Abstract
The expression of small, non-coding RNA, or microRNAs (miR), is frequently deregulated in human cancer, but how these pathways affect disease progression is still largely elusive. Here, we report on a microRNA, miR-296, which is progressively lost during tumor progression, and correlates with metastatic disease in colorectal, breast, lung, gastric, parathyroid, liver and bile ducts cancers. Functionally, miR-296 controls a global cell motility gene signature in epithelial cells by transcriptionally repressing the cell polarity-cell plasticity module, Scrib. In turn, loss of miR-296 causes aberrantly increased and mislocalized Scrib in human tumors, resulting in exaggerated random cell migration, and tumor cell invasiveness. Re-expression of miR-296 in MDA-MB231 cells inhibits tumor growth, in vivo. Finally, miR-296 or Scrib levels predict tumor relapse in hepatocellular carcinoma patients.
These data identify miR-296 as a global repressor of tumorigenicity, and uncover a previously unexplored exploitation of Scrib in tumor progression in humans.
Keywords: miR-296, Scribble, cell plasticity, tumor progression, metastases
Introduction
A network of small, non-coding RNA or microRNAs (miR) has attracted attention as regulators of gene expression (Bartel, 2004), influencing human diseases (Garzon et al., 2009). In cancer, the systematic cataloging of deregulated miR expression in transformed cells compared to normal tissues (Lu et al., 2005), has helped uncovering novel transcriptional networks relevant to cell invasion (Valastyan et al., 2009) and metastasis (Zhang et al., 2010), as main drivers of unfavorable disease outcome in humans (Calin and Croce, 2006).
In this context, perturbation of a normal apical-basal polarity in epithelial cells (Etienne-Manneville, 2008) is thought of as one of the earliest phenotypical changes in cellular transformation (Zhan et al., 2008; Ouyang et al., 2010), heralding invasive cancer (Hanahan and Weinberg, 2000). The transcriptional programs underlying this process are not completely understood, but so-called cell polarity-cell plasticity modules, for instance the mammalian homologs of Drosophila melanogaster Scribble (Scrib), atypical protein kinase C (aPKC) or Crumbs (Crb) play a key role in apical-basal polarity of epithelia, cell orientation (Etienne-Manneville, 2008), and directional cell movements (Petrie et al., 2009). In this context, loss of polarity proteins in Drosophila (Bilder et al., 2000), or mammalian cells (Etienne-Manneville, 2008), deregulates cell proliferation (Dow et al., 2008; Wu et al., 2010), survival (Zhan et al., 2008), epithelial-mesenchymal transition (Bahri et al., 2010), and random cell migration (Osmani et al., 2006), suggesting that these molecules may function in an evolutionary-conserved pathway of tumor suppression, in vivo (Humbert et al., 2008; Wu et al., 2010).
In this study, we aimed at identifying novel miR regulators of tumor progression conserved in animal models and in epithelial human cancers irrespective of the cancer origin. We found that miR-296, a miR previously associated with angiogenic endothelial cells (Wurdinger et al., 2008), or to prostate cancer (Wei et al., 2010), functions as a global repressor of tumorigenicity, and that this pathway, differently from previous models (Humbert et al., 2008; Wu et al., 2010), involves deregulation and subcellular mislocalization of Scrib in metastatic human cancer (Etienne-Manneville, 2008).
Results
Loss of miR-296 expression during tumorigenesis
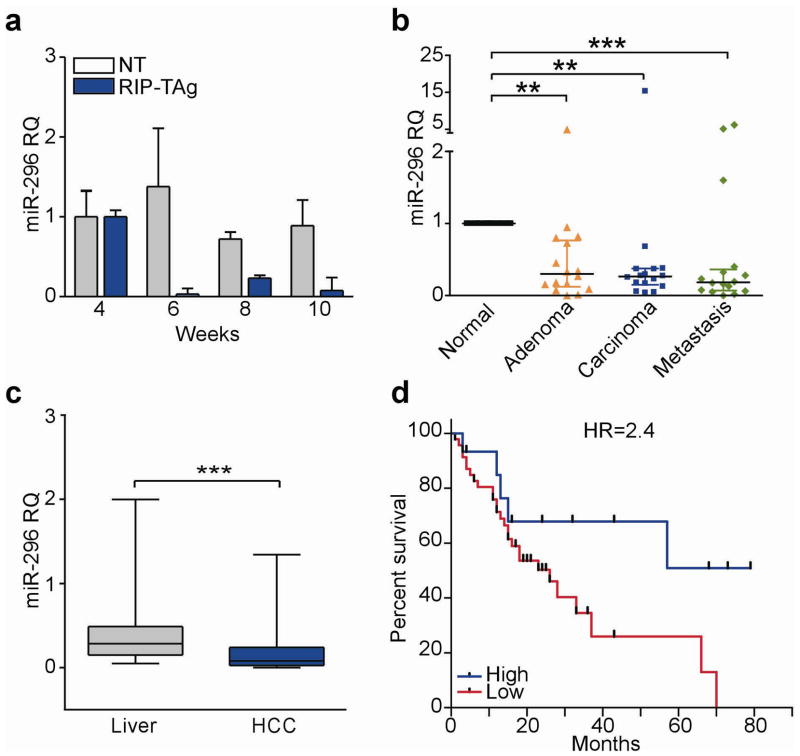
We began this study by profiling the expression of 132 orthologous miRs in microdissected pancreatic islets isolated from Rat Insulin Promoter-T Antigen (RIP-TAg) transgenic mice (Hanahan, 1988), an established model of tumor progression, suitable for miR analysis (Olson et al., 2009). Islets from RIP-TAg mice exhibited extensive changes in miR expression during oncogenic transformation (4–6 weeks) and evolution to invasive pancreatic islet cell carcinoma (10 weeks) (Supplementary Figure 1a). In this analysis, miR-296 was consistently downregulated throughout all stages of islet tumorigenesis, in vivo (Figure 1a). Conversely, miR-296 was expressed at comparable levels in non-transgenic (NT) islets at the same time intervals (Figure 1a). As control, oncogenic miRs, including miR-21 (Seike et al., 2009) (Supplementary Figure 1b), or miR-19a (Supplementary Figure 1c) (Olson et al., 2009), were up-regulated in transgenic islets, but not aged-matched controls. In addition, unrelated miRs, for instance miR-452 (Supplementary Figure 1d), or miR-216 (Supplementary Figure 1e) were indistinguishably expressed in non-transgenic or RIP-TAg pancreatic tissues.
Figure 1.
Loss of miR-296 expression in human cancer. (a) Pancreatic islets from non-transgenic (NT) or RIP-Tag islets were analyzed for miR-296 expression at the indicated time intervals by qPCR (RQ). Mean±s.d. of replicates. (b) Analysis of miR-296 expression in 16 patients with metastatic colon adenocarcinoma. Bars correspond to median expression value with interquartile range. Normal, matched normal mucosa (**, P<0.01 and ***, P<0.0001, Kruskal-Wallis test). (c) miR-296 levels in patients with hepatocellular carcinoma (HCC) or matched non-neoplastic liver (***, P<0.001, Wilcoxon signed-rank test). (d) Kaplan-Meier curves of relapse-free survival in HCC patients with high or low expression of miR-296. HR, hazard ratio (P=0.045, Log-rank test).
When analyzed in human samples, miR-296 levels were profoundly repressed in various cultured tumor cell lines representative of breast, lung, colorectal or liver adenocarcinoma, compared to normal mammary epithelial cells (HMEC) (Supplementary Figure 2a). Similarly, miR-296 expression was progressively lost in 86% of patient samples representative of the colorectal adenoma-to-carcinoma transition (13 out of 16 patients, P=0.0007, Table 1), compared to matched normal mucosa (Figure 1b). A comparable pattern was observed in disparate, genetically heterogeneous tumor types (Table 1), where miR-296 expression was frequently lost in primary (13 out of 20 patients, P<0.05), as well as metastatic lesions (15 out of 20 patients, P=0.009) (Table 1 and Supplementary Figure 2b). When correlated to disease outcome, loss of miR-296 levels in patients with hepatocellular carcinoma (HCC) (Augello et al., 2009), compared to matched normal liver (Figure 1c), was associated with considerably shortened disease-free survival (HR=2.4, Figure 1d).
Table 1.
Patients’ characteristics.
| Patient ID | Disease1 | Stage of Primary Tumor2 | Site of Distant Metastasis | miR-296 Level3 |
|---|---|---|---|---|
| Colon Ca1 | Colorectal Carcinoma § | D | Liver | 0.06 |
| Colon Ca2 | Colorectal Carcinoma § | C2 | Peritoneum | 0.01 |
| Colon Ca3 | Colorectal Carcinoma ‡ | D | Peritoneum | 0.0002 |
| Colon Ca4 | Colorectal Carcinoma ‡ | D | Liver | 0.32 |
| Colon Ca5 | Colorectal Carcinoma § | B2 | Liver | 0.27 |
| Colon Ca6 | Colorectal Carcinoma § | B2 | Liver | 5.13 |
| Colon Ca7 | Colorectal Carcinoma ‡ | D | Liver | 0.07 |
| Colon Ca8 | Colorectal Carcinoma § | D | Liver | 0.17 |
| Colon Ca9 | Colorectal Carcinoma ‡ | B2 | Liver | 6.18 |
| Colon Ca10 | Colorectal Carcinoma § | B2 | Liver | 0.19 |
| Colon Ca11 | Colorectal Carcinoma ‡ | C2 | Peritoneum | 0.23 |
| Colon Ca12 | Colorectal Carcinoma ‡ | B2 | Lung | 0.05 |
| Colon Ca13 | Colorectal Carcinoma ‡ | B2 | Liver | 0.16 |
| Colon Ca14 | Colorectal Carcinoma ‡ | B2 | Liver | 0.4 |
| Colon Ca15 | Colorectal Carcinoma ‡ | D | Liver | 0.13 |
| Colon Ca16 | Colorectal Carcinoma ‡ | B2 | Liver | 1.6 |
| BC1 | Breast Cancer | pT4,Nx,M0 | Skin | 0.02 |
| BC2 | Breast Cancer | pT2,N0,M0 | Skin | 0.15 |
| BC3 | Breast Cancer | pT1a,Nx,M0 | Skin | 0.42 |
| BC4 | Breast Cancer | pT2,N1,M0 | Uterus | 0.00039 |
| LC1 | NSCLC | pT4,N1,M1 | Pleura | 0.25 |
| LC2 | NSCLC | pT2,N0,M0 | Brain | 0.4 |
| LC3 | NSCLC | pT2,N1,M0 | Adrenal gland | 0.3 |
| LC4 | NSCLC | pT1,N0,M0 | Brain | 5.4 |
| GC1 | Gastric Cancer | pT2b,N3,M1 | Ileum | 1.1 |
| GC2 | Gastric Cancer | pT3,N1,M1 | Peritoneum | 13.9 |
| GC3 | Gastric Cancer | pT3,N3,M1 | Peritoneum | 0.19 |
| GC4 | Gastric Cancer | pT2b,N2,M1 | Liver | 0.4 |
| GC5 | Gastric Cancer | pT4,N2,M1 | Liver | 3.5 |
| GC6 | Gastric Cancer | pT3,N1,M0 | Peritoneum | 0.35 |
| GC7 | Gastric Cancer | pT3,N3,M1 | Peritoneum | 0.1 |
| GC8 | Gastric Cancer | pT4,N2,M1 | Liver | 0.26 |
| GC9 | Gastric Cancer | pT3,N1,M0 | Peritoneum | 0.25 |
| CC1 | Cholangiocarcinoma | pT3,N1,M1 | Liver | 0.11 |
| CC2 | Cholangiocarcinoma | pT1,N0,M0 | Liver | 0.08 |
| PC1 | Parathyroid Carcinoma | - | Lung | 0.09 |
§, mutated k-Ras (V12 or V13), ‡, wild-type k-Ras; NSCLC, non-small cell lung cancer-adenocarcinoma subtype; Breast Cancer, invasive ductal carcinoma hormones receptors-positive; Cholangiocarcinoma, extrahepatic carcinoma of the bile ducts.
Parathyroid carcinoma does not have a TNM staging.
miR-296 levels detected in distant metastatic lesion assuming that expression of matched normal counterpart is 1.
Regulation of tumor cell motility by miR-296
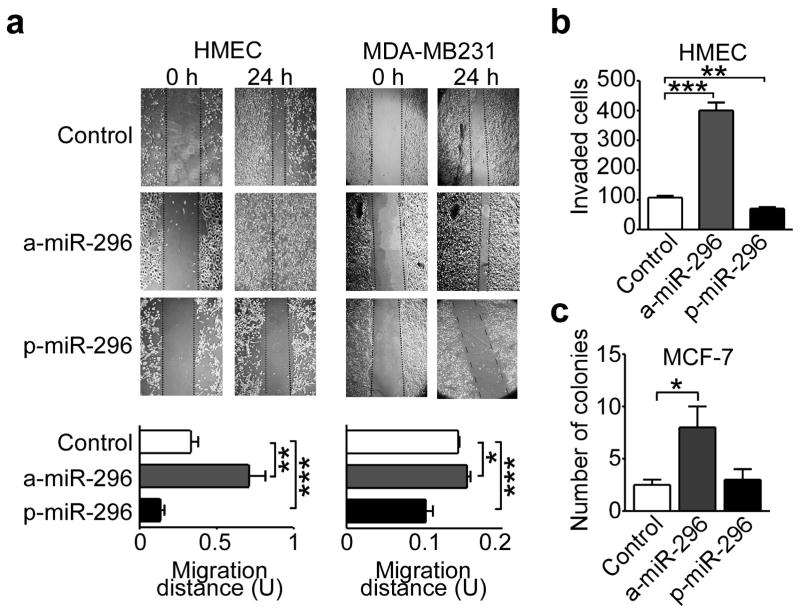
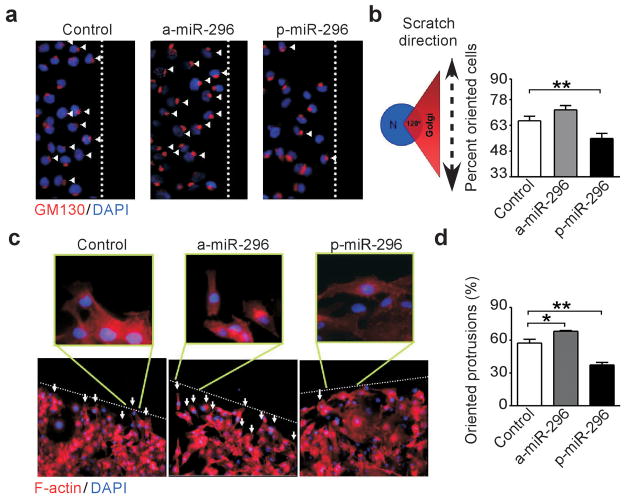
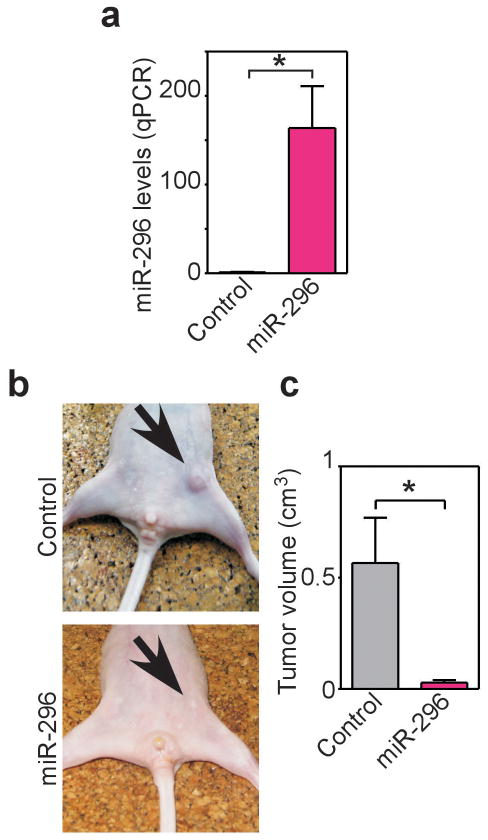
Manipulation of miR-296 levels by transfection of antagomir (a-miR-296, Supplementary Figure 3a), or, conversely, pre-miR-296 (p-miR-296, Supplementary Figure 3b) had no effect cell cycle transitions in various tumor types, by DNA content analysis and flow cytometry (Supplementary Figure 3c). Conversely, antagomir depletion of miR-296 potently stimulated the migration of HMEC or breast adenocarcinoma MDA-MB231 and MCF-7 cells in a wound healing assay, in vitro (Figure 2a and Supplementary Figure 4a). This was also associated with enhanced invasion of HMEC (Figure 2b) or MDA-MB231 cells (Supplementary Figure 4b,c) across Matrigel-coated inserts, using serum (Supplementary Figure 4b), or co-culture with primary carcinoma-isolated fibroblasts (Supplementary Figure 4c) as chemotrattactants. Cells depleted of miR-296 also exhibited increased colony formation in soft agar (Figure 2c, Supplementary Figure 4d), another marker of tumorigenicity. Reciprocally, forced expression of miR-296 by transfection of p-miR-296 repressed breast epithelial cell migration (Figure 2a), invasion (Figure 2b, Supplementary Figure 4b,c), colony formation in soft agar (Supplementary Figure 4d), and promoted anoikis in breast cancer cells (Supplementary Figure 4e,f). In these experiments, perturbation of miR-296 levels induced defects in cellular orientation, as judged by the position of the Golgi apparatus relative to the nucleus (Figure 3a,b), and affected the formation of polarized actin protrusions at the leading edge of directional cell migration (Figure 3c,d). Finally, we tested whether miR-296 re-expression in MDA-MB231 cells (Figure 4a) also inhibits tumor growth. We injected control miRNA and mir-296 transfected cells into the fourth mammary fat pads of immunocompromised mice and monitored tumor growth for three weeks (Figure 4b). Mice injected with miR-296 re-expressing cells resulted in reduced tumor growth compared with controls (Figure 4c).
Figure 2.
miR-296-regulation of cell migration and invasion. (a) The indicated cell types were transfected with antagomir-miR-296 (a-miR-296) or pre-miR-296 (p-miR-296) and analyzed in a wound healing assay after 24 h. Bottom, quantification of cell migration. Control, non-transfected. Scale bar, 100 μm (*, P=0.027; **, P =0.0011; ***, P <0.001). (b) HMEC were transfected as indicated in A, and analyzed for Matrigel invasion in a Transwell assay in the presence of serum (**, P=0.0098; *** P<0.001). (c) Breast adenocarcinoma MCF-7 cells were transfected as indicated and analyzed for colony formation in soft agar after 2 weeks (*, P=0.013). Values are mean ± s.e.m.
Figure 3.
miR-296 control of directional cell migration. (a) MDA-MB231 cells were transfected as indicated, and analyzed for Golgi staining (GM130) during directional cell migration. Dotted line, position of the wound. Arrowhead, oriented cell. Original magnification x200. (b) Quantification of directional cell migration in transfected MDA-MB231 cells. A cell was considered oriented when reactivity for GM130 fell within a 120° arc ahead of the nucleus. The baseline random orientation of the Golgi towards the wound edge was 33% (**, P=0.008). (c) MDA-MB231 cells were transfected as indicated and analyzed for F-actin immunostaining at the edge of directional migration (dotted line). Arrows indicate the formation of actin protrusion towards the direction of migration (enlarged areas). Original magnification x100, enlargements, x400. (d) Quantification of MDA-MB231 cells with oriented cell protrusions. Mean± s.e.m of replicates (*, P=0.03; **, P=0.009).
Figure 4.
miR-296 expression suppresses tumor growth in vivo. (a) MDA-MB231 cells were stably transfected with control miR or mir-296 and analyzed by real time PCR (qPCR). (b) The indicated transfected MDA-MB231 cells were injected s.c. in the mammary fat pad of immunocompromised mice, and tumor growth was visualized macroscopically after three weeks. (c) Quantification of tumor growth in control or miR-296-expressing MDA-MB231 cells. Mean±s.e.m with mice as the unit group.
miR-296 regulation of a cell polarity module
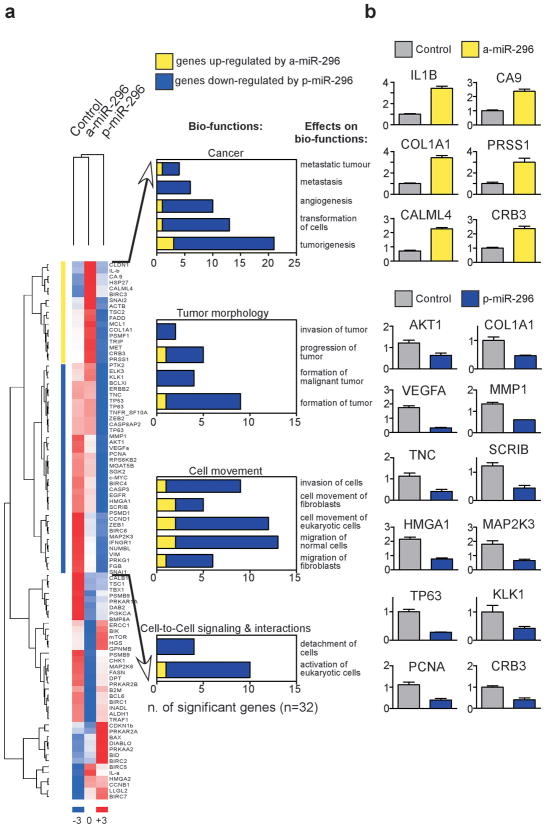
Consistent with the role of miRs in transcriptional control, changes in miR-296 levels had profound effects on the gene expression profile of tumor cells, as revealed by unsupervised clustering of candidate genes putatively modulated by miR-296 (Figure 5a). Out of 107 predicted candidate genes (Supplementary Table 1), potentially associated with miR-296 pathways (Supplementary Table 2), a 32-gene signature was validated by quantitative PCR in cells with acute changes in miR-296 expression (Figure 5a), comprising general regulators of tumor cell motility, migration, invasion, and metastasis (Figure 5b, Supplementary Table 1).
Figure 5.
mir-296 target gene signature in tumor cells. (a) Unsupervised clustering (dChip software) of eighty-nine candidate genes modulated by miR-296 expression in transfected MDA-MB231 cells (listed in Supplementary Table 1). Right, Candidate genes modulated by differential miR-296 expression levels with a >1.5 fold change (FC) cut-off were cataloged using Ingenuity Pathway software. (b) Validation of miR-296-regulated genes by qRT-PCR. Numbers in y-axis indicate relative quantification of target to TBP gene. Mean±s.d.
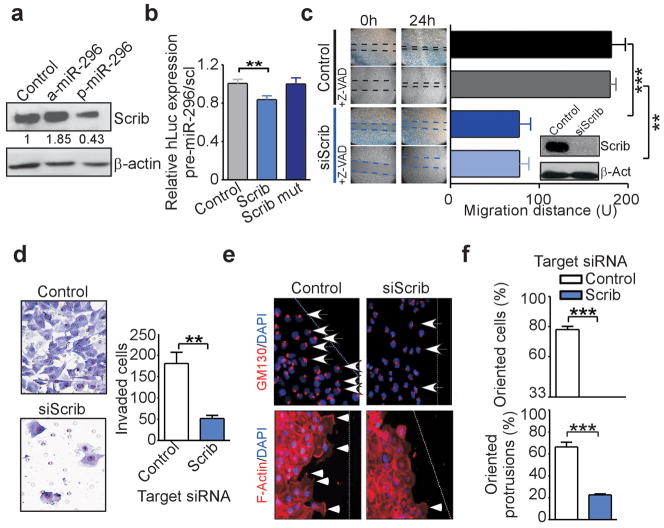
One of the target genes in the miR-296 gene signature was identified as the cell polarity-cell plasticity module, Scrib (Dow et al., 2007), an important regulator of apical-basal polarity and directional cell movement (Osmani et al., 2006). Accordingly, transfection of miR-296 suppressed Scrib protein expression in normal or transformed epithelial cells (Figure 6a and Supplementary Figure 5a), and significantly inhibited Scrib 3′ UTR activity in luciferase reporter assays (Figure 6b). In addition, mutagenesis of putative miR-296 binding site in Scrib identified by sequence analysis (Supplementary igure 5b) restored 3′ UTR activity to the levels of control transfectants (Figure 6b), validating Scrib as a target gene directly repressed by miR-296. Other putative or predicted target proteins as BMP8A (Supplementary Table 1) or HMGA1 (Wei et al., 2010), were not significantly modulated by miR-296 in our system (Supplementary Figure 5c).
Figure 6.
miR-296 regulation of Scrib cell polarity module. (a) MDA-MB231 cells were transfected as indicated and analyzed by Western blotting. Bands were quantified by the Image J program (b) HEK293 cells expressing wild type Scrib 3′ UTR reporter construct or Scrib 3′ UTR construct mutated in the miR-296 binding site (Scrib mut) were transfected with miR-296, and analyzed for luciferase activity. Mean± s.e.m. (n=5; **, P=0.007). (c) MDA-MB231 cells were transfected with non-targeting or Scrib-directed siRNA, and analyzed in a wound healing assay with or without the caspase inhibitor, zVAD-fmk. Right, quantification of cell migration (**, P=0.001; ***, P<0.001). Inset, immunoblotting for Scrib. βActin was a loading control. (d) MDA-MB231 cells were transfected with the indicated siRNA and analyzed for cell invasion. Right, quantification of cell invasion (**, P=0.0028). Mean±s.e.m (e) MDA-MB231 cells were transfected with the indicated siRNA, and analyzed for directional cell migration by Golgi (GM130, top) or F-actin (bottom) staining. Dotted line, position of the wound. Arrow, oriented cell. Arrowheads, formation of actin protrusion towards the direction of migration. Original magnification x200. (f) Quantification of oriented cells (top), or cells with oriented actin protrusion (bottom) in control or Scrib siRNA-silenced cultures (***, P<0.0001). Values are mean±s.e.m.
Functionally, depletion of Scrib by small interfering RNA (siRNA) inhibited migration of MDA-MB231 cells in a wound healing assay, in a response unaffected by the caspase inhibitor Z-VAD (Figure 6c). In addition, Scrib knockdown blocked tumor cell invasion across Matrigel-coated inserts (Figure 6d), induced anoikis (Supplementary Figure 5d,e), prevented a correct cellular orientation towards the leading edge of migration, and reduced the number of polarized actin protrusion at the leading edge of the cell (Figure 6e,f). In contrast, a control, non-targeting siRNA had no effect on cell migration (Figure 6c), invasion (Figure 6d), or requirements of cellular motility (Figure 6e,f).
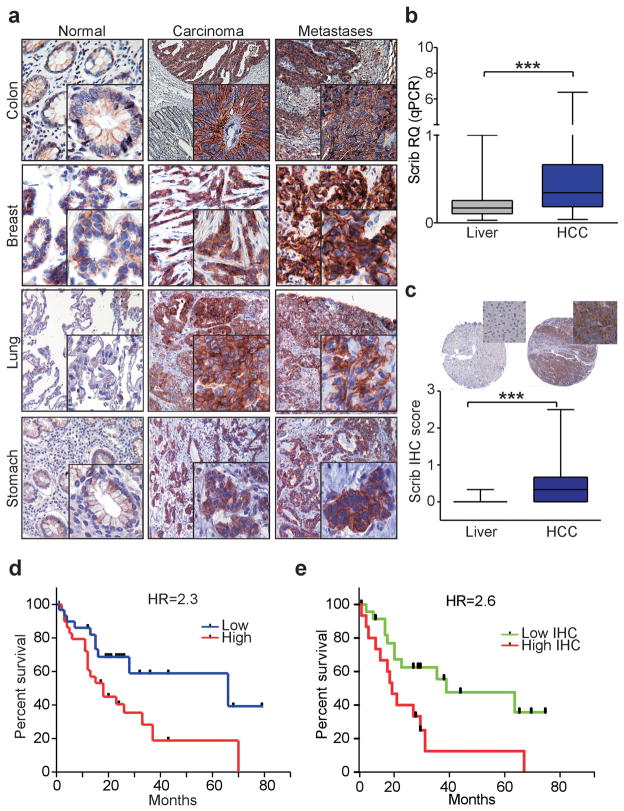
Deregulated Scrib expression in human tumors
Mirroring the loss of miR-296 expression in human cancer, Scrib levels were conversely upregulated in primary lesions and distant metastases of colon, lung, breast, and stomach cancer (n=18), as well as HCC (n=62) (Augello et al., 2009), compared with matched normal mucosa (Figure 7a, Supplementary Figure 6a). k-Ras mutations on codon 12, RasV12, or 13, RasV13, identified in six patients with colon cancer (40%), did not correlate with expression of miR-296 (P=0.8, Fisher exact test, Table 1), or its target, Scrib. Scrib protein levels were also increased in hyperplastic, angiogenic and invasive β-islets from RIP-TAg mice compared to control (4-weeks old RIP-Tag) or non-transgenic littermates (Supplementary Figure 6b). In immunohistochemical studies on tumor specimens, the subcellular localization of Scrib was also deregulated, changing from a predominantly membranous localization at cell-cell junctions, and characteristic of the normal mucosa, to a diffuse cytoplasmic staining in primary and metastatic tumor cells (Figure 7a and Supplementary Figure 6a).
Figure 7.
Deregulated expression of Scrib in human cancer. (a) Representative tissue samples of normal mucosa, adenocarcinoma or metastases from the indicated tumors were analyzed for expression of Scrib by immunohistochemistry. Original magnifications, 200x (normal mucosa), 100x (tumor lesions), and 400x (insets). Cases of HCC or matched normal liver were analyzed for expression of Scrib by qPCR (b) or immunohistochemistry (c). Cytoplasmic and membranous immunoreactivity was scored as negative (0), weak (1), moderate (2) or strong (3) by two authors independently. Original magnification x80 (tissue cores), and x400 (insets) (***, P<0.0001, Wilcoxon signed-rank test). (d) Kaplan-Meier curves of disease-free survival of HCC patients according to the miR-296 target expression. (P=0.025, Log-rank test). (e) Kaplan-Meier curves of relapse-free survival times of HCC patients with high or low Scrib immunohistochemical score (IHC score) (P=0.013, Log-rank test). HR, hazard ratio.
In terms of disease outcome, high levels of Scrib mRNA or protein expression in our series of HCC patients compared to normal liver (Figure 7b,c) significantly correlated with shortened overall survival (Figure 7d,e), thus mirroring the results with miR-296 expression in the same patient cohort (Figure 1d).
Discussion
In this study, we have shown that miR-296, a miR initially associated with angiogenic responses (Wurdinger et al., 2008), is frequently lost during tumor progression in a mouse model of disease, and in primary human specimens, correlating with worse. Functionally, loss of miR-296 deregulates a global cell polarity-cell plasticity module, resulting in aberrant over-expression and mislocalization of Scrib in the transformed cell population (Etienne-Manneville, 2008), and enhanced tumor cell migration and invasion. Conversely, forced re-expression of miR-296 in highly invasive model breast cancer cell types suppressed tumor growth, in vivo.
Although previously implicated in VEGFR2- or PDGFRβ-signaling (Wurdinger et al., 2008), anti-viral mechanisms (Pedersen et al., 2007), retinoic acid-mediated differentiation (Houbaviy et al., 2003; Tay et al., 2008), or to prostate cancer growth and invasion (Wei et al., 2010), a role of miR-296 in cancer has remained controversial, with studies alternatively reporting increased expression (Hong et al., 2010), or, conversely, downregulation (Yang et al., 2009; Wei et al., 2010) in primary patient series. Here, miR-296 was almost universally downregulated in cancer, compared to matched normal tissues, and often lost in metastatic lesions. This had profound effects on the transcriptional profile of transformed cells, unrestraining a distinctive “miR-296-loss” gene signature, enriched in multiple key regulators of cell motility, invasion and metastasis. A recent report (Wei et al., 2010) showed that the oncofetal factor HMGA1 is a direct target of miR-296 in prostate cancer. In our system HMGA1 was significantly down-modulated at mRNA levels in miR-296 re-expressing cells (Figure 4b), though its protein levels were not affected by the miR-296 modulation (Supplementary Figure 5c). Conversely, one of these molecules over-expressed in the miR-296 signature was identified as the cell polarity-cell plasticity module, Scrib (Etienne-Manneville, 2008), which controls apical-basal cell polarity (Bilder et al., 2000), cell orientation (Osmani et al., 2006), and directional cell movements (Petrie et al., 2009) via the assembly of multi-protein complexes at the leading edge of migrating cells (Assemat et al., 2008).
Although these properties, coupled with the pro-tumorigenic phenotype associated with Scrib loss-of-function (Wu et al., 2010; Nagasaka et al., 2010), suggested a role in tumor suppression (Humbert et al., 2008; Wu et al., 2010, Ouyang et al., 2010), our findings point to a far more complex scenario for this pathway in human cancer (Gardiol et al., 2006). Here, Scrib was found abundantly over-expressed in disparate primary and metastatic epithelial malignancies, correlating with worse patient outcome, and mechanistically linked to aberrantly increased tumor cell motility and invasion. Other data also support a causal role of Scrib in human tumorigenesis, as over-expression of Scrib has been observed in colorectal cancer patients, correlating with accumulation of cytoplasmic β catenin (Kamei et al., 2007), the Scrib locus on 8q24.3 is often amplified in breast and ovarian cancer (Kim et al., 2007; Naylor et al., 2005), and Scrib was identified as a mediator of HCC progression, in vivo (Woo et al., 2009).
In this context, it is possible that Scrib may alternatively function in tumor suppression, or, conversely, tumor progression depending on the specific cellular context, and in particular, the complement of genetic alterations acquired during disease progression. For instance, Scrib may function to maintain apical-basal polarity, thus opposing tumorigenicity (Humbert et al., 2008; Wu et al., 2010) at early phases of cellular transformation, whereas more advanced disease stages, characterized by loss of multiple checkpoint/tumor suppressor mechanisms, may exploit aberrantly over-expressed Scrib (Osmani et al., 2006) to enhance random, as opposed to directional cell migration (Petrie et al., 2009), and thus gaining a critical advantage towards metastatic potential (Hanahan and Weinberg, 2000). It is also possible that the subcellular mislocalization of Scrib in the cytosol of tumor cells, as characterized here at variance with its distribution at cell-cell junction, may contribute to this diversity of functions, and promote aberrant cell migration and invasion. This is consistent with previous observations, in which over-expression of Scrib in astrocytes perturbed apical-basal polarity and promoted the formation of random actin protrusion during cell migration (Osmani et al., 2006).
In summary, we identified miR-296 as a global regulator of cell migration, invasion, and tumorigenicity, via direct transcriptional repression of a Scrib cell polarity-cell plasticity module (Etienne-Manneville, 2008). These results uncover a more complex scenario than previously thought for how cell polarity-cell plasticity modules may be exploited during human tumorigenesis (Humbert et al., 2008), whereas the identification of a miR-296-Scrib signaling axis in tumor cell motility (Petrie et al., 2009) may open new venues for the development of anti-metastatic therapies in humans.
Materials and Methods
Sample collection and RNA purification
All experiments involving animals were approved by an Institutional Animal Care and Use Committee. Pancreatic β cells were obtained by laser-assisted microdissection (LMD; Leica Microsystems, Milan, Italy) from total pancreas of four groups of RIP-TAg transgenic mice (two animals per time point) at the following stages of β cell tumorigenesis: normal/transgenic β cells (4-weeks of age), hyperplastic islets (6-weeks of age), angiogenic switch/carcinoma in situ islets (8-weeks of age) and invasive β cell carcinoma (10-weeks of age). Total pancreas was surgically isolated, formalin-fixed (10% buffered formalin) and paraffin embedded (FFPE). As controls, non-transgenic mice were sacrificed at the same age and processed as above.
Epithelial cells from matched normal colonic mucosa, dysplastic adenoma, adenocarcinoma or corresponding metastasis were isolated by LMD from FFPE blocks of 16 patients with diagnosis of colorectal carcinoma. Epithelial cells from non-neoplastic, tumor or metastatic tissue samples were similarly isolated by LMD from patients with diagnosis of adenocarcinoma of the breast (n=4), lung (n=4), stomach (n=9), bile ducts (n=2), or parathyroid tumors (n=1). Patients’ characteristics are summarized in Table 1. Total RNA was isolated using MasterPure RNA Purification Kit (Epicentre Biotechnologies, Madison, WI, USA) following the manufacturer’s instructions and quantified spectrophotometrically.
Total RNA and TMA blocks were available from a series of 62 cases of hepatocellular carcinoma (HCC). Patients’ associated clinical follow-up data was described previously (Augello et al., 2009). Informed consent was obtained from all patients, and the study was approved by an Institutional Review Board of the University of Milan, School of Medicine.
miRNA detection
RT-PCR-based detection of 132 mature miRs and three mouse-specific reference snoRNAs (SNO 55, 412 and 142) was carried out from β cell samples isolated from RIP-TAg transgenic mice or control, non-transgenic mice using the microRNA Reverse Transcription Kit and gene-specific primers and probes (TaqMan MicroRNA Assays), according to the manufacturer’s specifications. All miRNAs were analyzed in duplicate, and instrument raw data (Ct values) were converted into miRNAs relative quantities (RQs) using the geometrical average of the three snoRNAs as normalization factor. For each miRNA, fold change ratios (FC) were calculated between 6, 8 and 10-week old RIP-TAg transgenic mice and 4-week old RIP-TAg animals. A FC=10 or 0.1 for over- or under-expression, respectively, was assigned as threshold for significant different expression. Significantly different miRs were analyzed in aged-matched non-transgenic mice as described above. The RNU48 was used as a reference snoRNA for human tissues or cell lines. For these experiments, miRNA expression relative to reference was calculated using the 2^(−ΔCt) formula. All reagents, primers, kits and instruments were obtained from Applied Biosystems (Life Technologies, Carlsbad, CA, USA).
k-Ras mutation analysis
Genomic DNA from tumor samples collected from 16 patients with diagnosis of metastatic colon adenocarcinoma was amplified with the following specific primers for k-Ras gene: k-Ras F: 5′-GTACTGGTGGAGTATTT-3′; k-Ras R: 5′-ATACAGCTAATTCAGAATCA-3′. Both strands of k-Ras amplicons were sequenced using BigDye terminators (Applied Biosystems, Life Technologies) and mutations occurring at codons 12 and 13 were identified.
Cells and cell culture conditions
Human colon carcinoma SW1116, hepatocellular carcinoma HepG2 and HUH-7, and breast adenocarcinoma MDA-MB231, MCF-7, SKBR3, and MDA-MB435 cells were purchased from the American Type Culture Collection (ATCC). Human embryonic kidney HEK293 cells were a generous gift from Dr. Locati (University of Milan). Normal human mammary epithelial cells (HMEC) were obtained from Gibco-Invitrogen (Life Technologies). Various cell types were seeded at 2×106 cells/well in 6-wells plates, and transfected with 100 pmol of pre-miR-296 (p-miR-296), antagomir-miR-296 (a-miR-296), pre-miR-1, pre-miR or Cy3-labeled anti-miR negative controls (Ambion, Life Technologies) in the presence of 5 μl Lipofectamine 2000 in 1 ml of OptiMem medium (Gibco-Invitrogen, Life Technologies). Stable miR-296 or control vector (GeneCopoeia, Rockville, MD, USA) expressing MDA-MB231 cells were generated after Lipofectamine 2000 transfection and puromycin (Sigma Aldrich, Milan, Italy) selection for one week. Then cells were trypsinized and plated without selection medium. MDA-MB231 with stable expression of miR-296 were validated by qPCR for recombinant miR expression and used for xenograft studies within passage four of culture.
Xenograft breast cancer model
Twelve female CD1 athymic mice were injected subcutaneously in the fourth mammary fat pad with 2×105 MDA-MB231 cells stably transfected with control miR or miR296 in a total volume of 100 μl of sterile PBS (six mice per condition). Tumor growth was monitored externally using vernier calipers and animals were euthanized after 3 weeks. Tumors were then formalin-fixed and paraffin embedded for histological examination. Tumor volume (mm3) was calculated using the formula: Volume = (width)2 × length/2.
Scrib siRNAs
MDA-MB231 cells were transfected with 100pmol of siGENOME or control Smart Pool siRNAs (cat. M-010500, D-001206 all from Dharmacon, Lafayette, CO, USA) and 5 μl of Lipofectamine 2000, as before. Subsequent to transfection, cells were maintained in complete medium for 24 or 48 and processed for individual experiments.
Cell cycle, apoptosis and cell viability assays
Cell cycle transitions and quantification of hypodiploid DNA content (i.e. sub-G1 cell fraction) were determined after 48 or 72 h in transfected cultures by propidium iodide staining and flow cytometry, as described (Romagnoli et al., 2008). Cells transfected with non-targeting constructs were used as unit sample.
Colony formation assay
MDA-MB231 or MCF-7 cells (5×103) were transfected with the various miRNA constructs, and seeded as a single-cell suspension in the presence of 1 ml of 0.7% top agar solution and 1% base agar solid layer after 48 h. Agar for both layers was dissolved in appropriate media supplemented with 10% FBS. Cell were cultured at 37°C, fed twice per week with complete culture medium, and plates were stained after 21 d with 0.5 ml of 0.05% Crystal Violet (Sigma-Aldrich) for 1h. Colonies with a diameter of at least 100 μm were counted by light microscopy and photographed.
Isolation of human breast cancer associated fibroblasts (CAFs)
We isolated tumor-associated fibroblasts from three patients surgically resected for breast cancer. Briefly the tissues were minced with blades and digested as previously described (Bauer et al., 2010). After isolation, CAFs were plated in cell culture dishes, cultured in 10%-FBS supplemented DMEM media (Gibco-Invitrogen, Life Technologies), and used for invasion assay of MDA-MB231 cells at the second and third splitting passage.
Motility and directional cell migration assays
Subconfluent (70%) cultures of MCF-7, MDA-MB231 or HMEC cells were transfected with the various constructs for 24–48 h. Wounds in the cell monolayer were created using a P200 micropipette tip, after which cells were washed in PBS and incubated in complete medium for 24 h. For migration assay, three random images (50x) were taken at the time of the scratch and after 24 h. The migration distance (units) was determined as reduction in the wound’s gap using NIH Image-J software. For directional cell migration, subconfluent monolayers of MDA-MB231 cells were transfected with the various constructs, scratched as above after 66 h or 42 h, respectively, and analyzed for Golgi orientation after 6 h, by immunofluorescence. The position of the Golgi relative to the nucleus and to the migration front was scored using Image-J software in cells at the leading edge, as described (Dow et al., 2007; Osmani et al., 2006; Phua et al., 2009). Golgi staining was performed using Alexa Fluor555-conjugated antibody to GM130 (BD Bioscience, Milan, Italy). The direction of stress fibers was assessed in the same experiments. F-actin was imaged by phalloidin-TRITC staining (Sigma-Aldrich) using an AxioImager Z1 microscope (Carl Zeiss, Göttingen, Germany) were mounted in DAPI I (Abbott, Abbott Park, IL, USA).
Invasion assay
MDA-MB231 or HMEC cells (5×104) transfected with the various constructs were seeded in serum-free medium in Matrigel-coated chambers (8.0 μm pores, BD Biosciences). Cells were allowed to migrate across coated inserts using serum-containing medium or CAFs as chemoattractant for 24 h. The cells on the apical surface of the insert were scraped off, and membranes with invaded cells were fixed in 100% methanol, stained with Toluidine blue (Sigma-Aldrich) and mounted on microscope slides. Invading cells were counted after photographing the membranes, and both the peripheral and the central areas were evaluated.
Anoikis assay
MCF-7 or MDA-MB231 cells were seeded (2×105) in poly-HEMA (20 mg/ml, Sigma-Aldrich)-coated 6-wells plates, after transfection with the various miRNA-mimics for 48 h or with the indicated siRNA for 24h. Cells were harvested after additional 24 h, and viability was assessed by propidium iodide content and flow cytometry (FACS). When PKH26 staining was performed (Sigma-Aldrich), 2×107 A549 or MDA-MB231 cells were labeled the day before transfection following manufacturer protocol.
miR-296 modulated genes analysis
The mRNA level of candidate genes (n=107, listed in Supplementary Table 1) was investigated in MDA-MB231 cells transfected with miR-296 or control constructs using TaqMan Gene Expression assays (Applied Biosystems, Life Technologies). TATA-binding protein (TBP) was used as housekeeping gene for target genes relative quantification (RQs) using the 2^(−ΔCt) formula. RQs were median-normalized and log2 transformed to generate an unsupervised hierarchical clustering. A 1.5 fold cutoff was used to select genes modulated by transfection of anti-miR-296 or pre-miR-296 relative to control, which were further analyzed using Ingenuity Pathway software. Functional profiling using Gene Ontology terms for biological process, molecular function and cellular component were conducted for miR-296 predicted targets identified by at least 4 algorithms (n=145) using GOTM tool (http://bioinfo.vanderbilt.edu/gotm). As reference gene set we used the human genome, and the top 10 functional categories (most significant p-values obtained using the hypergeometric test adjusted for multiple comparisons) with a minimum number of 4 required genes were identified (Supplementary Table 2).
Luciferase miRNA target reporter assay
Mutation in miR-296 seeding sequence within Scrib 3′UTR were created using QuikChangeII XL site directed mutagenesis kit (Stratagene, Agilent Technologies, Santa Clara, CA, USA). Vectors containing full length wild-type or mutated 3′UTR sequences of Scrib gene, or control sequence and dual luciferase reporter genes were purchased from GeneCopoeia. HEK293 cells were co-transfected as described above with 1 μg plasmid containing target gene 3′UTR or control sequence in the presence of 50 nM pre-miR-296, anti-miR-296, control oligonucleotides or unrelated miRNA constructs (anti-let-7g and pre-miR-1). Cells were lysed after 30 h and firefly to Renilla luciferase ratios was measured with a dual luciferase assay (Promega, Milan, Italy) using a Synergy 2 luminometer (BioTek, Winooski, VT, USA). Normalized luciferase ratios were determined for all combinations tested of miRNAs and vectors.
Immunoblotting, Immunohistochemistry and Immunofluorescence
For immunoblotting, aliquots of MDA-MB231, MCF-7, or HMEC cells were harvested 48 or 72 h post-transfection, and solubilized in 150 μl of RIPA buffer supplemented with 1X complete protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Cell lysates (50 μg) were separated by electrophoresis on 6% or 12% SDS-polyacrylamide gels, transferred to PVDF membranes (Millipore, Billerica, MA, USA), probed with 1 μg/μl of antibodies against Scrib (Santa Cruz Biotechnology, Santa Cruz, CA, USA), β-Actin (Sigma-Aldrich), BMP8A (Novus Biologicals Littleton, CO, USA), HMGA1 (LSBio Seattle, WA, USA) or cytokeratin AEI-AE3 (Dako, Milan, Italy), and reactive bands were visualized with ECL Plus reagents (GE Healthcare, Milan, Italy). CAFs were visualized by immunofluorescence staining with an antibody to vimentin (Dako). Immunohistochemical staining of FFPE tissue samples collected from patients with metastatic carcinomas or HCC, was carried out using an antibody to Scrib (10μg/μl, Santa Cruz Biotechnologies) for 1 h at 22°C followed by counterstaining with hematoxylin. As negative control, slides were incubated with dilution buffer instead of primary antibody. Slides were scored by light microscopy and photographed images were arranged with Photoshop for Windows. Cytoplasmatic and membranous Scrib immunoreactivity was scored as negative (0), weak (1), moderate (2) or strong (3) by two authors (GC and SB) independently.
Statistical analysis and clinical validation of miR-296 and Scrib
All experiments were performed in duplicate and performed at least three times. Differences among samples were analyzed using Student’s t-test, unless otherwise specified. The correlation between k-ras mutation status and miR-296 in colorectal carcinoma patients was determined using the Fisher’s exact test. For survival analysis, the Kaplan–Meier method was used. To explore a potential association between miR expression and metastatic relapse, patients were assigned to two groups depending on miR-296 relative expression in tumor versus the averaged non-neoplastic counterparts. A miR-296 ratio of ≤0.5 was used for the “Low” group, whereas a miR-296 value of >0.5 was used for the “High” group. To estimate the association of Scrib mRNA or protein expression to tumor recurrence, HCCs patients were categorized in two groups according to Scrib expression value as for the miR. The two-sided log-rank test was used to compare survival curves. Statistical analyses were performed using GraphPad Prism version 4, or Ministat 2.1 software. A P<0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments
Sources of Support for the present Investigation: This work was supported by grants from the Fondazione Invernizzi (to G.C.), Fondazione Berlucchi (to S.B.), and from National Institutes of Health grants HL54131, CA140043, CA78810 and CA118005 (to D.C.A).
We thank Riccardo Ghidoni, Michele Samaja and Massimo Locati for availability of laboratory equipment and reagents, Patrizia Doi and Delfina Tosi for technical help. A.F. is supported by a fellowship of the Doctorate School of Molecular Medicine at Università degli Studi di Milano.
Footnotes
Conflict of interest statement
The authors declare no competing interest to disclose.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Assemat E, Bazellieres E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochim Biophys Acta. 2008;1778:614–30. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- Augello C, Caruso L, Maggioni M, Donadon M, Montorsi M, Santambrogio R, et al. Inhibitors of apoptosis proteins (IAPs) expression and their prognostic significance in hepatocellular carcinoma. BMC Cancer. 2009;9:125. doi: 10.1186/1471-2407-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahri S, Wang S, Conder R, Choy J, Vlachos S, Dong K, et al. The leading edge during dorsal closure as a model for epithelial plasticity: Pak is required for recruitment of the Scribble complex and septate junction formation. Development. 2010;137:2023–32. doi: 10.1242/dev.045088. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bauer M, Su G, Casper C, He R, Rehrauer W, Friedl A. Heterogeneity of gene expression in stromal fibroblasts of human breast carcinomas and normal breast. Oncogene. 2010;9:1732–40. doi: 10.1038/onc.2009.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–6. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Dow LE, Kauffman JS, Caddy J, Zarbalis K, Peterson AS, Jane SM, et al. The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene. 2007;26:2272–82. doi: 10.1038/sj.onc.1210016. [DOI] [PubMed] [Google Scholar]
- Dow LE, Elsum IA, King CL, Kinross KM, Richardson HE, Humbert PO. Loss of human Scribble cooperates with H-Ras to promote cell invasion through deregulation of MAPK signalling. Oncogene. 2008;27:5988–6001. doi: 10.1038/onc.2008.219. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Polarity proteins in migration and invasion. Oncogene. 2008;27:6970–80. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]
- Gardiol D, Zacchi A, Petrera F, Stanta G, Banks L. Human discs large and scrib are localized at the same regions in colon mucosa and changes in their expression patterns are correlated with loss of tissue architecture during malignant progression. Int J Cancer. 2006;119:1285–90. doi: 10.1002/ijc.21982. [DOI] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Dissecting multistep tumorigenesis in transgenic mice. Annu Rev Genet. 1988;22:479–519. doi: 10.1146/annurev.ge.22.120188.002403. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hong L, Han Y, Zhang H, Li M, Gong T, Sun L, et al. The prognostic and chemotherapeutic value of miR-296 in esophageal squamous cell carcinoma. Ann Surg. 2010;251:1056–63. doi: 10.1097/SLA.0b013e3181dd4ea9. [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Humbert PO, Grzeschik NA, Brumby AM, Galea R, Elsum I, Richardson HE. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27:6888–907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- Kamei Y, Kito K, Takeuchi T, Imai Y, Murase R, Ueda N, et al. Human scribble accumulates in colorectal neoplasia in association with an altered distribution of beta-catenin. Hum Pathol. 2007;38:1273–81. doi: 10.1016/j.humpath.2007.01.026. [DOI] [PubMed] [Google Scholar]
- Kim SW, Kim JW, Kim YT, Kim JH, Kim S, Yoon BS, et al. Analysis of chromosomal changes in serous ovarian carcinoma using high-resolution array comparative genomic hybridization: Potential predictive markers of chemoresistant disease. Genes Chromosomes Cancer. 2007;46:1–9. doi: 10.1002/gcc.20384. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Nagasaka K, Pim D, Massimi P, Thomas M, Tomaić V, Subbaiah VK, et al. The cell polarity regulator hScrib controls ERK activation through a KIM site-dependent interaction. Oncogene. 2010;23:5311–21. doi: 10.1038/onc.2010.265. [DOI] [PubMed] [Google Scholar]
- Naylor TL, Greshock J, Wang Y, Colligon T, Yu QC, Clemmer V, et al. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005;7:R1186–98. doi: 10.1186/bcr1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–65. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani N, Vitale N, Borg JP, Etienne-Manneville S. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr Biol. 2006;16:2395–405. doi: 10.1016/j.cub.2006.10.026. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Zhan W, Dan L. hScrib, a human homolog of Drosophila neoplastic tumor suppressor, is involved in the progress of endometrial cancer. Oncol Res. 2010;18:593–9. doi: 10.3727/096504010x12767359114045. [DOI] [PubMed] [Google Scholar]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–22. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–49. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua DC, Humbert PO, Hunziker W. Vimentin regulates scribble activity by protecting it from proteasomal degradation. Mol Biol Cell. 2009;20:2841–55. doi: 10.1091/mbc.E08-02-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnoli S, Fasoli E, Vaira V, Falleni M, Pellegrini C, Catania A, et al. Identification of potential therapeutic targets in malignant mesothelioma using cell-cycle gene expression analysis. Am J Pathol. 2009;174:762–70. doi: 10.2353/ajpath.2009.080721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–46. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wei JJ, Wu X, Peng Y, Shi G, Olca B, Yang X, et al. Regulation of HMGA1 expression by microRNA296 affects prostate cancer growth and invasion. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-20-0229. [DOI] [PubMed] [Google Scholar]
- Woo HG, Park ES, Lee JS, Lee YH, Ishikawa T, Kim YJ, et al. Identification of potential driver genes in human liver carcinoma by genomewide screening. Cancer Res. 2009;69:4059–66. doi: 10.1158/0008-5472.CAN-09-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463:545–8. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–93. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Gu J, Wang KK, Zhang W, Xing J, Chen Z, et al. MicroRNA expression signatures in Barrett’s esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:5744–52. doi: 10.1158/1078-0432.CCR-09-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–78. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li Y, Lai M. The microRNA network and tumor metastasis. Oncogene. 2010;29:937–48. doi: 10.1038/onc.2009.406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.