Summary
Kinetochores link centromeric DNA to spindle microtubules and ensure faithful chromosome segregation during mitosis. In point-centromere yeasts, the CBF3 complex, Skp1:Ctf13:(Cep3)2:(Ndc10)2, recognizes a conserved centromeric DNA element through contacts made by Cep3 and Ndc10. We describe here the five-domain organization of Kluyveromyces lactis Ndc10 and the structure at 2.8 Å resolution of domains I–II (residues 1–402) bound to DNA. The structure resembles tyrosine DNA recombinases, although it lacks both endonuclease and ligase activities. Structural and biochemical data demonstrate that each subunit of the Ndc10 dimer binds a separate fragment of DNA, suggesting that Ndc10 stabilizes a DNA loop at the centromere. We describe in vitro association experiments showing that specific domains of Ndc10 interact with each of the known inner-kinetochore proteins or protein complexes in budding yeast. We propose that Ndc10 provides a central platform for inner-kinetochore assembly.
Introduction
Kinetochores are tightly regulated, multi-component complexes that assemble on centromeres and effect chromosome attachment to mitotic-spindle microtubules. They are the loci of force generation necessary to establish bipolar attachment of paired sister chromatids and the source of spindle checkpoint signals to prevent entry into anaphase until all chromatid pairs are correctly attached. The kinetochores of budding yeasts, with "point centromeres" of roughly 150 bp and a single microtubule attachment, appear to correspond to a single module of the multi-modular kinetochores in higher eukaryotes, which have "regional centromeres" of up to several Mb and 30–40 microtubule attachments1–4. Various lines of evidence -- e.g., protein sequence conservation5, quantitative fluorescence microscopy6, and the similarity of the CENP-A:H4:chaperone structures from point and regional centromeres7,8, strongly suggest conservation of basic kinetochore architecture across all eukaryotes.
An assembled kinetochore comprises a set of proteins that interact directly with centromeric DNA ("inner kinetochore" proteins), a set that contact the attached microtubule(s) ("outer kinetochore" proteins), and an intermediate array of linkers9. Proteins of the inner kinetochore conserved throughout evolution include a variant histone H3, known as Cse4 in budding yeast and as CENP-A in higher eukaryotes10–13, and a transcription-factor-like protein called Mif2 (CENP-C)10,14–17. The single-module kinetochore of S. cerevisiae assembles onto a ~150 bp centromere with three distinct DNA elements, designated centromere-determining element (CDE) I, II, and III2,18. CDEII wraps into a specialized nucleosome13,19,20, which appears to incorporate less than than the canonical 140 bp21,22. It is flanked in S. cerevisiae by Cbf1, a helix-loop-helix protein23, bound at CDEI, and by a four-protein complex called centromere-binding factor (CBF) 3, bound at CDEIII24–29. It is not yet clear to what extent contacts from these factors overlap the Cse4 nucleosome and hence exactly how much DNA winds onto the histone core. Neither Cbf1 nor CBF3 have obvious homologs in other eukaryotic cells, but CBF3 recruits a histone chaperone, Scm3, with a conserved domain and apparently conserved function21,30–33. Thus, the molecular mechanisms of kinetochore assembly appear to be very general, even if directed or regulated by somewhat different protein complexes in different organisms.
The components of CBF3 are a Skp1:Ctf13 heterodimer, a Cep3 homodimer, and an Ndc10 homodimer24–26,28,29,34. There is some in vitro evidence for a second Ndc10 homodimer assembling with the DNA-bound complex35. A Skp1:Ctf13:(Cep3)2 subassembly, which we will call here the "CBF3 core", is stable in vitro. The positions of various components with centromeric DNA have been mapped in vitro by DNA modification methods and in vivo by high-resolution chromatin immunoprecipitation (ChIP)19,35. Cep3 is a DNA-binding protein with an N-terminal, GAL4-like "Zn cluster"29. It binds DNA weakly but specifically, with contacts that include a CCG element on one side of an approximately palindromic sequence35. Ndc10 binds DNA strongly but nonspecifically (other than with a preference for A:T-rich sequences)36. The Skp1:Ctf13 heterodimer is probably a structural homolog of the higher eukaryotic Skp1:Skp2 complex, a component of the Skp1–Cul1–F-box protein (SCF) ubiquitin ligase37. What relationship, if any, its ubiquitin-ligase-like structure has to its organizational role in kinetochore assembly remains to be determined.
Although its initial characterization was as a component of CBF325,34, Ndc10 may also have centromere-independent functions. The chromosomal passenger complex (Ipl1–Sli15–Bir1–Nbl1 in yeast; Aurora B–INCENP–Survivin–Borealin in higher eukaryotes) and Ndc10 colocalize with interpolar microtubules at the spindle midzone in late anaphase38. The interaction of Ndc10 with the chromosomal passenger complex is through a contact with Bir139.
We report here an analysis of the structure and interactions of Ndc10 from Kluyveromyces lactis, a budding yeast with six point centromeric chromosomes40. The sequence characteristics of CDEI and CDEIII are conserved between K. lactis and S. cerevisiae, but the A:T-rich CDEII sequence is twice as long in the former (~160 bp) as in the latter (~80 bp)41. We find that the 85 kDa polypeptide chain of K. lactis Ndc10 has five domains, which we have defined either by direct structural analysis or by proteolytic dissection. We have mapped interactions of the Ndc10 domains with all the other known, DNA-proximal kinetochore components including Scm3, a Cse4 specific chaperone. The crystal structure of a large fragment comprising domains I and II in complex with DNA shows that the protein has an unexpected structural relationship to tyrosine DNA recombinases and that each subunit of an Ndc10 dimer is likely to bind a distinct DNA element, rather than closely spaced elements on a single DNA duplex. We have confirmed this latter conclusion in vitro. These results suggest that Ndc10 may create or stabilize a loop within centromeric DNA, and together with the data on Ndc10 interactions with other kinetochore proteins they suggest that Ndc10 is a central organizing hub of the inner kinetochore.
Results
Domains of K. lactis Ndc10
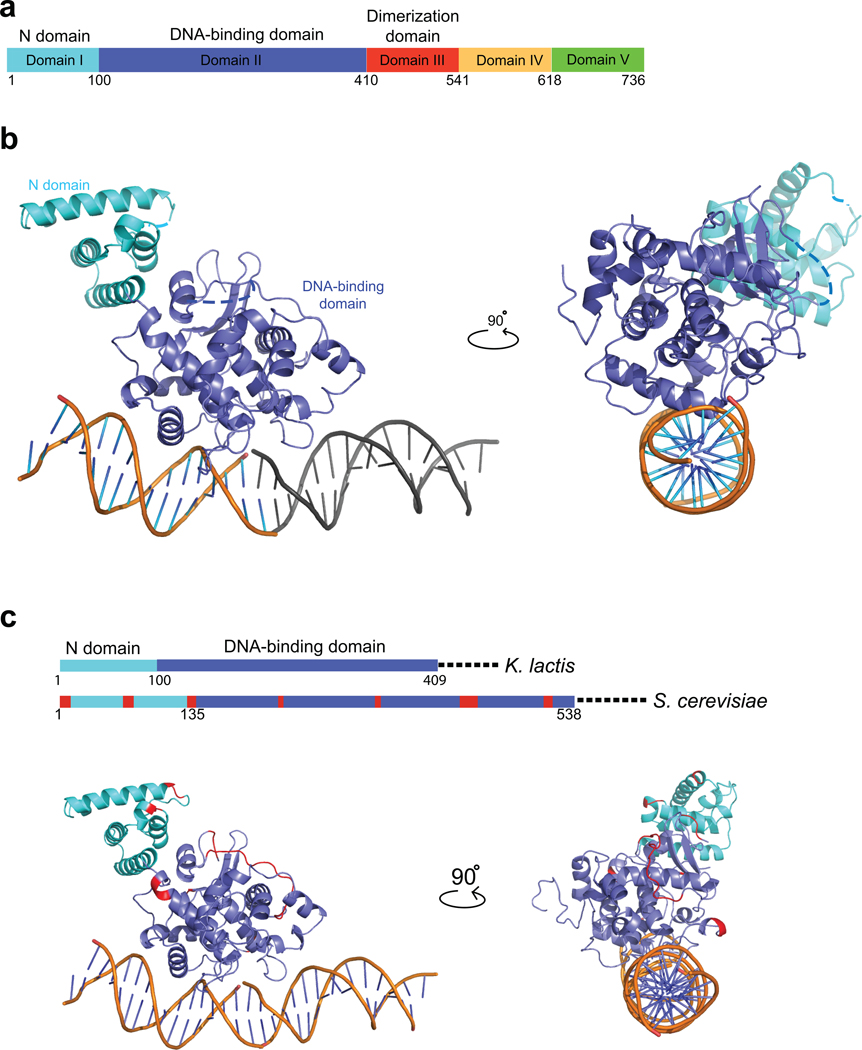
In an effort to overcome problems associated with bacterial expression of S. cerevisiae Ndc10, we cloned and expressed Ndc10 from other point-centromere yeasts. Ndc10 is highly diverged in these species, with the proteins from S. cerevisiae (956 amino acid residues), Ashbya gossypii (728), Kluyveromyces waltii (733), Saccharomyces kluyveri (875), Candida glabrata (906), and Kluyveromyces lactis (736) having between 25% and 32% pairwise identity. We expressed and purified the full-length K. lactis Ndc10, then carried out limited tryptic proteolysis to identify potential domain boundaries (Supplementary Fig. 1a); mass spectrometry showed that there are three positions of high trypsin sensitivity (C-terminal to Lys410, Lys541, Lys618), thereby defining four fragments that we designate as domains I–II (the further division is based on the three-dimensional structure; see below), III, IV, and V (Fig. 1a–c). In addition to the full-length protein, we purified fragments of K. lactis Ndc10 corresponding to DI–II (residues 1–403), DI–III (residues 1–534), and C-terminal fragments DIV–V (residues 552–736) and DV (residues 631–736). By sedimentation equilibrium analytical ultracentrifugation, DI–II is monomeric and DI–III is dimeric (Supplementary Fig. 1b,c), indicating that DIII is largely responsible for dimerization of the protein.
Figure 1. Domains of K. lactis Ndc10 and crystal structure of DI–II.
(a) Domain organization of Ndc10; numbers show residues at the domain boundaries derived either from limited proteolysis or from the crystal structure. (b) Structure of K. lactis Ndc10 (DI–II; 1–402) with 30 bp poly-dA:dT DNA. Domain I (N-domain, residues 1–100) is in cyan; domain II (DNA-binding domain, residues 101–402), in dark blue. Dashed lines represent disordered residues 36–39 and 283–292. A second, symmetry-related, 15 bp DNA fragment is shown in gray. The DNA has been modeled as polydA:dT (see text), with the sequence of 5’-TTAATTTATAAAATT-3’ (1–15) and 5’-AAATTTTATAAATTA-3’ (1’–15’), as indicated. (c) Sequence conservation of Ndc10 among point-centromere yeasts. Location of insertions (red) in S. cerevisiae Ndc10 DI–II with respect to K. lactis Ndc10 DI–II, shown both on a schematic representation of the primary sequence and on a ribbon representation of the structure. Illustration of all figures made with PYMOL (Delano Scientific, LLC).
DNA binding by Ndc10
The complete S. cerevisiae CBF3 complex -- Skp1:Ctf13, (Cep3)2, (Ndc10)2 -- covers about 57 bp of CDEIII35,42. In the absence of the specialized nucleosome, Ndc10 can also bind CDEII in vitro 36. We tested the affinity of full-length K. lactis Ndc10 for various lengths of the K. lactis CEN1 CDEIII and found that optimum binding required 25–30 bp of DNA (Supplementary Fig. 1d). We also tested sequence specificity using 30mers with different G:C content and found that binding was largely G:C-content and base-sequence independent (Supplementary Fig. 1e). We note that the Cep3 dimer probably occupies 25–30 bp at the "left-hand" end of CDEIII43 and that 25–30 bp is therefore just what we might expect if the Ndc10 dimer were to cover the rest of the 57 bp total.
We identified the domains of Ndc10 required for DNA binding, using an electrophoretic mobility shift assay (EMSA) and fluorescence polarization measurements with various fragments of the protein (Supplementary Fig. 1f,g). The DI–II fragment is the principal contributor to DNA binding; a DIV–V fragment also showed weak association with DNA. While isolated domain V does not bind CDEIII in our EMSA assay (Supplementary Fig. 1g), fluorescence polarization measurements indicate that the affinity of the DI–III fragment is slightly lower than that of the full-length protein, consistent with some contribution from the C-terminal region (Supplementary Fig. 1f).
Structure of a K. lactis Ndc10:DNA complex
We crystallized a complex of K. lactis Ndc10 DI–II (residues 1–403) with a 30 bp DNA fragment of K. lactis CEN1 CDEIII, and determined the structure to a resolution of 2.8 Å by single-wavelength anomalous-scattering (SAD) with SeMet-substituted protein (Fig. 1b, Table 1). The final model contains residues 1–402 of Ndc10, except for disordered loops from 36 to 39 and 283 to 292, and the DNA (built as poly-dA:dT, see below). The crystals are in space group C2221, with one Ndc10 subunit and 15 bp of DNA in the asymmetric unit. The DNA fragments stack end-to-end, parallel to the c-axis (which is equal in length to one 30bp DNA duplex). Because there is only half of the 30 bp DNA fragment in one asymmetric unit, the observed electron density is the two-fold average of the two halves of the DNA; we therefore modeled the DNA as poly-dA:dT. A single Ndc10 subunit contacts about 25 bp of DNA (consistent with the results in Supplementary Fig. 1d); overlapping contacts from adjacent Ndc10 subunits on opposite faces of each DNA account for the half-fragment asymmetric unit (Supplementary Fig. 1i).
Table 1.
Data collection and refinement statistics for K. lactis Ndc10 DI–II complexed with CDEIII DNA.
| Native 1 (C2221) | SAD (SeMet, C2221) | Native 2 (P212121) | |
|---|---|---|---|
| Data collection | |||
| Space group | C2221 | C2221 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 117.29, 147.51, 95.63 | 113.37, 148.20, 94.82 | 93.1, 99.7, 125.9 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
|
peak |
|||
| Wavelength | 0.9795 | 0.9795 | 0.9795 |
| Resolution (Å) | 50 – 2.8 (2.90-2.80)a | 50 – 3.0 (3.11-3.00) | 50 – 3.6 (3.73-3.60) |
| Rsym or Rmerge | 0.113 (0.717) | 0.127 (0.603) | 0.104 (0.912) |
| I / σ I | 23.8 (2.0) | 15.5 (2.0) | 6.4 (1.4) |
| Completeness (%) | 92.6 (74.2) | 91.7 (66.8) | 99 (99.9) |
| Redundancy | 14.7 | 5.2 | 3.4 |
| Refinement | |||
| Resolution (Å) | 50 – 2.8 | 50–3.6 | |
| No. reflections | 17663 | 13133 | |
| Rwork / Rfree | 19.3 / 25.1 | 28.6 / 33.1 | |
| No. atoms | |||
| Protein | 3196 | 6392 | |
| DNA | 615 | 1211 | |
| Water | 0 | 0 | |
| B-factors | |||
| Protein | 110.87 | 122.43 | |
| DNA | 159.20 | 142.26 | |
| Water | n/a | n/a | |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.005 | 0.009 | |
| Bond angles (°) | 1.147 | 1.466 |
Values in parentheses are for the highest-resolution shell.
Ndc10 (residues 1–402) contains two closely associated globular domains -- an N-domain (domain I, residues 1–100) and a DNA-binding domain (domain II, residues 101–402). The N-domain is a bundle of five α-helices, and the DNA-binding domain is largely helical with a small β-sheet. The domain I–domain II interface has well-packed hydrophobic residues, suggesting that the two domains constitute a single globular unit (Supplementary Fig. 1j). This conclusion is consistent with our failure to detect any tryptic cleavage at lysine 97, which lies just at the domain boundary.
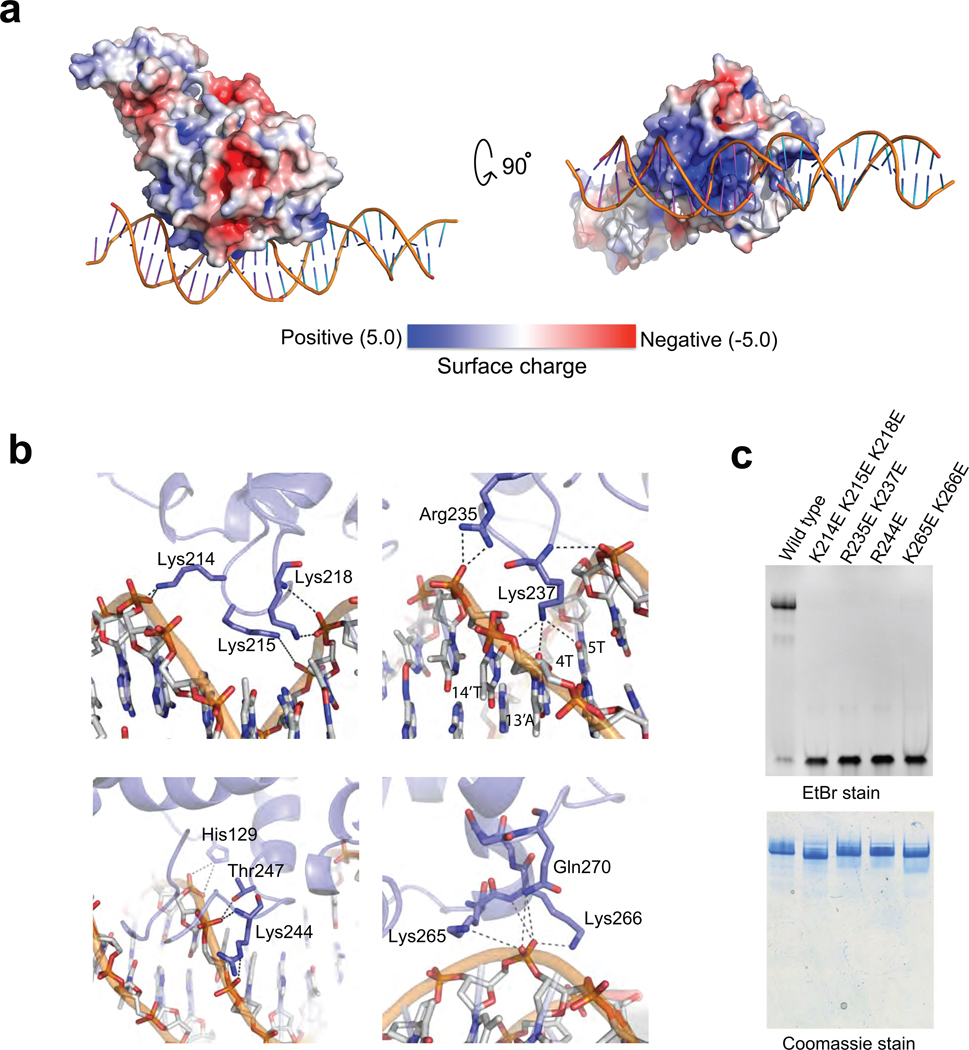
The contacts between Ndc10(1–402) and DNA are essentially all with the sugar-phosphate backbone, in keeping with the observed lack of sequence specificity (Supplementary Fig. 1e). The contact surface is entirely within domain II; a small crystal-packing contact from domain I (to another DNA segment) does not appear to be a functionally relevant interaction. The side chains of a number of residues (H129, K214, K215, K218, R235, K244, T247, K265, K266, Q270) have either ionic or hydrogen-bond interactions with DNA, and the main-chain amides of Lys218, Lys237, and Lys265 also donate hydrogen bonds to phosphate oxygens (Fig. 2b). These residues create three patches of positive electrostatic potential on the DNA-facing surface of domain II (Fig. 2a). Each patch contacts a neighboring segment of DNA backbone. The Lys237 side chain projects into the narrow minor groove, where it could donate hydrogen bonds to the minor-groove carbonyl groups of U or C at positions 4 and 14' or to carbonyl groups at positions 5 or 13'. It is therefore unlikely to dictate any base-pair preference beyond discriminating against G:C base pairs; the Ndc10-associated region of CDEIII is in any case quite A:T-rich. To verify the relevance of the observed contacts for DNA binding in solution, we mutated a number of the DNA contacting residues of Ndc10, and tested for DNA binding by EMSA: none of mutants we examined retained detectable affinity for DNA (Fig. 2c).
Figure 2. Surface charge distribution and DNA contacts of Ndc10 DI–II.
(a) Two views of the surface charge distribution of Ndc10 DI–II; bound DNA is shown in worm representation. (b) Sugar-phosphate backbone interactions. Residues involved in DNA contacts are labeled and shown in stick representation. (c) EMSA of wild-type and mutant Ndc10 (10% TBE acrylamide gel (w/v) stained sequentially with EtBr and Coomassie blue).
The S. cerevisiae Ndc10 polypeptide chain is 220 residues longer than that of K. lactis. The insertions are distributed into a number of discrete segments. Of the incremental residues, 129 are in domains I and II. As shown in Figure 1c, they form insertions into surface loops, none of them in DNA-contacting regions. The expansions in the S.cerevisiae Ndc10 sequence also tend to coincide with positions of poor conservation among the entire group of point-centromere yeast Ndc10 orthologs (Supplementary Fig. 2a,b).
Structural alignment with DNA recombinases
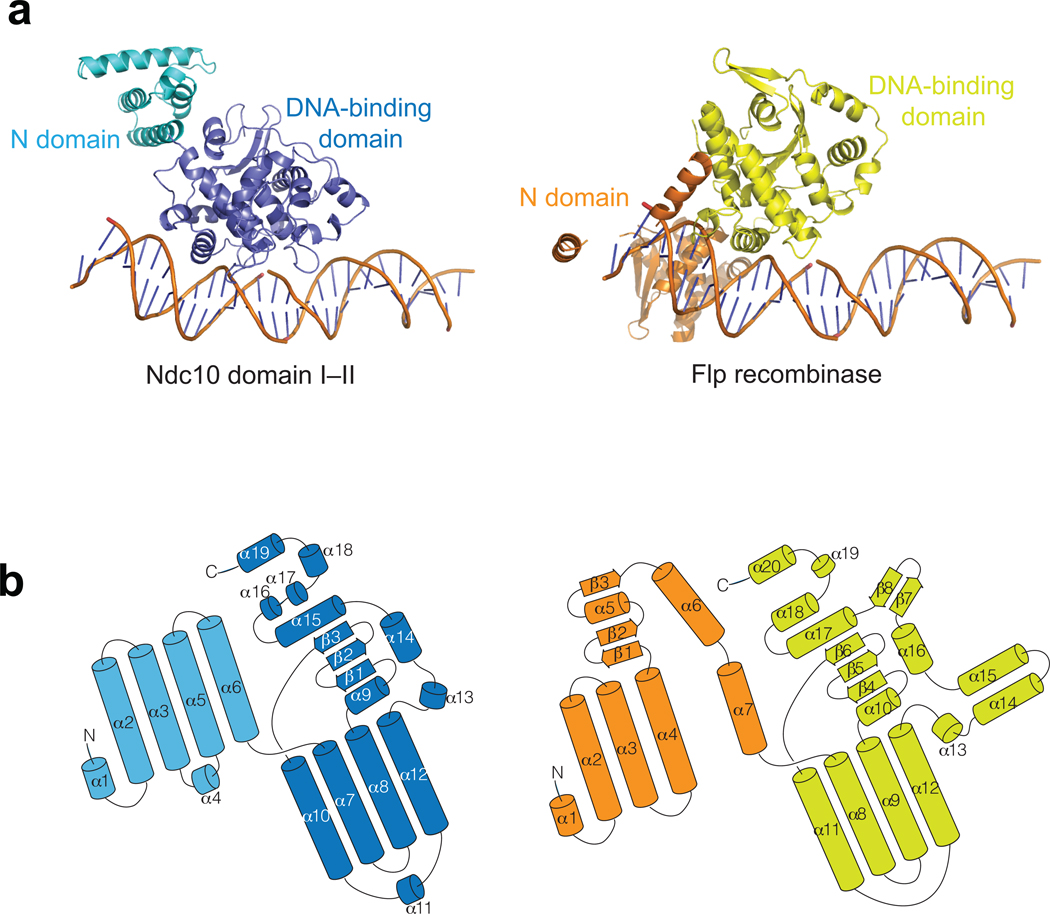
A search for three-dimensional structures related to Ndc10 using both a distance-matrix alignment program (Dali)44 and a secondary-structure matching program (SSM)45 yields the DNA recombinases Flp46 (PDB code: 1M6X, Fig. 3a,b) and Cre47 (PDB code: 1CRX), members of the λ-integrase family of tyrosine-based, site-specific DNA recombinases. Similarity of Ndc10 and λ-integrases is detectable from sequence relationships, but only after a large number of iterations of the program PSI-BLAST48. Also among PSI-BLAST-identified relatives are the crypton-encoded recombinase domains49, which in addition to the recombinase region, have a C-terminal domain related to Ndc10 domain V.
Figure 3. Structural alignment of K. lactis Ndc10 DI–II with Flp recombinases.
(a) Monomer structure of Flp (PDB ID: 1M6X) aligned with the K. lactis Ndc10 DI–II. The N-domain and the DNA binding domain of Flp recombinase are colored in orange and yellow, respectively. In Flp, the DNA structure of the Holliday junction was replaced by 30 bp CDEIII DNA for simple comparison. (b) Folding diagrams of K. lactis Ndc10 DI–II and Flp recombinase. Secondary-structure elements are labeled according to their position in the polypeptide chain; domains colored as in panel a.
Direct structure comparison shows that domains I and II of Ndc10 have essentially the same folds as do the corresponding domains of Cre and Flp, and when domain II of Ndc10 is superposed on domain II of either recombinase, the positions of bound DNA overlap (Supplementary Fig. 3a). Ndc10 has no catalytic activity, however, as it lacks the relevant active-site residues, nor does it have any base-sequence specificity (Supplementary Fig. 3b). The relative orientations of domains I and II are also different. Domains I in Cre and Flp wrap back against the DNA, whereas in Ndc10, domain I projects in the opposite direction; the well-packed hydrophobic interface between domains I and II in Ndc10 probably rules out any large-scale reorientation toward a configuration resembling the one in the recombinase-DNA complexes. The additional domains of Ndc10, which mediate dimerization and partner interactions, have no counterparts in Cre and Flp.
An Ndc10 dimer
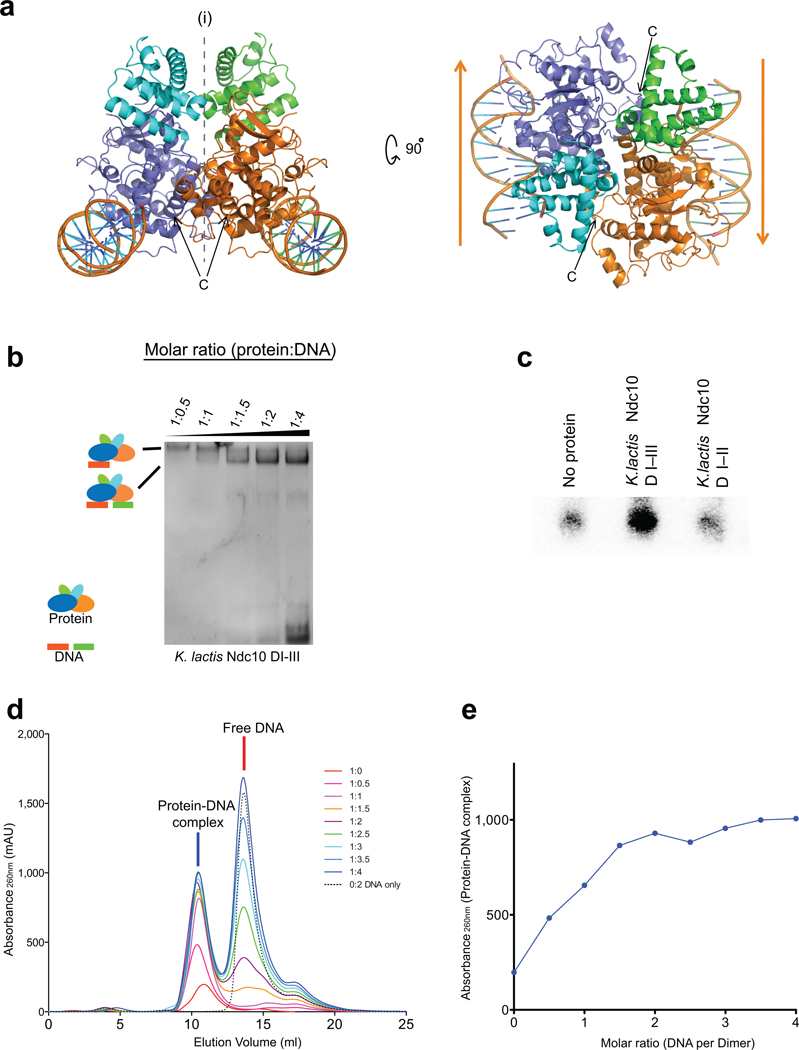
Ndc10 dimerization is largely enforced by domain III, as demonstrated by data described above (Supplementary Fig. 1b,c). Nonetheless, the twofold rotational axis along b in space group C2221 generates a relatively extended interface (~ 800 Å2) between two crystallographically related Ndc10 subunits (Fig. 4a, Supplementary Fig. 4a), suggesting that this contact may be present in the physiologically relevant dimer. Moreover, the C-terminal residues of the molecules related by this dyad are about 30 Å from each other, at one end of domain II, as might be expected if these residues were to connect into a pair of dimerized domains III. Supporting the relevance of this dimer interface is the presence of the same interface in a different crystal form of Ndc10(1–402) bound to a 30 bp DNA fragment. These crystals, in space group P212121, have an asymmetric unit that contains the Ndc10 dimer discussed above and two non-contiguous segments of 15 bp of DNA (Supplementary Fig. 4b,c) (see Supplementary Information). Thus, the stability of this interface is independent of crystal packing. In the following section, we describe experiments that support the dimer assignment, by showing that in solution, a dimer of Ndc10(1–534), which contains domains I–III, indeed binds two independent DNA segments. We note, however, that amino-acid residues that participate in the dimer interface are not well conserved among budding-yeast Ndc10 proteins and that domain III is clearly the principal determinant of dimerization.
Figure 4. Dimerization of K. lactis Ndc10 DI–III.
(a) Views of the likely Ndc10 DI–II dimer (symmetry axis along b in the C2221 space group). The subunits of the dimer contact different pseudocontinuous DNA duplexes. (b) EMSA of Ndc10 DI–III with increasing amounts of 30 bp CDEIII DNA. (c) DNA capture assay with two different labels. Either Ndc10 DI-III or Ndc10 DI-II was incubated with a mixture of equal amounts of biotinylated and unmodified CDEIII DNA, the including 32P-labeled product (10%). (d–e) Ratio of Ndc10 DI–III and CDEIII DNA determined by analytical size-exclusion chromatography.
Ndc10 dimer interacts with two independent DNA segments
To test whether an Ndc10 dimer binds a single extended site on one segment of DNA or more compact sites on each of two independent segments of DNA, we carried out EMSA experiments with 30bp CDEIII DNA and either dimeric Ndc10(1–534), containing domains I–III, or monomeric Ndc10(1–403), containing domains I and II only. We monitored the DNA mobility shifts as a function of DNA concentration, keeping the protein concentration fixed. If an Ndc10(1–534) dimer binds two independent 30bp DNA fragments non-cooperatively (as the flexible linkage of the dimerization domain suggests), then as the DNA concentration increases, an initially observed band, corresponding to a single 30 bp DNA duplex shifted by an Ndc10 dimer, will decrease in strength, in favor of a more rapidly migrating band, corresponding to binding of a second DNA fragment. The Ndc10(1–403) monomer should yield only a single shifted band at all DNA concentrations. The results shown in Figure 4b are fully consistent with these predictions.
To confirm these results by an alternative method, we tested whether an Ndc10 dimer can bind two differentially labeled DNA fragments in solution (Fig. 4c). We incubated a mixture of biotin-modified CDEIII DNA and 32P-labeled CDEIII DNA, at equal concentrations, with either dimeric Ndc10(1–534) or monomeric Ndc10(1–403). After affinity pull-down with streptavidin-coated resin, the reaction with dimeric Ndc10(1–534) yielded substantial amounts of 32P-labeled DNA in the bead eluate; the reaction with monomeric Ndc10(1–403) yielded no more label than a control containing no protein.
We also measured the ratio of Ndc10 to bound CDEIII in solution by two other methods: size-exclusion chromatography and isothermal titration calorimetry (ITC). In the former experiments, a fixed quantity of Ndc10 dimer (1–534) was incubated with 30bp CDEIII DNA in various molar ratios, then applied to an analytical size-exclusion column. The amounts of protein-DNA complex and unbound free DNA were monitored by absorption at 260 nm. Consistent with our other experiments, the binding capacity of the protein saturated a 1:2 molar ratio of protein-dimer:DNA; peaks corresponding to unbound DNA appeared when larger amounts of DNA were used (Fig. 4d,e). In the ITC measurements (Supplementary Fig. 5), Ndc10 monomer (1–403) bound a 30 bp DNA fragment in a 1:1 molar ratio; Ndc10 dimer (1–534) bound the same fragment in about a 1:2 molar ratio (Ndc10 dimer:DNA). These results further substantiate our conclusion that one Ndc10 dimer binds two separate DNA fragments in solution.
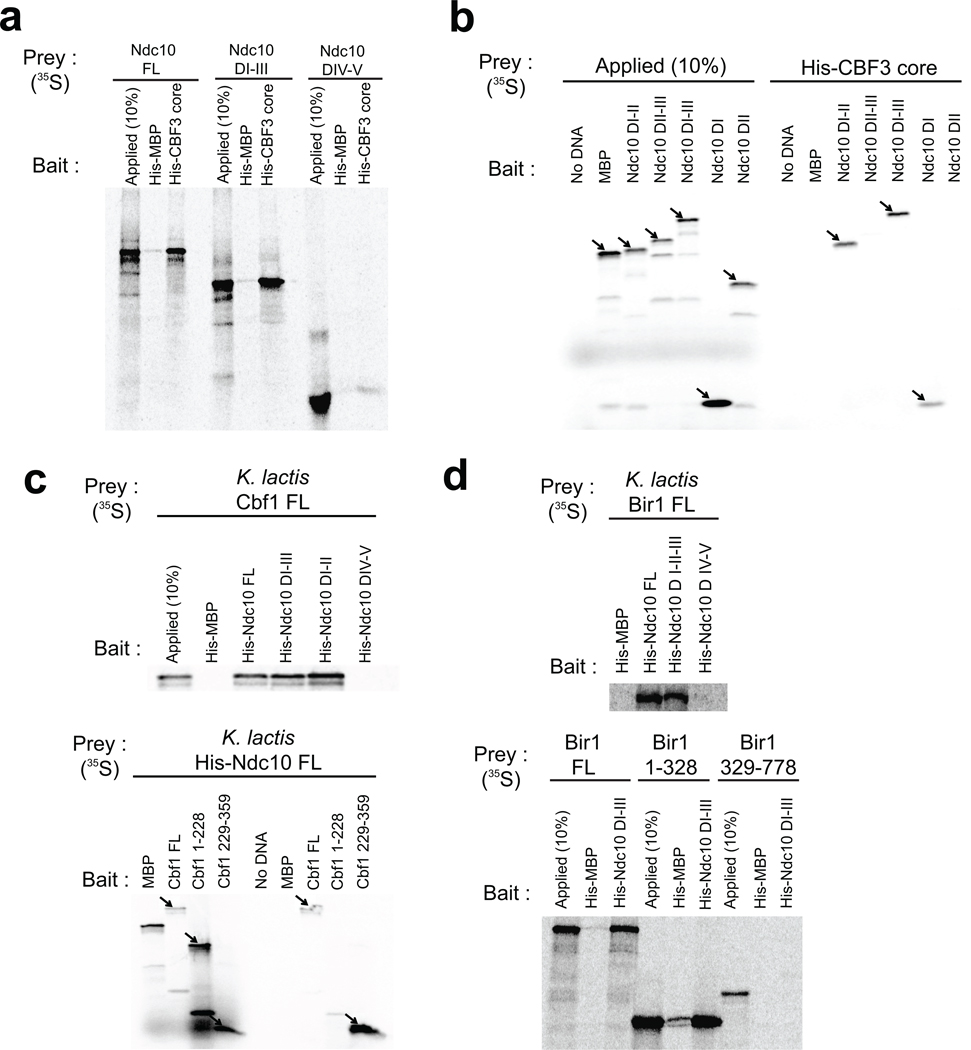
Interactions with other kinetochore components
Identification of structurally meaningful domain boundaries has allowed us to carry out interaction experiments with well defined domains of Ndc10. We performed pull-down experiments with His-tagged, biochemically well-characterized proteins or protein complexes as bait and in vitro translated, 35S-labeled proteins as prey. In these experiments, we used the K. lactis homologs of all the proteins in question; previous results with yeast-two-hybrid methods on some of the S. cerevisiae counterparts support our findings39,50.
CBF3 core: His-tagged CBF3 core (Skp1:Ctf13:(Cep3)2; His-tag on Cep3) pulled down full-length Ndc10, a domain I–III fragment, and a domain I fragment, but not fragments that lacked domain I (Fig. 5a,b). The critical contact with the CBF3 core thus appears to be in domain I. A reversed experiment, using His-tagged, full-length K. lactis Ndc10 and in vitro translated CBF3 core components, yielded a weak interaction with Cep3 and no interactions with the remaining two components (data not shown). Stronger Ndc10 association with Cep3 might therefore require the complete CBF3 core complex.
Figure 5. Interactions of Ndc10-associated proteins or protein complexes in the inner kinetochore.
(a–d) Ni2+ affinity pull-down of 35S-labeled, in vitro translated prey proteins with purified, His-tagged bait protein, analyzed by SDS-PAGE and visualized by phosphoimaging. Each panel includes a lane loaded with 10% of the in vitro translation reaction mixture (to monitor extent of synthesis) and either in vitro translated maltose-binding protein (MBP) as a prey or purified His-tagged MBP as a bait (negative controls).
Cbf1: The DNA-binding region of Cbf1, residues 229–359, a basic-helix-loop-helix-zipper (bHLHZ) element, is at the C-terminus of the molecule; residues 1–228 are probably largely unstructured. The data in Figure 5c show that the bHLHZ region of Cbf1 interacts with Ndc10 DI–II. Analytical size-exclusion chromatography with purified recombinant proteins confirms that Cbf1 associates with the CBF3 holocomplex but not with the CBF3 core (Supplementary Fig. 6).
Bir1: CBF3 recruits the chromosomal passenger complex (in yeast, Ipl1, Bir1, and Sli15) through an interaction between Ndc10 and Bir139. This association may have a centromere-independent function, as Ndc10 and the chromosomal passenger complex move together to the spindle midzone in late anaphase38. We carried out pull-down experiments with in vitro translated K. lactis Bir1 and His-tagged K. lactis Ndc10 and found that domains I–III of Ndc10 were sufficient for association with Bir1 and that the N-terminal half of Bir1 (residues 1–328) bears the Ndc10 target site (Fig. 5d).
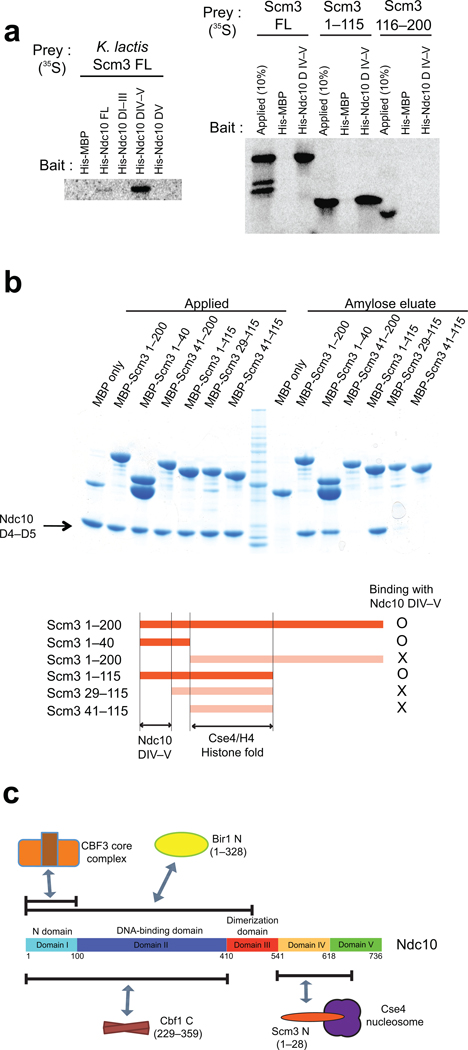
N-terminal region of Scm3 interacts with Ndc10 domain IV
The 200-residue protein, Scm3, binds Ndc10 and links CBF3 with the Cse4-containing nucleosome51. In yeast with regional centromeres, which lack a clearly identified CBF3 equivalent, orthologs of Scm3 bear additional domains appended to the C-terminus, which may have DNA-recognition properties of their own52. The Scm3 ortholog in higher eukaryotes, HJURP30,31, is likewise a larger protein with an N-terminal Scm3-like region33. The structures of mammalian HJURP:CENP-A:H4 and yeast Scm3:Cse4:H4 show conserved recognition of the centromeric histone complex by its chaperone7,8. We find that an N-terminal, 115-residue fragment of Scm3 (which includes the Cse4 binding domain, residues 41–115)7 interacts with a DIV–V fragment of Ndc10, but not with domain V alone (Fig. 6a). Amylose pull-down experiments with various MBP-tagged Scm3 fragments show that residues 1–28 are sufficient for Ndc10 binding (Fig. 6b). Size exclusion chromatography confirms that a fragment containing residues 1–115 of Scm3, but not the fragment spanning residues 29–115, forms a stable complex with Ndc10 domain IV–V (Supplementary Fig. 7).
Figure 6. Interaction of Ndc10 Domain IV–V with N-terminal Scm3.
(a) Ni2+ affinity pull-down of 35S-labeled, in vitro translated Ndc10 proteins with purified, His-tagged Scm3 proteins, analyzed by SDS-PAGE and visualized by phosphoimaging. (b) In vitro amylose pull down of purified MBP tagged Scm3 proteins with Ndc10 domain IV–V. (c) Schematic overview of domain association of K. lactis Ndc10 with other kinetochore proteins. Ndc10 DI interacts with CBF3 core; Ndc10 DI–II, with Cbf1 (229–359) and Bir1p (1–328). Scm3 (1–28) associates with Ndc10 DIV–V but not with DV. Interaction of Cbf1 with Ndc10 was confirmed by analytical size-exclusion chromatography with purified proteins (Supplementary Fig. 6).
Discussion
Ndc10 is a multi-domain, dimeric protein. We have located boundaries that define five distinct domains, through use of limited proteolysis and by determining a high-resolution structure of the fragment comprising residues 1–402. Domains I and II form a DNA-binding module; domain III stabilizes the dimer; all of the five domains (with the possible exception of domain V) interact with other kinetochore components. Pull-down experiments suggest that there are detectable contacts with the CBF3 core, Scm3, and Cbf1 -- in short, with all the other known DNA-contacting or nucleosome-contacting proteins in the yeast inner kinetochore (Fig. 6c). Thus, Ndc10 appears to be a central scaffold for inner kinetochore assembly.
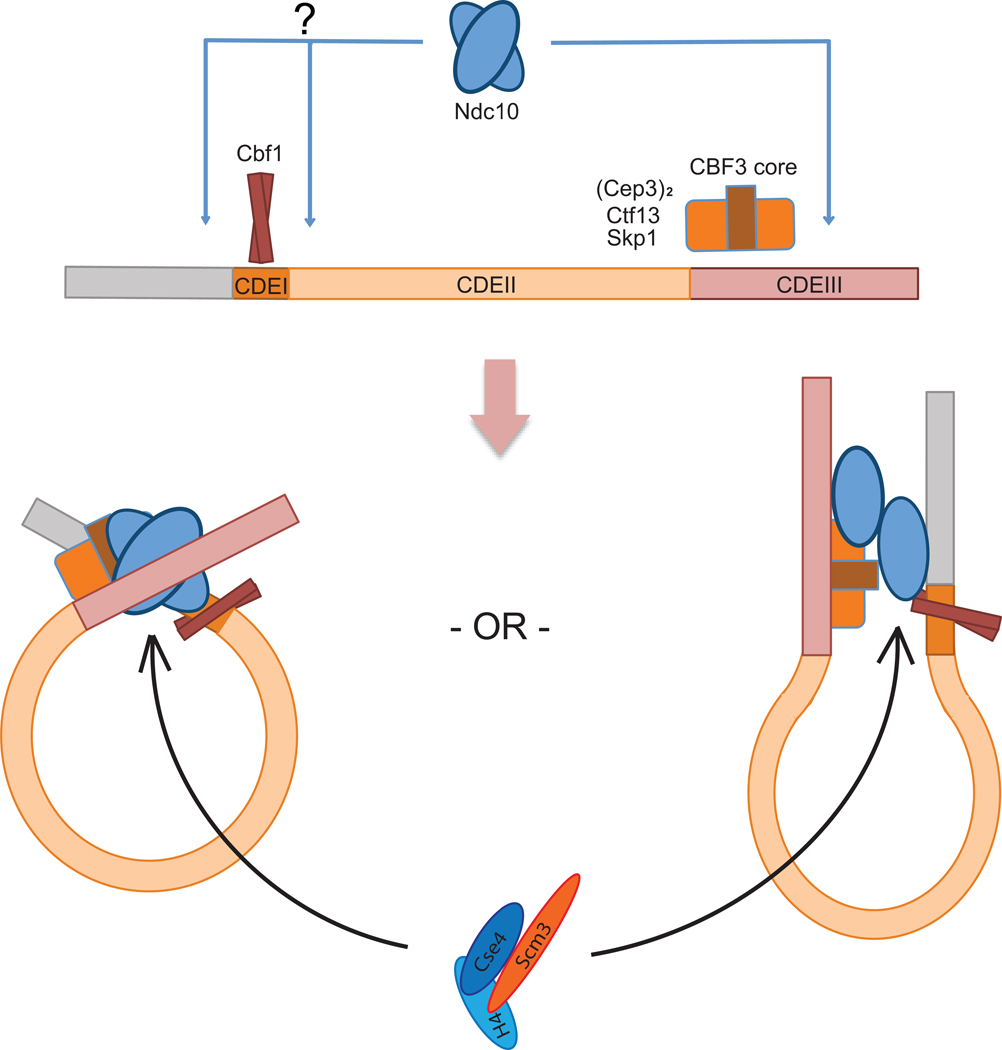
The crystal structure of DNA-bound Ndc10(1–402) suggests that the two subunits in a single Ndc10 dimer bind independent DNA fragments, rather than adjacent sites along a single fragment. Our experiments on DNA-binding in solution confirm this inference. For assembly of a kinetochore in vivo, we can entertain three possibilities. One is that an Ndc10 dimer bridges between sister chromatids. Because sister kinetochores can be widely separated even before anaphase onset, such a state could only occur during a limited part of the cell cycle. A second possibility is that one of the two DNA contacts remains unsatisfied -- for example during all but a period during which sister centromeres might indeed be adjacent. A third possibility -- the one that impresses us as most likely -- is that centromeric DNA is looped and that Ndc10 contacts both the well-characterized site in CDEIII and a site (perhaps less precisely defined) upstream of CDEI or downstream of CDEIII (Fig. 7).
Figure 7. Schematic model of Ndc10 interactions on budding yeast centromeres.
Cbf1 and CBF3 core recognize CDEI and CDEIII, respectively. Ndc10 does not have sequence-specific DNA contacts, but it binds in defined register through its interactions with Cbf1 and CBF3 core. We propose that these contacts bring CDEI and CDEIII together to form a loop. Two potential loop configurations are shown. The Scm3:Cse4:H4 heterotrimeric complex can be recruited through Scm3–Ndc10 interaction.
A further conclusion from the crystal structure is that the fragment containing domains I and II closely resembles tyrosine recombinases, but with no endonuclease activity. The complex common fold of these proteins is not one that seems likely to have arisen independently, and the local structural relationships between Ndc10 and the Cre and Flp recombinases are quite extensive (Fig. 4, Supplementary Fig. 4). Moreover, the homologous surfaces of domain II contact DNA. We suggest that the two groups of proteins are distant evolutionary cousins, but the functional implications, if any, of the homology between Ndc10 and recombinases are not apparent from the information currently in hand.
The notion that Ndc10 binding might introduce or stabilize a loop in yeast centromeric DNA, suggested by the finding that the partners in an Ndc10 dimer bind two distinct DNA sites rather than one extended site, is consistent with our interaction data from pull-down experiments with in vitro translated targets. Ndc10, which contacts DNA at the 3' edge of the consensus centromere35, also contacts Cbf1, which binds at the 5' edge (Fig. 7). Moreover, incorporation of Ndc10 at centromeric DNA will recruit the Scm3:Cse4:H4 complex through the Ndc10–Scm3 interaction (Fig. 6b, Fig. 7). Between the Ndc10 footprint in CDEIII and DNA-bound Cbf1 lies CDEII, which is coiled around the Cse4-containing nucleosome20. In K. lactis and A. gossypii, CDEII is exactly twice the length (80 bp) of CDEII in S. cerevisiae5 and most other budding yeasts for which genome sequences are known, and it is reasonable to suppose that the double-length CDEII will incorporate two CenH3-containing nucleosomes or CDEII is wrapped twice, rather than once as in S. cerevisiae. In either case, this wrapping will bring CDEI and CDEIII close enough together that each region can bind one subunit of an Ndc10 dimer. The interaction we have detected between Cbf1, the CDEI-binding component, and Ndc10 supports the idea that Ndc10 may form a bridge between juxtaposed CDEI and CDEIII sequences. A related model for DNA looping has been proposed previously, based on experiments showing that centromeric DNA loops in vivo and that this looping depends on the function of Ndc1053. This model is also supported by the nearly bimodal distribution across CDEI, II, and III of Ndc10 binding detected by high-resolution ChIP experiments19.
Our recently reported structure of an Scm3:Cse4:H4 complex shows that a fragment containing residues 41–115 of Scm3 interacts tightly with Cse4 and H4, and stabilizes the Cse4:H4 dimer in solution7. Ndc10 can then recruit such a complex through the Scm3 N-terminal segment (residues 1–28), which does not overlap the Cse4:H4 binding region (Fig. 6b). Multiple alignment of Scm3 sequences from point-centromeric yeasts identifies two conserved regions, corresponding to the regions that bind Ndc10 and Cse4, respectively (Supplementary Fig. 8). A crystal structure of the HJURP:CENP-A:H4 complex superposes well on that of the Scm3:Cse4:H4 complex (rmsd = 0.7 Å for Cα atoms of 176 overlapping residues). Recognition and recruitment of CENP-A:H4 by its chaperone are thus the same for both point and regional centromeres7,8.
The structure and interaction data reported here suggest that Ndc10 contributes to the formation and stability of a compact structure for the inner kinetochore. Ndc10 is a platform for initial kinetochore assembly. By recruiting the Scm3:Cse4:H4 complex into centromeres, it initiates formation of an elaborate superstructure. Because the specialized nucleosome is a universally conserved architectural feature of any kinetochore, whatever the length and organization of its centromere, the way in which Ndc10 scaffolds centromere compaction in budding yeast is likely to have counterparts in all other eukaryotes.
Supplementary Material
Acknowledgments
We thank the staff at the Advanced Photon Source NE-CAT beamlines for advice and assistance with data collection and interpretation, David King, HHMI Mass Spectrometry Facility, UC Berkeley, for mass spectrometry, and Kevin Corbett and Peter Sorger, for helpful discussions. This research was supported by the Howard Hughes Medical Institute.
Footnotes
Accession codes
Coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank with accession codes 3SQI (C2221 form), 3T79 (P212121 form).
Author Contributions
U-S.C. designed and performed experiments, determined and refined the structures, analyzed data and wrote the manuscript; S.C.H directed the project, analyzed data and wrote the manuscript.
References
- 1.Clarke L, Baum MP. Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences. Mol. Cell Biol. 1990;10:1863–1872. doi: 10.1128/mcb.10.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 3.Murphy TD, Karpen GH. Localization of centromere function in a Drosophila minichromosome. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willard HF. Centromeres of mammalian chromosomes. Trends Genet. 1990;6:410–416. doi: 10.1016/0168-9525(90)90302-m. [DOI] [PubMed] [Google Scholar]
- 5.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joglekar AP, et al. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc. Natl. Acad. Sci. USA. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu H, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- 10.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 11.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 12.Palmer DK, O'Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 14.Brown MT. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 1995;160:111–116. doi: 10.1016/0378-1119(95)00163-z. [DOI] [PubMed] [Google Scholar]
- 15.Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitoh H, et al. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 17.Yang CH, Tomkiel J, Saitoh H, Johnson DH, Earnshaw WC. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 19.Cohen RL, et al. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 2008;19:4480–4491. doi: 10.1091/mbc.E08-03-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole HA, Howard BH, Clark DJ. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc. Natl. Acad. Sci. USA. 2011 doi: 10.1073/pnas.1104978108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 22.Dechassa ML, et al. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat. Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellor J, Rathjen J, Jiang W, Barnes CA, Dowell SJ. DNA binding of CPF1 is required for optimal centromere function but not for maintaining methionine prototrophy in yeast. Nucleic Acids Res. 1991;19:2961–2969. doi: 10.1093/nar/19.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doheny KF, et al. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyman AA, Middleton K, Centola M, Mitchison TJ, Carbon J. Microtubule-motor activity of a yeast centromere-binding protein complex. Nature. 1992;359:533–536. doi: 10.1038/359533a0. [DOI] [PubMed] [Google Scholar]
- 27.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 28.Russell ID, Grancell AS, Sorger PK. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 1999;145:933–950. doi: 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strunnikov AV, Kingsbury J, Koshland D. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J. Cell Biol. 1995;128:749–760. doi: 10.1083/jcb.128.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 31.Foltz DR, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato T, et al. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang W, Lechner J, Carbon J. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J. Cell Biol. 1993;121:513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J. Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol. Biol. Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaplan KB, Hyman AA, Sorger PK. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 38.Bouck DC, Bloom KS. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA. 2005;102:5408–5413. doi: 10.1073/pnas.0405925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon HJ, Carbon J. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. USA. 1999;96:13208–13213. doi: 10.1073/pnas.96.23.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heus JJ, Zonneveld BJ, Steensma HY, Van den Berg JA. Centromeric DNA of Kluyveromyces lactis. Curr. Genet. 1990;18:517–522. doi: 10.1007/BF00327022. [DOI] [PubMed] [Google Scholar]
- 41.Heus JJ, Zonneveld BJ, de Steensma HY, van den Berg JA. The consensus sequence of Kluyveromyces lactis centromeres shows homology to functional centromeric DNA from Saccharomyces cerevisiae. Mol. Gen. Genet. 1993;236:355–362. doi: 10.1007/BF00277133. [DOI] [PubMed] [Google Scholar]
- 42.Sorger PK, et al. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc. Natl. Acad. Sci. USA. 1995;92:12026–12030. doi: 10.1073/pnas.92.26.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bellizzi JJ, 3rd, Sorger PK, Harrison SC. Crystal structure of the yeast inner kinetochore subunit Cep3p. Structure. 2007;15:1422–1430. doi: 10.1016/j.str.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Chapter 5. Curr. Protoc. Bioinformatics. 2006:5. doi: 10.1002/0471250953.bi0505s14. Unit 5. [DOI] [PubMed] [Google Scholar]
- 45.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 46.Conway AB, Chen Y, Rice PA. Structural plasticity of the Flp-Holliday junction complex. J. Mol. Biol. 2003;326:425–434. doi: 10.1016/s0022-2836(02)01370-0. [DOI] [PubMed] [Google Scholar]
- 47.Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodwin TJ, Butler MI, Poulter RT. Cryptons: a group of tyrosine-recombinase-encoding DNA transposons from pathogenic fungi. Microbiology. 2003;149:3099–3109. doi: 10.1099/mic.0.26529-0. [DOI] [PubMed] [Google Scholar]
- 50.Stoyan T, Carbon J. Inner kinetochore of the pathogenic yeast Candida glabrata. Eukaryot. Cell. 2004;3:1154–1163. doi: 10.1128/EC.3.5.1154-1163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camahort R, et al. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 52.Aravind L, Iyer LM, Wu C. Domain architectures of the Scm3p protein provide insights into centromere function and evolution. Cell Cycle. 2007;6:2511–2515. doi: 10.4161/cc.6.20.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeh E, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.