also origin selection and usage.
Comparison of genome-wide replication timing profiles from established Drosophila cell lines revealed that ∼20% of autosomal sequences have different times of replication in different cell types (Schwaiger et al., 2009). Additionally, comparing regions of the genome that display cell-type differences with respect to replication timing confirms correlations between replication timing and gene expression. It still remains unclear whether increased gene expression actively promotes early replication, or transcription and replication are governed by a common chromatin state.
In Drosophila, males have only one copy of the X chromosome. This necessitates the need to increase gene expression from the single X chromosome, a process termed dosage compensation. Early experiments analyzing the replication patterns of Drosophila salivary gland polytene chromosomes by tritium labeling revealed that the X chromosomes of males completed replication earlier than the female X chromosomes, whereas autosomes replicated with similar kinetics (Berendes, 1966). Consistent with these observations, analysis of the dosage-compensated X chromosome from a male-derived cell line revealed that the male X replicates almost entirely in early S phase, whereas the X chromosomes from a female-derived cell line have early and late replication profiles similar to those of autosomes (Schwaiger et al., 2009). Interestingly, however, dosage compensation-dependent upregulation of transcription cannot explain the shift to early replication of the male X chromosome, because non-transcribed regions are also early replicating. Furthermore, histone H4 lysine16 (H4K16) acetylation profiles, which reflect transcriptionally active chromatin, were found to correlate very well with early replication of the male X and early replicating regions of autosomes even in regions of no transcription (Schwaiger et al., 2009). Therefore, these results favor a model in which chromatin accessibility, not transcription per se, dictates replication timing.
Comparison of multiple transformed human cell lines has demonstrated that independent cell lines of the same lineage display a remarkable similarity in replication timing profiles (Hansen et al., 2010). By contrast, ∼50% of the genome displays cell type-specific differences in replication timing profiles (Hansen et al., 2010). Similar to observations in human cell lines, analysis of 22 cell lines derived from different stages of mouse development indicated that ∼50% of the genome displays differences in replication timing (Hiratani et al., 2010). Although useful, comparing transformed non-isogenic cell lines that have been independently cultured for many years has limitations when assessing how differentiation affects processes such as replication timing. However, replication timing profiles of independently derived mouse embryonic stem cell (mESC) lines, separated for over 15 years in culture were shown to be nearly identical, suggesting that replication timing is an extremely stable property, at least in mESCs (Hiratani et al., 2008).
Cells in culture that can be triggered to differentiate provide useful models for understanding how replication is affected during development. Multiple independent studies using both mouse and human ESCs and various cell types derived from them have begun to define the effect of differentiation on replication timing. In both human and mouse cells, the size of replication domains ranges from ∼200 kb to many megabytes, and these are consolidated into fewer and larger domains during differentiation. The molecular events responsible for these changes are yet to be established (Desprat et al., 2009; Hiratani et al., 2008; Pope et al., 2010). Replication of many early and late domains appears to be coordinated, meaning multiple domains fire at very precise times during S phase. Monitoring changes in replication timing as a function of differentiation in a mouse ESC line and in a single differentiated neural precursor cell type derived from that line revealed that ∼20% of the genome is subject to changes in replication timing (Hiratani et al., 2008). This might be a more accurate percentage than that derived from comparing independently derived differentiated cell lines.
Given the vast array of cell types present in a metazoan organism, more cell types will have to be examined to determine the full impact that differentiation has on the replication program. Most importantly, future studies need to focus on individual replication origins. This is because multiple potential origins of replication have been shown to cluster within zones (Vaughn et al., 1990), and replication-timing experiments lack the resolution to distinguish individual origin-firing events (Fig. 2). For example, insertion of a single transgene into a region of late replication is sufficient to change the replication timing of that region from late to early (Lin et al., 2003). Therefore, relatively small changes on a genetic and/or epigenetic level can dramatically change the properties of a much larger replication domain.
Origin firing is modulated by differentiation
Characterization of individual replication origins within a chromosomal domain has revealed that origin firing is itself regulated during differentiation. This was first observed at the II/9A locus of the fly Sciara coprophila, which undergoes developmental programmed changes in gene copy number (Lunyak et al., 2002). Mapping of replication origins by nascent-strand analysis within the 120 kb HoxB locus in undifferentiated mouse P19 cells (see Glossary, Box 1) identified at least five origins of replication (Grégoire et al., 2006). Retinoic acid treatment causes P19 cells to differentiate and induce gene expression from the HoxB locus. Origins of replication are silenced during differentiation with the exception of the Hoxb1 origin, which becomes the single dominant replication origin within this domain. Furthermore, by assaying transcript levels at various time points during retinoic acid treatment, kinetic analysis revealed that the Hoxb1 origin is not affected by transcription and that restriction of origin usage in this locus upon differentiation is correlated with histone acetylation (Grégoire et al., 2006). These results contrast with the positive correlations between origin activation, transcription and histone acetylation seen in genome-wide studies using cell culture. They reveal the importance of studying individual replication origins to address the mechanisms that regulate origin firing and the potential for diverse regulatory schemes for modulation of individual origin usage.
Powerful single-molecule analysis of replicated DNA (SMARD; Box 2) has also been used to study the properties of individual replication origins upon differentiation (Norio et al., 2005). The benefit of SMARD analysis is that replication initiation and fork elongation can be monitored on individual chromatin fibers. In primary mouse ESCs and non-B cell lines, the Igh locus is silent and replication initiates from a zone ∼80 kb downstream of the locus. A single unidirectional replication fork travels ∼400 kb and is responsible for replicating the entire Igh locus. However, in both pro- and pre-B cell lines in which the locus is expressed, multiple origins of replication become active within the Igh locus, demonstrating that individual replication origins are regulated during differentiation (Norio et al., 2005).
Regions in which a unidirectional replication fork emanating from an early domain is responsible for replicating a large origin-less region often separate early and late replicated domains and are termed temporal transition regions (Ermakova et al., 1999). Elegant single molecule analysis of the Igh locus confirmed the existence of such domains, which have the potential to be regions of genomic instability (Norio et al., 2005; Watanabe et al., 2004). Furthermore, because temporal transition regions are a product of differences in replication timing, particularly replication timing influenced by development, temporal transition regions have the potential to be developmentally augmented regions of genomic instability.
SMARD analysis of the replication program of the POU5F1 locus in human cells also revealed a change in both origin firing and replication fork progression as a function of differentiation (Schultz et al., 2010). In multiple ESC lines, the pattern of origin usage and replication fork progression is similar within the POU5F1 locus. By contrast, analysis of a differentiated cell line as well as a multipotent neuronal rosette cell line derived from one of the ESC lines used in this study revealed a change in origin usage and directionality of fork progression within the POU5F1 region (Schultz et al., 2010).
Modulation of origin firing during differentiation has recently been shown to have a profound effect on genomic stability. Common fragile sites are chromosomal regions susceptible to breakage upon replication stress (Durkin and Glover, 2007). Recent work has demonstrated that the fragility at these sites results from the combined lack of initiation sites within these regions and their late-replicating nature (Letessier et al., 2011). Furthermore, changes in origin usage in multiple cell types can result in cell type-specific differences in chromosomal fragility (Letessier et al., 2011). These results suggest that changes in the DNA replication program, which occur during the course of differentiation, could ultimately be responsible for the tissue and/or cell type-specific genomic instability associated with tumor cell progression.
Differential replication in development
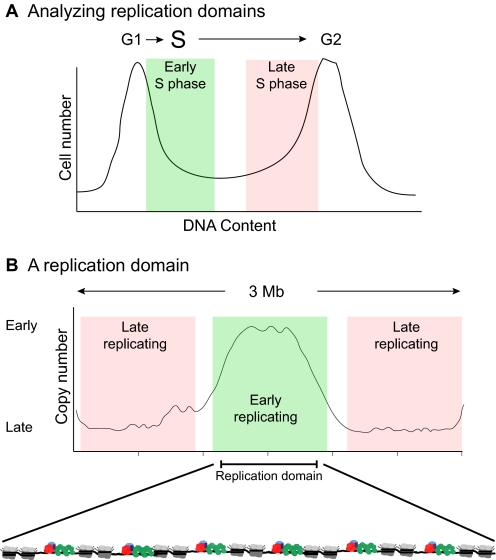
Alterations in the cell cycle and DNA replication programs are often utilized during organismal development to ensure proper tissue function. One variant cell cycle is the endo cycle (see Glossary, Box 1) in which repeated S-G cycles occur in the absence of mitosis. This results in polyploid cells, or cells with increased genome content (Fig. 3) (Edgar and Orr-Weaver, 2001; Lee et al., 2009).
Fig. 3.
Differential DNA replication. (A) A mitotic cycle with four distinct phases: G1, S, G2 and M phase. (B) By contrast, the endo cycle consists of repeated rounds of S and G phases with no intervening mitoses, resulting in polyploid cells with increased genome content per cell. (C) Developmentally programmed gene amplification can occur through repeated rounds of origin firing followed by bidirectional replication fork progression, resulting in gradients of copy number over a 100 kb domain. The example shown depicts four rounds of origin firing. Arrows at top indicate direction of fork progression. (D) Schematic of the copy number differences associated with under-replication. Arrows at top indicate direction of fork progression relative to the maximally under-replicated region. Under-replicated domains within euchromatin range from 100 to 400 kb.
Polyploidy (see Glossary, Box 1) is common in plant and animal development and is generally thought to be a developmental strategy to produce large, highly metabolically active cells. In mammals, polyploidy is essential for the development of the placenta. In the mouse, genetic ablation of the S phase cyclin, cyclin E, results in embryonic lethality (Geng et al., 2003; Parisi et al., 2003) due to a block in the endo cycle of the placental giant trophoblast cells, resulting in their failure to become polyploid. The giant trophoblast cells attain ploidy levels up to 1000C in rodents, and their large size might facilitate a maternal-fetal barrier. Megakaryocytes, the large precursor cells from which blood platelets are derived, also are polyploid (Nguyen and Ravid, 2010). The large size of polyploid megakaryocytes is required for sufficient cytoplasm to bud off adequate numbers of platelets. In Drosophila, nearly all larval tissues and numerous adult tissues have increased ploidy. Most of these cell types are polytene (see Glossary, Box 1), meaning that the replicated sister chromatid copies are held in tight association with one another. Blocking polyploidization in Drosophila inhibits cell and larval growth and perturbs proper tissue function (Edgar and Orr-Weaver, 2001).
Although polyploid cells have increased genome content per cell, gene copy number is not always uniform throughout the genome (Fig. 3). For example, programmed gene amplification (see Glossary, Box 1) in Drosophila ovarian follicle cells and the Sciara salivary glands results in an increase in genomic copy number by repeated rounds of origin firing at specific loci (Claycomb and Orr-Weaver, 2005). This process is essential to the function of these cells during development. Polyploid cells can also have regions of reduced copy number relative to overall ploidy, a phenomenon termed under-replication (see Glossary, Box 1). The function of under-replication during development still remains unclear. However, the identification of under-replicated regions by genome-wide techniques has demonstrated tissue-specific control of under-replication and has provided insight into the mechanisms that result in the repression of DNA replication in these regions.
The developmental control of under-replication
In Drosophila, it has long been appreciated that heterochromatic regions of the genome are under-replicated with respect to overall ploidy (Spradling and Orr-Weaver, 1987). Additionally, a small number of sites distributed along the euchromatic arms in the larval salivary gland and larval fat body have been shown at the molecular level to be under-replicated (Belyaeva et al., 1998; Marchetti et al., 2003). Using genome-wide array-based comparative genomic hybridization (aCGH; see Box 2) it has now been demonstrated that under-replication is prevalent throughout the euchromatic arms of the Drosophila genome (Nordman et al., 2011). Comparing the copy number profiles of DNA isolated from larval salivary gland, larval midgut and the larval fat body tissues revealed that differential replication displays tissue specificity. The mechanisms responsible for these tissue-specific differences in DNA copy number are not clear, but differences in ploidy cannot account for them (Nordman et al., 2011). This would suggest that an active, developmentally controlled mechanism exists to regulate differential replication. Whether this is at the level of chromatin structure or regulation of DNA replication factors remains to be determined as does the role played by differential replication during organismal development. Additionally, regions of under-replication provide a powerful tool for investigating the molecular mechanisms that control DNA copy number during development.
DNA under-replication in Drosophila is dependent on the Suppressor of Under-Replicaton gene, SuUR (Belyaeva et al., 1998). Genome-wide aCGH experiments have demonstrated that all regions of under-replication become fully replicated if SuUR is not functional (Nordman et al., 2011). Little is known about how SUUR functions to repress DNA replication, but its N terminus shows homology to the SWI/SNF family of chromatin remodelers and, therefore, it could act by influencing chromatin structure (Makunin et al., 2002). SUUR has been shown to be a component of a newly identified repressive form of chromatin in Drosophila termed BLACK chromatin (different chromatin states were named with color names for simplicity), which in cell culture is generally late replicating and repressed for transcription (Filion et al., 2010).
To determine whether SUUR prevents replication by inhibiting origin function within regions of under-replication, ORC binding was mapped genome-wide in wild-type and SuUR mutant larval salivary glands. The frequency of ORC binding sites was reduced significantly within regions of under-replication compared with fully replicated regions of the genome (Sher et al., 2012).
Remarkably, in the SuUR mutant, although copy number is completely restored to the normally under-replicated regions, they remain devoid of ORC binding, showing a distribution of ORC binding virtually identical to that in wild type. This result indicates that SUUR does not affect ORC binding, but rather could inhibit replication forks from entering regions of under-replication. Using a model system to study replication fork progression, it was shown that replication forks travel a greater distance in the same developmental time frame in SuUR mutant cells compared with wild-type cells (Sher et al., 2012). Therefore, these results strongly indicate that developmentally programmed changes in gene copy number can be achieved by regulation of fork progression.
During S phase of a mitotic cell cycle, multiple mechanisms exist to ensure that the genome is replicated once per cell cycle (Arias and Walter, 2007). However, during the S phases of the endo cycle, both heterochromatic and euchromatic sequences are under-replicated, suggesting that the checkpoints that regulate proper copy number in mitotic cells are inactive in endo cycling cells (Lilly and Spradling, 1996). In support of this idea, it has been demonstrated that the boundary between fully replicated euchromatin and under-replicated heterochromatin accumulates DNA damage (Hong et al., 2007). Furthermore, endo cycling cells are unable to induce apoptosis following genotoxic stress, owing to the inability of these cells to induce the apoptotic genes that would be expressed in mitotic cells following genotoxic stress (Mehrotra et al., 2008). The mechanism(s) responsible for silencing the checkpoint activity in endo cycling cells is currently unknown. Additionally, these studies raise the possibility that the response of a cell to DNA damage and replication stress can be influenced by its developmental state, which has broad implications for cell-type specific genomic instability and tumor formation.
Developmentally regulated gene amplification
Developmentally programmed gene amplification is another form of differential DNA replication necessary for proper tissue function (Claycomb and Orr-Weaver, 2005). The follicle cells of the Drosophila ovary form an epithelial layer around the developing oocyte. After a series of mitotic divisions, follicle cells enter the endo cycle and increase their overall ploidy to 16C. Follicle cells then exit the endo cycle and enter into a developmentally regulated gene amplification program, in which six sites are repeatedly amplified, forming so-called amplicons (see Glossary, Box 1), by a ‘re-replication’-based mechanism resulting in ∼100 kb gradients of copy number (Fig. 3) (Claycomb et al., 2004; Kim et al., 2011). The mechanism by which most replication origins are silenced but six origins are selected as amplification sites is unknown. Follicle cell amplicons provide a powerful tool to study metazoan origin regulation, because changes in gene copy number are precisely timed during development. Thus, both initiation of replication and replication fork progression can be individually assessed during development.
The identification of all the amplicons in follicle cells has permitted the integrative analysis of genome-wide ORC binding, histone acetylation, and transcription levels (Kim et al., 2011). Amplification was previously shown to be dependent on histone acetylation, a property correlated with ORC binding and origin activation in many genome-wide studies (Aggarwal and Calvi, 2004; Hartl et al., 2007). Histone acetylation was assayed both genome wide and specifically within amplification regions revealing that histone acetylation is found at some sites of amplification, but not all. These results demonstrate that histone acetylation is not a prerequisite for origin firing (Kim et al., 2011).
Follicle cell gene amplification is a developmental strategy that increases the amount of DNA template to support the high level of transcription necessary to produce sufficient chorion (see Glossary, Box 1) proteins in a narrow developmental window (Spradling and Mahowald, 1980). Although this strategy has been well characterized for a small number of genes involved in chorion synthesis, the relationship between gene amplification and transcription had not been analyzed genome wide. To understand this relationship better, whole-transcriptome profiling was performed on amplifying follicle cells and gene expression levels were compared with copy number changes (Kim et al., 2011). As expected, many of the most highly expressed genes are located within the two main chorion gene clusters. Many robustly transcribed genes lie outside the amplicons, however. In addition, genes located within amplified regions are not always highly expressed (Kim et al., 2011). This could be important for understanding the significance of amplified genomic regions that are prevalent in cancer cells. The discovery that gene expression is not always correlated with copy number in Drosophila amplicons is an important consideration when assessing how copy number variation might affect gene expression in cancer cells.
Conclusions
It is now clear that the DNA replication program is modulated as a function of development. Numerous genome-wide studies and studies of individual replication origins have established a correlation between origin efficiency and chromatin state (Ding and MacAlpine, 2011). It remains to be determined whether the changes in the replication program that occur during development reflect an active mechanism to maximize the efficiency of DNA replication and/or genomic stability during development, or if changes in the DNA replication program are a passive reflection of developmentally regulated changes in chromatin state. Research in model systems such as Drosophila has clearly demonstrated that developmentally programmed changes in DNA replication are necessary for proper cell function. How common programmed changes in DNA replication are in development remain to be determined.
Recent studies addressing the cause of MGS demonstrated that failure to initiate properly DNA replication can have specific effects on human development (Bicknell et al., 2011a; Bicknell et al., 2011b; Guernsey et al., 2011). But, why are certain tissues and/or cell types so susceptible to defects in DNA replication? A simple hypothesis is that the most highly proliferative cell types would be the most vulnerable to alterations in the DNA replication program. The phenotypes associated with MGS suggest that this is unlikely to be the case. Therefore, a major focus in addressing the relationship between DNA replication and development will be to understand why certain cell types and tissues are affected by alterations in the DNA replication program. Only then will we be able to gauge the impact programmed DNA replication has on proper organismal development.
Defects in proper DNA replication have long been associated with cancer. For example, gene amplification and polyploidy are associated with numerous tumor types, but the primary defects responsible for generating gene amplification have been difficult to identify (Albertson, 2006; Beroukhim et al., 2010). Recent work has demonstrated directly that loss of DNA replication control can initiate the amplification process (Green et al., 2010). Furthermore, defects in proper replication licensing result in increased levels of chromosomal instability and spontaneous tumor formation (Beroukhim et al., 2010; Kawabata et al., 2011). Thus, proper DNA replication is not only essential for proper organismal development but also to prevent the onset of cancer.
Acknowledgments
We thank Mary Gehring, Jane Kim, Gerry Fink, Angelika Amon and Steve Bell for constructive comments on the manuscript. We are grateful to Tom DiCesare for technical assistance in figure preparation.
Footnotes
Funding
J.N. is a Howard Hughes Medical Institute (HHMI) Fellow of the Damon Runyon Cancer Research Foundation and has received support from a Margaret and Herman Sokol postdoctoral award. This work was supported by the National Institutes of Health (NIH) and an American Cancer Society Research Professorship to T.L.O.-W. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
References
- Aggarwal B. D., Calvi B. R. (2004). Chromatin regulates origin activity in Drosophila follicle cells. Nature 430, 372–376 [DOI] [PubMed] [Google Scholar]
- Albertson D. G. (2006). Gene amplification in cancer. Trends Genet. 22, 447–455 [DOI] [PubMed] [Google Scholar]
- Araki H. (2010). Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr. Opin. Cell Biol. 22, 766–771 [DOI] [PubMed] [Google Scholar]
- Arias E. E., Walter J. C. (2007). Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21, 497–518 [DOI] [PubMed] [Google Scholar]
- Bell S. P., Dutta A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374 [DOI] [PubMed] [Google Scholar]
- Belyaeva E. S., Zhimulev I. F., Volkova E. I., Alekseyenko A. A., Moshkin Y. M., Koryakov D. E. (1998). Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc. Natl. Acad. Sci. USA 95, 7532–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendes H. D. (1966). Differential replication of male and female X-chromosomes in Drosophila. Chromosoma 20, 32–43 [Google Scholar]
- Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J. S., Dobson J., Urashima M., et al. (2010). The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell L. S., Bongers E. M., Leitch A., Brown S., Schoots J., Harley M. E., Aftimos S., Al-Aama J. Y., Bober M., Brown P. A., et al. (2011a). Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 43, 356–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell L. S., Walker S., Klingseisen A., Stiff T., Leitch A., Kerzendorfer C., Martin C. A., Yeyati P., Al Sanna N., Bober M., et al. (2011b). Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat. Genet. 43, 350–355 [DOI] [PubMed] [Google Scholar]
- Blumenthal A. B., Kriegstein H. J., Hogness D. S. (1974). The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb. Symp. Quant. Biol. 38, 205–223 [DOI] [PubMed] [Google Scholar]
- Boos D., Sanchez-Pulido L., Rappas M., Pearl L. H., Oliver A. W., Ponting C. P., Diffley J. F. (2011). Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr. Biol. 21, 1152–1157 [DOI] [PubMed] [Google Scholar]
- Boynton S., Tully T. (1992). latheo, a new gene involved in associative learning and memory in Drosophila melanogaster, identified from P element mutagenesis. Genetics 131, 655–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoret J. C., Meisch F., Hassan-Zadeh V., Luyten I., Guillet C., Duret L., Quesneville H., Prioleau M. N. (2008). Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc. Natl. Acad. Sci. USA 105, 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C., Coulombe P., Vigneron A., Stanojcic S., Ganier O., Peiffer I., Rivals E., Puy A., Laurent-Chabalier S., Desprat R., et al. (2011). Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 21, 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Bell S. P. (2011). CDK prevents Mcm2-7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes Dev. 25, 363–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J. M., Orr-Weaver T. L. (2005). Developmental gene amplification: insights into DNA replication and gene expression. Trends Genet. 21, 149–162 [DOI] [PubMed] [Google Scholar]
- Claycomb J. M., Benasutti M., Bosco G., Fenger D. D., Orr-Weaver T. L. (2004). Gene amplification as a developmental strategy: isolation of two developmental amplicons in Drosophila. Dev. Cell 6, 145–155 [DOI] [PubMed] [Google Scholar]
- Davidson I. F., Li A., Blow J. J. (2006). Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol. Cell 24, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprat R., Thierry-Mieg D., Lailler N., Lajugie J., Schildkraut C., Thierry-Mieg J., Bouhassira E. E. (2009). Predictable dynamic program of timing of DNA replication in human cells. Genome Res. 19, 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q., MacAlpine D. M. (2011). Defining the replication program through the chromatin landscape. Crit. Rev. Biochem. Mol. Biol. 46, 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin S. G., Glover T. W. (2007). Chromosome fragile sites. Annu. Rev.Genet. 41, 169–192 [DOI] [PubMed] [Google Scholar]
- Eaton M. L., Prinz J. A., MacAlpine H. K., Tretyakov G., Kharchenko P. V., MacAlpine D. M. (2011). Chromatin signatures of the Drosophila replication program. Genome Res. 21, 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B. A., Orr-Weaver T. L. (2001). Endoreplication cell cycles: more for less. Cell 105, 297–306 [DOI] [PubMed] [Google Scholar]
- Ermakova O. V., Nguyen L. H., Little R. D., Chevillard C., Riblet R., Ashouian N., Birshtein B. K., Schildkraut C. L. (1999). Evidence that a single replication fork proceeds from early to late replicating domains in the IgH locus in a non-B cell line. Mol. Cell 3, 321–330 [DOI] [PubMed] [Google Scholar]
- Farkash-Amar S., Lipson D., Polten A., Goren A., Helmstetter C., Yakhini Z., Simon I. (2008). Global organization of replication time zones of the mouse genome. Genome Res. 18, 1562–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., Ward L. D., Brugman W., de Castro I. J., Kerkhoven R. M., Bussemaker H. J., et al. (2010). Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143, 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y., Yu Q., Sicinska E., Das M., Schneider J. E., Bhattacharya S., Rideout W. M., Bronson R. T., Gardner H., Sicinski P. (2003). Cyclin E ablation in the mouse. Cell 114, 431–443 [DOI] [PubMed] [Google Scholar]
- Gilbert D. M. (2002). Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol. 14, 377–383 [DOI] [PubMed] [Google Scholar]
- Gilbert D. M. (2010). Evaluating genome-scale approaches to eukaryotic DNA replication. Nat. Rev. Genet. 11, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göndör A., Ohlsson R. (2009). Replication timing and epigenetic reprogramming of gene expression: a two-way relationship? Nat. Rev. Genet. 10, 269–276 [DOI] [PubMed] [Google Scholar]
- Gorlin R. J. (1992). Microtia, absent patellae, short stature, micrognathia syndrome. J. Med. Genet. 29, 516–517 [PMC free article] [PubMed] [Google Scholar]
- Green B. M., Finn K. J., Li J. J. (2010). Loss of DNA replication control is a potent inducer of gene amplification. Science 329, 943–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grégoire D., Brodolin K., Méchali M. (2006). HoxB domain induction silences DNA replication origins in the locus and specifies a single origin at its boundary. EMBO Rep. 7, 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guernsey D. L., Matsuoka M., Jiang H., Evans S., Macgillivray C., Nightingale M., Perry S., Ferguson M., LeBlanc M., Paquette J., et al. (2011). Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat. Genet. 43, 360–364 [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Thomas S., Sandstrom R., Canfield T. K., Thurman R. E., Weaver M., Dorschner M. O., Gartler S. M., Stamatoyannopoulos J. A. (2010). Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl. Acad. Sci. USA 107, 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl T., Boswell C., Orr-Weaver T. L., Bosco G. (2007). Developmentally regulated histone modifications in Drosophila follicle cells: initiation of gene amplification is associated with histone H3 and H4 hyperacetylation and H1 phosphorylation. Chromosoma 116, 197–214 [DOI] [PubMed] [Google Scholar]
- Havens C. G., Walter J. C. (2009). Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 35, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A. S., Prasanth S. G., Siddiqui K., Stillman B. (2009). Orc1 controls centriole and centrosome copy number in human cells. Science 323, 789–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I., Ryba T., Itoh M., Yokochi T., Schwaiger M., Chang C. W., Lyou Y., Townes T. M., Schübeler D., Gilbert D. M. (2008). Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 6, e245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I., Ryba T., Itoh M., Rathjen J., Kulik M., Papp B., Fussner E., Bazett-Jones D. P., Plath K., Dalton S., et al. (2010). Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 20, 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong A., Narbonne-Reveau K., Riesgo-Escovar J., Fu H., Aladjem M. I., Lilly M. A. (2007). The cyclin-dependent kinase inhibitor Dacapo promotes replication licensing during Drosophila endocycles. EMBO J. 26, 2071–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Mechali M. (1993). Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 12, 4511–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I., Petojevic T., Pesavento J. J., Botchan M. R. (2010). Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 37, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N., Taylor C. M., Malhotra A., Dutta A. (2010). Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol. Biol. Cell 21, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T., Luebben S. W., Yamaguchi S., Ilves I., Matise I., Buske T., Botchan M. R., Shima N. (2011). Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell 41, 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. C., Nordman J., Xie F., Kashevsky H., Eng T., Li S., Macalpine D. M., Orr-Weaver T. L. (2011). Integrative analysis of gene amplification in Drosophila follicle cells: parameters of origin activation and repression. Genes Dev. 25, 1384–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S., Shimamoto A., Goto M., Miller R. W., Smithson W. A., Lindor N. M., Furuichi Y. (1999). Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 22, 82–84 [DOI] [PubMed] [Google Scholar]
- Larizza L., Roversi G., Volpi L. (2010). Rothmund-Thomson syndrome. Orphanet J. Rare Dis. 5, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. O., Davidson J. M., Duronio R. J. (2009). Endoreplication: polyploidy with purpose. Genes Dev. 23, 2461–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier A., Millot G. A., Koundrioukoff S., Lachages A. M., Vogt N., Hansen R. S., Malfoy B., Brison O., Debatisse M. (2011). Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470, 120–123 [DOI] [PubMed] [Google Scholar]
- Lilly M. A., Spradling A. C. (1996). The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10, 2514–2526 [DOI] [PubMed] [Google Scholar]
- Lim J. W., Hummert P., Mills J. C., Kroll K. L. (2011). Geminin cooperates with Polycomb to restrain multi-lineage commitment in the early embryo. Development 138, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. M., Fu H., Martinovsky M., Bouhassira E., Aladjem M. I. (2003). Dynamic alterations of replication timing in mammalian cells. Curr. Biol. 13, 1019–1028 [DOI] [PubMed] [Google Scholar]
- Lunyak V. V., Ezrokhi M., Smith H. S., Gerbi S. A. (2002). Developmental changes in the Sciara II/9A initiation zone for DNA replication. Mol. Cell. Biol. 22, 8426–8437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalpine D. M., Rodríguez H. K., Bell S. P. (2004). Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 18, 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine H. K., Gordân R., Powell S. K., Hartemink A. J., MacAlpine D. M. (2010). Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 20, 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makunin I. V., Volkova E. I., Belyaeva E. S., Nabirochkina E. N., Pirrotta V., Zhimulev I. F. (2002). The Drosophila suppressor of underreplication protein binds to late-replicating regions of polytene chromosomes. Genetics 160, 1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti M., Fanti L., Berloco M., Pimpinelli S. (2003). Differential expression of the Drosophila BX-C in polytene chromosomes in cells of larval fat bodies: a cytological approach to identifying in vivo targets of the homeotic Ubx, Abd-A and Abd-B proteins. Development 130, 3683–3689 [DOI] [PubMed] [Google Scholar]
- Martin M. M., Ryan M., Kim R., Zakas A. L., Fu H., Lin C. M., Reinhold W. C., Davis S. R., Bilke S., Liu H., et al. (2011). Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 21, 1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry T. J., Kirschner M. W. (1998). Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043–1053 [DOI] [PubMed] [Google Scholar]
- Mehrotra S., Maqbool S. B., Kolpakas A., Murnen K., Calvi B. R. (2008). Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes Dev. 22, 3158–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner L. D., Crawford E. L., Hamlin J. L. (2006). Isolating apparently pure libraries of replication origins from complex genomes. Mol. Cell 21, 719–726 [DOI] [PubMed] [Google Scholar]
- Mesner L. D., Valsakumar V., Karnani N., Dutta A., Hamlin J. L., Bekiranov S. (2011). Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res. 21, 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer S. E., Lewis P. W., Botchan M. R. (2006). Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 103, 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. G., Ravid K. (2010). Polyploidy: mechanisms and cancer promotion in hematopoietic and other cells. Adv. Exp. Med. Biol. 676, 105–122 [DOI] [PubMed] [Google Scholar]
- Nordman J., Li S., Eng T., Macalpine D., Orr-Weaver T. L. (2011). Developmental control of the DNA replication and transcription programs. Genome Res. 21, 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P., Kosiyatrakul S., Yang Q., Guan Z., Brown N. M., Thomas S., Riblet R., Schildkraut C. L. (2005). Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol. Cell 20, 575–587 [DOI] [PubMed] [Google Scholar]
- Parisi T., Beck A. R., Rougier N., McNeil T., Lucian L., Werb Z., Amati B. (2003). Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 22, 4794–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto S., Quintana D. G., Smith P., Mihalek R. M., Hou Z. H., Boynton S., Jones C. J., Hendricks M., Velinzon K., Wohlschlegel J. A., et al. (1999). latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron 23, 45–54 [DOI] [PubMed] [Google Scholar]
- Pope B. D., Hiratani I., Gilbert D. M. (2010). Domain-wide regulation of DNA replication timing during mammalian development. Chromosome Res. 18, 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell J. C., Fan A., Chan C., Francis L. I., Heller R. C., Galani K., Bell S. P. (2010). Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol. Cell 40, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D., Beuron F., Tolun G., Griffith J. D., Morris E. P., Diffley J. F. (2009). Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 139, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Gilbert D. M. (2007). The many faces of the origin recognition complex. Curr. Opin. Cell Biol. 19, 337–343 [DOI] [PubMed] [Google Scholar]
- Schübeler D., Scalzo D., Kooperberg C., van Steensel B., Delrow J., Groudine M. (2002). Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet. 32, 438–442 [DOI] [PubMed] [Google Scholar]
- Schultz S. S., Desbordes S. C., Du Z., Kosiyatrakul S., Lipchina I., Studer L., Schildkraut C. L. (2010). Single-molecule analysis reveals changes in the DNA replication program for the POU5F1 locus upon human embryonic stem cell differentiation. Mol. Cell. Biol. 30, 4521–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M., Stadler M. B., Bell O., Kohler H., Oakeley E. J., Schübeler D. (2009). Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev. 23, 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J., Díaz-Uriarte R., Apedaile A., Huntley D., Brockdorff N., Gómez M. (2009). Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 5, e1000446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher N., Bell G. W., Li S., Nordman J., Eng T., Eaton M. L., MacAlpine D. M., Orr-Weaver T. L. (2012). Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y. J., Stillman B. (2010). The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A., Orr-Weaver T. (1987). Regulation of DNA replication during Drosophila development. Annu. Rev. Genet. 21, 373–403 [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Mahowald A. P. (1980). Amplification of genes for chorion proteins during oogenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 77, 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. (2007). CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445, 328–332 [DOI] [PubMed] [Google Scholar]
- Vaughn J. P., Dijkwel P. A., Hamlin J. L. (1990). Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell 61, 1075–1087 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Ikemura T., Sugimura H. (2004). Amplicons on human chromosome 11q are located in the early/late-switch regions of replication timing. Genomics 84, 796–805 [DOI] [PubMed] [Google Scholar]
- Wohlschlegel J. A., Dwyer B. T., Dhar S. K., Cvetic C., Walter J. C., Dutta A. (2000). Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290, 2309–2312 [DOI] [PubMed] [Google Scholar]
- Yang V. S., Carter S. A., Hyland S. J., Tachibana-Konwalski K., Laskey R. A., Gonzalez M. A. (2011). Geminin escapes degradation in G1 of mouse pluripotent cells and mediates the expression of Oct4, Sox2, and Nanog. Curr. Biol. 21, 692–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Diffley J. F. (2007). Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445, 281–285 [DOI] [PubMed] [Google Scholar]