Abstract
Molecular investigations of riboswitches bound to small-molecule effectors have produced a wealth of information on how these molecules achieve high affinity and specificity for a target ligand. X-ray crystal structures have been determined for the ligand-free state for representatives of the preQ1-I, SAM-I, lysine, and glycine aptamer classes. These structures in conjunction with complimentary techniques, such as in-line probing, NMR spectroscopy, Förster resonance energy transfer, small-angle scattering, and computational simulations, have demonstrated that riboswitches adopt multiple conformations in the absence of ligand. Despite a number of investigations that support ligand-dependent folding, mounting evidence suggests that free-state riboswitches interact with their effectors in sub-populations of largely pre-folded states as embodied by the principle of conformational selection, which has been documented extensively for protein-mediated ligand interactions. Fundamental riboswitch investigations of the bound and free states have advanced our understanding of RNA folding, ligand recognition, and how these factors culminate in communication between an aptamer and its expression platform. An understanding of these topics is essential to comprehend riboswitch gene regulation at the molecular level, which has already provided a basis to understand the mechanism of action of natural antimicrobials.
Keywords: Riboswitch, Ligand-Free State, RNA Structure, RNA Folding, X-Ray Crystallography, Gene Regulation, Conformational Selection, Antimicrobial Target
Introduction
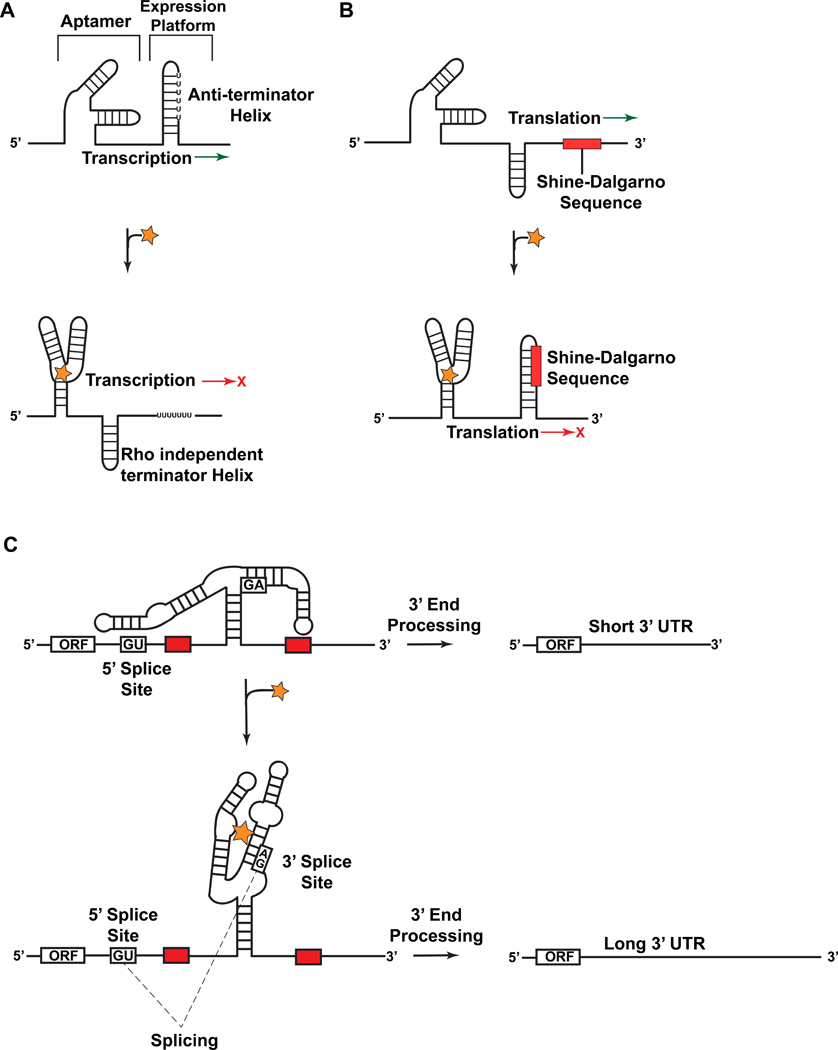
Riboswitches detect small molecules leading to conformational changes at the messenger RNA level that sequester or expose regulatory sequences that control gene expression1,2. Such structural modulation can have the effect of activating or attenuating transcription (Figure 1(a)) or translation (Figure 1(b))3, or it can change the availability of splice-site sequences required for pre-mRNA processing (Figure 1(c)) [reviewed in Watcher4]. This regulatory paradigm is distributed widely in the biosphere as illustrated by the strong showing of riboswitches in the mRNA of eubacteria3, as well as the transcripts from archaea5, fungi6, plants7–9 and algae10, where riboswitches are present to a lesser extent. The success of riboswitches is due not only to their economical architecture (ranging from 30-nt to 200-nt) but also because of their ability to function without proteins or external factors11–13.
Figure 1.
Representative mechanisms of action by which riboswitches regulate gene expression. (a) Schematic depiction of transcriptional repression by a bacterial riboswitch. In the ligand-free state the aptamer is not stably folded and favors anti-terminator helix formation, thus allowing transcription. Upon binding ligand (star) the folded structure predominates and a rho-independent terminator helix forms, thus attenuating transcription. (b) A schematic depiction of translational regulation by a bacterial riboswitch. In the ligand-free state the aptamer is not stably folded and favors an exposed Shine-Dalgarno (SD) sequence favoring translation. Upon ligand binding the folded structure predominates and the SD sequence is sequestered, thus attenuating translation. (c) A schematic depiction of splicing regulation by the THIC TPP riboswitch from Arabidopsis thaliana. When TPP levels are low the aptamer base pairs with a sequence used in 5´-splice-site selection. This bypasses splicing and necessitates use of a 3´-end processing signal (red rectangle) that yields a short 3’-UTR associated with high protein expression levels. When TPP levels are high the aptamer binds TPP during transcription and exposes the 5´-splice site. The ligand-bound aptamer results in splicing whereby the upstream 3´-end processing site is removed, which necessitates the use of an inferior downstream processing site. This results in a longer, unstable mRNA that produces lower quantities of THIC protein.
Riboswitches are encoded within the transcripts they regulate, which often include genes that metabolize or transport amino acids, enzymatic cofactors, metal ions, nucleobases, or second messengers3,12. The ligands of riboswitches (Table 1) are frequently metabolic end-points of a specific pathway that is regulated by a riboswitch whose classification is based on the effector that it responds to12. The affinity of most riboswitches for their cognate ligand resides in the middle to lower nanomolar range, thereby providing high sensitivity as well as the capacity to respond over a broad dynamic range14–16.
Table 1.
Known Riboswitch Structures with Representative Ligand Bound and Free States
| Riboswitch | Ligand | Free- State |
Organism or Source | PDB Code and Select References |
||
|---|---|---|---|---|---|---|
| Purine Class | ||||||
| Guanine | Hypoxanthine | B. subtilis | 1U8D59 | |||
| Guanine | Guanine | B. subtilis | 1Y2760 | |||
| Adenine | Adenine | V. vulnificus | 1Y2660 | |||
| PreQ1 | PreQ0, PreQ1 | yes | T. tengcongensis | 3GCA61, 3Q50, 3Q5146 | ||
| PreQ1 | PreQ1 | B. subtilis | 3FU263, 2L1V62 | |||
| SAM Class | ||||||
| SAM-I | SAM | T. tengcongensis | 2GIS47, 3IQN45 | |||
| SAM-I (A94G) | - | yes | T. tengcongensis | 3IQP45 | ||
| SAM-I | SAM | B. Subtilis | 3NPB50 | |||
| SAM-II | SAM | Sargasso Sea metagenome | 2QWY51 | |||
| SAM-III | SAM | E. faecalis | 3E5C55 | |||
| Amino Acid Classes | ||||||
| Lysine | Lysine | yes | T. maritima | 3D0U, 3D0X23, 3DIL, 3DIS44 | ||
| Glycine (VCII) | Glycine | yes | V. cholerae | 3OWW, 3OXO43 | ||
| Others (Bound State Only) | ||||||
| glmS | GlcN6P | T. tengcongensis | 2Z75 | |||
| FMN | FMN | F. nucleatum | 3F2Q | |||
| M-box | Mg(II) | B. subtilis | 2QBZ | |||
| SAH | SAH | R. solanacearum | 3NPQ | |||
| TPP | TPP | A. thaliana | 2CKY | |||
| TPP | TPP | E. coli | 2GDI | |||
| c-di-GMP | c-di-GMP | V. cholerae | 3MXH | |||
Riboswitches themselves are organized functionally into a high-affinity aptamer that binds ligand and a downstream expression platform3 (Figure 1). Aptamers tend to be conserved for different genes of the same species as well as spanning multiple species. The expression platform is often less conserved in terms of sequence, spatial relationship with the aptamer, and its genetic control mechanism3. Effector recognition is accompanied generally by conformational rearrangements that attenuate or activate expression at an adjacent open reading frame. Transcriptional control may entail structural changes that shift the equilibrium of hairpin loops (Figure 1(a)). In the majority of cases, ligand binding results in stabilization of a rho-independent terminator helix that arrests transcription (figure 1(a)), although there are cases where the terminator helix is stable in the absence of ligand and binding results in stabilization of an anti-terminator helix favoring transcription (not shown). By contrast, translational attenuation involves conformational changes that restrict access to the ribosome-binding site (RBS) (Figure 1(b)), which must hybridize with the 3´-end of the 30S ribosome to initiate decoding17. In higher organisms, riboswitches influence splice-site selection by ligand sensing that alters the accessibility of 5´- or 3´-splice sites, as well as the branch point adenosine. This can lead to alternative splicing and decreased mRNA stability (Figure 1(c))4.
As many as 4% of bacterial genes are controlled by riboswitches18. The susceptibility of specific riboswitches to antimicrobials has garnered considerable attention19–29 due to the emergence of drug-resistant opportunistic pathogens. The analysis of riboswitch structure and function has provided useful insights into the mechanism of action of ligand binding, thereby facilitating the design of antibacterial compounds, which holds promise for therapeutic development30,31. The untapped potential of riboswitches as antimicrobial targets is underscored by the existence of more than 20 classes of riboswitches that recognize 15 discrete small molecules12,32. In several cases multiple, structurally distinct riboswitches respond to the same ligand. For example, there are seven S-adenosylmethionine (SAM) riboswitches5,33–36 (reviewed in Batey37), two preQ1 riboswitches38,39, and two c-di-GMP riboswitches40,41.
At present, nearly a dozen distinct classes of riboswitch aptamers have been solved in complex with their cognate ligands (Table 1)12.,32,42,43. In addition to understanding the molecular basis for ligand recognition, an equally important question is how conformational changes in the aptamer and expression platforms lead to gene regulation. To elucidate this area it is important to investigate bound as well as free-state riboswitches. Visualization of these states is of special importance because they provide a unique opportunity to locate atomic-scale conformational changes resulting from ligand recognition that lead to gene regulation. However, the aptamer and expression platform are separated frequently to allow formation of the mutually exclusive structures that are necessary for gene regulation. This aspect of riboswitch organization can complicate structural approaches such as crystallography. Nonetheless, the structures of four riboswitches have been reported in the ligand-free state (Table 1). Three represent transcriptional riboswitches in which part of the expression platform is missing23,43–45. The fourth is a translational riboswitch that includes the aptamer as well as a portion of the expression platform46. Despite these advances, free-state crystal structures cannot provide a comprehensive picture of the thermodynamic ensemble45,46 but they do expand our understanding of the regulatory process and provide a focus for experimentation.
Herein, we emphasize the known three-dimensional structures of free-state riboswitches (Table 1). We compare both free and bound states to learn how ligand binding culminates in gene regulation. Whenever possible, structural observations are tempered by biochemical or biophysical analyses to provide a broader perspective. In addition, we discuss aspects of riboswitch structure and function that would benefit from further experimentation, and make suggestions on topics that require additional exploration.
RIBOSWITCHES THAT BIND S-ADENOSYL METHIONINE (SAM)
SAM-I riboswitch
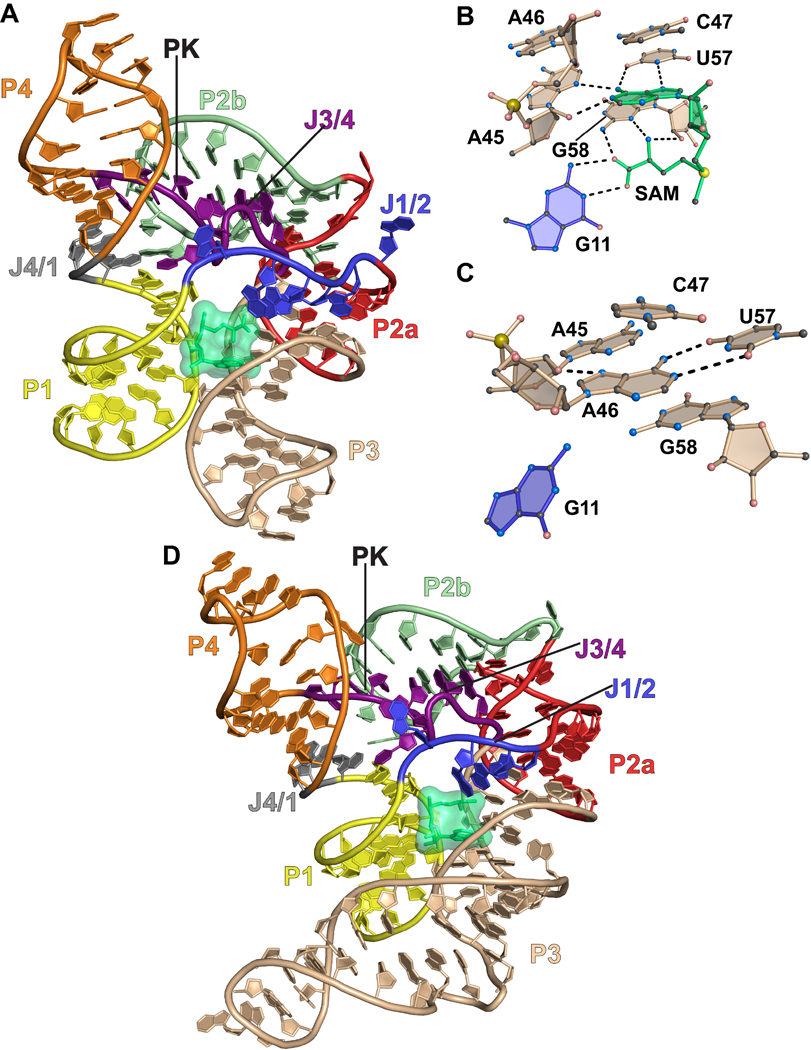
The structure of a SAM-I (class 1) riboswitch from the hotspring bacterium Thermoanaerobacter tengcongensis was determined in the presence of the methyl-group donor SAM47. In the ligand-bound state, the SAM-I riboswitch has two sets of co-axially stacked helices (Figure 2(a))47. SAM binds at the interface of P1 and P3 (Figures 2(a) and 2(b)), which generates a ligand-specific recognition pocket. The adenine ring of SAM stacks on base C47 of the aptamer (Figure 2(a) and 2(b)). Groups N7 and N6 from the Hoogsteen edge of the SAM adenosyl ring pair with N3 and O4 of aptamer base U57, respectively (Figure 2(b)). The Watson-Crick or WC face of the SAM adenosyl ring pairs with the sugar edge of aptamer base A45. Specifically, N1 of the SAM base accepts a hydrogen bond from the O2’ of aptamer ribose A45, whereas the N6 of the SAM adenine base donates a hydrogen bond to N3 of A45. The WC face of G11 forms hydrogen bonds with the carboxyl group of the methionine moiety. Increased specificity is achieved via hydrogen bonds between the carboxyl group of SAM and the N2 amine of G58. Similarly, the amino group of SAM forms hydrogen bonds with the sugar edge of G58 where it interacts with the base N3 and ribose O2’ groups. Finally, the amino group of SAM points into its own adenine ring to stabilize its conformation via a cation-π interaction.
Figure 2.
Cartoon and ball-and-stick diagrams of the SAM-I riboswitch aptamer domains. (a) Cartoon depiction of the T. tengcongensis SAM-I riboswitch structure in the ligand-bound state. SAM is depicted as space-filling surface (green); PK is the abbreviation for pseudoknot. A common color code is used in all figures that corresponds to the pairing (P) and joining (J) regions. Dashed black lines show putative hydrogen bonds. The coordinates are from PDB entry 2GIS. (b) The SAM ligand binding site in the bound state derived from (a). (c) The SAM binding site in the ligand-free state. The coordinates are from PDB entry 3IQP. (d) Cartoon diagram of the tertiary structure of the B. subtilis SAM-I riboswitch from PDB entry 3NPB.
Recently a crystal structure of the SAM-I riboswitch was determined in the ligand-free state. In a supporting analysis, the structural heterogeneity of the aptamer was characterized in the SAM-free state45. The free-state structure exhibited only local rearrangements at the site of SAM binding with a root-mean-square displacement (rmsd) of 0.12 Å over all RNA atoms compared to the bound state. The free-state structure also revealed that A46 occupies the binding site in place of SAM. In particular A46 forms a WC pair with nearby base U57, which is highly conserved and involved in SAM recognition (Figure 2(d)). In addition, the N7 group of A46 interacts with the 2´-OH group of A45, which is also involved in SAM binding. The observation that A46 occludes the binding pocket suggested that the observed free-state structure was not representative of the complete thermodynamic ensemble45. Instead, this “closed” conformation was hypothesized to be one of many possible with the likelihood that A46 interconverts from closed to open states to accommodate SAM binding.
To evaluate the possibility that open and closed conformational states of the SAM-I aptamer co-exist in the ligand-free state complementary methods were utilized. One method used to analyze the apo and bound riboswitch in solution was selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE). SHAPE reports on secondary structure of an RNA by treating the folded RNA with N-methylisatoic anhydride (NMIA), an agent that acylates the 2’-OH of flexible nucleotides – i.e. those not forming stable base pairs48. A primer extension reaction is run and the products are separated by electrophoresis. Acylated sites are detected as premature stops. SHAPE analysis in the absence of SAM at 10 mM Mg2+ concentrations demonstrated the presence of secondary-structure corresponding to both the bound and free-state45. Small angle X-ray scattering (SAXS) performed on the aptamer in 7.6 mM and 10.0 mM Mg2+ in the SAM-bound state indicated that the experimental scattering profiles fit closely to that calculated from the bound-state crystal structure as indicated by a χ2 value of 1.6. In contrast, the scattering profile in the absence of SAM did not resemble that of the bound aptamer based on a χ2 value of 5.1. To deconvolute the free-state scattering profile a conformational diversity pool was generated comprising nearly 103 structures. Using a genetic algorithm, a series of 13 structures were identified that fit the experimental scattering profile with a χ2 value of 0.8345. Among these structures, there were substantial differences in the distance between P1 and P3, as well as the orientation of the P4-PK-P2a/b subdomain.
The resulting structural conformations showed eleven open and two closed A46 conformations for the binding pocket. To explore the energetic differences between these states, replica-exchange molecular dynamics was employed using structures of the ligand-bound and the SAM-free state as starting models. Two free-energy minima were observed that included the “closed” conformation with A46 occupying the binding pocket, and a “primed” conformation resembling the SAM-bound state that is competent to bind ligand. Although the closed conformation is more stable, energetic differences between the respective states were not prohibitive for interconversion. This result when combined with the chemical modification analysis of A45 and A46 suggests that the free-state aptamer comprises an ensemble of structures in closed and primed conformations. This result is suggestive of a conformational selection model for ligand recognition (Sidebar 1).
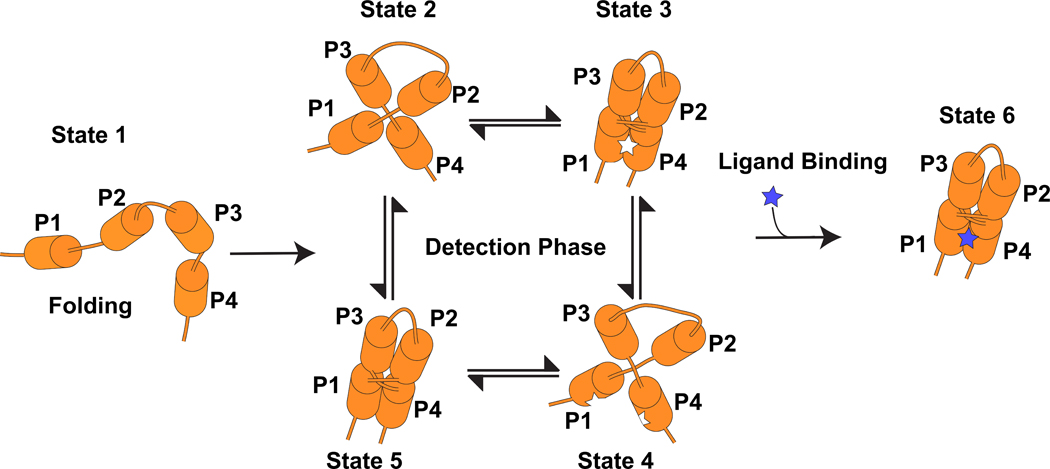
SIDEBAR 1.
A cartoon illustrating conformational selection by a riboswitch, which is a prominent mode of ligand recognition by proteins49. In state 1 only secondary structures are formed. In states 2–5, tertiary structure has formed but the ligand is not bound. The aptamer exists as an equilibrium of conformational states with both local changes to the ligand-binding pocket, as well as long-range tertiary interactions between different pairing regions. Only conformations with a fully formed, unblocked binding sites are receptive to ligand binding. Upon binding the aptamer adopts a stable conformation (state 6) that effects expression of the associated mRNA.
Since SAM is only able to bind when A46 is not occupying the binding site the kinetics of the closed-to-open transition can alter the on-rate of ligand binding. Thus far nearly a dozen SAM-I riboswitches have been identified in Bacillus subtilis transcripts and these require different concentrations of SAM to achieve half-maximal transcription termination16. Sequence variations provide a plausible mechanism to alter tertiary interactions that differentiate riboswitch responses at the same ligand concentration45. These observations emphasize the nuanced relationship between RNA folding, ligand sensing and transcriptional control within the same riboswitch class.
We now turn our attention to differences in SAM-I riboswitches between genera. In the case of the SAM-I riboswitch, structural and biochemical investigations have been conducted on riboswitch aptamers from the thermophilic bacterium T. tengcongensis, as well as the mesophile B. subtilis50. Crystal structures of the T. tengcongensis and B. subtilis aptamers showed reasonable agreement with an rmsd of 2.5 Å, with the most overlap at the SAM binding site50. The structure of the B. subtilis SAM-I aptamer was probed using SHAPE50. Like that of T. tengcongensis, the B. subtilis aptamer also appears to be well organized in the ligand-free state with J1/J2, J3/J4, and nucleotides A46 and A47 – involved directly in SAM binding – show greater protection in the ligand-bound state. P1, P2, P3, and P4 show nearly the same protection pattern in the presence and absence of SAM50. The results suggest that folding of the free-state aptamer is a trait shared by thermophiles and mesophiles.
SAM-II riboswitch
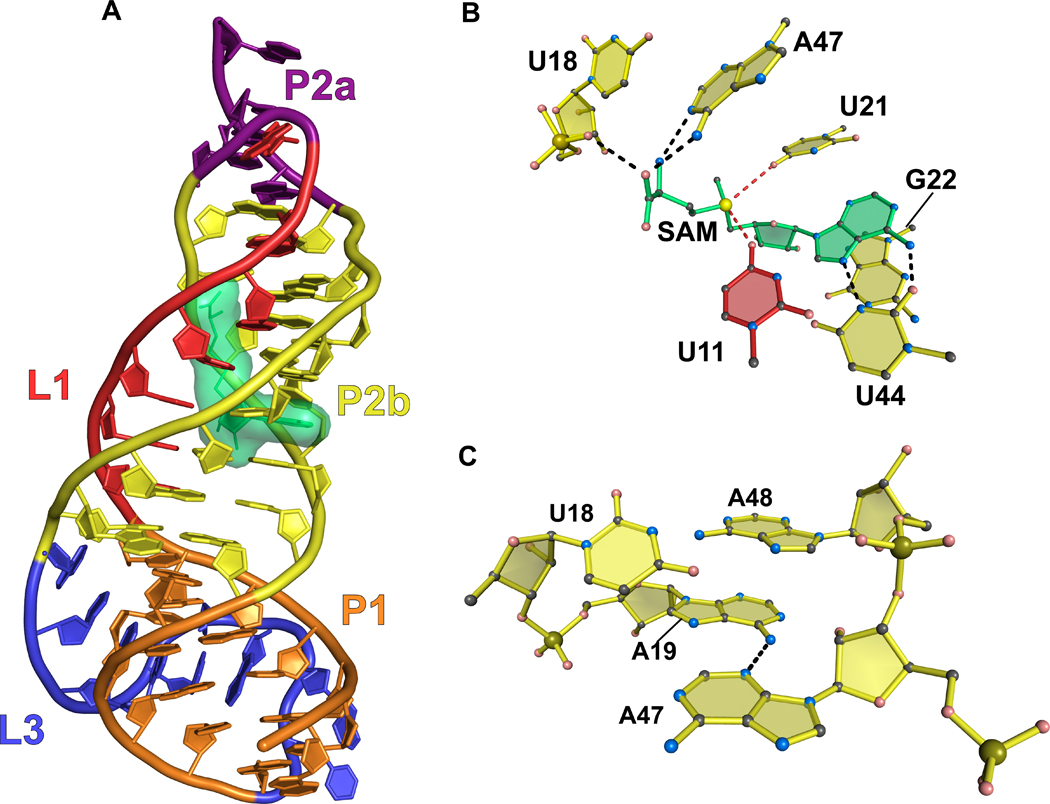
A second class of SAM-binding riboswitch called class 2 was identified in proteobacteria. SAM-II shows no homology with SAM-I in terms of its overall fold but utilizes similar ligand interaction traits. The riboswitch forms an H-type pseudoknot comprising three stacked helices (Figure 3(a))51. L1 interacts with the major groove of P2a/b and L3 interacts with the minor groove of P1. SAM is recognized by hydrogen bonds to its respective methionyl and adenosyl moieties. On the methionine side, the carboxyl group donates and receives hydrogen bonds from the nonbridging oxygen of the U18 phosphate and the N6 amino of A47, respectively (Figure 3(b)). The positively charged methylsulfonium ion of SAM forms electrostatic interactions with the N4 keto group of both U11 and U21. Like SAM-I, the adenine ring of SAM forms a Hoogsteen interaction with U44 that is part of a base triple that includes U10 (not shown).
Figure 3.
Cartoon and ball-and-stick diagrams of the metX SAM-II riboswitch from the Sargasso Sea metagenome. (a) Cartoon depicting the tertiary structure of the SAM-II riboswitch in the bound state. The coordinates are from PDB entry 2QWY. (b) Ball-and-stick diagram of the structure in (a) showing interactions with SAM; red dashed lines indicate putative electrostatic interactions. (c) Close up view of the interaction between A19 and A47 showing the potential for chemical modification. (See the text for details).
Chemical modification was used to monitor SAM-II conformational changes that occur in the presence and absence of ligand. NMIA modifies accessible 2´-OH groups, whereas DMS modifies accessible and unpaired adenine N1 and the N3 groups of cytosine bases. The P1 helix showed no significant change in reactivity in the presence or absence of SAM, and L3 showed only modest decreases in reactivity. This suggested that these structures are stable in the presence or absence of ligand. The SAM-binding site formed by P2b and L1 (Figure 3(a)) showed marked decreases in NMIA reactivity, indicating that SAM facilitates the formation of a stable binding pocket. A19 (Figure 3(c)), whose N6 amino interacts with A47 when SAM is bound, showed increased reactivity with DMS in the presence of SAM because its WC face is exposed upon binding ligand. Overall, these data indicate that the SAM-II riboswitch, like SAM-I, is significantly structured in the ligand-free state although the SAM-binding site is not formed fully. These observations are complemented by a recent biophysical analysis of the SAM-II riboswitch that show that the dynamic, unliganded state is predisposed to binding ligand52.
SAM-III (SMK) riboswitch
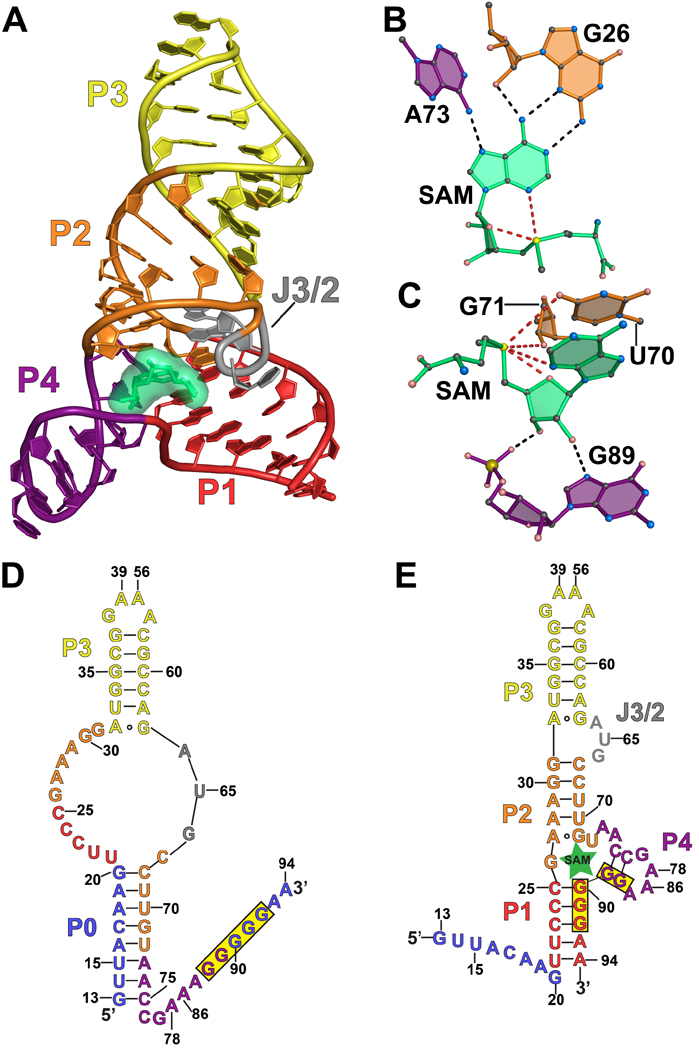
The SAM-III (SMK) riboswitch regulates translation of the bacterial metK gene. The SAM-III riboswitch has been shown to be a reversible riboswitch capable of binding, releasing, and then rebinding the SAM ligand multiple times during the lifetime of the metK transcript53. Recent experiments have framed its mode of ligand binding in the context of conformational selection54. A co-crystal structure of the Enterococcus faecalis SMK riboswitch bound to SAM has been solved revealing a Y-shaped fold comprising four helices with co-axial stacking (Figure 4(a))55. SAM binds at a three-way junction formed by helices P1, P2 and P4. The RBS is sequestered in P1 and P4, restricting its ability to pair with the anti-RBS, thus representing an example of direct translational attenuation3. SAM is recognized by the aptamer through interaction with its adenosyl and methionyl groups. The adenosyl moiety pairs with the sugar edge and minor grove of aptamer base G26 (Figure 4(b)). In particular, N6 of SAM hydrogen bonds to the 2´-OH and N3 groups of G26 whereas the N1 of SAM pairs to the N2 amino group of G26. The N7 of the adenosyl group accepts a hydrogen bond from the N6 exocyclic amine of A73. Both ribose hydroxyl groups of SAM form hydrogen bonds with G89 at N7 and a nonbridging oxygen (Figure 4(c)). The positively charged methylsulfonium group forms electrostatic interactions with the O2’ of G71 and the O4 keto of U70.
Figure 4.
Cartoon, ball-and-stick, and secondary structure diagrams of the SAM-III (SMK) riboswitch from E. faecalis. (a) Cartoon diagram showing the tertiary structure of the SMK riboswitch in the ligand-bound state. The coordinates are from PDB entry 3E5C. (b) Ball-and-stick diagram of the structure in (a) showing interactions between SAM and nucleotides A73 and G26 involved in SAM binding. (c) Additional interactions for the structure in (a) between SAM and nucleotides that stabilize ligand binding. (d) Secondary structure of the E. faecalis SMK riboswitch when SAM is absent. Nucleotides in blue are present in SMK59 and absent in SMK5154; the RBS is highlighted in yellow. (e) Secondary structure of the E. faecalis SMK riboswitch when ligand is present. Nucleotides in blue are present in SMK59 and absent in SMK5154.
In the absence of SAM, a new aptamer helix designated P0 was predicted to form35, thereby disrupting P1, P2 and P4, and freeing the RBS sequence to interact with the anti-RBS (Figure 4(d)). NMR experiments confirmed the formation of P0 and the loss of P1, P2 and P4 in a 59-nt E. faecalis SMK riboswitch – dubbed SMK59 – observed in the absence of SAM54. When 8-nt at the 5’ end that help compose P0 are removed (Figure 4(e)) – called SMK51 – the structure is nearly the same in the unbound and bound states, although the structure is less stable when SAM is absent. Formation of P0 and contacts required for binding SAM are mutually exclusive, so sampling a ‘primed’ state that resembles the bound conformation, as observed for the SAM-I riboswitch, seems most probable. Analysis by isothermal titration calorimetry (ITC) showed that SMK51 – which is locked in the primed conformation – has a KD 6-fold lower than the 59-nt riboswitch indicating that in the absence of SAM the ensemble populates only 20% in the primed SAM-binding conformation, while the remaining 80% is in the so-called ‘ISO’, non-binding-competent conformation. These data indicate that the SMK riboswitch exists primarily in a conformation incompetent to bind ligand but samples a primed conformation in the free state that is competent to bind ligand. Further evidence for an equilibrium between the ISO and primed conformations was provided by SHAPE experiments performed on apo SAM-III, which were consistent with the secondary structure identified by NMR56. Disruption of P0 by substitution of poly(A) in the pairing region, or overstabilization of P4, resulted in stabilization of the primed state and caused a reporter gene under control of the riboswitch to be constitutively off. Furthermore, both stabilizing P4 and substituting poly(A) in P0 resulted in a 5-fold decrease in the KD of SAM binding. Further evidence for the presence of the primed state in vivo in the wildtype riboswitch is the low free energy change of 3.2 kcal/mol between the ISO and primed conformations as determined by secondary structure prediction software. This indicates that 0.5% of ligand-free riboswitches are in the primed conformation in vivo and represents one of the strongest cases for conformational selection in the riboswitch field.
PURINE RIBOSWITCHES
Four classes of riboswitches recognize purine ligands including guanine, adenine, 2’-deoxyguanosine, and preQ1. Whereas the guanine, adenine, and dG riboswitches show high levels of sequence conservation57, the preQ1 riboswitches show no conservation between its two classes or to other purine binders58. As such, folding and ligand recognition by the preQ1 riboswitch classes appears to be a case of convergent evolution39. Structures have been determined for the guanine59,60, adenine60, and preQ1-I riboswitches46,61–63 (Table 1).
Adenine and Guanine riboswitches
X-ray co-crystal structures have been determined for the guanine riboswitch bound to hypoxanthine59 and guanine60, as well as the adenine riboswitch bound to adenine60. The structures are highly homologous on a tertiary level and comprise two co-axially-stacked helices. These regions form the purine-binding site at a three-way helical junction involving P3 (Figures 5(a) and 5(b)). The guanine riboswitch structure was derived from the xpt gene of B. subtilis, which attenuates transcription. The adenine riboswitch structure was derived from the add gene of Vibrio vulnificus, which causes translational activation upon ligand binding.
Figure 5.
Cartoon, ball-and-stick, and secondary structure diagrams of the adenine and guanine riboswitches. (a) Cartoon diagram showing the tertiary structure of the B. subtilis xpt guanine binding riboswitch. The coordinates are from PDB entry 1Y27. (b) Cartoon diagram showing the tertiary structure of the V. vulnificus add adenine riboswitch. The coordinates are from PDB entry 1Y26. (c) Ball-and-stick diagram of the structure in (a) depicting interactions between guanine and the riboswitch. (d) Ball-and-stick diagram of the structure in (b) depicting interactions between adenine and the riboswitch. (e) Secondary structure diagram of the pbuE adenine riboswitch in the bound state. Nucleotides that were mutated to disrupt loop-loop interactions are highlighted in green as described in the text and ref.64.
Recognition of guanine occurs by a WC base pair to C74 of the aptamer (Figure 5(c)). The minor groove edge of guanine interacts with the WC face of nearby base U51, which hydrogen bonds to N2, N3 and N9 of the ligand, and with U47 which also forms a hydrogen bond with N9 of guanine; N7 of guanine accepts a hydrogen bond from O2’ of U22. Remarkably, the adenine riboswitch forms similar interactions but C74 is replaced by U74. This substitution allows ligand-specific WC base pairing with adenine rather than guanine. Although adenine has no N2 amine, a mode of minor-groove ligand recognition occurs that is comparable to the guanine riboswitch whereby adenine pairs with U51 (Figure 5(d)). U47 and U22 again form hydrogen bonds with N9 and N7, respectively. Upon binding, these riboswitches sequester ~98% of the ligand from solvent.
The role of distal elements in ligand binding was explored in the context of the transcriptional pbuE adenine riboswitch (Figure 5(e))64. The base analogue 2-aminopurine (2AP) allowed probing of the fold since this molecule undergoes a change in quantum yield upon binding RNA, and is able to bind in a manner comparable to the adenine ligand. Binding analysis revealed an apparent KD for 2AP of 354 ± 17 nM. Further probing revealed that ligand binding was disrupted significantly as a result of P2 mutations U27G and G28C or the P3 mutants C51G, C52G, or G53U (Figure 5(e)). This was hypothesized to be the result of changes in loop-loop interactions that normally occur between these nucleotides to maintain structural stability64. Probing with T1 RNase was used to determine if the loop-loop mediated interactions between P2 and P3 occurred in the absence of ligand. The results revealed that in the absence of ligand or Mg2+ T1 cleaves after G28 and G29 in P2 and after G53 in P3 (Figure 5(e)). Addition of 10 mM Mg2+ prevented cleavage, indicating a loop-loop interaction between L2 and L3 that blocks T1 RNase access. Addition of adenine or 2AP did not alter cleavage of these bases, confirming that ligand binding is not required for the loop-loop interaction.
The interaction between L2 and L3 was investigated further by ensemble FRET64. The VV1 adenine riboswitch, which shows high levels of conservation with the pbuE riboswitch, was labeled on L2 and L3 at positions that form loop-loop interactions. High FRET efficiency was seen in the absence of adenine, demonstrating significant folding in the absence of ligand. However, the concentration of Mg2+ required to achieve half-maximal fluorescence was 2-fold lower in the presence of adenine indicating that adenine assists tertiary folding. Single-molecule (sm)FRET on the VV1 aptamer revealed a folding intermediate with a FRET efficiency intermediate between unfolded and folded states64. The transition among these states increased to the folded conformation with greater Mg2+. Addition of adenine increased the rate of folding and decreased the rate of unfolding. Overall, the smFRET experiments demonstrate the value of the single molecule approaches to identify folding intermediates. A future challenge will be to characterize folding in the context of larger transcripts in the presence of RNA polymerase or the translation initiation complex.
Fluorescence experiments on the B. subtilis xpt guanine riboswitch provide further evidence of riboswitch folding in the absence of ligand65. Riboswitches with labels on P1 and P2, P1 and P3, and P2 and P3 were generated, and bulk and smFRET measurements were made at 0 or 500 nM guanine with 0 to 100 mM magnesium. Consistent with the data for the adenine riboswitch, addition of magnesium is sufficient for prefolding with formation of the loop-loop interaction between L2 and L3 as well as the coaxial stack of P1 and P3. Addition of saturating guanine results in a higher FRET efficiency between P1 and P2 at physiological magnesium concentrations, consistent with ligand-induced structural changes that result in gene regulation. Independent evidence from high-resolution NMR also supports prefolding for this riboswitch66.
PreQ1 Riboswitch
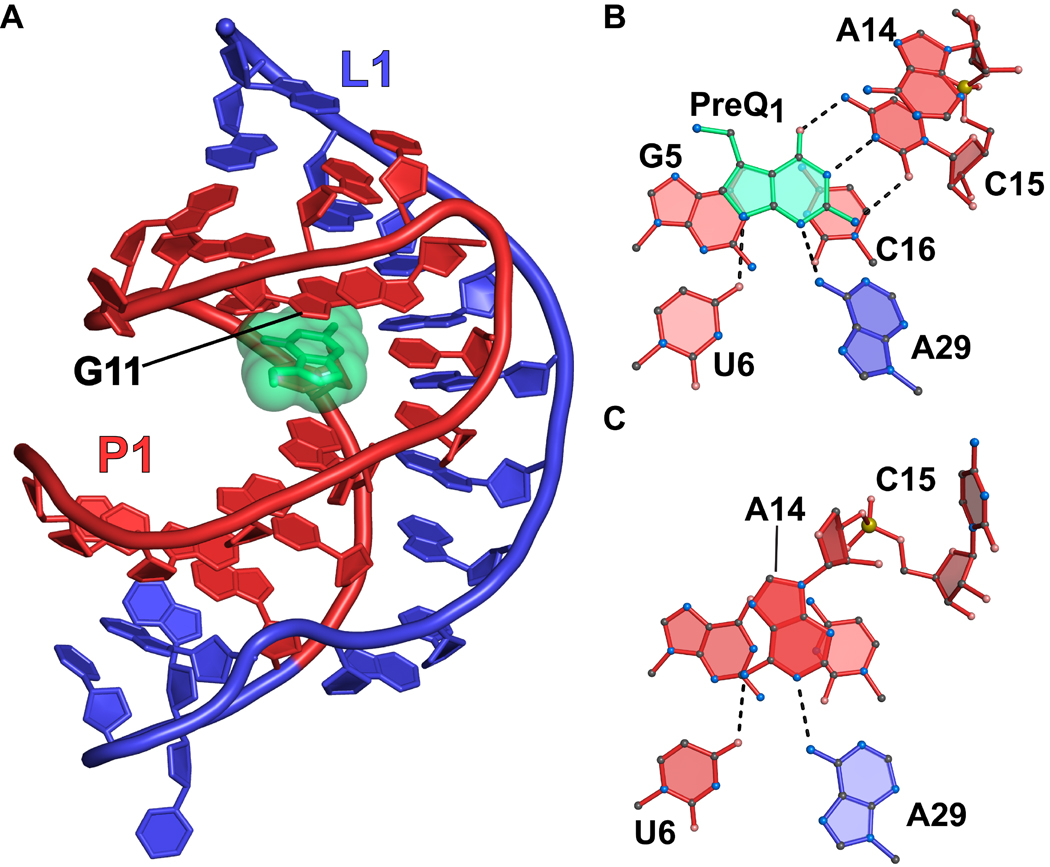
The preQ1-I riboswitch has the smallest aptamer identified to date at a diminutive 34 nucleotides67. Three-dimensional structures of the riboswitch have been determined in complex with preQ1 46,62,63, preQ0 61, and in a ligand-free state46. In the folded state, the aptamer forms an H-type pseudoknot in which the stem-loop poised atop the P1 helix interacts with bases in the A-rich aptamer tail (Figure 6(a)). PreQ1 is recognized by hydrogen bonding to the WC face of conserved base C15 (Figure 6(b)). Additional interactions with conserved bases occur between N6 of A29 to N3 of preQ1, and O4 of U6 to N9 of preQ1 (Figure 6(b)). When bound to the aptamer, preQ1 completes a base stacking interaction that connects G11 at the “ceiling” of the pocket to P1 bases G5-C16 at the aptamer “floor” (Figures 6(a) and 6(b)).
Figure 6.
Cartoon and ball-and-stick diagrams of the T. tengcongensis preQ1-I riboswitch. (a) Cartoon diagram showing the tertiary structure of the riboswitch. The A-rich pseudoknotted tail is depicted in blue. The coordinates are from PDB entry 3Q50. (b) Ball-and-stick diagram of the structure in (a) depicting interactions between preQ1 and the aptamer. (c) The preQ1-binding site in the ligand-free state from PDB entry 3Q51.
An NMR analysis of the B. subtilis queC preQ1-I transcriptional riboswitch was conducted in the ligand-bound and free states62. In the absence of ligand, an alternate helix called P1A was identified in lieu of P1 and the A-rich tail did not form a pseudoknot. Folding of the Fusobacterium nucleatum preQ1-I transcriptional riboswitch aptamer and expression platform were also interrogated by NMR68,69. The results showed that the expression platform could adopt one of two mutually exclusive secondary structures favoring either an anti-terminator or a terminator helix. In the absence of ligand imino proton NMR suggested the expression platform existed as a 50:50 distribution between the anti-terminator and terminator conformations69. This was corroborated by 19F NMR in which U37 was replaced by 5-fluoro-uridine; this position is single-stranded in the terminator helix but double-stranded in the anti-terminator helix. The 19F label revealed that the anti-terminator and terminator helices were present in equilibrium with equal amounts of each. The addition of preQ1 shifted the equilibrium to 80% terminator helix.
Given the ligand-dependent folding in the B. subtilis and F. nucleatum analyses, the recent crystal structure of the T. tengcongensis aptamer in the ligand-free state was somewhat unexpected46. A comparison of the preQ1-bound and ligand-free states showed an all-atom rmsd of 1.7 Å with the most significant rearrangement at A14. This base flanks the ligand-binding pocket in the bound state where it pairs with G11 (Figure 6(b)). A14 moves 7.5 Å away from G11 in the free state into the location occupied by preQ1 (Figure 6(c)). Moreover, A14 hydrogen bonds to U6 and A29, which form part of the preQ1 recognition network46. Base C15 of the aptamer, which WC pairs with the guanine-like face of preQ1, rotates away from the binding pocket (Figure 6(b) versus 6(c)). Because crystal structures are subject to packing interactions, the compactness and degree of folding of the T. tengcongensis aptamer were assessed in solution using SAXS. The preQ1-bound and free-state aptamers – identical to those used in crystallization – were equally compact in solution with radius of gyration (RG) values of 16.6 ± 0.02 Å and 17.0 ± 0.02 Å, respectively; these values were comparable to those of the bound and free-state crystal structure whose RG values were 16.9 Å and 17.0 Å, respectively46. Additional evidence for ligand-independent compaction for the T. tengcongensis aptamer came from estimation of the maximum intraparticle distance, rMax. Again both preQ1 bound and free-state samples exhibited similar rMax values of 51 ± 5 Å and 57 ± 6 Å, respectively. The rMax values from bound and free-state crystal structures were 50 Å and 52 Å, respectively, which are comparable to SAXS-based values measured for the preQ1-bound state. Control SAXS measurements were conducted on the F. nucleatum riboswitch. In the preQ1-bound state, the F. nucleatum aptamer exhibited RG and rMax values of 19.3 ± 0.03 Å and 64 ± 6 Å. By contrast, the ligand-free state showed values of 31.0 ± 0.07 Å and 107 ± 11 Å. This corroborates prior ligand-dependent folding observations for the F. nucleatum aptamer68,69 and such findings are consistent with ligand-dependent folding by the B. subtilis aptamer62. These results stand in contrast to those of the T. tengcongensis preQ1 aptamer, which appears equally compact in the ligand bound and free states.
To evaluate whether the crystal structure of the free-state T. tengcongensis aptamer and its compactness in solution are representative of the predominant solution conformation, the observed and calculated scattering profiles were compared. The preQ1-bound crystal structure agreed relatively well with the bound-state SAXS data producing a χ2 fit of 1.7. By contrast, the free-state crystal structure was a poor match with the free-state SAXS data, as indicated by a χ2 value of 5.846. One possible reason for this discrepancy is the crystal packing experienced by the free-state molecule. Overall this result and the observation that A14 “closes” the preQ1-binding pocket in the free state (Figure 6(c)) suggest that the free-state aptamer must also form an “open” conformation receptive to ligand binding. Such results appear comparable to the SAM-I aptamer45, although the latter work is notable for its characterization of the solution ensemble by a variety of complementary techniques.
AMINO ACID BINDING RIBOSWITCHES
Lysine Riboswitch
Riboswitches that regulate lysine biosynthesis genes have been identified in both Gram-positive and Gram-negative bacteria19,70,71. The Gram-positive riboswitches regulate genes by premature transcription termination, whereas Gram-negative varieties utilize translational repression70,71. These riboswitches bind L-lysine and the apparent KD for the B. subtilis lysC riboswitch was determined to be approximately 1 µM for a 179 nucleotide construct (nucleotides 25–205 of the lysC 5’-UTR) that composes only the aptamer domain. When nucleotides 1–315 of the lysC 5’-UTR were included the KD increased to 500 µM19. In vitro transcription showed that between 3 µM70 and 10 µM19 L-lysine was required to achieve 75% transcription termination.
Despite its relatively modest affinity, the lysine riboswitch shows exceptional discrimination against lysine analogues with nearly undetectable binding to D-lysine and ornithine19.The lysine precursor diaminopimelate is converted to lysine by the action of the lysA gene product, which is also regulated by a lysine riboswitch. This molecule differs from lysine only by the presence of a single carboxyl group, which renders it incapable of inducing transcription termination19,70.
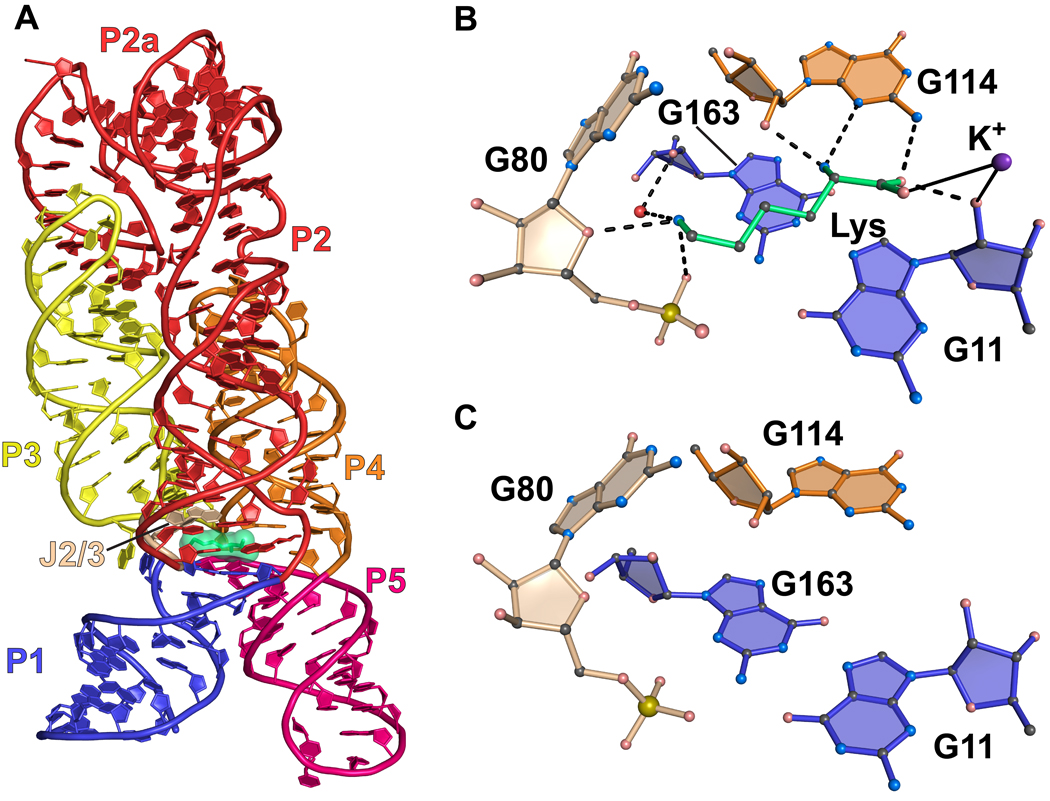
The structure of the lysine riboswitch from Thermotoga maritima has been determined in both the ligand-bound and free states23,44. The global fold comprises five main helices with collinear stacking, which establishes the lysine-binding pocket at the confluence of a multi-helix junction (Figure 7(a)). The mode of lysine recognition is multifold and utilizes hydrogen bonding as well as shape complementarity. At the acid end of lysine, one carboxylic oxygen accepts hydrogen bonds from both the N2 amino group of G114, as well as the ribosyl O2’ from G11 (Figure 7(b)). The other carboxylate oxygen coordinates a K+ ion that also interacts with the O2’ of G11. The α-amino group of lysine hydrogen bonds to the O2’ and N3 imino groups of G114 at the sugar edge of the nucleoside. At the basic end, the ε-amino group of lysine hydrogen bonds to the O4’ ribosyl oxygen of G80, as well its non-bridging oxygen (Figure 7(b)). A water molecule also mediates specific contacts between the ε-amino group and the RNA aptamer. The prominent electrostatic interactions with the ion and the phosphate backbone are notable affinity determinants.
Figure 7.
Cartoon and ball-and-stick diagrams of the T. maritima lysine riboswitch. (a) Cartoon diagram depicting the tertiary structure of the lysine riboswitch in the bound state. The coordinates are from PDB entry 3DIL. (b) Ball-and-stick diagram of the structure in (a) depicting hydrogen bonds with the riboswitch and coordination with a potassium ion; water is shown as a red sphere. The coordinates were derived from PDB entry 3DIL (c) The lysine binding site in the ligand-free state riboswitch. No significant conformational changes are observed compared to the structure in (b). The coordinates were derived from PDB entry 3DIS.
Free-state structures of the lysine riboswitch showed only minor changes compared to their respective bound states with typical rmsd values of 0.35 Å; this differs from the larger rearrangements seen in the preQ1 and SAM-I free states. The lysine-binding pocket is in a distinctly ‘closed’ state, comparable to the SAM-I and preQ-I aptamers. However, the lysine pocket is not occluded by an aptamer base (Figure 7(c)), and the empty amino acid binding site is well-formed but inaccessible to solvent in free-state crystal structures23,44. This observation is corroborated by SHAPE analysis of both the B. subtilis and T. maritima lysine riboswitches. These results revealed only small, local changes in secondary structure upon addition of lysine at sub-physiological Mg2+ concentrations ranging from 0.2 to 0.5 mM with the most significant changes in J2/3, P5 and P123. SAXS was employed to assess the extent of structural changes under various conditions for the B. subtilis aptamer. At 5 mM Mg2+ the structure of the riboswitch was substantially compact with an RG of 32.0 ± 0.1 Å. The addition of 2 mM lysine had no apparent affect as revealed by an RG of 31.5 ± 0.1 Å23. These results mirror observations for the SAM-I and translational preQ1 riboswitches.
Formation of the P1 helix of the lysine riboswitch (Figure 7(a)) promotes folding of a terminator structure that simultaneously abrogates formation of an adjacent anti-terminator helix (e.g., Figure 1(a)). To probe the stability of the P1 helix under biologically relevant conditions, the effects of increasing P1 length were examined in the context of the B. subtilis lysC riboswitch. Extension of P1 was expected to increase helix stability in the absence of lysine, which was then measured by use of in vitro transcription72. The results revealed significantly higher premature transcription termination in the absence of lysine as compared to wildtype. FRET was also conducted to examine folding transitions in the B. subtilis lysC riboswitch72. Fluorophore pairs were placed at locations on P1 and P5 (Figure 7(a)), and folding was analyzed at various Mg2+ concentrations in the presence and absence of lysine. The results indicated that half-maximal FRET efficiency in the absence of lysine occurred in 0.5 mM Mg2+, whereas addition of 5 mM lysine produced half maximal FRET efficiency in 0.05 mM Mg2+. Overall, the results suggest that the lysine riboswitch can pre-fold in the free state, but lysine stabilizes the tertiary structure. This fortification is important in the context of RNA polymerase, which is likely to shift the RNA folding equilibrium to the unfolded state.
Glycine Riboswitch
Computational analysis of bacterial genomes revealed the presence of two putative riboswitch classes called gcvT type I and type II, which were identified upstream of genes responsible for glycine metabolism73. Glycine riboswitches activate gene expression upon binding ligand and most operate at the level of transcription27. Further analysis on a pair of homologous aptamers upstream of the VC1422 gene from Vibrio cholerae showed that the type I (designated VCI) and II (designated VCII) riboswitches actually compose a single riboswitch with tandem aptamers and a single expression platform. Each aptamer binds one glycine molecule but does so in a cooperative manner, as indicated by a Hill coefficient of 1.6. For comparison, VCII alone binds glycine with a Hill coefficient of 0.9774,75. The cooperative nature of ligand binding was confirmed by in vitro transcription assays with B. subtilis gcvT. Both type I and type II aptamers showed high specificity for glycine but were unable to bind alanine or serine74.
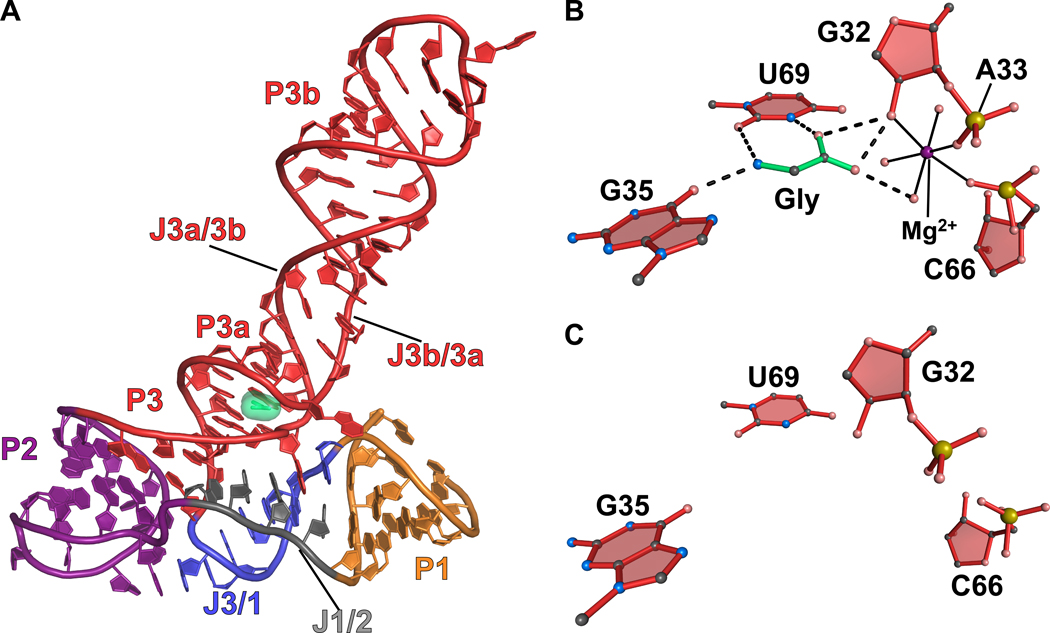
A structure of the VCII aptamer domain was determined to 2.85 Å resolution in the glycine-bound state43. The VCII aptamer forms a three-way helical junction with glycine bound just above the junction (Figure 8(a)). Glycine is recognized by hydrogen bonds between its amino group and keto groups O6 of G35 and O2 of U69 (Figure 8(b)). The glycine carboxyl group hydrogen bonds via one of its oxygens to the N3 imine of U69 and O2’ of the G32 ribose. The other oxygen forms hydrogen bonds with O2’ of G32 and water in the hydration sphere of Mg2+. This magnesium neutralizes the negative charge of the carboxylate group of the glycine ligand and forms inner-sphere contacts to the O2’ hydroxyl of G32 and non-bridging oxygens of C66 and A33 (Figure 8(b)). A second magnesium ion present at the ligand binding site forms interactions between water in its hydration sphere and the nonbbridging oxygens of G35 and A34, as well as the O2’ hydroxyl of A81 (not shown). This magnesium also repels the positively charged ammonium group of glycine.
Figure 8.
Cartoon and ball-and-stick diagrams of the V. cholerae VCII glycine riboswitch. (a) Cartoon diagram depicting the tertiary structure of the glycine VCII riboswitch in the ligand-bound state. The coordinates are from PDB entry 3OWW. (b) Ball-and-stick diagram of the structure in (a) depicting hydrogen bonds between the riboswitch and glycine. (c) The VCII glycine-binding site from a ligand-free crystal structure of the VCII glycine riboswitch. There are no substantial conformational changes relative to the bound structure. The coordinates were derived from PDB entry 3OX0.
The VCII aptamer was also determined in the glycine-free state to a resolution of 3.05 Å43. The ligand-free aptamer showed only modest rearrangements compared to the bound state with an all-atom rmsd of 0.96 Å. The Mg2+ that coordinated glycine was absent, resulting in a more open binding pocket due to rearrangement of the nucleotides that form the ion’s outer coordination sphere. As suggested for other ligand-free riboswitches, this structure most likely represents a single low-energy conformation present in the solution ensemble. Notably, the free-state glycine riboswitch was crystallized in the presence of 80 mM Mg2+, which likely contributed significantly in the stabilization of a bound-state-like conformation. A longer P1 helix was utilized also to stabilize the structure43.
The tandem VCI-II aptamers have been investigated using a variety of methods including SAXS and hydroxy-radical footprinting75. SAXS data indicated that the VCI-II riboswitch undergoes significant folding in the presence of Mg2+ with no glycine present. Compaction of the riboswitch was observed in which the RG decreases from 55 Å in the absence of Mg2+ to 45 Å when 10 mM Mg2+ is added. Further compaction to 39 Å is observed upon addition of 10 mM glycine. These observations provide independent confirmation that the VCI-II riboswitch adopts a compact conformation in the absence of ligand that is suggestive of a partly folded free state. When examined in light of the crystal structure, the results suggest that the glycine aptamer may favor a low-energy solution conformation predisposed to glycine binding, and is therefore poised to undergo gene activation when triggered by metabolite. Further experiments are required to understand how chemical binding is communicated to the expression platform and the allosteric basis for glycine signaling at the molecular level.
Concluding Remarks
Structural investigations of riboswitches have helped to uncover the molecular basis for ligand affinity and specificity. However, because riboswitch-mediated gene regulation entails formation of RNA conformations that are mutually exclusive, greater emphasis should be placed on the analysis of free-state riboswitch structures. At present, our understanding of how riboswitches fold in the free state is poor despite the importance of this state in ligand recognition, ligand-binding kinetics, and kinetic control of gene regulation. Although the kinetics of ligand association and dissociation are determined routinely, such measurements often take place in the absence of the expression platform and without RNA polymerase or the translation initiation machinery. These important factors will impact significantly RNA folding events and necessarily gene regulation. The most sophisticated biophysical analyses indicate that riboswitches exist as an ensemble of conformations45,52,54,64–66,69 and hint at use of conformational selection for ligand recognition (Sidebar 1). To advance the field, greater strides must be taken to explore such mechanisms, and current analysis suggest this must be done on a case-by-case basis. As a practical goal, knowledge of riboswitch mechanisms of action has helped targeting by small molecules (reviewed in Mulhbacher et. al.76). The emergence of drug resistant organisms with widespread riboswitch utilization suggests ample opportunities exist to bring new therapeutics to the forefront of medicine.
Acknowledgment
We thank members of the Wedekind lab for support and helpful discussions. This work was funded in part by NIH grants GM063162 and RR026501 awarded to J.E.W.
Footnotes
Further Reading/Resources
Rfam rfam.sanger.ac.uk (A database comprising a collection of RNA families derived from multiple sequence alignments, consensus secondary structures and covariance analysis).
Protein Data Bank www.pdb.org (A database comprising experimentally-derived atomic coordinates and structure-factor amplitudes that includes riboswitch structures).
References
- 1.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 2.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 3.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wachter A. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 2010;7:67–76. doi: 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 7.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3' end processing of mRNAs. Plant Cell. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci U S A. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochim Biophys Acta. 2009;1789:584–591. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henkin TM. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith KD, Lipchock SV, Livingston AL, Shanahan CA, Strobel SA. Structural and Biochemical Determinants of Ligand Binding by the c-di-GMP Riboswitch. Biochemistry. 2010;49:7351–7359. doi: 10.1021/bi100671e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 15.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shine J, Dalgarno L. Terminal-sequence analysis of bacterial ribosomal RNA. Correlation between the 3'-terminal-polypyrimidine sequence of 16-S RNA and translational specificity of the ribosome. Eur J Biochem. 1975;57:221–230. doi: 10.1111/j.1432-1033.1975.tb02294.x. [DOI] [PubMed] [Google Scholar]
- 18.Irnov, Kertsburg A, Winkler WC. Genetic control by cis-acting regulatory RNAs in Bacillus subtilis: general principles and prospects for discovery. Cold Spring Harb Symp Quant Biol. 2006;71:239–249. doi: 10.1101/sqb.2006.71.021. [DOI] [PubMed] [Google Scholar]
- 19.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards TE, Ferre-D'Amare AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 25.Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6:187–194. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ataide SF, Wilson SN, Dang S, Rogers TE, Roy B, Banerjee R, Henkin TM, Ibba M. Mechanisms of resistance to an amino acid antibiotic that targets translation. ACS Chem Biol. 2007;2:819–827. doi: 10.1021/cb7002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serganov A, Patel DJ. Amino acid recognition and gene regulation by riboswitches. Biochim Biophys Acta. 2009;1789:592–611. doi: 10.1016/j.bbagrm.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du Q, Wang H, Xie J. Thiamin (vitamin B1) biosynthesis and regulation: a rich source of antimicrobial drug targets? Int J Biol Sci. 2011;7:41–52. doi: 10.7150/ijbs.7.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thore S, Frick C, Ban N. Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. J Am Chem Soc. 2008;130:8116–8117. doi: 10.1021/ja801708e. [DOI] [PubMed] [Google Scholar]
- 30.Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000865. e1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR. Design and antimicrobial action of purine analogues that bind Guanine riboswitches. ACS Chem Biol. 2009;4:915–927. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serganov A. Determination of riboswitch structures: light at the end of the tunnel? RNA Biol. 2010;7:98–103. doi: 10.4161/rna.7.1.10756. [DOI] [PubMed] [Google Scholar]
- 33.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 34.Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 36.Poiata E, Meyer MM, Ames TD, Breaker RR. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA. 2009;15:2046–2056. doi: 10.1261/rna.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batey RT. Wiley Interdisciplinary Reviews: RNA. Volume 2. John Wiley and Sons LTD; 2011. Recognition of S-adenosylmethionine by riboswitches; pp. 199–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth A, Winkler WC, Regulski EE, Lee BW, Lim J, Jona I, Barrick JE, Ritwik A, Kim JN, Welz R, et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat Struct Mol Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 39.Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA. 2008;14:685–695. doi: 10.1261/rna.937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards AL, Reyes FE, Heroux A, Batey RT. Structural basis for recognition of S-adenosylhomocysteine by riboswitches. RNA. 2010;16:2144–2155. doi: 10.1261/rna.2341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenkins JL, Krucinska J, McCarty RM, Bandarian V, Wedekind JE. Comparison of a PreQ1 Riboswitch Aptamer in Metabolite-bound and Free States with Implications for Gene Regulation. J Biol Chem. 2011;286:24626–24637. doi: 10.1074/jbc.M111.230375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 48.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 49.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu C, Ding F, Chowdhury A, Pradhan V, Tomsic J, Holmes WM, Henkin TM, Ke A. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J Mol Biol. 2010;404:803–818. doi: 10.1016/j.jmb.2010.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 52.Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nat Chem Biol. 2011;7:393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 53.Smith AM, Fuchs RT, Grundy FJ, Henkin TM. The SAM-responsive S(MK) box is a reversible riboswitch. Mol Microbiol. 2010;78:1393–1402. doi: 10.1111/j.1365-2958.2010.07410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson RC, Smith AM, Fuchs RT, Kleckner IR, Henkin TM, Foster MP. Tuning riboswitch regulation through conformational selection. J Mol Biol. 2011;405:926–938. doi: 10.1016/j.jmb.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, Ke A. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol. 2008;15:1076–1083. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu C, Smith AM, Ding F, Chowdhury A, Henkin TM, Ke A. Variable Sequences outside the Sam-Binding Core Critically Influence the Conformational Dynamics of the SAM-III/ SMK Box Riboswitch. J Mol Biol. 2011;409:786–799. doi: 10.1016/j.jmb.2011.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2'-deoxyguanosine. Proc Natl Acad Sci U S A. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim JN, Breaker RR. Purine sensing by riboswitches. Biol Cell. 2008;100:1–11. doi: 10.1042/BC20070088. [DOI] [PubMed] [Google Scholar]
- 59.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 60.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J Biol Chem. 2009;284:11012–11016. doi: 10.1074/jbc.C900024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang M, Peterson R, Feigon J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 63.Klein DJ, Edwards TE, Ferre-D'Amare AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 65.Brenner MD, Scanlan MS, Nahas MK, Ha T, Silverman SK. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry. 2010;49:1596–1605. doi: 10.1021/bi9019912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. Interplay of 'induced fit' and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liberman JA, Wedekind JE. Base ionization and ligand binding: how small ribozymes and riboswitches gain a foothold in a protein world. Curr Opin Struct Biol. 2011;21:327–334. doi: 10.1016/j.sbi.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rieder U, Lang K, Kreutz C, Polacek N, Micura R. Evidence for pseudoknot formation of class I preQ1 riboswitch aptamers. Chembiochem. 2009;10:1141–1144. doi: 10.1002/cbic.200900155. [DOI] [PubMed] [Google Scholar]
- 69.Rieder U, Kreutz C, Micura R. Folding of a transcriptionally acting preQ1 riboswitch. Proc Natl Acad Sci U S A. 2010;107:10804–10809. doi: 10.1073/pnas.0914925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grundy FJ, Lehman SC, Henkin TM. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci U S A. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res. 2003;31:6748–6757. doi: 10.1093/nar/gkg900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blouin S, Chinnappan R, Lafontaine DA. Folding of the lysine riboswitch: importance of peripheral elements for transcriptional regulation. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci U S A. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 75.Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mulhbacher J, St-Pierre P, Lafontaine DA. Therapeutic applications of ribozymes and riboswitches. Curr Opin Pharmacol. 2010;10:551–556. doi: 10.1016/j.coph.2010.07.002. [DOI] [PubMed] [Google Scholar]