Abstract
Secreted protein, acidic and rich in cysteine (SPARC, also known as osteonectin or BM-40) is a glycoprotein component of the extracellular matrix that has been reported to be involved with a variety of cellular processes. Although SPARC expression levels are frequently altered in a variety of tumor types, the exact implications of deregulated SPARC expression — whether it promotes, inhibits or has no effect on tumor progression — have remained unclear. Our recent gene expression analyses have shown that SPARC is significantly downregulated in highly metastatic human prostate cancer cells. To test the role of endogenous SPARC in tumorigenesis directly, we examined cancer progression and metastasis in SPARC+/− and SPARC−/− mice using two separate transgenic mouse tumor models: transgenic adenocarcinoma of the mouse prostate (TRAMP) and murine mammary tumor virus-polyoma middle T (MMTV-PyMT). Surprisingly, in both instances, we found that loss of SPARC had no significant effects on tumor initiation, progression or metastasis. Tumor angiogenesis and collagen deposition were also largely unaffected. Our results indicate that, although differential SPARC expression may be a useful marker of aggressive, metastasis-prone tumors, loss of SPARC is not sufficient either to promote or to inhibit cancer progression in two spontaneous mouse tumor models.
Keywords: SPARC, prostate cancer, breast cancer, metastasis, TRAMP, PyMT-MMTV
INTRODUCTION
Secreted protein, acidic and rich in cysteine (SPARC) is an extracellular, calcium-binding glycoprotein that functions as a modulator of cell-matrix interactions [1, 2]. SPARC is widely expressed during development and, later in the adult, is highly expressed in tissues that undergo rapid turnover of matrix, such as the gut and bone [3]. SPARC has also been observed to be upregulated in instances that involve cell migration, such as during tissue remodeling, wound healing and angiogenesis [4].
SPARC is generally thought to function as a de-adhesive molecule [5, 6]. Purified SPARC protein added exogenously to cells in vitro has been reported to induce disassembly of cellular focal adhesions and reorganization of stress fibers [6], which may promote an “intermediate” state of adhesion that favors cell migration [5]. This is likely mediated, at least in part, through the ability of SPARC to bind various matrix substrates such as collagen and laminin [6, 7]; through interactions with cellular receptors and cytokines [8, 9]; and through induction and/or activation of pro-migratory factors such as matrix metalloproteinases (MMPs) and TGF-β [10–12]. Importantly, SPARC has also been reported to be critical for the proper assembly of collagen fibers, and SPARC-deficient mice have been observed to exhibit a variety of subtle phenotypes which are thought to stem from defective matrix assembly [2].
Given the varied effects of SPARC on cell adhesion and matrix integrity, it is perhaps not surprising that SPARC expression is frequently altered in a variety of tumors [13–17]. In addition, experimental manipulation of SPARC expression in tumor cell lines has yielded findings that suggest that SPARC promotes an invasive phenotype in prostate cancer and melanoma [16, 18–21]. However, these results are contrasted by reports that SPARC may also suppress tumor progression in ovarian, breast, colon and immortalized human kidney cells [22–25]. Interestingly, enhanced subcutaneous growth has been observed when un-manipulated lymphoma, lung and pancreatic tumor cells were implanted into SPARC−/− mice, as compared with wild-type animals [7, 26]. However, evidence that the opposite can be true — reduced mammary tumor growth in SPARC-deficient mice — has also been reported [27].
These seemingly contradictory results might be explained by the variety of factors that are thought to influence the activity of, as well as the cellular response to, SPARC. Perhaps most notably, as SPARC is known to be proteolytically processed by matrix metalloproteinases [28], the protease profile of the tumor microenvironment could potentially impact the activity of this protein [23]. In addition, the local composition of matrix molecules and the availability of cytokines are also likely to be important, as are the identities of the responding cell types. Thus, given the complexity of these systems, it is possible that SPARC may enhance tumor growth in certain experimental contexts but have opposite or no effect in others.
An important point that needs to be considered is that, in most cases, studies examining the effects of SPARC on tumorigenesis have relied either on in vitro experiments or on implantation of already-transformed cells into immunocompromised or SPARC-deficient animals. While significant findings can be gleaned from these experiments, spontaneous models of tumorigenesis, which recapitulate all the steps of cancer progression from tumor initiation to metastasis, are generally regarded as more accurate representations of tumorigenesis. Indeed, recent studies have reported that SPARC−/− mice appear resistant to UV irradiation-induced squamous cell carcinomas and APCMin/+-induced spontaneous intestinal adenomas, relative to SPARC+/+ animals [29, 30]; however, as mentioned previously, the effects of SPARC may be organ/cell-type specific, and therefore these findings need to be extended to different spontaneous models of tumorigenesis.
Our interest in SPARC grew out of our gene expression analyses of variably metastatic human prostate cancer cell lines, which indicated that SPARC expression was specifically downregulated in highly metastatic cells [31]. This finding was concordant with human clinical gene expression data which showed that reduced SPARC expression was associated with an aggressive prostate and breast cancer phenotype. To test the effects of SPARC in spontaneous models of tumorigenesis, we used SPARC−/− and SPARC+/− mice to examine tumor progression and metastasis in two transgenic models of prostate and breast cancer. In both instances, we observed that loss of endogenous SPARC expression had no significant impact on tumorigenesis. Our findings indicate that the effects of SPARC may depend not only on the cell type(s) being tested but also on the experimental setting. Although SPARC expression levels may be prognostic of tumor severity and/or aggressiveness, our results argue that loss of SPARC, by itself, neither directly promotes nor inhibits spontaneous prostate or breast cancer progression.
MATERIALS AND METHODS
Tumor Models
Surgical orthotopic implantation (SOI) was performed, as previously described [32, 33]. Variant subclones of the human prostate cancer cell line PC-3 (American Type Culture Collection, Manassas, VA [34]) were derived by isolating metastatic cells from CD-1 immunodeficient mice (Charles River Laboratories, Wilmington, MA) that had been implanted by SOI and by expanding these cell in vitro [31]. The PC3-pMicro-1 cell line was first derived by passaging PC-3 cells in vivo using SOI. Re-implantation of pMicro-1 cells by SOI led to the subsequent isolation of poorly metastatic PC3-#78 and high metastatic PC3-#82 cells. SPARC−/− mice, which were originally generated by Norose et al., were obtained from Dr. E. H. Sage and were in a mixed genetic background [35]. TRAMP mice, originally generated by Greenberg et al., were obtained from Dr. Ailin Bai (MIT) and were in a pure C57BL/6 background [36]. MMTV-PyMT mice, originally generated by Muller et al., were obtained from Dr. Lei Xu (MIT) and were in an FVB background [37]. The following cross was performed to generate the majority of mice used in the TRAMP prostate studies: TRAMP+/− ; SPARC+/− x SPARC−/−. The following cross was performed to generate the majority of mice used in the MMTV-PyMT mammary studies: PyMT-MMTV+/− ; SPARC+/− x SPARC−/− (for both TRAMP and MMTV-PyMT, the oncogenic transgene was carried as a single copy in either parent, who was either SPARC+/− or SPARC−/−). Thus, only SPARC+/− or SPARC−/− mice were generated (as littermates or cousins) for either analysis. In our spontaneous prostate cancer studies, 26-week-old TRAMP mice were sacrificed, and their urogenital systems were dissected, weighed and fixed in zinc (Becton Dickinson, San Diego, CA). Lymph node metastases were identified by the gross expansion of the draining lumbar/para-aortic lymph nodes, and, in some marginal cases, were confirmed by histology. In our spontaneous breast cancer studies, 10-week-old MMTV-PyMT mice were checked twice weekly for the appearance of tumor nodules in the mammary fat pads. About four weeks after the initial date of tumor detection, mice were sacrificed, and all tumors and fat pads were dissected, individually weighed and fixed in formalin. Tumor mass was then assessed either as the sum of the masses of all tumors from each mouse (Figure 4B), or as the sum of the masses of all tumors removed from each set of mammary glands found in each physiological quadrant of every mouse (upper left quadrant, upper right quadrant, lower left quadrant, lower right quadrant; Figure 4C). Lungs were also removed at the time of sacrifice, and metastases were counted under a dissecting microscope. All animal experiments were performed in accordance with the animal care guidelines established by the MIT Division of Comparative Medicine.
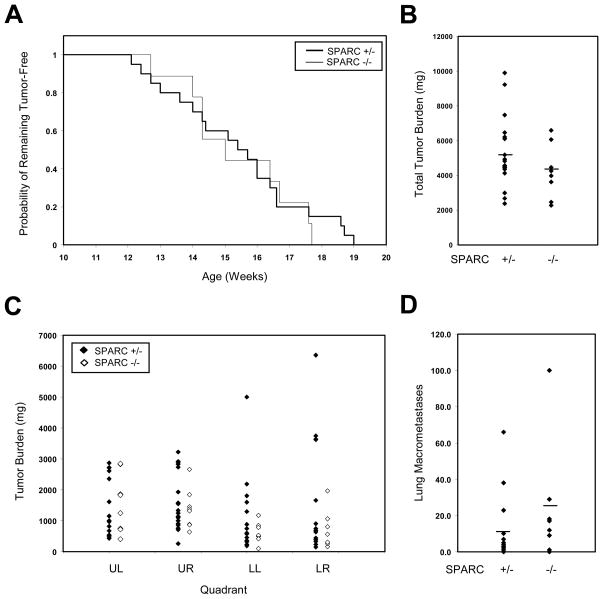
Figure 4. Absence of SPARC does not affect breast cancer initiation, progression or metastasis.
(A) The time to tumor initiation for MMTV-PyMT tumors did not differ between genotypes. (B) Average total tumor burden, ~4 weeks after the initial date of tumor initiation, also did not vary between the two cohorts (p = 0.2). (C) When tumors were sub-divided by quadrant (UL, upper left; UR, upper right; LL, lower left; LR, lower right), average masses did not differ between SPARC+/− and SPARC−/− tumors. (D) The average number of lung metastases observed per animal was not significantly changed between genotypes (p = 0.17).
Histology, Immunohistochemistry and Western Blot
For TRAMP mice, prostates fixed in zinc were separated into ventral and dorsolateral lobes, and each of these was sectioned and stained by hematoxylin and eosin. The sections were each blindly scored by a pathologist (R.T.B.) on a scale of 1–6 (with ‘1’ representing normal prostate and ‘6’ representing undifferentiated prostate adenocarcinoma), according to the system devised by Hurwitz et al [38]. Each section was awarded two grades: a highest grade for the area of most severe pathology, and a predominant grade for the most common degree of pathology observed on the section. An overall highest grade was also assigned for each prostate, identical to the highest grade awarded to any constituent lobe. For MMTV-PyMT mice, formalin-fixed mammary carcinomas were sectioned and stained by hematoxylin and eosin. In all cases, lobular carcinomas were observed. Masson’s trichrome staining was performed, as described [39], on Grade 4-matched prostate cancer sections and on size-matched mammary tumors (~200–300 mg) for each genotype. A single section from at least six independent animals was stained for each organ and for each genotype. For immunohistochemistry, Grade 4 prostate sections were microwaved in BD Retrievagen buffer (Becton Dickinson), probed with rat anti-mouse CD34 antibody (clone RAM34, 1:25; BD Pharmingen, San Diego, CA), detected with biotinylated rabbit anti-rat immunoglobulin (1:200; Vector Labs, Burlingame, CA), and stained with Vectastain ABC kit (Vector Labs). Four SPARC+/− and six SPARC−/− mice bearing Grade 4 dorsal prostate lesions were blindly scored for CD34+ vessels in representative low-power fields under a 10X objective. For Western blot, total prostate or mammary lysates were extracted from high grade tumors using Laemmli SDS loading buffer containing 5% β-mercaptoethanol and resolved by 15% or 18% SDS-PAGE gel. Membranes were incubated overnight with chicken anti-SPARC antibody (ab14071; 1:500; Abcam, Cambridge, MA), goat anti-SPARC antibody (AF942; 1:500; R&D Systems, Minneapolis, MN) and/or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody (1:5000; Chemicon, Temecula, CA).
Gene Expression Studies, Bioinformatics and Statistics
Gene expression analyses on PC-3 prostate cancer cells were performed as previously described [31]. Briefly, total RNA from subcutaneous tumors was converted into cRNA and hybridized onto human U133A chips (Affymetrix, Santa Clara, CA). Data were analyzed by dChip software (Harvard Medical School, Boston, MA) [40] and deposited in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo; accession no. GSE7930). Quantitative PCR validation of SPARC expression was performed on total RNA extracted from tumors and tissue culture cells using the RNeasy kit (Qiagen, Valencia, CA). Total RNA was reverse-transcribed into cDNA using TaqMan reverse transcription reagent (Applied Biosystems, Branchburg, NJ), and the cDNA was amplified for qPCR using the SYBR Green PCR amplification kit (Applied Biosystems). Human SPARC message levels were normalized to those of human GAPDH internal controls. Primer sets used for SPARC amplification were as follows: 5′-GAGAGCGCGCTCTGCCTGCCG-3′ (forward); 5′-CACCACCTCTGTCTCATCAGGC-3′ (reverse). Primer sets for GAPDH were as follows: 5′-GGAAGGTGAAGGTCGGAGTC-3′ (forward); 5′-CTGGAAGATGGTGATGGGATTTC-3′ (reverse). Clinical prostate cancer gene expression data-sets were obtained from Oncomine 3.0 and analyzed at www.oncomine.org [41]. The clinical breast cancer data-set originally obtained by van’t Veer et al. [42], as well as the accompanying support information, were downloaded from www.nature.com and from http://www.rii.com/publications/2002/vantveer.html. Normalized SPARC expression data were rank ordered and matched with patient metastatic outcome. Log-rank test of the Kaplan-Meier plots was performed using S-Plus software (Insightful, Seattle, WA). For other statistical analyses, unpaired Student’s t-test was performed at http://www.physics.csbsju.edu/stats/Index.html. Chi-square analyses were performed at http://www.psych.ku.edu/preacher/.
RESULTS
We isolated variably metastatic subclones of the human prostate cancer cell line PC-3 by repeated passage of these cells in vivo, using a xenotransplant mouse tumor model known as surgical orthotopic implantation (SOI). As has been described previously, SOI is a two-step procedure where subcutaneous tumors are first grown in immunodeficient mice [32, 33]. Small fragments of these tumors are then used as solid graft material for implantation into the prostates (the orthotopic site) of additional immunodeficient animals. This approach has several advantages over other xenotransplant techniques for modeling prostate cancer but, most notably, the primary tumors metastasize locally to lymph nodes and also distally to the lungs via systemic circulation [43]. By isolating and culturing metastases that arose from this model system (see Materials and Methods for further details), we previously derived a series of cell lines that varied in metastatic potential when implanted by SOI: poorly metastatic PC3-#78 cells, moderately metastatic PC3-pMicro1 cells and highly metastatic PC3-#82 cells [31].
We next performed gene expression analyses on subcutaneous tumors derived from these three cell lines, as has also been previously described [31]. Our results indicated that SPARC was highly expressed in poorly metastatic PC3-#78 cells, significantly downregulated in highly metastatic PC3-#82 cells, and expressed at intermediate levels in PC3-pMicro1 cells (Figure 1A). We subsequently confirmed these findings by performing quantitative PCR using cDNAs derived from additional subcutaneous tumors and also from tissue culture cells (Figure 1A).
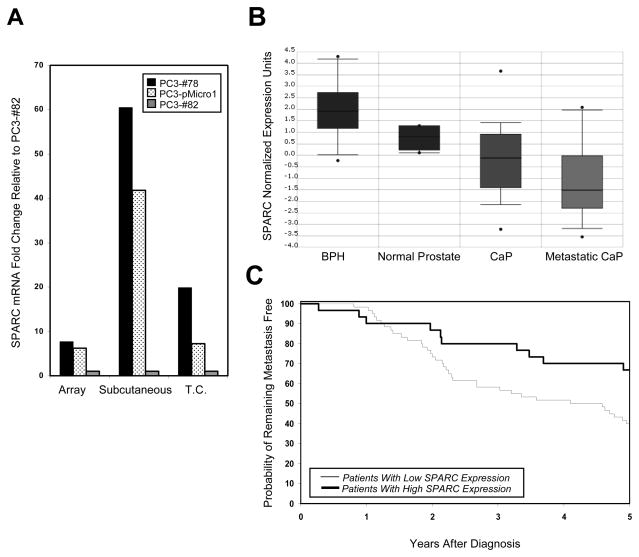
Figure 1. SPARC expression is downregulated in aggressive prostate and breast tumors.
(A) SPARC expression levels, as assessed by gene expression analyses on tumors (“Array”), and by quantitative PCR on tumors (“Subcutaneous”) and on tissue culture cells (“Tissue Culture”), are shown for poorly metastatic PC3-#78 cells (black bars), moderately metastatic PC3-pMicro1 cells (dotted bars) and highly metastatic PC3-#82 cells (gray bars). Within each set of comparisons, the fold change in SPARC expression was expressed relative to SPARC expression in PC3-#82 cells, which was set to 1. (B) Analysis of data originally obtained by Dhanasekaran et al. [44], revealed that SPARC is highly expressed in benign prostate hyperplasia (BPH) and normal prostate tissue, and significantly downregulated in both prostate cancer (CaP) and metastatic CaP (p < 0.01 for CaP or metastatic CaP versus BPH or normal prostate tissue). (C) Analysis of data obtained by van’t Veer et al. [42], revealed that breast cancer patients with low SPARC expression exhibited increased likelihood of developing metastases within 5 years of diagnosis, relative to patients with higher SPARC expression (p = 0.026 by log rank test).
Our results suggested that SPARC may function as a suppressor of prostate cancer progression and that downregulation of SPARC gene expression may enhance tumor aggressiveness. We found that this hypothesis was supported by results obtained from several studies that had examined gene expression in human clinical prostate cancer and which were compiled on the ONCOMINE database. Among them, we found that, in a study performed by Dhanasekaran et al. [44], SPARC expression was significantly downregulated in human prostate carcinomas relative to normal prostate tissue or benign prostate hyperplasia (Figure 1B, p < 0.01), and that SPARC was further downregulated in metastatic prostate cancer. Additional gene expression studies performed on clinical prostate samples by Lapointe et al., and Welsch et al., were also found to yield concordant results [45, 46].
Using gene expression data originally obtained by van’t Veer et al., for human clinical breast cancer [42], Smid et al., noted that downregulation of SPARC expression was significantly associated with tumors harboring the BRCA1 mutation (data not shown) [47]. Interestingly, after analyzing the same data-set, we found that 33% of patients bearing breast tumors with high SPARC expression developed metastases within 5 years of diagnosis (Figure 1C, n = 10 patients out of a total of 30). In contrast, 60% of patients with low SPARC expression possessed metastases within the same time period (n = 36 patients out of a total of 60; p = 0.026 by log rank test). Thus, these data, together with those for prostate cancer, indicated that downregulation of SPARC may either be causal for, or associated with, an aggressive tumor phenotype. However, it should be noted that these gene expression studies do not distinguish whether alterations in SPARC expression occurred within the tumor epithelium and/or in the surrounding stroma.
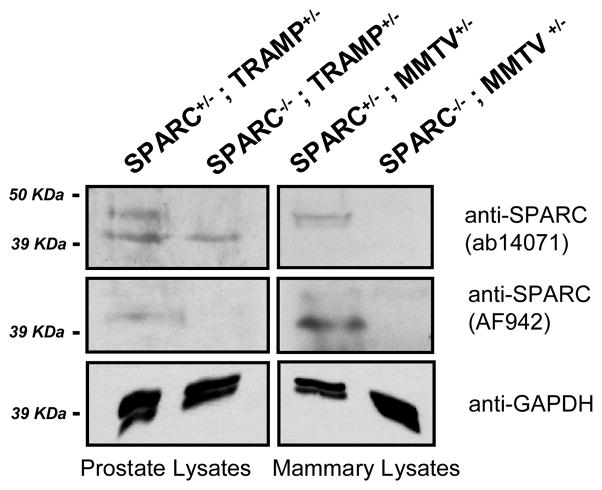
To test the role of SPARC in spontaneous tumor progression, we obtained SPARC−/− mice (gift of E. H. Sage; originally generated by Norose et al.) [35]. For the most part, these animals are healthy and viable, exhibit subtle phenotypes, and are not pre-disposed to forming tumors [2]. Therefore, we crossed SPARC-deficient mice into two spontaneous, viral-oncogene-driven models of prostate and breast cancer: transgenic adenocarcinoma of the mouse prostate (TRAMP) [36] and murine mammary tumor virus-polyoma middle T (MMTV-PyMT) [37]. To confirm loss of SPARC expression in knock-out animals, we extracted total protein from high grade SPARC+/− and SPARC−/− TRAMP prostate and MMTV-PyMT mammary tumors, and performed Western blot analysis for SPARC. In both tumor types, SPARC was detected in tumors derived from SPARC+/− mice, but not in those from SPARC−/− animals, as expected (Figure 2).
Figure 2. SPARC protein expression in spontaneous prostate and mammary tumors derived from SPARC−/− and SPARC+/− animals.
Total lysates from TRAMP prostate and MMTV-PyMT mammary tumors were probed for SPARC using two different antibodies. Top and middle panels, SPARC is present as a ~45 kDa protein in SPARC+/−, but not SPARC−/−, tissues. The lower bands in the top panel for the TRAMP samples are non-specific bands detected by the ab14071 SPARC antibody but not by the AF942 SPARC antibody (middle). Bottom, GAPDH loading controls are shown for all samples.
For our prostate cancer studies, we compared 26-week-old SPARC−/− and SPARC+/− mice that were either TRAMP+ or TRAMP−. In the case of TRAMP− animals, urogenital mass did not differ significantly between SPARC−/− and SPARC+/− mice (Figure 3A), and prostate abnormalities were not observed (data not shown). While TRAMP+ mice exhibited an expansion of prostate and seminal vesicle mass, there was again no difference in overall urogenital mass between SPARC−/− and SPARC+/− animals (Figure 3A). Almost all TRAMP+ mice in the study had developed prostate cancer at the time of analysis, and neither the incidence of palpable carcinomas (35% versus 28% of animals) nor of macroscopic lumbar/para-aortic lymph node metastases (25% versus 21% of animals) differed significantly between SPARC+/− and SPARC−/− mice, respectively (Figure 3B).
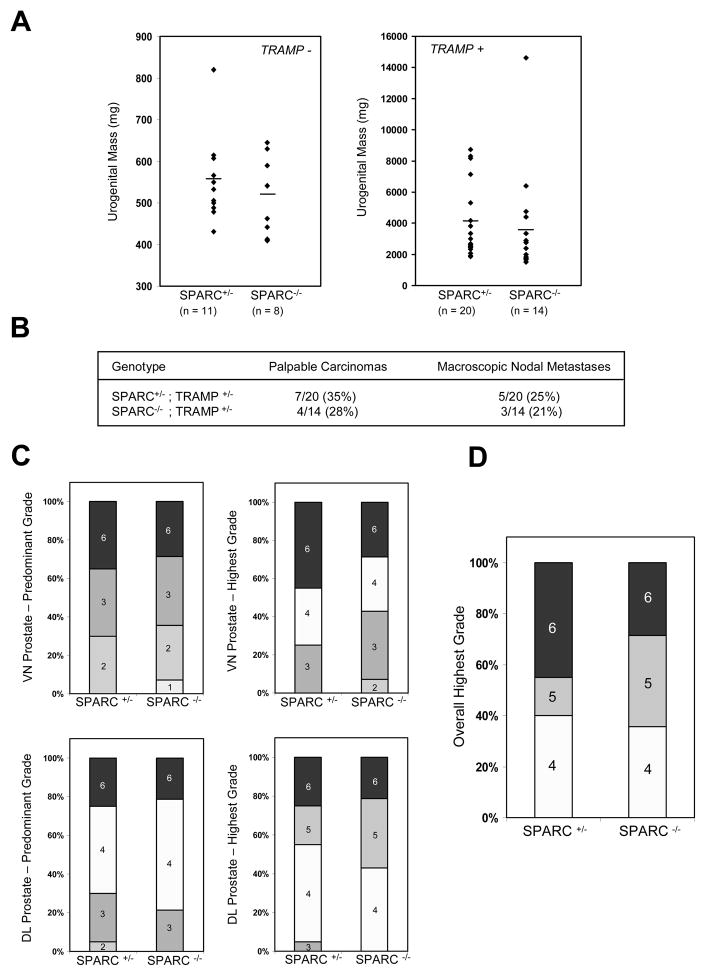
Figure 3. Absence of SPARC does not affect prostate cancer progression or metastasis.
(A) Average urogenital masses of TRAMP− (left, p = 0.44) or TRAMP+ animals (right, p = 0.75) did not differ between genotypes. (B) The incidence of palpable prostate carcinomas and macroscopic para-aortic/lumbar lymph node metastases did not differ between the two cohorts. (C) Disease progression also did not differ between SPARC+/− and SPARC−/− tumors, as assessed by histological grading of the individual dorsolateral (DL) and ventral (VN) prostate lobes. (D) When an overall highest grade was assigned to each prostate sample (identical to the highest grade given to any constituent lobe), no significant differences were observed between genotypes in the distribution of grades awarded.
To assess disease progression at the histological level, we separated and sectioned the ventral (VN) and dorsolateral (DL) lobes of each prostate, and graded the samples based on the scoring method originally described by Hurwitz et al. (see Materials and Methods) [38]. Each section was assigned a predominant grade and also a highest grade, both ranging from a score of ‘1’ for normal prostate to ‘6’ for undifferentiated adenocarcinoma. In all cases, no significant differences in tumor progression were observed between SPARC+/− and SPARC−/− mice (Figure 3C). When a single highest grade was assigned to each prostate (identical to the highest grade given to any constituent lobe), 45% of SPARC+/− mice developed the most severe grade of prostate cancer (Grade 6), compared to 29% of SPARC−/− mice; however, these differences were not statistically significant (Figure 3D, p = 0.33 by Chi-Square test). In addition, the distribution of lower graded tumors was not significantly changed between genotypes.
As mentioned previously, we also used SPARC-deficient animals to assess the role of this protein in spontaneous breast cancer progression. SPARC+/− and SPARC−/− mice expressing PyMT in the mammary fat pads were examined twice weekly beginning at 10 weeks of age for the presence of tumor nodules. The first date of tumor detection was noted, and this served as a rough estimate of time to tumor initiation. All mice were sacrificed approximately four weeks after the initial date of detection, and the total tumor mass for each mouse was assessed, which served as an estimation of the rate of growth. Macroscopic lung metastases were also quantitated at the time of sacrifice.
Using these criteria to measure time to tumor initiation, rate of tumor growth and metastasis, we found no significant differences in cancer progression between SPARC+/− and SPARC−/− mice. The average time to tumor initiation for both cohorts was 15.4 weeks, and all mice bore tumors by 19 weeks of age (Figure 4A). The mean tumor mass of SPARC+/− mice was 5.3 grams, compared with 4.2 grams for SPARC−/− mice (Figure 4B, p = 0.2). Also, no significant differences in mass were observed when tumors were subdivided by quadrant and compared by genotype (Figure 4C). Lastly, SPARC+/− mice bore an average of 9.6 metastatic lung nodules at the time of sacrifice, compared with 23.3 nodules in SPARC−/− animals (Figure 4D, p = 0.17). These differences, however, were largely due to a single outlier SPARC−/− mouse with > 100 lung metastases, and, aside from this animal, the rest of the SPARC−/− cohort (n = 7) averaged 12.3 metastases.
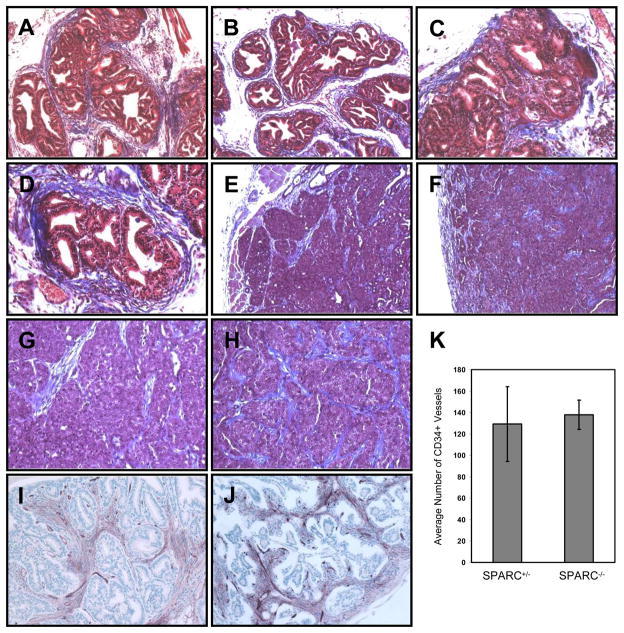
SPARC has been observed in some studies either to enhance or inhibit tumorigenesis by affecting the integrity of the surrounding extracellular matrix, particularly of collagen fibers [7, 25–27]. We therefore performed Masson’s Trichrome staining on both TRAMP prostate and MMTV-PyMT mammary tumors to visualize the collagen networks encapsulating the tumors. In the prostate, collagen staining was observed at the periphery of the acinar ductal structures that contained cancerous lesions (Figure 5A–D). For both SPARC+/− and SPARC−/− tumors, staining was heterogeneous, with some acini surrounded by a fibrous, collagen-rich network, and others possessing little matrix. In mammary tumors, collagen staining was detected at the periphery and also within connective tissue septa located at the interior (Figure 5E–H). However, for both tumor types, we found no readily observable differences in either collagen staining intensity or localization within the tumor, as assessed by trichrome staining. Tumor angiogenesis was also largely unchanged between genotypes (p = 0.80), as observed by staining for the blood vessel-specific marker, CD34 (Figure 5I–K).
Figure 5. Collagen deposition and angiogenesis were largely unaffected by loss of SPARC.
Masson’s trichrome staining was performed on TRAMP prostate cancer lesions (A–D) and on MMTV-PyMT mammary carcinomas (E–H). 10X magnification of images is shown in A, B, E and F; 20X magnification of images is shown in C, D, G and H. CD34 staining of blood vessels at 20X magnification is shown for prostate lesions from SPARC+/− (I) and SPARC−/− (J) animals, and the results are quantitated in (K).
DISCUSSION
Previous studies have reported that SPARC can affect several aspects of the tumorigenic process, including matrix assembly, angiogenesis, proliferation, apoptosis and invasion [2]. Most notably, SPARC is thought to enhance the integrity of collagen matrices which encapsulate tumors. This encapsulation has been postulated to act as a physical restraint that limits tumor size (thus serving a tumor suppressive function) [7, 26], but it may also prevent leukocytes from infiltrating into the tumor, as has been observed for mammary carcinomas [27]. In addition, recent work has suggested that SPARC may inhibit TGF-β-mediated fibroblast activation, as assessed by expression of smooth muscle actin [48]. Although the exact implications of this remain unclear, activated fibroblasts are thought to promote tumor progression by secreting cytokines and matrix molecules that could potentially affect tumor cell proliferation and angiogenesis [49].
It is interesting to note that expression of SPARC is frequently altered in a variety of human cancers, and there has also been experimental evidence that this protein can both promote and inhibit tumorigenesis. In light of these conflicting results, the effects of SPARC have been interpreted as being cell type- and/or tumor microenvironment-specific, and may depend upon the local availability of MMPs and other proteolytic enzymes which are known to process SPARC post-translationally [23, 27]. For instance, plasmin-mediated cleavage has been reported to convert SPARC from an anti-angiogenic molecule into one that favors blood vessel growth [28]. Thus, when considering the effects of SPARC on tumorigenesis, several factors need to be taken into account, including the degree to which SPARC is processed, the stromal context in which SPARC and its fragments are present, and also the cell types that ultimately respond to this molecule. Clearly, this is a complicated situation — one that becomes even more complex if the varying effects attributed to SPARC synergize or counter-balance one another during tumor progression.
Although numerous reports have implicated SPARC in tumorigenesis, most of these studies used tumor cell transplant experiments. We therefore evaluated the role of endogenous SPARC in spontaneous tumor progression for prostate and breast cancer. Surprisingly, in both systems, we observed no significant differences in tumor initiation, progression or metastasis between SPARC+/− and SPARC−/− mice. Collagen deposition and angiogenesis were also largely unaffected.
There are several possible explanations for why our results are discrepant with those previously reported by others. Foremost among these is our use of spontaneous tumor models to test the effects of SPARC, which is an approach that differs drastically from xenotransplant systems in that SPARC is deleted both in the tumor and the stroma, and that most of the steps of tumorigenesis are recapitulated in vivo. Thus, it is likely that a variety of factors may have differed between our experimental system and those of others, including immune response, tumor matrix composition and the local protease profile of the tumor microenvironment. Our study also differed in that we compared SPARC+/− and SPARC−/− mice in our tumor experiments, whereas others utilized SPARC+/+ versus SPARC−/− animal comparisons [7, 27, 29, 30]. It is currently unclear whether SPARC+/− mice may be, to some degree, haplo-insufficient, although this is unlikely, given that SPARC−/− mice are healthy and viable, and manifest only subtle phenotypes [2]. In addition, while some studies have suggested that SPARC may act as a chemotactic factor for renal and prostate cancer cells, particularly to bone [18, 21], spontaneous tumors in mice metastasize rarely, if at all, to bone. Therefore, our experiments do not rule out the possibility that SPARC could affect osseous metastasis, a process that has been better modeled using xenotransplant approaches such as intracardiac injection of tumor cells [50]. Finally, although SPARC has been reported to affect matrix deposition and integrity in tumors [7, 25–27], we observed no readily apparent differences in collagen matrix deposition between SPARC+/− and SPARC−/− tumors. Whether the integrity of this matrix was affected remains to be tested.
While our results suggest that loss of SPARC, by itself, is insufficient to promote or inhibit spontaneous prostate and breast cancer progression, our findings do not exclude the possibility that SPARC is important for other tumor types. For instance, SPARC-deficient mice have been reported to be resistant to intestinal adenomas and squamous cell carcinomas [29, 30]. In addition, in spite of our animal model results, SPARC expression may still prove useful as a metric for assessing tumor aggressiveness. Indeed, our interest in SPARC originally grew out of our observation that its expression is significantly downregulated in highly metastatic prostate cancer cells, as well as in several studies of human clinical prostate cancer. We have also found that reduced SPARC expression is predictive of metastatic disease in human breast cancer patients. Thus, while our results indicate that loss of SPARC does not contribute causally to prostate or breast cancer in the mouse models tested, its association with disease progression may, nonetheless, make it a potentially useful biomarker.
Acknowledgments
We are very grateful to Dr. E. Helene Sage and Sarah Funk (Benaroya Research Institute, Seattle, WA) for SPARC-deficient mice; Dr. Lei Xu (MIT) for MMTV-PyMT mice; Dr. Ailin Bai (MIT) for TRAMP mice; Jane Trevithick for technical assistance; and the MIT Division of Comparative Medicine for animal maintenance. This work was supported by grants from the NIH (RO1CA17007); from the Virginia and D. K. Ludwig Fund for Cancer Research; from the Prostate Cancer Foundation; from the National Cancer Institute’s Integrative Cancer Biology Program (U54-CA112967); and from the Howard Hughes Medical Institute, of which R.O.H. is an Investigator. S.Y.W. was further supported by an NIGMS Predoctoral Training Grant to the MIT Biology Department and by a David H. Koch Research Fellowship from the Center for Cancer Research.
Abbreviations
- SPARC
secreted protein, acidic and rich in cystein
- TRAMP
transgenic adenocarcinoma of the mouse prostate
- MMTV-PyMT
murine mammary tumor virus-polyoma middle T
- DL
dorsolateral prostate
- VN
ventral prostate
- SOI
surgical orthotopic implantation
References
- 1.Brekken RA, Sage EH. SPARC, a matricellular protein: at the crossroads of cell-matrix communication. Matrix Biol. 2001;19:816–827. doi: 10.1016/s0945-053x(00)00133-5. [DOI] [PubMed] [Google Scholar]
- 2.Framson PE, Sage EH. SPARC and tumor growth: where the seed meets the soil? J Cell Biochem. 2004;92:679–690. doi: 10.1002/jcb.20091. [DOI] [PubMed] [Google Scholar]
- 3.Bradshaw AD, Sage EH. SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J Clin Invest. 2001;107:1049–1054. doi: 10.1172/JCI12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradshaw AD, Reed MJ, Sage EH. SPARC-null mice exhibit accelerated cutaneous wound closure. J Histochem Cytochem. 2002;50:1–10. doi: 10.1177/002215540205000101. [DOI] [PubMed] [Google Scholar]
- 5.Murphy-Ullrich JE. The de-adhesive activity of matricellular proteins: is intermediate cell adhesion an adaptive state? J Clin Invest. 2001;107:785–790. doi: 10.1172/JCI12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sage H, Vernon RB, Funk SE, Everitt EA, Angello J. SPARC, a secreted protein associated with cellular proliferation, inhibits cell spreading in vitro and exhibits Ca+2-dependent binding to the extracellular matrix. J Cell Biol. 1989;109:341–356. doi: 10.1083/jcb.109.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brekken RA, et al. Enhanced growth of tumors in SPARC null mice is associated with changes in the ECM. J Clin Invest. 2003;111:487–495. doi: 10.1172/JCI16804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motamed K, Sage EH. SPARC inhibits endothelial cell adhesion but not proliferation through a tyrosine phosphorylation-dependent pathway. J Cell Biochem. 1998;70:543–552. doi: 10.1002/(sici)1097-4644(19980915)70:4<543::aid-jcb10>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 9.Kupprion C, Motamed K, Sage EH. SPARC (BM-40, osteonectin) inhibits the mitogenic effect of vascular endothelial growth factor on microvascular endothelial cells. J Biol Cell. 1998;273:29635–29640. doi: 10.1074/jbc.273.45.29635. [DOI] [PubMed] [Google Scholar]
- 10.Schiemann BJ, Neil JR, Schiemann WP. SPARC inhibits epithelial cell proliferation in part through stimulation of the transforming growth factor-beta-signaling system. Mol Biol Cell. 2003;14:3977–3988. doi: 10.1091/mbc.E03-01-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tremble PM, Lane TF, Sage EH, Werb Z. SPARC, a secreted protein associated with morphogenesis and tissue remodeling, induces expression of metalloproteinases in fibroblasts through a novel extracellular matrix-dependent pathway. J Cell Biol. 1993;121:1433–1444. doi: 10.1083/jcb.121.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shankavaram UT, DeWitt DL, Funk SE, Sage EH, Wahl LM. Regulation of human monocyte matrix metalloproteinases by SPARC. J Cell Physiol. 1997;173:327–334. doi: 10.1002/(SICI)1097-4652(199712)173:3<327::AID-JCP4>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 13.Wang CS, Lin KH, Chen SL, Chan YF, Hsueh S. Overexpression of SPARC gene in human gastric carcinoma and its clinic-pathologic significance. Br J Cancer. 2004;91:1924–1930. doi: 10.1038/sj.bjc.6602213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin D, et al. Novel markers for poor prognosis in head and neck cancer. Int J Cancer. 2005;113:789–797. doi: 10.1002/ijc.20608. [DOI] [PubMed] [Google Scholar]
- 15.Yamanaka M, et al. Analysis of the gene expression of SPARC and its prognostic value for bladder cancer. J Urol. 2001;166:2495–2499. [PubMed] [Google Scholar]
- 16.Thomas R, True LD, Bassuk JA, Lange PH, Vessella RL. Differential expression of osteonectin/SPARC during human prostate cancer progression. Clin Cancer Res. 2000;6:1140–1149. [PubMed] [Google Scholar]
- 17.Bail BL, et al. Osteonectin/SPARC is overexpressed in human hepatocellular carcinoma. J Pathol. 1999;189:46–52. doi: 10.1002/(SICI)1096-9896(199909)189:1<46::AID-PATH392>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 18.De S, et al. Molecular pathway for cancer metastasis to bone. J Biol Chem. 2003;278:39044–39050. doi: 10.1074/jbc.M304494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ledda MF, et al. Suppression of SPARC expression by antisense RNA abrogates the tumorigenicity of human melanoma cells. Nat Med. 1997;3:171–176. doi: 10.1038/nm0297-171. [DOI] [PubMed] [Google Scholar]
- 20.Robert G, et al. SPARC represses E-cadherin and induces mesenchymal transition during melanoma development. Cancer Res. 2006;66:7516–7523. doi: 10.1158/0008-5472.CAN-05-3189. [DOI] [PubMed] [Google Scholar]
- 21.Jacob K, Webber M, Benayahu D, Kleinman HK. Osteonectin promotes prostate cancer cell migration and invasion: a possible mechanism for metastasis to bone. Cancer Res. 1999;59:4453–4457. [PubMed] [Google Scholar]
- 22.Yiu GK, et al. SPARC (secreted protein acidic and rich in cysteine) induces apoptosis in ovarian cancer cells. Am J Pathol. 2001;159:609–622. doi: 10.1016/S0002-9440(10)61732-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koblinski JE, et al. Endogenous osteonectin/SPARC/BM-40 expression inhibits MDA-MB-231 breast cancer cell metastasis. Cancer Res. 2005;65:7370–7377. doi: 10.1158/0008-5472.CAN-05-0807. [DOI] [PubMed] [Google Scholar]
- 24.Tai IT, Dai M, Owen DA, Chen LB. Genome-wide expression analysis of therapy-resistant tumors reveals SPARC as a novel target for cancer therapy. J Clin Invest. 2005;115:1492–1502. doi: 10.1172/JCI23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chlenski A, et al. SPARC expression is associated with impaired tumor growth, inhibited angiogenesis and changes in the extracellular matrix. Int J Cancer. 2006;118:310–316. doi: 10.1002/ijc.21357. [DOI] [PubMed] [Google Scholar]
- 26.Puolakkainen PA, Brekken RA, Muneer S, Sage EH. Enhanced growth of pancreatic tumors in SPARC-null mice is associated with decreased deposition of extracellular matrix and reduced tumor cell apoptosis. Mol Cancer Res. 2004;2:215–224. [PubMed] [Google Scholar]
- 27.Sangaletti S, Stoppacciaro A, Guiducci C, Torrisi MR, Colombo MP. Leukocytes, rather than tumor-produced SPARC, determines stroma and collagen type IV deposition in mammary carcinoma. J Exp Med. 2003;198:1475–1485. doi: 10.1084/jem.20030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iruela-Arispe ML. Expression of SPARC during development of the chicken chorioallantoic membrane: evidence for regulated proteolysis in vivo. Mol Biol Cell. 1995;6:327–343. doi: 10.1091/mbc.6.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aycock RL, Bradshaw AC, Sage EH, Starcher B. Development of UV-induced squamous cell carcinomas is suppressed in the absence of SPARC. J Invest Dermatol. 2004;123:592–599. doi: 10.1111/j.0022-202X.2004.23316.x. [DOI] [PubMed] [Google Scholar]
- 30.Sansom OJ, Mansergh F, Evans M, Wilkins J, Clarke A. Deficiency of SPARC suppresses intestinal tumourigenesis in APCMin/+ mice. Gut. 2007 doi: 10.1136/gut.2006.116921. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong SY, et al. Protein 4. 1B suppresses prostate cancer progression and metastasis. Proc Natl Acad Sci USA. 2007;104:12784–12789. doi: 10.1073/pnas.0705499104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang XH, et al. Improved metastatic animal model of human prostate carcinoma using surgical orthotopic implantation (SOI) Anticancer Res. 1999;19:4199–4202. [PubMed] [Google Scholar]
- 33.An Z, Wang X, Geller J, Moossa AR, Hoffman RM. Surgical orthotopic implantation allows high lung and lymph node metastatic expression of human prostate carcinoma cell line PC-3 in nude mice. Prostate. 1998;34:169–174. doi: 10.1002/(sici)1097-0045(19980215)34:3<169::aid-pros3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 34.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3) Invest Urol. 1979;17:16–23. [PubMed] [Google Scholar]
- 35.Norose K, et al. SPARC deficiency leads to early-onset cataractogenesis. Invest Ophthalmol Vis Sci. 1998;39:2674–2680. [PubMed] [Google Scholar]
- 36.Greenberg NM, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci USA. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurwitz AA, Foster BA, Allison JP, Greenberg NM, Kwon ED. Current Protocols in Immunology. Wiley; New York: 2001. The TRAMP mouse as a model for prostate cancer. [DOI] [PubMed] [Google Scholar]
- 39.Armed Forces Institute of Pathology Staff, Prophet EB. Afip Laboratory Methods in Histotechnology. American Registry of Pathology; Washington: 1992. [Google Scholar]
- 40.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:0032.1–0032.11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–535. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 43.Wong SY, et al. Tumor-secreted vascular endothelial growth factor-C is necessary for prostate cancer lymphangiogenesis, but lymphangiogenesis is unnecessary for lymph node metastasis. Cancer Res. 2005;65:9789–9798. doi: 10.1158/0008-5472.CAN-05-0901. [DOI] [PubMed] [Google Scholar]
- 44.Dhanasekaran SM, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 45.Lapointe J, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welsh JB, et al. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- 47.Smid M, Dorssers LC, Jenster G. Venn Mapping: clustering of heterologous microarray data based on the number of co-occurring differentially expressed genes. Bioinformatics. 2003;19:2065–2071. doi: 10.1093/bioinformatics/btg282. [DOI] [PubMed] [Google Scholar]
- 48.Chlenski A, et al. SPARC enhances tumor stroma formation and prevents fibroblast activation. Oncogene. 2007;26:4513–4522. doi: 10.1038/sj.onc.1210247. [DOI] [PubMed] [Google Scholar]
- 49.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;6:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]