Abstract
Snail1 is a central regulator of epithelial cell adhesion and movement in epithelial-to-mesenchymal transitions (EMTs) during embryo development; a process reactivated during cancer metastasis. While induction of Snail1 transcription precedes EMT induction, post-translational regulation of Snail1 is also critical for determining Snail1's protein level, subcellular localization, and capacity to induce EMT. To identify novel post-translational regulators of Snail1, we developed a live cell, bioluminescence-based screen. From a human kinome RNAi screen, we have identified Lats2 kinase as a novel regulator of Snail1 protein level, subcellular localization, and thus, activity. We show that Lats2 interacts with Snail1 and directly phosphorylates Snail1 at residue T203. This occurs in the nucleus and serves to retain Snail1 in the nucleus thereby enhancing its stability. Lats2 was found to positively influence cellular EMT and tumour cell invasion, in a Snail1-dependent manner. Indeed during TGFβ-induced EMT Lats2 is activated and Snail1 phosphorylated at T203. Analysis in mouse and zebrafish embryo development confirms that Lats2 acts as a positive modulator of Snail1 protein level and potentiates its in vivo EMT activity.
Keywords: breast cancer, Lats2, Snail1
Introduction
Epithelial–mesenchymal transition (EMT) is critical for many developmental events such as gastrulation, neural crest migration, cardiac valve formation, and oral palate fusion (Thiery et al, 2009). Many parallels between developmental EMT and cancer metastasis have been observed, leading to the current view that EMT could directly contribute to cancer metastasis by affecting localized adenoma tumour cell de-adhesion and subsequent invasion through basement membrane and migration through the extracellular matrix (ECM) to intravasate into the blood stream or lymphatics (Kalluri and Weinberg, 2009). The EMT process is thought to influence the arrest in cell proliferation that occurs as cells differentiate (Barrallo-Gimeno and Nieto, 2005) and enhance cell survival (Vega et al, 2004). EMT is also thought to contribute to the resistance of tumour cells to therapy and the production of cancer stem cells (CSCs; Kalluri and Weinberg, 2009; Thiery et al, 2009).
EMT describes a dynamic and reversible morphogenetic program. Expression of epithelial genes, such as E-cadherin, is directly repressed while mesenchymal genes, such as Vimentin, are indirectly induced. Morphologically, this results in loss of epithelial phenotype and acquisition of spindle-shaped mesenchymal cell phenotype and function. Although multiple environmental stimuli (e.g., TGFβ, FGF, hypoxia, and Notch1) can induce EMT their disparate intracellular signalling pathways tend to ultimately converge upon a limited group of transcriptional repressors that are considered to be the inducers of EMT. Central among these, in both development and cancer metastasis, is the Zinc-finger protein Snail1.
Normal epithelia do not express Snail1; however, its transcription is induced in response to environmental EMT signals. Snail1 mRNA and protein levels can be regulated in discoordinate manner and although Snail1 protein is found in the cytosol and nucleus it needs to be nuclear to induce EMT. These observation lead to the identification of post-transcriptional regulation as being a critical factor influencing total cellular Snail1 protein level or stability, Snail1 subcellular distribution, and Snail1 function (Dominguez et al, 2003; Zhou et al, 2004; Peinado et al, 2005; Yook et al, 2006; MacPherson et al, 2010). Snail1 shuttles between the nucleus and cytoplasm in a regulated manner that is not completely understood (Zhou et al, 2004; Yook et al, 2006). Total cellular Snail1 protein is a highly unstable protein. In the nucleus, its turnover is decreased while in the cytosol it is rapidly degraded by proteasomes (Zhou et al, 2004). Post-translational phosphorylation, ubiquitination, and lysine oxidation all influence Snail1 protein stability, subcellular localization, and activity. Snail1 phosphorylation by GSK3β, for example, affects its nuclear to cytosol transport and cytosolic degradation (Zhou et al, 2004; Yook et al, 2006). Although GSK3β likely phosphorylates Snail1 in the nucleus leading to its nuclear export signals mediating Snail1 nuclear import and, or retention are not known.
In various cells, we noted a differential response in Snail1 protein level to inhibition of GSK3β versus inhibition of proteasome action. Moreover, in some cells inhibition of GSK3β had little effect upon Snail1 protein levels even though proteasome inhibition did. These results suggested that other cellular proteins or pathways could influence Snail1 protein level possibly post-translationally. If one was able to identify these cellular proteins or signalling pathways and discern how they affect Snail1 protein level and function this could identify new cellular targets more amenable to the development of drugs that could attenuate Snail1-induced EMT, and thus, potentially inhibit cancer metastasis. Towards this end, we developed a bioluminescence cell-based assay that reflects Snail1 protein stability. This provided us with a method to screen for positive and negative post-translational regulators of total cellular Snail1 protein level. We performed a human kinome RNAi screen. This screen identified the protein kinase Lats2, a putative tumour suppressor, as stabilizing cellular Snail1 protein levels post-translationally. Lats2 does so by interacting with Snail1 in cells and directly phosphorylating Snail1 at T203 in response to multiple signals that activate Lats2, including TGFβ-induced EMT. Phosphorylation of Snail1 at T203, by Lats2, occurs in the nucleus and serves to retain Snail1 in the nucleus thereby enhancing protein stability and function. In cells, we found that Lats2 can affect TGFβ-induced EMT and that this effect depends upon the presence of Snail1. Lats2 potentiates Snail1 EMT-inducing function in zebrafish and mouse embryo development, as well as Snail1's capacity to regulate tumour cell invasion/migration. In aggressive breast tumour cells, Lats2 levels were found to be increased and influence invasion and or migration of these tumour cells in 3D collagen gels. These findings indicate that Lats2 potentiates Snail1 function and suggest that this kinase, previously described as a tumour suppressor, may also exhibit tumour-promoting activity.
Results
Development of a cell-based screen to identify post-translational modifiers of Snail1 protein stability
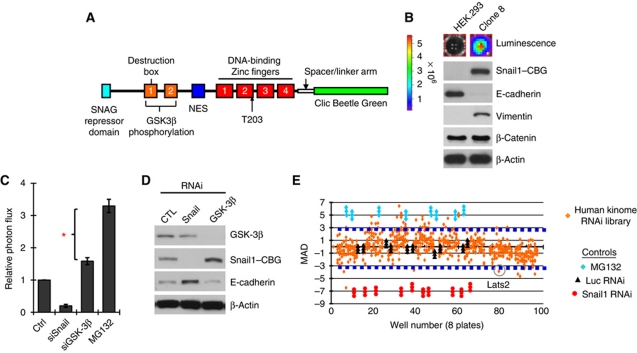
To identify novel post-translational modifiers of Snail1 protein level, we developed a cell-based bioluminescent assay amenable to high-throughput screening. First, we created a plasmid expressing human Snail1 fused to the bioluminescent peptide Clic Beetle Green (Snail1–CBG) (Figure 1A). To minimize potential CBG interference with DNA binding by the C-terminal Zinc fingers of human Snail1 a flexible poly-Alanine spacer arm separated the C-terminus of Snail1 and CBG. CBG was placed at the C-terminus of Snail1 so as to not interfere with its N-terminal SNAG repressor domain; a domain required for Snail1 activity (Ayyanathan et al, 2007; Langer et al, 2008). A CMV promoter controlled transcription of this Snail1–CBG transgene.
Figure 1.
A screen for post-translational modifiers of Snail1 protein stability. (A) Stick figure representation of human Snail1-Clic Beetle Green (CBG) bioluminescent plasmid. NES: nuclear export signal region. (B) Bioluminescence and western blot of Snail1–CBG expressing HEK293 (clone #8) or untransfected control HEK293 cells, for the indicated proteins (right). (C) Bioluminescence of HEK293.Sn-CBG clone 8 transfected with Snail1 RNAi, GSK3β RNAi, or treated with the proteosome inhibitor MG132. Results are presented as bioluminescence relative to control untreated cells (set at an arbitrary value of 1.0). *Identifies the difference in Snail1 stabilizing effect between inhibition of GSK3β and inhibition of proteasome function. (D) Western blot analysis for the EMT marker E-cadherin (i.e., a Snail1 target gene) in HEK293.Sn-CBG clone 8 cells following transfection with control Luc RNAi, Snail1 RNAi, or GSK3β RNAi. (E) A human kinome RNAi screen (Qiagen) for proteins that stabilize or destabilize Snail1 protein level, as described in Materials and methods. Individual RNAi values are presented as Median Average Deviation (MAD) bioluminescence from the median of the complete library and Luc control RNAi. Control for maximum stabilization is MG1323 treatment (blue diamonds); for maximum destabilization is Snail RNAi (red circles); and for RNAi control is Luciferase RNAi (black triangles). The RNAi library results are in triplicate for each RNAi (orange diamonds). There were two RNAi's per kinase in the library. The blue broken lines identify ±3 MAD.
Multiple HEK293 cells clones stably expressing Snail1–CBG were generated and characterized for functionality of the Snail1–CBG fusion protein. One, clone 8, was selected for subsequent analyses. Parental HEK293 cells do not express detectable Snail1 protein. In clone 8, an appropriately sized Snail1–CBG fusion protein was present, exhibited robust bioluminescence, and did not affect expression of endogenous Snail1 protein (Figure 1B; Supplementary Figure S1A). Snail1–CBG, like wt Snail1, induced cellular EMT as evidenced by a morphological change from epithelial-to-mesenchymal phenotype (Supplementary Figure S1B), suppression of epithelial E-cadherin expression, and induction of mesenchymal Vimentin and N-cadherin expression (Figure 1B; and data not shown). Immunofluorescence analysis demonstrated that Snail1–CBG was largely confined to the nucleus (Supplementary Figure S1C).
EMT is often reversible and RNAi-mediated depletion of exogenous Snail1–CBG in clone 8 resulted in decreased Snail1–CBG bioluminescence, decreased Snail1–CBG protein level, re-expression of E-cadherin, and a return to epithelial cell morphology (Figure 1C and D, and data not shown). To ensure that Snail1–CBG protein was regulated in a manner similar to endogenous Snail1, clone 8 cells were treated with the proteosome inhibitor MG132 or GSK3β was inhibited by RNAi depletion and/or LiCl treatment. In all situations, Snail1–CBG bioluminescence and protein level increased (Figure 1C and D, and not shown). Importantly, inhibition of GSK3β alone was not equivalent to proteosome inhibition (Figure 1C), suggesting that cellular proteins or signalling pathways other than GSK3β influence cellular Snail1 protein level.
A human kinome RNAi screen identified Lats2 as stabilizing Snail1 protein level post-translationally
Since post-translational phosphorylation of Snail1 has been shown to be important for controlling its cellular protein level, subcellular localization, and function (Dominguez et al, 2003; Zhou et al, 2004), we performed an RNAi screen with the Qiagen RNAi human kinome library to identify protein kinases that influence Snail1 protein level, both positively or negatively. To define the range of effects possible in our screen, proteosome inhibition (MG132) was used to define the maximal stabilizing effect on Snail1–CBG levels and Snail1 RNAi was used to define the maximal destabilizing effect (Figure 1E). Non-specific Luciferase RNAi was used as a control for RNAi transfection. Each 96-well library plate contained 15 control wells (4 proteosome inhibition, 4 Snail1 RNAi, 4 Luc RNAi, and 3 untransfected). For a plate to be considered valid for analysis controls must fall within their prescribed range, as set by a master control plate. Change in cell numbers, due to targeting of cell-cycle or apoptosis regulators, was controlled for by measuring resazurin dye uptake immediately after bioluminescent determination and normalizing bioluminescent values to viable cell number. Final bioluminescence photon values are presented as the median of absolute deviation (MAD) relative to non-specific Luc RNAi controls and median value of all RNAi in the library (Zhang et al, 2006, 2008b; Figure 1E). Samples more than +3 from the median (i.e., Snail1 stabilized) or less than −3 from the median (i.e., Snail1 destabilized) were selected for further study. Of 694 protein kinases screened 34 fell into these two groups. Two of these, PAK1 and p70 S6 kinase, have already been shown to influence Snail1 protein stability (Yang et al, 2005; Pon et al, 2008).
Confirmatory secondary studies included: (1) absence of any effect of candidate RNAi upon CMV promoter using HEK293 cells expressing CMV-CBG and (2) demonstration that new siRNA oligonucleotides (different from those present in the library) or lentiviruses expressing shRNAi recapitulated the effect observed by library candidate siRNAi. One of the confirmed positives was Lats2.
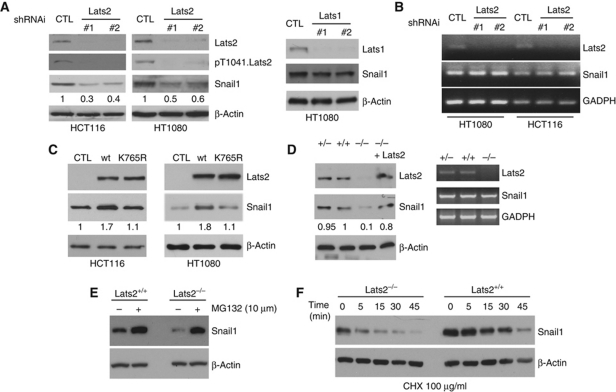
RNAi-mediated depletion of endogenous Lats2 in clone 8 (overexpressing exogenous Snail1–CBG), using non-Qiagen library RNAi oligos resulted in decreased Snail1–CBG protein level and bioluminescence without affecting Snail1 mRNA levels (Supplementary Figure S1D–F). Depletion of related Lats1 kinase did not affect the level of Snail1 protein (Figure 3D). Lentiviruses expressing Lats2 shRNAi also reduced Snail1–CBG protein level in these cells (Supplementary Figure S1G). Although normal epithelial cells do not express Snail1 invasive carcinoma cells such as the colon cancer cell line HCT116 do (Figure 2A). Moreover, since mesenchymal cells express Snail1 and its regulation and function in these cells may be distinct from cells undergoing EMT we also determined the effect of Lats2 depletion upon Snail1 protein level in the mesenchymal fibrosarcoma cell line HT1080. In both HCT116 and HT1080 cells, low level of active Lats2 was present in cells in their basal state (i.e., proliferating in serum-containing cultures), as indicated by the presence of pT1041.Lats2 (Figures 2A and 4A; Ikeda et al, 2009). RNAi-mediated depletion of Lats2, but not related Lats1, in both cell types resulted in decreased Snail1 protein level (Figure 2A) without affecting the level of Snail1 mRNA (Figure 2B). When Lats2 was overexpressed in these same cells Snail1 protein level was increased and this effect required active enzyme as overexpression of a kinase-inactive mutant of Lats2 (K765R) did not alter Snail1 protein level (Figure 2C; Supplementary Figure S1H).
Figure 2.
Presence of Lats2 protein stabilizes Snail1 protein level without affecting Snail1 transcription. Western blots (A) and RT–PCR analysis (B) for indicated proteins or mRNA in Lats2-depleted colon cancer HCT116 or mesenchymal HT1080 cells or Lats1-depleted HT1080 cells using two different shRNAi-containing lentiviruses against each or control luciferase shRNAi (shCTL). (C) Western blots for the indicated proteins in HCT116 and HT1080 cells transfected with wt or kinase-inactive (K765R) Lats2. (D) Western blot (left panel) or RT–PCR (right panel) for indicated protein or mRNA in Lats2+/+, +/−, and −/− MEFs and Lats2−/− MEFs rescued by Lats2 re-expression. (E) Western blot comparing Snail1 protein level in indicated MEFs treated with the proteasome inhibitor MG132. (F) Snail1 protein stability in Lats2 wt (+/+) and null (−/−) MEFs. Protein translation was inhibited by pretreatment with cycloheximide (CHX). Snail1 protein level relative to control cells (arbitrarily set as 1.0) (A, C) or wt MEFs (D) is listed.
In Lats2−/− MEFs, basal Snail1 protein level was dramatically decreased without any change in Snail1 mRNA level (Figure 2D). Importantly, re-expression of Lats2 in Lats2−/− MEFs, or inhibition of proteasome function in Lats2−/− cells, restored cellular Snail1 protein level to that seen in wt MEFs (Figure 2D and E). The protein half-life of Snail1 was also decreased in Lats2−/− MEFs (Figure 2F). In sum these confirmatory studies, including cells expressing endogenous Snail1, demonstrated that the presence of Lats2 kinase influenced the total cellular level of Snail1 protein and that this occurred at the level of post-translational regulation.
Lats2 interacts with and phosphorylates Snail1 at T203 to increase cellular levels of Snail1 protein
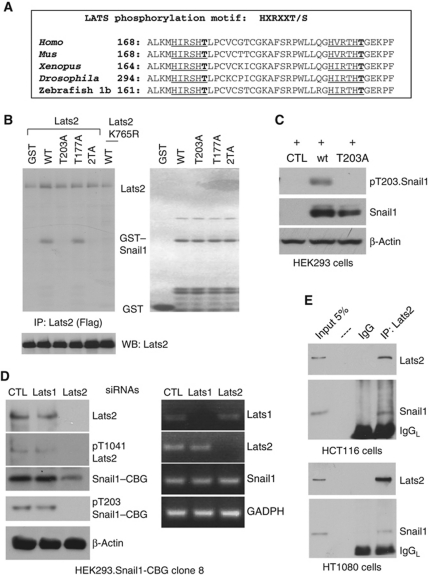
Lats2 kinase could affect Snail1 protein level either directly (phosphorylation) or indirectly by influencing components of some upstream signalling pathway that itself affects Snail1 stability. To determine if Lats2 might directly phosphorylate Snail1 protein sequence of Snail1 from multiple organisms was analysed for the presence of consensus Lats2 phosphorylation sites (Zhao et al, 2007). This identified two highly conserved, potential Lats2 phosphorylation sites at T177 and T203 (human Snail1) (Figure 3A). To determine if these could be phosphorylated by Lats2, we immunoprecipitated Flag–Lats2, or kinase-inactive Flag–Lats2 (K765R), from transfected HEK293 cells and performed in vitro kinase assays using purified GST–Snail1 or GST–Snail1 phosphorylation site mutants (T to A) as exogenous substrate. In vitro, Lats2 was found to specifically phosphorylate human Snail1 at T203 but not at T177 (Figure 3B).
Figure 3.
Lats2 phosphorylates Snail1 at T203 in vitro and in cells. (A) Peptide alignment of Snail1 from various organisms. Putative Lats2 phosphorylation sites are underlined. (B) Flag–Lats2 (lanes 1–5) or kinase-inactive Lats2 (K765R) (lane 6) was immunoprecipitated from transfected HEK293 cells, washed, and an in vitro kinase assay performed using purified GST (lane 1), GST-Snail1 (lane 2), GST-T203A.Snail1 (lane 3), GST-T177A.Snail1 (lane 4), GST-T177A; T203A.Snail1 (2TA, lane 5) and GST-Snail1 (lane 6) as exogenous substrate. Left panel: autoradiograph; right panel: Coomassie-stained gel. The left lower panel is a western blot for amount of immunoprecipitated Lats2. (C) A human anti-Snail1 pT203-containing peptide antibody (pT203.Snail1) specifically recognizes T203 phosphorylated Snail. Western blot for indicated proteins in HEK293 cells transfected with empty plasmid (CTL), wt Snail1, or T203A.Snail1. (D) HEK293 cells containing Snail1–CBG (clone #8) were infected with control (CTL: Luciferase siRNA oligos), Lats1, or Lats2 siRNA oligos. Extracts from a confluent plate of cells were immunoblotted (left panel) or RT–PCR performed (right panel) for indicated proteins or mRNA, respectively. (E) Lats2 associates with Snail1 in cells. Endogenous Lats2 was immunoprecipitated from HCT116 cells (upper panel) or HT1080 cells (lower panel) and bound products immunoblotted for Lats2 and Snail1. Input controls (5% of extract used for IP) are in the first lane.
To determine whether T203 of Snail1 was phosphorylated in cells two approaches were used. First, Flag-tagged Snail1 was immunoprecipitated from cells that had been treated with nocodazole, a manipulation (mitotic injury) that has been shown to activate Lats2 kinase (Aylon et al, 2006), and analysed by nano-LC-MS. This identified a prominent phosphopeptide 201THPTGEKPFSCPHCSR215 (Supplementary Figure S2A and B). Next, we generated a phospho-specific antibody to a pT203-containing human Snail1 peptide. This antibody detected a band migrating at the molecular size of Snail1 in extracts from HEK293 cells transfected with wt Snail1 but not in cells transfected with T203A.Snail1 (Figure 3C). In clone 8 cells (HEK293 cells containing Snail1–CBG used in the screen), pT203.Snail1–CBG was detected while RNAi depletion of Lats2, but not Lats1, abrogated detection of pT203.Snail1–CBG without affecting Snail1–CBG mRNA levels (Figure 3D). In basal clone 8 cells, low level of active Lats2 was present, as detected by a pT1041.Lats2 antibody (Figure 3D).
Lats2 and Snail1 were also found to associate (co-immunoprecipitate) in carcinoma HCT116 and mesenchymal fibrosarcoma HT1080 cells, which endogenously express each protein (Figure 3E). This association depended upon Lats2 kinase activity as Lats2 (K765R) did not associate with Snail1 (Supplementary Figure S3A); however, their association did not require phosphorylation of T203 as T203A.Snail1 still associated with Lats2, as well as wt Snail1 did (Supplementary Figure S3A). In interaction mapping experiments, it was found that an N-terminal region, aa 10–40, of Snail1 directed its association with Lats2 (Supplementary Figure S3B). This region is well removed from the DNA-binding C-terminal Zinc fingers region.
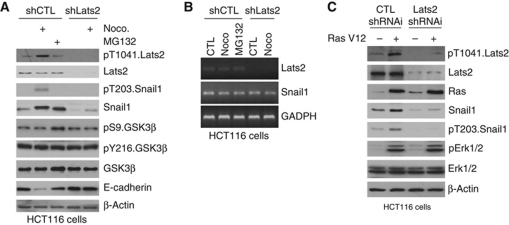
Lats2 kinase translocates from centrosomes to the nucleus in response to mitotic or oncogenic stress and results in increased level of Lats2 protein (Aylon et al, 2006, 2009); however, whether these same signals also activate Lats2 kinase activity is not known. Activation of the Hippo pathway, a critical pathway regulating organ size, does lead to activation of Lats2 kinase, however (Chan et al, 2005). When mitotic stress was induced in HCT116 cells, by treating with nocodazole to inhibit centrosome and mitotic apparatus function, low basal Lats2 activity was increased as determined by western blotting with a pT1041.Lats2 antibody that identifies active Lats2 (Ikeda et al, 2009; Figure 4A). Snail1 was phosphorylated at T203, as determined by western blotting with a pT203.Snail1 antibody and mobility shift of Snail1 protein on SDS–PAGE (Figure 4A; Supplementary Figure S2C), and total cellular level of Snail1 protein increased (Figure 4A; Supplementary Figure S2C) in response to nocodazole treatment. There was no change in the level of Snail1 mRNA in response to nocodazole treatment (Figure 4B). These effects upon Snail1 were not observed when Lats2 was shRNAi depleted (Figure 4A). Nocodazole-induced pT203.Snail1 and the decrease in mobility of Snail1 on SDS–PAGE was lost following phosphatase treatment of cell extracts (Supplementary Figure S2C). As a further control for Lats2 specificity of the nocodazole treatment effects upon Snail1 GSK3β activity or level, a known regulator of Snail1 stability (Zhou et al, 2004) did not to change in response nocodazole treatment (Figure 4A). To determine if Lats2 phosphorylation of Snail1, in response to mitotic stress, was associated with alterations in Snail1 function we asked whether E-cadherin level (a transcriptional target of Snail1) changed in nocodazole-treated cells. E-cadherin level decreased and this required the presence of Lats2 (Figure 4A). Nocodazole treatment of wt MEFs also resulted in Snail1 phosphorylation and increased total cellular protein level (Supplementary Figure S3A). This required the presence of Lats2 as in Lats2−/− MEFs nocodazole treatment did not stabilize Snail1 protein level (Supplementary Figure S3A). Finally, T203A.Snail1 protein half-life was found to be four-fold less than wt Snail1 in transfected HEK293 cells (Supplementary Figure S2D). In sum, these data indicated that mitotic stress activated Lats2 kinase and this resulted in the phosphorylation of Snail1, at T203, and the post-translational stabilization of total cellular Snail1 protein level.
Figure 4.
Activation of Lats2, by mitotic stress or oncogenic stress, phosphorylates Snail1 at T203 and stabilizes total cellular Snail1 protein level. (A, B) HCT116 cells were transduced with control (CTL) or Lats2 shRNA lentiviruses and then untreated, treated with nocodazole (noco) or the proteasome inhibitor MG132. Western blot with the indicated antibodies was performed (A) or RT–PCR for the indicated mRNA was performed (B). (C) HCT116 cells were infected with H-RasV12 expressing lentivirus (+) or control empty lentivirus (−) and western blot performed with the indicated antibodies.
To determine whether oncogenic stress signal affected Snail1, in a Lats2-dependent manner, we overexpressed RasV12 in HCT116 cells. Even though HCT116 cells contain oncogenic Ras mutations (Dunn et al, 2011) when H-RasV12 was overexpressed Lats2 was further activated, Snail1 phosphorylated at T203, and total cellular Snail1 protein level increased (Figure 4C). This action of overexpressed RasV12 required Lats2 as in HCT116 cells depleted of Lats2 Snail1 was neither phosphorylated nor stabilized (Figure 4C). Similar results were observed when RasV12 was overexpressed in wt and Lats2−/− MEFs (Supplementary Figure S3B).
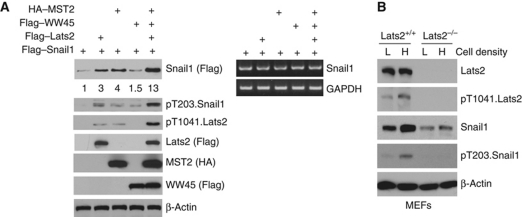
To determine if Hippo pathway activation of Lats2 kinase affected Snail1 T203 phosphorylation and stabilization we used two approaches. First, we transfected HEK293 cells with the mammalian Hippo kinase Mst2, the upstream kinase in the Hippo pathway responsible for Lats2 activation (Chan et al, 2005), and its adapter protein WW45 in the presence or absence of exogenous Lats2 and determined whether Snail1 protein level changed. Transfection of only Mst2 and WW45 resulted in endogenous Lats2 activation, phosphorylation of Snail1 at T203 and increase of total cellular Snail1 protein (Figure 5A). When Lats2 was co-transfected with Mst2 and WW45 the level of activated Lats2, pT203.Snail1, and total cellular Snail1 protein was dramatically increased without any change in Snail1 mRNA level (Figure 5A).
Figure 5.
Activation of Lats2 by the Hippo pathway phosphorylates Snail1 on T203 and stabilizes total cellular Snail1 protein. (A) HEK293 cells were transfected with epitope-tagged plasmids expressing the indicated cDNAs (HA–Mst2, Flag–WW45, Flag–Lats2, and Flag–Snail1). Western blot (left panel) or RT–PCR (right panel) was performed with the indicated antibodies or for the indicated mRNA, respectively. (B) WT (+/+) or Lats2 null (−/−) MEFs were grown to a low density (L: <50% confluent) or high density (H: confluent) and western blot with the indicated antibodies performed.
A physiological activator of the endogenous Hippo pathway is cell–cell contact growth inhibition (Zhao et al, 2007). Since invasive carcinoma cell lines (e.g., HCT116 cells) are not contact growth inhibited, we asked whether contact growth inhibition of non-transformed MEFs affected Snail1 protein level and whether this was Lats2 dependent. MEFs were used in this experiment, as untransformed normal epithelial cells, do not express Snail1. When wt MEFs were grown at high-density (growth arrested) Lats2 was activated, Snail1 phosphorylated at T203, and total cellular Snail1 protein level increased (Figure 5B). However, in proliferating low-density wt MEFs these effects were not observed (Figure 5B). The effect of MEF cell density upon total cellular Snail1 protein level was dependent upon the presence of Lats2 as in Lats2−/− MEFs grown to confluence Snail1 protein level did not change nor was Snail1 phosphorylated at T203 (Figure 5B).
In sum, in response to three different signals that activate Lats2 kinase (nocodazole-induced mitotic stress, RasV12-induced oncogenic stress, and the Hippo pathway), Snail1 was phosphorylated at T203 and total cellular Snail1 protein level stabilized post-transcriptionally.
Lats2 phosphorylates Snail1 in the nucleus and serves to retain Snail1 in the nucleus
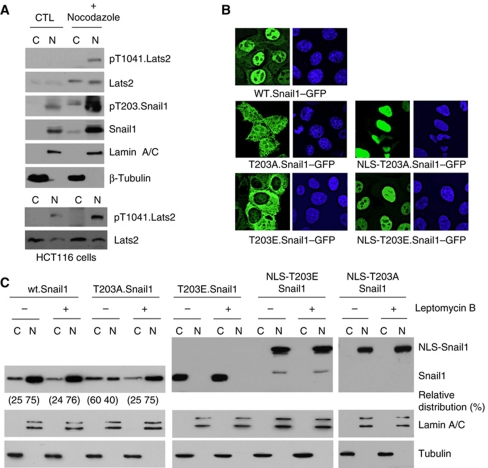
Lats2 localizes to centrosomes and cytosol yet injury to the mitotic apparatus or oncogenic stress results in Lats2 translocation to the nucleus (Aylon et al, 2006, 2009). Furthermore, when Snail1 is in the nucleus it's turnover is decreased (i.e., stabilized) whereas it is rapidly degraded when cytosolic (Zhou et al, 2004; Yook et al, 2006). Therefore, we asked whether phosphorylation of Snail1 by Lats2 might occur in the nucleus and prevent its nuclear-cytosolic transport thereby leading to its stabilization. To test this, HCT116 cells were subjected to nocodazole treatment and fractionated into cytosolic and nuclear fractions. Active Lats2 was only detected in the nucleus, despite total Lats2 protein being equally distributed between cytosol and nucleus (Figure 6A). Correspondingly, increased pT203.Snail1 and Snail1 protein were also predominantly nuclear (Figure 6A). A low level of active Lats2 and pT203.Snail1 was also present in untreated cells and these were also exclusively nuclear (Figure 6A).
Figure 6.
Phosphorylation of Snail1 at T203 occurs in the nucleus and affects Snail1 nuclear retention. (A) HCT116 cells were untreated (CTL) or treated with nocodazole and cytosolic (C) and nuclear (N) fractions prepared. Western blot with the indicated antibodies was performed on each fraction. Lamin A/C served as a nuclear fraction control and β-tubulin served as a cytosolic control. The lower two panels are a repeat experiment of the upper two lanes but overexposed. (B) Immunofluorescent analyses of MCF10A cells transfected with the indicated Snail1 mutant protein fused to GFP. DAPI staining was used to identify nuclei. NLS: nuclear export signal. (C) Same cells as in (B) were treated (+) or not (−) with Leptomycin B to inhibit nuclear export. Cells were fractionated into nuclear (N) and cytosolic (C) fractions and each fraction western blotted for the indicated proteins. The relative distribution of Snail1 between the nucleus and cytosol is shown (%).
To determine whether the presence of T203 in Snail1 was critical for its nuclear retention, we expressed wt Snail1, the T203A mutant, or the phospho-mimetic mutant T203E in the breast epithelial cells MCF10A and determined and contrasted the subcellular distribution of the various Snail1 mutants by immunofluorescence analysis and western blotting of nuclear and cytosolic fractions. MCF10A cells were chosen, as they do not express endogenous Snail1. WT Snail1 was predominantly nuclear whereas T203A.Snail1 was equally distributed between the nucleus and cytoplasm and T203E.Snail1 was exclusively cytosolic (Figure 6B and C). This suggested that phosphorylation of Snail1 at T203 could affect nuclear entry, nuclear retention, or both. To distinguish between these possibilities, cells were treated with Leptomycin B (LMB), an inhibitor of Crm-mediated nuclear export prior to analysis. Treatment of cells with LMB resulted in nuclear accumulation of T203A.Snail1 while T203E-Snail1 was unresponsive to LMB treatment and remained cytosolic (Figure 6C).
These results indicated that cytosolic phosphorylation of Snail1 at T203 was not required for its nuclear entry, possibly T203 phosphorylation of Snail1 might even inhibit its nuclear entry. Rather T203 phosphorylation likely occurred in the nucleus and contributed to Snail1 retention therein. Consistent with this possibility the capacity of T203E.Snail1 to associate with nuclear pore importins, which are required for Snail1 nuclear entry (Mingot et al, 2009), was found to be severely attenuated compared with wt Snail1 or T203A-Snail1 (Supplementary Figure S4C).
Lats2 enhances EMT in a Snail1-dependent manner
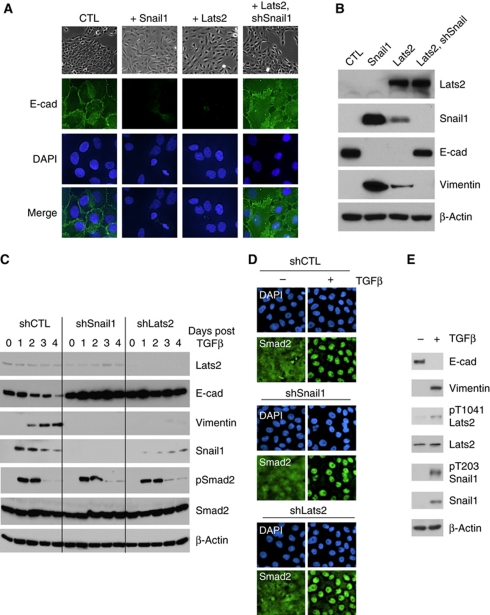
One of the central functions of Snail1 during development and pathologic processes, such as cancer metastasis, is to induce EMTs (Thiery et al, 2009). The breast epithelial cells MCF10A cells, in particular, have been extensively utilized for studies of cellular EMT. MCF10A cells undergo EMT, in response to TGFβ1, by activating expression of Snail1. MCF10A cells also undergo TGFβ-independent EMT when EMT inducers, such as Snail1, are overexpressed (Maeda et al, 2005; Overholtzer et al, 2006). To determine whether Snail1 phosphorylation by Lats2 influenced Snail1's capacity to induce EMT, we first asked whether Lats2 affected EMT and if so whether this was Snail1 dependent. As expected, overexpression of Snail1 in confluent MCF10A cells resulted in EMT, as evidenced by change in cellular morphology, loss of epithelial E-cadherin expression, and expression of mesenchymal Vimentin (Figure 7A and B). Overexpression of Lats2 in these cells also resulted in EMT and this was dependent upon the presence of Snail1, as when Snail1 was shRNAi depleted in Lats2-overexpressing MCF10A cells EMT did not occur (Figure 7A and B).
Figure 7.
Lats2 influences Snail1-dependent cellular EMT. (A, B) MCF10A cells were transfected with an empty control vector (CTL), Snail1, Lats2, or Lats2 and an shRNAi against Snail1. Cells were grown to confluence and analysed for EMT changes. (A) Upper set of panels is phase images of cell morphology and lower panels are immunofluorescent analysis for E-cadherin and DAPI staining. (B) Western blot for the indicated markers of EMT: epithelial E-cadherin and mesenchymal Vimentin. (C) MCF10A cells were transduced with control (CTL: luciferase shRNAi), Snail1 or Lats2 shRNA lentiviruses. Following selection in puromycin, cells were grown to confluence and then treated with TGFβ1 (2 ng/ml) for 8 h, washed, fresh media without TGFβ added and cells cultured for 4 days. Western blot of extracts from cells at each of 4 days was performed using the indicated antibodies. (D) Immunofluorescence analysis of Smad2 subcellular distribution on the same set of cells as in (C) at day 0 (−) and day 4 after 8 h treatment with TGFβ (+). DAPI staining was used to identify nuclei. (E) Western blot analysis, with the indicated antibodies, of untreated confluent parental MCF10A cells (−) or parental MCF10A cells 4 days after treatment with TGFβ (+) to induce EMT.
When Lats2 was shRNAi depleted in MCF10A cells EMT, in response to TGFβ, was significantly inhibited but not completely blocked (Figure 7C). Control shRNAi-treated MCF10A cells readily underwent EMT in response to TGFβ, while Snail1-depleted cells did not (Figure 7C). This effect of Lats2 depletion was not due to a loss of TGFβ responsiveness, as Smad2 was phosphorylated and accumulated in the nucleus of Snail1- and Lats2-depleted MCF10A cells exposed to TGFβ (Figure 7D). Furthermore, TGFβ-induced EMT of parental MCF10A resulted in activation of Lats2 and phosphorylation of Snail1 at T203 (Figure 7E).
While overexpression of wt Snail1 induced EMT in MCF10A cells (Figure 7A; Supplementary Figure S5A and B), overexpression of T203A.Snail1 or T203E.Snail1 cells did not (Supplementary Figure S5A and B). This could be explained by the inability of T203E.Snail1 or T203A.Snail to enter or accumulate, respectively, in the nucleus (see Figure 6) and not because they were inactive as EMT inducers. To test this possibility, we forced their nuclear entry and accumulation in MCF10A cells by fusing a strong nuclear localization signal to their N-terminus (NLS-T203E/A.Snail1). Control immunofluorescent and subcellular fractionation analyses revealed that NLS-T203E.Snail1 and NLS-T203A.Snail1 indeed entered and accumulated in the nucleus (i.e., were retained; Figure 6B and C). In contrast to T203E.Snail1 or T203A.Snail1, both NLS-T203E.Snail1 and NLS-T203A.Snail1 induced robust EMT of MCF10A cells (Supplementary Figure S5A and B). This result indicated that T203E and T203A mutations do not abrogate Snail1 function; rather they do not induce EMT because they do not enter or are not retained in the nucleus. Furthermore, it suggested that T203 phosphorylation of Snail1 likely occurs in the nucleus to enhance its nuclear retention and that T203 phosphorylation is not essential for Snail1 transcriptional function.
Snail1 also has other cellular functions, such as inhibition of cell-cycle progression and proliferation, enhanced cell survival, and stimulation of cell motility (Vega et al, 2004; Barrallo-Gimeno and Nieto, 2005). In MCF10A cells, expression of wt Snail1 inhibited cell proliferation (Supplementary Figure S6A), induced G1/S cell-cycle arrest (Supplementary Figure S6B), prevented cell death in response to TNFα (Supplementary Figure S6C) and serum withdrawal (Supplementary Figure S6D), and stimulated increased cell motility (Supplementary Figure S6E). In contrast, T203A.Snail1 was attenuated in its ability to perform all these cellular functions (Supplementary Figure S6A–E).
Lats2 levels are increased in aggressive breast cancer cells and influence their invasive/migratory capacity
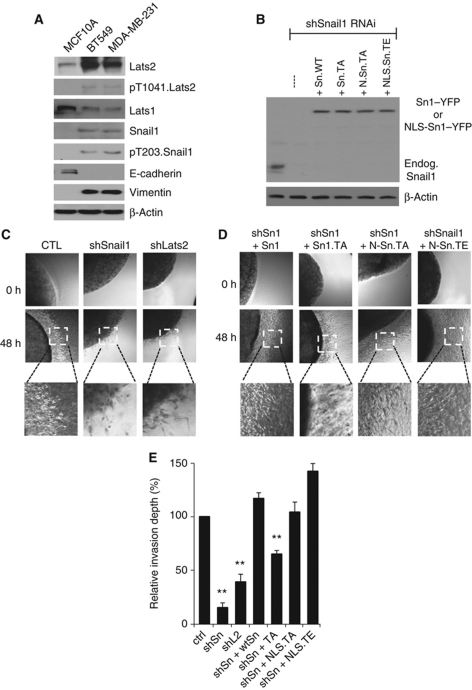
To determine if Lats2 influenced Snail1 function in cancer progression, we analysed two aggressive (i.e., metastatic) human breast cancer cell lines with characteristic EMT features: MDA-MB-231 and BT549, and compared these with non-tumourigenic MCF10A cells. Relative to MCF10A cells in both MDA-MB-231 and BT549 cells, total cellular Lats2 protein level was increased and there was a low level of Lats2 kinase activity present (pT1041.Lats2 antibody; Figure 8A). Snail1 was phosphorylated at T203 and total cellular Snail1 protein level was increased (Figure 8A).
Figure 8.
Metastatic breast cancer cells contain increased amounts of Lats2 and in these cells Lats2 affects their invasive capacity. (A) Western blot analysis with the indicated antibodies of cell lysates from the non-transformed and non-tumourigenic human breast epithelial cell line MCF10A and two tumourigenic and metastatic human breast cancer cell lines MDA-MB-231 and BT549. (B) A Snail1 antibody western blot analysis of MDA-MB-231 cells transduced with lentiviruses expressing both a Snail1 shRNAi and various YFP-tagged, RNAi-resistant Snail1 mutants, as indicated. TA: T203A mutation; TE: T203E mutation; N or NLS: nuclear localization signal. (C, D) MDA-MB-231 cells as described in (B) were aggregated and placed in a 3D collagen I gel (2 mg/ml). Phase image from a representative imbedded cell aggregate shows the distance of invasion/migration of cells at 48 h. Magnified boxes show presence of cells leaving the aggregate. Results from multiple aggregates (@10) per well from multiple experiments (3) are quantified in (E) and depict the distance cells migrated centrifugally from the aggregates, relative to control cells arbitrarily set at 100%. Data are represented as mean±s.d. **Indicates P<0.01.
To determine whether the increased Lats2 level present in MDA-MB-231 cells influenced Snail1 cancer function, such as tumour cell invasion/migration, we asked whether depletion of Lats2 affected the ability of MDA-MB-231 cells to invade/migrate in 3D collagen I gels. MDA-MB-231 cells require Snail1 to invade through the basement membrane and migrate through the ECM in vivo (Ota et al, 2009). In control experiments, depletion of Snail11 in MDA-MB-231 (Supplementary Figure S7A) indeed inhibited their invasion/migration in 3D collagen I gels (Figure 8C and E). When Lats2, but not Lats1, was RNAi depleted in MDA-MB-231 cells this resulted in reduced total cellular Snail1 protein level (Supplementary Figure S7A) and inhibited the invasion/migration of these cells in 3D collagen I gels (Figure 8C and E).
Next, we asked whether phosphorylation at T203 of Snail1 in MDA-MB-231 cells was required for their invasion/migration in collagen I gels. To do so, we made use of a dual cassette lentivirus that shRNAi depletes endogenous Snail1 while concurrently expressing in the same cell an RNAi-resistant YFP-tagged Snail1 isoform at levels that approximate the level of endogenous Snail1 (Figure 8B; Feng et al, 2010). In control experiments, rescue with RNAI-resistant wt Snail1 restored collagen I invasion/migration of Snail1-depleted MDA-MB-231 cells (Figure 8D and E). Expression of RNAi-resistant T203A.Snail1 provided a partial rescue, while expression of either RNAi-resistant NLS-T203A.Snail1 or NLS-T203E.Snail1 completely rescued or enhanced collagen I invasion/migration (Figure 8D and E).
In sum these results indicate that invasive, metastatic breast cancer cell lines contain increased level of Lats2 and that this increased Lats2 influences the invasive capacity of these cells. Phosphorylation of Snail1 at T203 also appears to be critical for the invasive potential of MDA-MB-231 cells. Furthermore, forced nuclear entry of T203A.Snail1 and T203E.Snail1 indicated that they were functional in mediating tumour cell invasion, and lends further support to the contention that phosphorylation of Snail1 by Lats2 serves to retain Snail1 in the nucleus and thereby enhance its function.
Lats2 potentiates Snail1 function in vivo
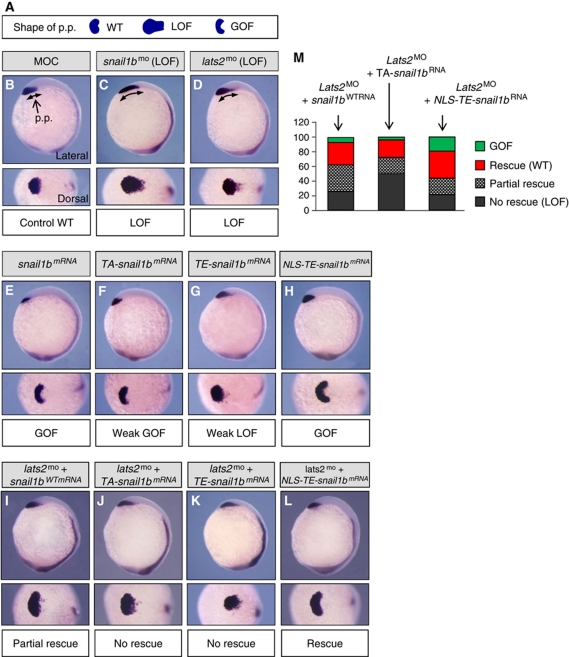
To determine whether Lats2 modulates Snail1 activity in vivo we examined zebrafish embryo development, as the phenotype of Snail1 gain and loss of function have been well described at gastrulation stages (Blanco et al, 2007). Loss of function was obtained after embryo injection with morpholino antisense oligonucleotides at the one cell stage (morphant embryos). There are two snail1 genes in the zebrafish embryo and interestingly, the reported phenotype of lats2 morphant embryos at gastrulation (Chen et al, 2009) appears to be similar to that of snail1b morphants when analysed at the tailbud stage. A side-by-side comparison of embryos defective in snail1b or lats2 function confirmed the similarity of the two phenotypes (Figure 9B–D and quantified in Supplementary Table 1). In particular, snail1b morphants showed a defective migration of the axial mesendoderm (Figure 9C), as assessed by the shape of the most anterior embryonic structure, the prechordal plate (p.p.) (Blanco et al, 2007). The p.p. was visualized by expression of the hgg1 gene (hatching gland gene 1), and a similar defective shape was also observed after lats2 morpholino injection (Figure 9D). Compared with snail1b morpholino, lats2 morpholino injections produced consistently weaker phenotypes, as would be expected if Lats2 was an in vivo upstream modulator of Snail1b function.
Figure 9.
Snail1 rescues the Last2 morpholino defect in migration of the mesoderm during zebrafish embryo development. Zebrafish embryos at the one cell stage were injected with indicated morpholinos, mRNA, or combination of morpholino and mRNA as indicated above each panel. All were analysed at the end of gastrulation (tailbud stage). Prechordal plate (p.p.) migration was visualized by expression of hatching gland gene hgg1. The arrows indicate the distance occupied by the p.p. along the anteroposterior axis. (A) A diagram showing the shapes of the p.p. under different conditions is shown: WT: wild type; LOF: loss of function; GOF: gain of function. The phenotype associated with the specified injections is indicated below each panel (C–L). Human Snail1 is equivalent to zebrafish snail1b. Human Snail1 T203 is equivalent to zebrafish snail1b T196. MOC is a control, scrambled morpholino. TA: T196A; TE: T196E; NLS: nuclear localization signal. (M) Snail1b rescues the lats2MO phenotype. The percentage of phenotypes (LOF: no rescue; partial rescue; WT: rescue; GOF: gain-of-function) obtained after injection of the corresponding mRNAs with lats2MO, relative to embryos injected with lats2MO alone. Pictures (I, J, L) show representative embryos of each condition. Conditions: (I) lats2MO+snail1b WT mRNA n=85 (GOF: 7%; rescue: 31%; partial rescue 37%; no rescue: 25%). (J) lats2MO+TA-snail1b n=91 (GOF: 4%; rescue: 24%; partial rescue: 22%; no rescue: 50%). (L) lats2MO+NLS-TE-snail1b n=88 (GOF: 18%; rescue: 38%; partial rescue: 23%; no rescue: 21%). GOF phenotype: similar to snail1b overexpression (E). Rescue: similar to the wild-type condition (B). Partial rescue: intermediate phenotype between lats2MO (D) and the wild-type condition (B). No rescue: similar to lats2MO (D).
It is worth noting that although lats1 morphant embryos also show mesodermal defects (Chen et al, 2009), the main effect is in the convergence movements, and the embryos are much wider than their wild-type counterpart. This phenotype is very different from that observed in both lats2 and snail1b morphants. The similarity in lats2 and snail1b defective phenotypes is compatible with Lats2 interacting with Snail1 in the mouse and with snail1b in the fish as the endogenous expression patterns indicate that Lats2 and Snail transcripts colocalize in several tissues at the appropriate stages. In the fish, both lats1 and lats2 are ubiquitously expressed at early stages and at stages equivalent to embryonic days 7–9 in the mouse. Lats2 but not Lats1 is expressed in the neural crest, a predominant location of Snail1 expression. In the mouse embryo, Lats1 is mainly expressed in ectodermal tissues while Lats2, like Snail1, is mainly expressed in mesodermal tissues (McPherson et al, 2004).
Overexpression in the fish embryo is usually obtained after mRNA injection. Injection of snail1b mRNA resulted in the opposite phenotype to that of morpholino injection, an increase in axial mesendoderm migration assessed by the anterior compression of the p.p. (Blanco et al, 2007; Figure 9E and quantified in Supplementary Table 1). In embryos injected with T196A.snail1b mRNA (TA-snail1b), a partial increase in axial mesendoderm was observed (Figure 9F). If, as seen in cell lines, Snail1 function is potentiated upon nuclear phosphorylation at T203 in vivo, then possibly fish embryos overexpressing the equivalent version of the zebrafish snail1 gene (T196E) in the nucleus would show a clear gain-of-function phenotype. T196E.snail1b mRNA (TE-snail1b) injected embryos actually inhibited axial mesendoderm migration (Figure 9G), possibly reflecting its inability to enter the nucleus and thus accumulation in the cytosol. However, injection with NLS-TE.snail1b mRNA dramatically increased axial mesendoderm migration (Figure 9H) consistent with the TA and TE mutants being active but restricted in their nuclear accumulation or entry, respectively, and that phosphorylation of snail1b in the nucleus potentiates Snail function.
Importantly, NLS-TE-snail1b injection rescued the lats2 morphant axial mesendoderm migration defect (Figure 9L and quantified in Figure 9M). WT snail1b and TA-snail1 mRNA partially rescued the lats2 morphant phenotype (Figure 9I and J, and quantified in Figure 9M). WT snail1b gave a better rescue than TA-snail1b. TE-snail1b mRNAs did not rescue lats2 morphant phenotype at all (Figure 9K).
In conclusion, Last2 contributes to the migration of the mesoderm during zebrafish embryo development, a process that requires EMT and is regulated by Snail1 in different organisms.
Lats2−/− mouse embryos die around E12.5 with CNS developmental defects and are smaller in size (Yabuta et al, 2007). Snail1−/− embryos are resorbed around E8.5 and although mesoderm forms it is disorganized and E-cadherin expression fails to be downregulated as occur in wt embryos at a similar stage (Carver et al, 2001). Therefore, we asked whether gastrulating E7.5 Lats2−/− embryos contain less Snail1 protein and correspondingly higher E-cadherin levels, relative to wt controls, as would be predicted from our biochemical and cellular studies. Western blots of dissected E7.5 embryos extracts revealed that Snail1 protein level was reduced in embryos deficient in Lats2 while E-cadherin level was increased (Supplementary Figure S7B) without significant change in Snail1 mRNA level (Supplementary Figure S7C).
Discussion
Post-translational modification of Snail1 is a critical regulator of its total cellular protein level, subcellular localization, and function (Dominguez et al, 2003; Zhou et al, 2004; Peinado et al, 2005; Yook et al, 2006). Through a screen to identify novel post-translational modifiers of Snail1 protein stability we identified the tumour suppressor kinase Lats2. Lats2 phosphorylates Snail1 in the nucleus leading to its nuclear retention and stabilization. As such Lats2 influences various Snail1 cellular functions, such as EMT induction, growth arrest, survival, and cell migration. During embryonic development, mouse embryos lacking Lats2 have less Snail1 protein and correspondingly higher E-cadherin protein levels. In developing zebrafish embryos, Lats2 contributes to the migration of the mesoderm, a process that requires EMT and is regulated by Snail1. Finally, analysis of a limited set of invasive/metastatic human breast cancer cell lines with features of EMT revealed that they express high levels of Lats2 that was required to support Snail1-mediated invasion of these cells in 3D collagen I gels.
Our data suggest the following model for how Lats2 influences Snail1 level and potentiates its activity. Following activation, Lats2 interacts with Snail1 and directly phosphorylates it at residue T203 in the nucleus, which serves to retain Snail1 in the nucleus where it is stable and functional as a transcription factor.
Three lines of evidence support the contention that Lats2 phosphorylates Snail1 in the nucleus. First, nocodazole-induced mitotic stress activates Lats2 kinase activity and results in Lats2 nuclear translocation (Aylon et al, 2006). Under these conditions, Snail1 phosphorylation at T203 is enhanced and pT203.Snail1 and increased Snail1 protein are predominantly found in the nucleus (Figure 6). Second, a mutant form of Snail1, T203A, which cannot be phosphorylated by Lats2 is inhibited in its EMT and tumour invasion functions (but not completely inactive) and found predominantly in the cytosol. Although it can associate with nuclear pore importin proteins and enter the nucleus it is not retained. However, when T203A.Snail1 nuclear entry and retention is forced by attaching a strong NLS it is functional to induce EMT and facilitate tumour cell invasion, suggesting that phosphorylation of Snail1 at T203A serves primarily to enhance nuclear Snail1 level and not its transcriptional repressor activity. Third, the phospho-mimetic T203E.Snail1 mutant is exclusively cytosolic and rapidly degraded. It does not associate with nuclear importins nor does it enter the nucleus even when nuclear export is inhibited. T203E.Snail1 does not induce EMT or tumour cell invasion. The cellular behaviour of this mutant suggests that pT203 phosphorylation in the cytosol could inhibit Snail1 nuclear entry and function. When nuclear entry and retention of T203E.Snail1 is forced in cells by attaching a strong NLS it now induces EMT and tumour cell invasion. Moreover, when injected into zebrafish embryos it exhibits a gain-of-function snail1b migratory phenotype during gastrulation and rescues both snail1b and lats2 morphant zebrafish phenotype. Thus, it is likely the inability of these mutants either to be retained in the nucleus (T203A) or to enter the nucleus (T203E) that diminishes their function. Precisely, how T203 phosphorylation of Snail1 results in its nuclear accumulation is unclear. Possibilities include phosphorylation at T203 alters Snail1 binding with the nuclear export machinery or with some resident nuclear proteins or chromatin structures.
A recent analysis of Snail1 phosphorylation in cells did not identify T203 as being phosphorylated (MacPherson et al, 2010). Although we used similar cell types (HEK293), in the latter study cells were not stimulated prior to analysis, whereas we activated Lats2, by treating cells with nocodazole prior to phosphopeptide isolation and analysis or prior to pT203.Snail1 antibody western blotting. This could suggest that T203 phosphorylation of Snail1 by Lats2 is tightly regulated, particularly during embryonic development. Precisely, how Lats2 is activated during embryonic development remains to be determined.
Mammalian Lats kinases were initially identified as putative tumour suppressors (Warts or wts) in a Drosophila screen for regulators of organ size (Justice et al, 1995; Xu et al, 1995). Subsequent studies revealed that mammalian Lats1 and 2, and Drosophila wts are central components of the Hippo signalling pathway that regulate cell number and thus organ size (Zeng and Hong, 2008). Activation of the Hippo pathway leads to activation of Lats kinases, which in turn phosphorylate and limit the transcriptional co-activators YAP and TAZ (Zeng and Hong, 2008). YAP and TAZ overexpression can induce EMT in epithelial cells (Overholtzer et al, 2006; Lei et al, 2008). Since Lats inhibits YAP and TAZ, by sequestering them in the cytoplasm together with Smad3, it leads to the suppression of EMT (Varelas et al, 2010). We now identify Snail1 as a substrate of Lats2 kinase, but not of Lats1 kinase. Manipulation of the Hippo (Mst1/2) kinase, by overexpression or cell–cell contact results in Lats2 activation (Chan et al, 2005) and Snail1 phosphorylation and stabilization. But, we found that overexpression of Lats2 induced cellular EMT, in a Snail1-dependent manner, while reducing Lats2 level inhibits EMT. This raises the possibility that under certain circumstances Lats2 may promote EMT and tumour progression through its effects on Snail1. In support of a possible role for Lats2 in tumour progression, we found that two aggressive breast cancer cell lines express high levels of Lats2, and when reduced this inhibited the invasion/migration of MDA-MB-231 breast cancer cells in 3D collagen gels, much like depletion of Snail1 does in these cells.
How then could Lats inhibit EMT on the one hand (Varelas et al, 2010) and be a positive modulator in other circumstances? A plausible explanation for this apparent opposing view could be that Snail1 is a nuclear target of Lats2 whereas the Hippo components YAP and TAZ are cytoplasmic targets (Hao et al, 2008; Varelas et al, 2010; Zhao et al, 2010). Thus, it could be possible that a stimulus leading to Lats2 activation could generate two opposing responses depending on the compartmentalization of Lats2 kinase activity and particular substrate in the cytoplasm or nucleus.
It is also worth noting that there may be distinct functions or substrates for Lats1 and Lats2. We find that Lats1 does not phosphorylate Snail1 or affect Snail1 cellular protein level. Recent studies in MCF10A cells have shown that Lats1, but not Lats2, suppresses YAP function (Zhang et al, 2008a), raising the intriguing possibility that Lats2 function in MCF10A cells is primarily via its phosphorylation of Snail1 and not through inhibition of YAP or TAZ. Moreover, the ability of the two Lats kinases to suppress TGFβ signals, and thus EMT, also differs with Lats1 being much more effective (Varelas et al, 2010). The phenotype of Lats1- and Lats2-deficient mice is also distinct (McPherson et al, 2004; Yabuta et al, 2007; Visser and Yang, 2010), and only for Lats1 null mice has in vivo tumour suppressive activity been observed (St John et al, 1999).
Our data suggest that Lats2, through its effect upon Snail1, may positively affect EMT independent of its effects upon YAP or TAZ. In summary, we propose that Lats2, in addition to behaving as a tumour suppressor, may also have a tumour-promoting activity not only via modulation of mutant p53 (Aylon et al, 2006, 2009, 2010), but also as a positive regulator of Snail1-mediated EMT and survival.
Materials and methods
Cell lines and cell culture conditions
HEK293, HCT116, HT1080, MDCK, BT549, MDA-MB-231, and MCF10A cells were obtained from ATCC (Manassas, VA), Lats2 MEFs (Yabuta et al, 2007) and maintained in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 1 mM L-glutamine and 100 units/ml of penicillin, 100 μg/ml of streptomycin. MCF-10A cells were cultured as described (Debnath et al, 2003). For HEK293 cells stably overexpressing Snail–CBG, cells were transfected with pEN1-Snail–CBG using Lipofectamine 2000 (Invitrogen) and selected with 1000 μg/ml G418. Single clones were isolated by limiting dilution. 3D cyst cultures of individual MCF-10A cells in type I collagen gels were carried out as described (O’Brien et al, 2001). Culture medium was changed every 2–3 days and cells grown for 12 days until cysts with lumens were formed.
Antibodies and chemical inhibitors
Total Lats2, phospho-Lats2 T1041, and phospho-Lats2 S871 are described in Yabuta et al (2007). Rabbit polyclonal Lats2 antibody was from Bethyl Laboratories (Montgomery, TX). The Snail1 monoclonal antibody, clone 17EC3, was provided by I Virtanen (Helsinki, Finland) (Franci et al, 2006). Snail (L70G2), GSK-3β, and YAP antibodies were from Cell Signaling Technology (Beverly, MA). β-Actin, β-tubulin, Flag (M2 monoclonal), HA antibodies were from Sigma (St Louis, MO). E-cadherin and Vimentin antibodies were from BD Biosciences (San Jose, CA). Lamin A/C, β-catenin, and p21 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against importin α, β, and transportin have been described previously (Mingot et al, 2009). H-Ras and Smad2 antibody were from Santa Cruz Biotechnology. Phospho-Smad2 (Ser465/467) antibody was from Cell Signaling Technology. GSK-3β (pY216) antibody was from BD Biosciences. MG132, Nocodazole, and LMB were from Sigma.
Plasmids
p2 × Flag–CMV2–YAP was provided by M Sudol (Mount Sinai School of Medicine, New York, NY); pCMV-HA-MST2 and pME18SFL-WW45 were provided by K Guan (UCSD, San Diego, CA); pGL2-E-cadherin luciferase has been previously described (Ayyanathan et al, 2007); human Snai1l cDNA and pcDNA3.1-GFP–Snail1 were provided by M Hung (M.D. Anderson, Houston, TX); pBabe-puro and pGEX-6P-3 (GE Healthcare). The pEN1-Snail1–CBG construct was made by replacing β-catenin with human Snail1 cDNA in pβ-cat-CBG (Naik and Piwnica-Worms, 2007). Mutations in Snail1 and Lats2 were introduced using the Quick-Change site-directed mutagenesis kit (Stratagene). pLKO.1 lentiviral vectors containing shRNAs were from the Washington University Genome Center. Coding DNAs for Danio rerio snail1b were cloned as an EcoRI fragment in pCS2+ or CS2+NLS MT (Rupp et al, 1994) that provides an SV40 Large T antigen NLS followed by six myc epitope tags. For E. coli expression, Snail1 and mutants were subcloned into pNzztev80 (Mingot et al, 2009), which provides an N-terminal ZZ tag and a TEV protease cleavage site, or pGEX. All PCR-amplified products were verified by DNA sequencing. pBabe-myc–Lats2 was made by subcloning lats2 cDNA into pBabe-puro plasmid. Flag-tagged Snail1 mutant constructs were subcloned into pCMV14–3 × Flag. pBabe-H-Ras plasmid was supplied by Dr J Weber (Washington University, St Louis, MO, USA).
siRNA kinome screen
The siRNA screen was performed as previously described (Naik and Piwnica-Worms, 2007) with minor modifications. Briefly, 7000 cells/well were seeded in 96-well plate 1 day before transfection. Forward transfection was performed with a 96 multichannel head on the FX liquid handler, adding 200 μl/well of media-complexed Lipofectamine 2000 (Invitrogen) to 20 pmol of the combined Kinase siRNA Set V2.0 2 pools (targeting 691 genes, Qiagen). Experimental siRNA controls were arrayed in columns 2–11 of each plate and individual controls placed manually in columns 1 and 12. After an siRNA/Lipofectamine 2000 incubation (20 min at room temperature), 50 μl of the complexed siRNA was added to each well of a triplicate cell plate set using the FX liquid handler, yielding a final concentration of ∼5 pmol siRNA/well. Seventy-two hours after transfection, luminescent signal was measured in ultrasensitive detection mode on an EnVision plate reader (Perkin-Elmer). Cell viability was then determined as conversion of resazurin dye (Sigma R7017) (final concentration 44 μM) to resorufin after 4 h incubation at 37°C, monitored at 590 nm on a FLUOstar OPTIMA fluorescence reader (BMG Labtech) excited at 544 nm. For each gene, the robust Z-scores (MAD) for all siRNAs (three independent plates) were averaged. Targets with an ∣MAD∣ of at least three are selected as potential hits for further identification (Zhang et al, 2008b).
Supplementary Material
Acknowledgments
This work was supported by NIH Grants P30 DK52574, P30 CA91842, P41 RR00954, and UL1 RR024992 to the Proteomics Core at the Siteman Cancer Center of Washington University and P50CA94056 to the Imaging Core of the Siteman Cancer Center at Washington University. NIH grants CA85839 and GM080673 to GDL. Grants from Ministerio de Ciencia e Innovación (BFU2008-01042; CONSOLIDER-INGENIO 2010 CSD2007-00017 and CSD2007-00023) and Generalitat Valenciana (Prometeo 2008/049) to MAN.
Author contributions: KZ, ER-A, NY, RJO, and JM conceived aspects of the experimental design, performed experiments, and interpreted the data. GL, HN, and MAN conceived experiments, interpreted results, and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M (2006) A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev 20: 2687–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Ofir-Rosenfeld Y, Yabuta N, Lapi E, Nojima H, Lu X, Oren M (2010) The Lats2 tumor suppressor augments p53-mediated apoptosis by promoting the nuclear proapoptotic function of ASPP1. Genes Dev 24: 2420–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Yabuta N, Besserglick H, Buganim Y, Rotter V, Nojima H, Oren M (2009) Silencing of the Lats2 tumor suppressor overrides a p53-dependent oncogenic stress checkpoint and enables mutant H-Ras-driven cell transformation. Oncogene 28: 4469–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanathan K, Peng H, Hou Z, Fredericks WJ, Goyal RK, Langer EM, Longmore GD, Rauscher FJ III (2007) The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic Shuttling. Cancer Res 67: 9097–9106 [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA (2005) The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132: 3151–3161 [DOI] [PubMed] [Google Scholar]
- Blanco MJ, Barrallo-Gimeno A, Acloque H, Reyes AE, Tada M, Allende ML, Mayor R, Nieto MA (2007) Snail1a and Snail1b cooperate in the anterior migration of the axial mesendoderm in the zebrafish embryo. Development 134: 4073–4081 [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T (2001) The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol 21: 8184–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH (2005) The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24: 2076–2086 [DOI] [PubMed] [Google Scholar]
- Chen CH, Sun YH, Pei DS, Zhu ZY (2009) Comparative expression of zebrafish lats1 and lats2 and their implication in gastrulation movements. Dev Dyn 238: 2850–2859 [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS (2003) Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30: 256–268 [DOI] [PubMed] [Google Scholar]
- Dominguez D, Montserrat-Sentis B, Virgos-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J, Franci C, Garcia de Herreros A (2003) Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol Cell Biol 23: 5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EF, Iida M, Myers RA, Campbell DA, Hintz KA, Armstrong EA, Li C, Wheeler DL (2011) Dasatinib sensitizes KRAS mutant colorectal tumours to cetuximab. Oncogene 30: 561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Nie L, Thakur MD, Su Q, Chi Z, Zhao Y, Longmore GD (2010) A multifunctional lentiviral-based gene knockdown with concurrent rescue that controls for off-target effects of RNAi. Genomics Proteomics Bioinformatics 8: 238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franci C, Takkunen M, Dave N, Alameda F, Gomez S, Rodriguez R, Escriva M, Montserrat-Sentis B, Baro T, Garrido M, Bonilla F, Virtanen I, García de Herreros A (2006) Expression of Snail protein in tumor-stroma interface. Oncogene 25: 5134–5144 [DOI] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X (2008) Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem 283: 5496–5509 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Kawata A, Nishikawa M, Tateishi Y, Yamaguchi M, Nakagawa K, Hirabayashi S, Bao Y, Hidaka S, Hirata Y, Hata Y (2009) Hippo pathway-dependent and -independent roles of RASSF6. Sci Signal 2: ra59. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9: 534–546 [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA (2009) The basics of epithelial-mesenchymal transition. J Clin Invest 119: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer EM, Feng Y, Zhaoyuan H, Rauscher FJ III, Kroll KL, Longmore GD (2008) Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell 14: 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL (2008) TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol 28: 2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson MR, Molina P, Souchelnytskyi S, Wernstedt C, Martin-Perez J, Portillo F, Cano A (2010) Phosphorylation of serine 11 and serine 92 as new positive regulators of human Snail1 function: potential involvement of casein kinase-2 and the cAMP-activated kinase protein kinase A. Mol Biol Cell 21: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Johnson KR, Wheelock MJ (2005) Cadherin switching: essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J Cell Sci 118: 873–887 [DOI] [PubMed] [Google Scholar]
- McPherson JP, Tamblyn L, Elia A, Migon E, Shehabeldin A, Matysiak-Zablocki E, Lemmers B, Salmena L, Hakem A, Fish J, Kassam F, Squire J, Bruneau BG, Hande MP, Hakem R (2004) Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J 23: 3677–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot JM, Vega S, Maestro B, Sanz JM, Nieto MA (2009) Characterization of Snail nuclear import pathways as representatives of C2H2 zinc finger transcription factors. J Cell Sci 122: 1452–1460 [DOI] [PubMed] [Google Scholar]
- Naik S, Piwnica-Worms D (2007) Real-time imaging of beta-catenin dynamics in cells and living mice. Proc Natl Acad Sci USA 104: 17465–17470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien LE, Jou TS, Pollack AL, Zhang Q, Hansen SH, Yurchenco P, Mostov KE (2001) Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly. Nat Cell Biol 3: 831–838 [DOI] [PubMed] [Google Scholar]
- Ota I, Li XY, Hu Y, Weiss SJ (2009) Induction of a MT1-MMP and MT2-MMP-dependent basement membrane transmigration program in cancer cells by Snail1. Proc Natl Acad Sci USA 106: 20318–20323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA (2006) Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA 103: 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Del Carmen Iglesias-de la Cruz M, Olmeda D, Csiszar K, Fong KS, Vega S, Nieto MA, Cano A, Portillo F (2005) A molecular role for lysyl oxidase-like 2 enzyme in snail regulation and tumor progression. EMBO J 24: 3446–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon YL, Zhou HY, Cheung AN, Ngan HY, Wong AS (2008) p70 S6 kinase promotes epithelial to mesenchymal transition through snail induction in ovarian cancer cells. Cancer Res 68: 6524–6532 [DOI] [PubMed] [Google Scholar]
- Rupp RA, Snider L, Weintraub H (1994) Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev 8: 1311–1323 [DOI] [PubMed] [Google Scholar]
- St John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG, Parlow AF, McGrath J, Xu T (1999) Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet 21: 182–186 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL (2010) The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell 19: 831–844 [DOI] [PubMed] [Google Scholar]
- Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA (2004) Snail blocks the cell cycle and confers resistance to cell death. Genes Dev 18: 1131–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser S, Yang X (2010) LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle 9: 3892–3903 [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W (1995) Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Yabuta N, Okada N, Ito A, Hosomi T, Nishihara S, Sasayama Y, Fujimori A, Okuzaki D, Zhao H, Ikawa M, Okabe M, Nojima H (2007) Lats2 is an essential mitotic regulator required for the coordination of cell division. J Biol Chem 282: 19259–19271 [DOI] [PubMed] [Google Scholar]
- Yang Z, Rayala S, Nguyen D, Vadlamudi RK, Chen S, Kumar R (2005) Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail's subcellular localization and functions. Cancer Res 65: 3179–3184 [DOI] [PubMed] [Google Scholar]
- Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, Cha SY, Ryu JK, Choi YJ, Kim J, Fearon ER, Weiss SJ (2006) A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol 8: 1398–1406 [DOI] [PubMed] [Google Scholar]
- Zeng Q, Hong W (2008) The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell 13: 188–192 [DOI] [PubMed] [Google Scholar]
- Zhang J, Smolen GA, Haber DA (2008a) Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res 68: 2789–2794 [DOI] [PubMed] [Google Scholar]
- Zhang XD, Kuan PF, Ferrer M, Shu X, Liu YC, Gates AT, Kunapuli P, Stec EM, Xu M, Marine SD, Holder DJ, Strulovici B, Heyse JF, Espeseth AS (2008b) Hit selection with false discovery rate control in genome-scale RNAi screens. Nucleic Acids Res 36: 4667–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Yang XC, Chung N, Gates A, Stec E, Kunapuli P, Holder DJ, Ferrer M, Espeseth AS (2006) Robust statistical methods for hit selection in RNA interference high-throughput screening experiments. Pharmacogenomics 7: 299–309 [DOI] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL (2010) A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev 24: 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC (2004) Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol 6: 931–940 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.