Abstract
SET8 is implicated in transcriptional regulation, heterochromatin formation, genomic stability, cell-cycle progression, and development. As such, it is predicted that SET8 might be involved in the development and progression of tumour. However, whether and how SET8 might be implicated in tumourigenesis is currently unknown. Here, we report that SET8 is physically associated with TWIST, a master regulator of epithelial–mesenchymal transition (EMT). We demonstrated that SET8 and TWIST are functionally interdependent in promoting EMT and enhancing the invasive potential of breast cancer cells in vitro and in vivo. We showed that SET8 acts as a dual epigenetic modifier on the promoters of the TWIST target genes E-cadherin and N-cadherin via its H4K20 monomethylation activity. Significantly, in breast carcinoma samples, SET8 expression is positively correlated with metastasis and the expression of TWIST and N-cadherin and negatively correlated with E-cadherin. Together, our experiments revealed a novel role for SET8 in tumour invasion and metastasis and provide a molecular mechanism underlying TWIST-promoted EMT, suggesting SET8 as a potential target for intervention of the metastasis of breast cancer.
Keywords: breast cancer, EMT, metastasis, SET8, TWIST
Introduction
It is abundantly clear now that epigenetic dysregulation of histone modifications, which is imparted by epigenetic modulators and other factors that mediate the installation, removal, and/or interpretation of the modifications, actively contributes to human cancer (Minucci and Pelicci, 2006; Chi et al, 2010). Recently, biological effects of histone methylation/demethylation at distinct lysine residues have received considerable attentions, and emerging data link aberrant histone methylation and/or demethylation events to tumourigenesis (Krivtsov and Armstrong, 2007; Chi et al, 2010). For example, it has been reported that the mixed lineage leukaemia (MLL) family of histone methyltransferases is involved in human myeloid and lymphoblastic leukaemia (Krivtsov and Armstrong, 2007); enhancer of zeste homologue 2 (EZH2), a histone H3 lysine K27 (H3K27)-specific methyltransferase, promotes the proliferation, invasion, and angiogenesis of epithelial cancer cells, and is predictive of poor clinical outcome (Richter et al, 2009; Lu et al, 2010); the jumonji family of lysine demethylases, including JARID1A, JARID1B, and JARID1C, have been implicated in leukaemia, prostate cancer, and renal carcinoma (Chi et al, 2010); and recently, we reported that lysine-specific histone demethylase 1 (LSD1), an H3K4 mono- and di-methylation (H3K4me1/2)-specific demethylase, suppresses the invasiveness and metastasis of breast cancer cells (Wang et al, 2009b).
SET8 (also known as PR-Set7/9, SETD8, KMT5A), a member of the SET domain-containing methyltransferase family (Fang et al, 2002; Nishioka et al, 2002) specifically targeting H4K20 for monomethylation, has been implicated in a diverse array of biological processes, such as controlling gene transcription (Congdon et al, 2010; Li et al, 2011b), modulating replication origins (Tardat et al, 2010), maintaining genome integrity (Houston et al, 2008; Oda et al, 2009), and regulating cell-cycle progression and development (Jorgensen et al, 2007; Oda et al, 2009; Abbas et al, 2010; Centore et al, 2010; Wu et al, 2010), through its histone monomethylating activity. Interestingly, SET8 has been reported to be involved in both activation and repression of transcription. For example, it has been shown that SET8-mediated H4K20me1 is associated with a reader/effector L3MBTL1 to create a transcriptionally repressive status of chromatin (Nishioka et al, 2002; Trojer et al, 2007) and is thus considered as a transcription repression mark (Kalakonda et al, 2008); however, it has also been observed that SET8/H4K20me1 is enriched in the promoter and coding regions of transcriptionally active genes (Talasz et al, 2005; Li et al, 2011a) and mediates the transcriptional activation of Wnt target genes (Li et al, 2011b). Moreover, in addition to histone modifications, SET8 has been shown to be able to monomethylate non-histone protein p53 at lysine 382 (p53K382me1) and to repress its related proapoptotic and cell-cycle arrest functions (Shi et al, 2007). The involvement of SET8 in cell proliferation and genome stability and the importance of its non-histone target p53 in cell-cycle control predict its possible role in tumourigenesis. However, whether and how SET8 is involved in oncogenesis is currently not known.
Tumour metastasis is the major cause of death in cancer patients. To achieve a metastatic process, tumour cells must experience multiple and sequential steps: cell motility, tissue invasion, intravasation, extravasation, and final colonization and proliferation of tumour cells at new sites (Yang et al, 2004; Joyce and Pollard, 2009; Klein, 2009; Nguyen et al, 2009; Nicoloso et al, 2009; Polyak and Weinberg, 2009; Psaila and Lyden, 2009). Each of these steps is regulated by a panel of signalling pathways. Epithelial–mesenchymal transition (EMT) is believed to be the initial step of tumour metastasis, during which epithelial cells shed their differentiated characteristics, including cell–cell adhesion planar, apical-basal polarity as well as immobility, and acquire mesenchymal features, such as motility, invasiveness, and a heightened resistance to apoptosis. EMT program is triggered by a diverse set of elements including growth factors (Oft et al, 1998; Acevedo et al, 2007), transcription factors such as TWIST (Yang et al, 2004), FOXC2 (Mani et al, 2007), SNAIL (Kudo-Saito et al, 2009), HIF1α (Yang et al, 2008), and NF-κB (Radisky and Bissell, 2007), as well as genetic or epigenetic alterations (Wang et al, 2009b). Despite considerable research efforts on EMT program, the molecular pathway underlying this process is still not totally understood.
TWIST (also known as TWIST1), a highly conserved basic helix-loop-helix transcriptional factor, plays a pivotal role in tumour metastasis by promoting EMT (Yang et al, 2004; Taube et al, 2010). Ectopic expression of TWIST results in both morphological and molecular alterations in the expression of particular proteins, such as downregulation of epithelial protein markers (e.g., E-cadherin, α-catenin, β-catenin, and γ-catenin) as well as upregulation of mesenchymal markers (e.g., N-cadherin, fibronectin, vimentin, and SM-actin; Yang et al, 2004; Taube et al, 2010). In addition, as a transcription factor, it is believed that TWIST regulates transcriptional activity through homo- or hetero-dimerization (Castanon et al, 2001), thereby conferring its dual mode of transcriptional output, activation or repression (Yang et al, 2004; Alexander et al, 2006; Cheng et al, 2007; Fu et al, 2011). For instance, TWIST represses E-cadherin transcription (Yang et al, 2004; Fu et al, 2011) on the one hand, but, on the other hand, upregulates the expression of N-cadherin (Alexander et al, 2006), YB-1 (Shiota et al, 2008), and AKT2 (Cheng et al, 2007), eventually leading to tumour cell EMT and metastasis. In spite of these, the molecular events leading to TWIST-facilitated EMT, especially its dual transcriptional mode, are still not fully understood.
In the present work, we found that SET8 physically interacts with TWIST in vivo. We showed that SET8 acts as a dual epigenetic modifier on the promoter of E-cadherin and N-cadherin through its H4K20 monomethylation activity. We demonstrated that TWIST and SET8 cooperate to regulate the expression of E-cadherin and N-cadherin. We showed that SET8 and TWIST interplay to promote EMT and invasion of breast cancer cells, and that SET8 expression is positively correlated with N-cadherin and TWIST expression and negatively correlated with E-cadherin expression in breast carcinoma samples.
Results
SET8 is physically associated with TWIST
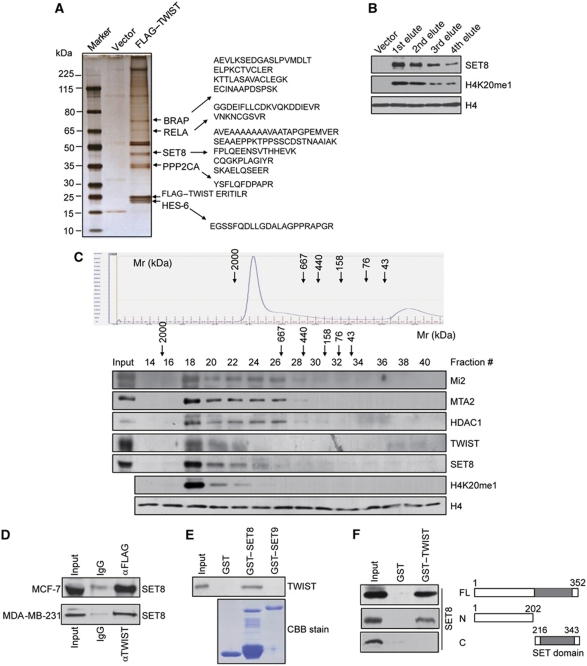
Tumour metastasis has received extensive research efforts for its association with high cancer mortality, and EMT is believed to represent a critical step in tumour metastasis. In an effort to better understand the regulatory mechanisms underlying EMT, we employed affinity purification and mass spectrometry (MS) to identify proteins that are associated with TWIST, a major inducer in EMT and tumour metastasis. In these experiments, FLAG-tagged TWIST (FLAG–TWIST) was stably expressed in breast cancer cell line MCF-7. Whole cell extracts were prepared and subjected to affinity purification using an anti-FLAG affinity gel. MS analysis indicates that TWIST was co-purified with a number of proteins including BRAP (BRCA1-associated protein), RELA (a subunit of NF-κB), PPP2CA (Protein phosphatase 2, catalytic subunit, alpha isozyme), and HES-6 (Figure 1A). Interestingly, five peptides matching the H4K20-specific histone methyltransferase SET8 were also identified in the TWIST complex (Figure 1A). When incubated with recombinant histone octamers, the eluted TWIST complex indeed exhibited a monomethyltransferase activity towards H4K20, supporting the existence of SET8 in the TWIST complex (Figure 1B).
Figure 1.
SET8 is physically associated with TWIST. (A) Mass spectrometry analysis of TWIST-associated proteins. Whole cellular extracts from FLAG–TWIST stably expressing MCF-7 cells were subjected to affinity purification with anti-FLAG antibody that was immobilized on the agarose beads. The purified protein complex was resolved on SDS–PAGE and silver stained, and the bands were retrieved and analysed by mass spectrometry. (B) Western blotting examination of SET8 protein level and its H4K20 monomethyltransferase activity in the FLAG–TWIST-purified fractions using the indicated antibodies. (C) Co-fractionation of TWIST and SET8 complex by FPLC. MCF-7 cell nuclear proteins were extracted, concentrated, and then 6 mg of nuclear extract was fractionated on Superose 6 size exclusion columns. Chromatographic elution profiles and immunoblotting analysis of the chromatographic fractions are shown. The elution positions of calibration proteins with known molecular masses (kDa) are indicated, and an equal volume from each fraction was analysed. (D) SET8 interacts with TWIST in vivo. Whole cell lysates from FLAG–TWIST stably expressing MCF-7 cells or from MDA-MB-231 cells were prepared and immunoprecipitation was performed with anti-FLAG or anti-TWIST followed by immunoblotting with anti-SET8. (E) SET8 interacts directly with TWIST in vitro. GST pull-down assays were performed with the indicated GST-fused SET8 or SET9 protein and in vitro transcribed/translated TWIST. Coomassie brilliant blue (CBB) staining of the GST-fused proteins was shown. (F) Mapping the interface of SET8 interacting with TWIST by GST pull-down experiments using GST-fused TWIST and in vitro transcribed/translated full-length SET8 or its deletion constructs.
To further validate the presence of SET8 in the TWIST complex in vivo, MCF-7 cell nuclear extracts were examined by protein fractionation experiments using fast protein liquid chromatography (FPLC) with Superose 6 columns and a high salt extraction and size exclusion approach. Native TWIST and SET8 were eluted with an apparent molecular mass much greater than that of the monomeric protein; TWIST and SET8 immunoreactivity were detected in chromatographic fractions from the Superose 6 column with a relatively symmetrical peak centred between 667 and 2000 kDa and were accompanied by histone H4K20 monomethylation activity, as assayed by incubating these fractions with recombinant histone octamers and then immunoblotted with anti-H4K20me1 (Figure 1C). In addition, it has been reported that TWIST directly interacts with multiple components of the Mi2/NuRD/MTA2 complex (Fu et al, 2011). Indeed, the elution pattern of TWIST and SET8 in our experiments largely overlapped with that of the NuRD complex proteins including Mi2, MTA2, and HDAC1 (Figure 1C).
To further support the in vivo interaction of SET8 and TWIST, total protein extracts from FLAG–TWIST-expressing MCF-7 cells were prepared and co-immunoprecipitation experiments were performed with specific antibodies against target proteins. Immunoprecipitation (IP) with anti-FLAG followed by immunoblotting (IB) with the anti-SET8 indicated that SET8 is co-immunoprecipitated with TWIST (Figure 1D). This interaction is also confirmed with endogenous proteins in MDA-MB-231 cells (Figure 1D). To test whether TWIST and SET8 are able to interact in vitro, glutathione S-transferase (GST) pull-down assay was carried out using GST-fused SET8 construct and in vitro transcribed/translated TWIST. The results revealed that SET8 interacted with TWIST in vitro, whereas another SET domain-containing histone methyltransferase SET9 had no interaction with TWIST (Figure 1E). In an effort to map the interaction interface of SET8 with TWIST, GST pull-down assays were performed with GST-fused TWIST and in vitro transcribed/translated full-length SET8, SET8 N-terminal fragment (1–202 aa), and C-terminal SET domain (216–343 aa). The results showed that the SET8 N-terminal fragment is essential for the interaction of SET8 with TWIST (Figure 1F).
Functional interplay between SET8 and TWIST in promoting EMT in breast cancer cells
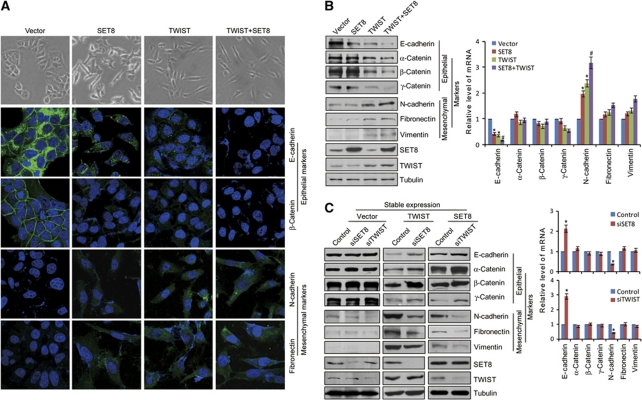
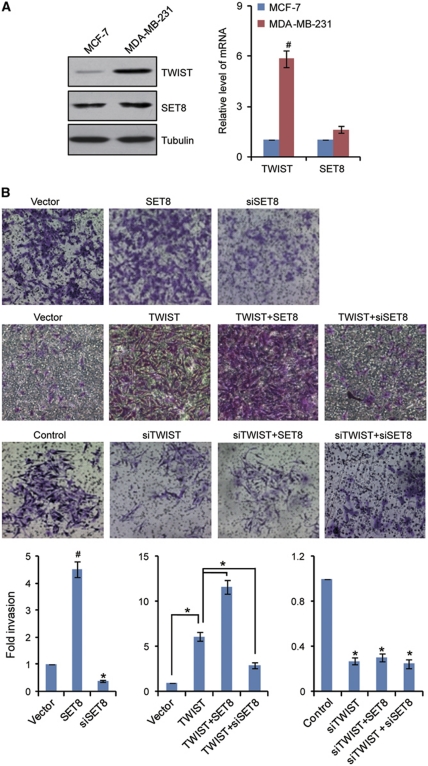
As stated above, TWIST is considered to be a master regulator of EMT (Yang et al, 2004; Horikawa et al, 2007). The interaction of SET8 with TWIST suggests that SET8 might also be involved in tumour EMT and metastasis. In order to further support the physical interaction and explore the functional connection between TWIST and SET8, we next investigated what role, if any, SET8 might play in the EMT stage of breast cancer metastasis. To this end, the morphological alterations and epithelial or mesenchymal marker changes in SET8- or/and TWIST-expressing MCF-7 cells were assessed by microscopy and western blotting, respectively. As shown in Figure 2A, while control MCF-7 cells maintained organized cell–cell adhesion and cell polarity, overexpression of TWIST or/and SET8 in MCF-7 cells led to loss of cell–cell contacts; these cells became scattering and their cobble stone-like appearance was replaced by a spindle-like, fibroblastic morphology (Figure 2A), which represents morphological changes of EMT. Consistently, we found that overexpression of SET8 or TWIST alone resulted in reduction of epithelial protein markers (E-cadherin, α-catenin, β-catenin, and γ-catenin) and induction of mesenchymal protein markers (N-cadherin, fibronectin, and vimentin), and co-expression of SET8 with TWIST led to a more dramatic change in expression of these proteins (Figure 2B). In agreement with these observations, immunofluorescent microscopy showed a reduction or loss of E-cadherin and β-catenin staining from cell membrane in the TWIST- or/and SET8-expressing MCF-7 cells compared with their strong membrane staining in control cells, whereas mesenchymal markers, including N-cadherin and fibronectin, exhibited a reverse trend (Figure 2A). Additionally, knockdown of SET8 in the cells stably expressing TWIST resulted in an elevation (or derepression) of the epithelial markers and a decrease (or deactivation) of the mesenchymal markers (Figure 2C). Analogously, knockdown of TWIST in SET8-expressing MCF-7 cells was associated with an evident derepression of the epithelial markers and a significant deactivation in the mesenchymal marker (Figure 2C). Together, these experiments indicate that SET8 and TWIST are functionally interdependent and act in a cooperative fashion to promote EMT. Examination of the expression of endogenous SET8 and TWIST in non-metastatic cell line MCF-7 and metastatic cell line MDA-MB-231 by western blotting and quantitative real-time PCR (qPCR) indicated that the expression of both protein and mRNA of TWIST is high in MDA-MB-231 and low in MCF-7 cells while SET8 has no significant changes in both cell lines (Figure 3A). This could contribute to the marginal changes in EMT markers in blank vector-transfected MCF-7 cells with SET8 or TWIST knockdown (Figure 2C).
Figure 2.
Functional interplay of SET8 and TWIST in promoting EMT of breast cancer cells. (A) SET8 cooperates with TWIST to induce morphological changes in MCF-7 cells. MCF-7 cells were transfected with vector, SET8 or/and TWIST, and the morphological alterations of MCF-7 cells were observed by phase-contrast microscopy. Immunofluorescence staining of epithelial (E-cadherin and β-catenin) and mesenchymal (N-cadherin and fibronectin) markers was visualized by confocal microscopy (Green). DAPI staining was included to visualize the cell nucleus (Blue). (B) Immunoblotting examination of the epithelial and mesenchymal proteins (left) and qPCR analyses of their mRNA expression (right) in MCF-7 cells overexpressing SET8 or/and TWIST. (C) SET8 or TWIST was silenced in vector, TWIST, or SET8 stably transfected MCF-7 cells, and the epithelial and mesenchymal markers were measured by immunoblotting (left) and qPCR (right). Each bar indicates mean±s.d. of three independent experiments. P-values were determined by Student's t-test. *P<0.05; #P<0.01.
Figure 3.
Functional interdependence of SET8 and TWIST in enhancing the invasive potential of breast cancer cells. (A) The endogenous protein or mRNA expression of SET8 and TWIST was examined by western blotting (left) or qPCR (right). (B) MDA-MB-231 cells were transfected with empty vector, TWIST or SET8 expression construct, control siRNA, TWIST or SET8 siRNA. After 48 h of transfection, cells were starved for 18 h before cell invasion assays were performed using Matrigel transwell filters. The invaded cells were stained and counted. The images represent one field under microscopy in each group. Each bar indicates mean±s.d. of a representative experiment performed in triplicate. P-values were determined by Student's t-test. *P<0.05; #P<0.01.
Functional interdependence of SET8 and TWIST in enhancing the invasive potential of breast cancer cells
In order to further support the SET8-promoted EMT and to explore the role of SET8 in tumour invasion, SET8 or/and TWIST was overexpressed or/and silenced in MDA-MB-231 cells, and the gain-of-function or loss-of-function of SET8 on the invasive potential of these cells was investigated using transwell invasion assays. The results indicated that SET8 overexpression resulted in elevated cell invasion of MDA-MB-231 cells by 4.5-fold and knockdown of SET8 expression led to a 2.5-fold decrease in cell invasion of these cells (Figure 3B). Moreover, SET8 could further enhance TWIST-promoted invasion of MDA-MB-231 cells, whereas SET8 knockdown rendered an impaired TWIST-promoted invasion (Figure 3B). On the other hand, in TWIST-depleted MDA-MB-231 cells, neither overexpression nor knockdown of SET8 was able to affect the invasive potential of these cells (Figure 3B). This is similar to MCF-7 cells that exhibit very low level of TWIST expression and low metastatic potential (Figure 3A). Collectively, these results indicate that SET8 and TWIST are functionally interdependent in promoting cell invasion.
SET8-mediated H4K20me1 has a dual function in TWIST-regulated gene expression
As mentioned above, SET8 is a member of methyltransferase family that specifically targets H4K20 for monomethylation (Fang et al, 2002; Nishioka et al, 2002) and is implicated in regulating either activation (Talasz et al, 2005; Wakabayashi et al, 2009; Li et al, 2011b) or repression (Nishioka et al, 2002; Kalakonda et al, 2008) of gene transcription. In order to explore the mechanistic insight into the interplay between SET8 and TWIST in facilitating EMT and cell invasion, we first hypothesized, based on the physical interaction between SET8 and TWIST and the report that SET8 is able to, in addition to H4K20, also methylate non-histone proteins (Shi et al, 2007), that SET8 might target TWIST for methylation. However, extensive efforts using MS and western blotting to identify TWIST methylation by SET8 yielded negative results. Therefore, we went to the conventional gene regulation model in which the transcription factor TWIST might require/recruit SET8 to the promoters of its target genes to regulate their transcription. As shown in Figure 2B (right), SET8 or/and TWIST suppressed E-cadherin mRNA expression and enhanced N-cadherin mRNA expression, whereas had limited effects on the expression of other markers, suggesting that E-cadherin and N-cadherin may be transcriptionally regulated by SET8 and TWIST. qPCR analyses through loss-of-function of SET8 or TWIST confirmed the results; depletion of SET8 or TWIST in TWIST or SET8 stably expressing MCF-7 cells led to a significant increase in E-cadherin mRNA level and a sharp decrease in N-cadherin gene expression (Figure 2C, right). Together, these results are consistent with the reports that E-cadherin and N-cadherin are the direct targets of TWIST (Yang et al, 2004; Alexander et al, 2006) and suggested that both E-cadherin and N-cadherin are regulated by TWIST and SET8 at the transcriptional level.
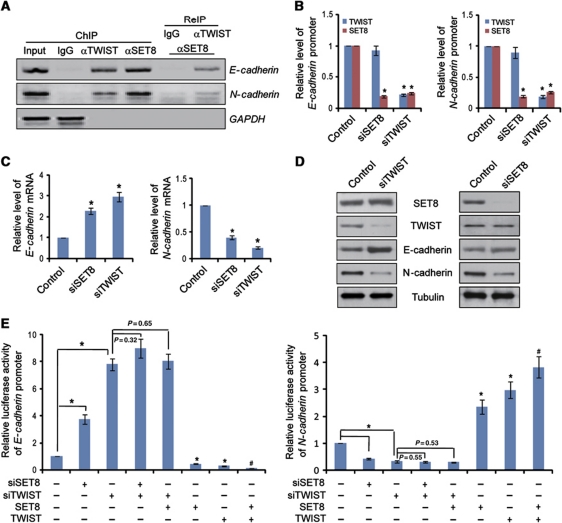
We then analysed the binding mode of TWIST and SET8 on the promoters of E-cadherin and N-cadherin genes. In these experiments, MDA-MB-231 cells were harvested for chromatin immunoprecipitation (ChIP) analysis, and the results indicated that SET8 was not only detected on the promoter of E-cadherin gene, but also on that of N-cadherin gene (Figure 4A). Furthermore, ChIP/re-immunoprecipitation (ChIP/ReIP) experiments validated co-occupancy of SET8 and TWIST on the E-cadherin and N-cadherin promoters (Figure 4A). Since the promoter recruitment of SET8 and TWIST is consistent with E-cadherin and N-cadherin expression patterns, it appeared that SET8 functions in a dual mode in TWIST-regulated gene expression.
Figure 4.
SET8 cooperates with TWIST to regulate E-cadherin and N-cadherin transcription. (A) Recruitment of SET8 and TWIST on E-cadherin and N-cadherin promoters. ChIP and ChIP/ReIP experiments were performed in MDA-MB-231 cells with indicated antibodies. (B) SET8 is recruited by TWIST to the E-cadherin or N-cadherin promoter. SET8 or TWIST was silenced individually in MDA-MB-231 cells; qChIP assays were performed with indicated antibodies. (C) SET8 or TWIST silencing led to increase or decrease in E-cadherin or N-cadherin mRNA level measured by qPCR. (D) SET8 or TWIST depletion led to an elevation or a reduction in E-cadherin or N-cadherin protein level, respectively, as measured by western blotting. (E) SET8 interplays with TWIST to regulate E-cadherin promoter- or N-cadherin promoter-driven luciferase activity. MDA-MB-231 cells were transfected with promoter luciferase constructs together with SET8 or/and TWIST expression or silencing molecules. Luciferase activities were measured and normalized to those of Renilla. Each bar indicates mean±s.d. of three independent experiments. P-values were determined by Student's t-test. *P<0.05; #P<0.01.
To further understand the molecular interdependence of SET8 and TWIST on E-cadherin and N-cadherin promoters, SET8 or TWIST was silenced individually in MDA-MB-231 cells and quantitative ChIP (qChIP) assays were performed. The results demonstrated that TWIST was still associated with the E-cadherin and N-cadherin promoters upon SET8 knockdown, whereas TWIST silencing abolished the occupancy of SET8 on these promoters, suggesting that SET8 is recruited by TWIST on E-cadherin and N-cadherin promoters (Figure 4B). Consistent with the promoter occupancy, in SET8-depleted MDA-MB-231 cells, the mRNA (Figure 4C) and protein (Figure 4D) expression of E-cadherin and N-cadherin increased and decreased, respectively, further supporting the notion that SET8 is required for trans-activation or trans-repression of TWIST target genes. Clearly, the dual regulatory function of TWIST and SET8 is interdependent, at least on E-cadherin and N-cadherin promoters.
To further support the argument that TWIST and SET8 are functionally interdependent, reporter activity assays were carried out in MDA-MB-231 cells with E-cadherin or N-cadherin promoter-driven luciferase reporter under overexpression or depletion of SET8. These experiments indicated that SET8 overexpression or knockdown resulted in repressed or enhanced E-cadherin reporter activity, respectively, whereas its overexpression or silencing led to activation or repression of the N-cadherin reporter activity, respectively (Figure 4E). However, in TWIST-depleted cells, neither overexpression nor depletion of SET8 had a significant effect on E-cadherin or N-cadherin promoter activity (Figure 4E).
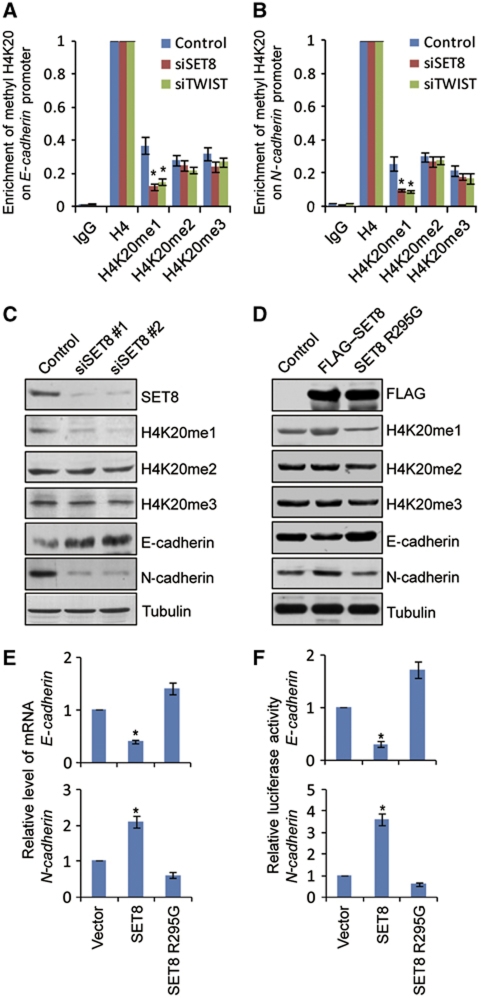
We next analysed the methylation status of histone H4K20 on E-cadherin and N-cadherin promoters. It has been reported that histone H4K20 can be mono-, di-, or tri-methylated by SET8 (Fang et al, 2002; Nishioka et al, 2002), NSD1 (Rayasam et al, 2003), Suv4-20h1/KMT5B and Suv4-20h2/KMT5C (Schotta et al, 2004), respectively. qChIPs in MDA-MB-231 cells indicated that H4K20me1, H4K20me2, and H4K20me3 were all detected on E-cadherin and N-cadherin promoters. To examine whether or not H420me1 was catalysed by SET8 on these promoters, endogenous SET8 expression was knocked down in MDA-MB-231 cells and the methylation status of H4K20 on E-cadherin and N-cadherin promoters were measured by qChIP. The results indicated that SET8 knockdown was associated with a sharp reduction of H4K20me1 as well as a slight decrease in H4K20me2 and H4K20me3 on both E-cadherin (Figure 5A) and N-cadherin promoters (Figure 5B). Interestingly, TWIST silencing also led to a marked decrease of H4K20me1 on E-cadherin and N-cadherin promoters (Figure 5A and B), further supporting the argument that SET8 is recruited by TWIST to the E-cadherin and N-cadherin promoters for monomethylating H4K20. Meanwhile, the promoter occupancy of H4K20me1 was associated with the expression tendency of E-cadherin and N-cadherin (Figure 5C and E). Collectively, these experiments indicate that SET8 is recruited by TWIST to E-cadherin promoter to repress its transcription and to N-cadherin promoter to activate its transcription through its H4K20 monomethylation activity. In support of this argument, we created catalytically inactive SET8 via R295G point mutation within the SET domain (Nishioka et al, 2002). Ectopic expression of the inactive SET8 mutant had a dominant-negative role in H4K20 monomethylation in vivo (Figure 5D), and this was accompanied by transcriptional derepression of E-cadherin and deactivation of N-cadherin (Figure 5E), which is also confirmed by the E-cadherin or N-cadherin promoter-driven luciferase activity (Figure 5F).
Figure 5.
SET8-directed H4K20me1 is a dual epigenetic mark on the E-cadherin and N-Cadherin promoters. SET8 recruitment on E-cadherin (A) or N-cadherin (B) promoter is associated with the monomethylation status of histone H4K20. After transfection of MDA-MB-231 cells with siRNAs of control, TWIST or SET8 for 72 h, qChIP assays were performed with mono-, di-, and tri-methylated H4K20-specific antibodies with anti-H4 normalized as 100%. (C) MDA-MB-231 cells were treated with control or SET8 siRNAs, and western blotting analysis was performed with antibodies against the indicated proteins. (D) Western blotting was conducted in MDA-MB-231 cells overexpressing the FLAG-tagged vector (control), FLAG–SET8 construct or a dominant-negative FLAG-tagged SET8 R295G point mutant (SET8 R295G). (E) Catalytically inactive SET8 R295G mutant resulted in derepression or deactivation of E-cadherin or N-cadherin transcription, respectively, as measured by qPCR. (F) Catalytically inactive SET8 R295G mutant resulted in derepression or deactivation of E-cadherin or N-cadherin promoter activity. Each bar indicates mean±s.d. of three independent experiments. P-values were determined by Student's t-test. *P<0.05.
SET8 cooperates with TWIST to promote breast cancer metastasis in vivo
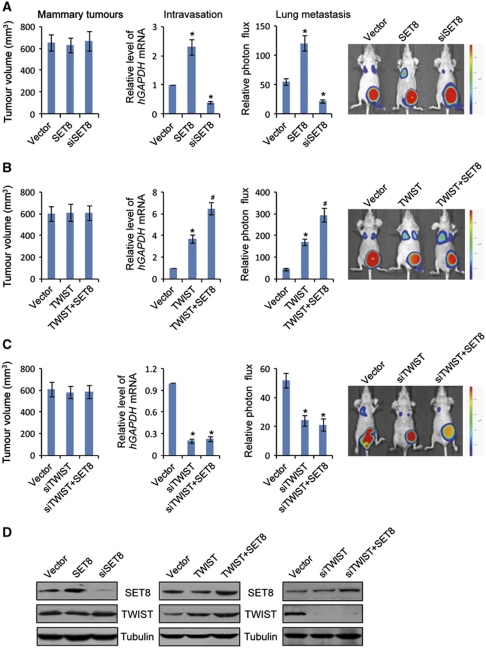
To further support the functional interdependence of SET8 and TWIST and to investigate their possible roles in breast cancer metastasis in vivo, MDA-MB-231 cells that had been engineered to stably express firefly luciferase (MDA-MB-231-Luc-D3H2LN, Xenogen Corporation) were infected with retroviruses or/and lentiviruses carrying empty vector, TWIST or SET8 expression construct, control shRNA, TWIST or SET8 shRNA. The effect of the gain-of-function and loss-of-function of SET8 or/and TWIST was assessed in immunocompromised female BALB/c mice by orthotopic implantation of MDA-MB-231-Luc-D3H2LN cells into the left abdominal mammary fat pad. The growth/dissemination of tumours was monitored weekly by bioluminescence imaging with IVIS imaging system (Xenogen Corporation). A metastatic event was defined as any detectable luciferase signal above background and away from the primary tumour site. The results showed that, while either overexpression or depletion of SET8 or/and TWIST had limited effects on the primary tumour growth (Figure 6A–C, left), the cell intravasation and spontaneous lung metastasis were enhanced in mice carrying MDA-MB-231-Luc-D3H2LN tumours with SET8 or/and TWIST overexpression and were suppressed in mice carrying MDA-MB-231-Luc-D3H2LN tumours with SET8 or TWIST depletion, as assessed by real-time RT–PCR analysis of the relative level of human GAPDH expression to murine β2-microglobulin in blood samples (Figure 6A–C, middle) or by bioluminescence imaging quantifying photon flux (Figure 6A–C, right). Collectively, these experiments support the notion that SET8 and TWIST cooperate to increase the metastatic potential of breast cancer cells in vivo.
Figure 6.
SET8 cooperates with TWIST to promote breast cancer metastatic potential in orthotopic mouse model. (A) The effect of SET8 gain-of-function or loss-of-function on spontaneous lung metastasis of orthotopic breast cancer cells. (B, C) Cooperation of SET8 and TWIST on spontaneous lung metastasis of orthotopic breast cancer cells. MDA-MB-231-Luc-D3H2LN cells were co-infected with retroviruses and lentiviruses carrying empty, SET8 or/and TWIST expression vector, and control shRNA, SET8 or TWIST shRNA-containing vector. These cells were inoculated into the left abdominal mammary fat pad (2 × 106 cells) of 6-week-old immunocompromised female BALB/c mice. Tumour size was measured on day 42 (mammary tumours, n=10). The presence of circulating tumour cells (intravasation, n=10) was assessed by qPCR of human GAPDH expression relative to murine β2-microglobulin in 1 ml of mouse blood perfusate. Lung metastases were quantified using bioluminescence imaging (n=10) after 6 weeks of initial implantation, and representative in vivo bioluminescent images are shown. (D) Examination of SET8 and/or TWIST overexpression and/or knockdown in the cell used in animal experiments by IB. In all panels, each bar indicates mean±s.d. P-values were determined by Student's t-test. *P<0.05; #P<0.01.
Positive association of SET8 expression with metastasis in human breast tumours
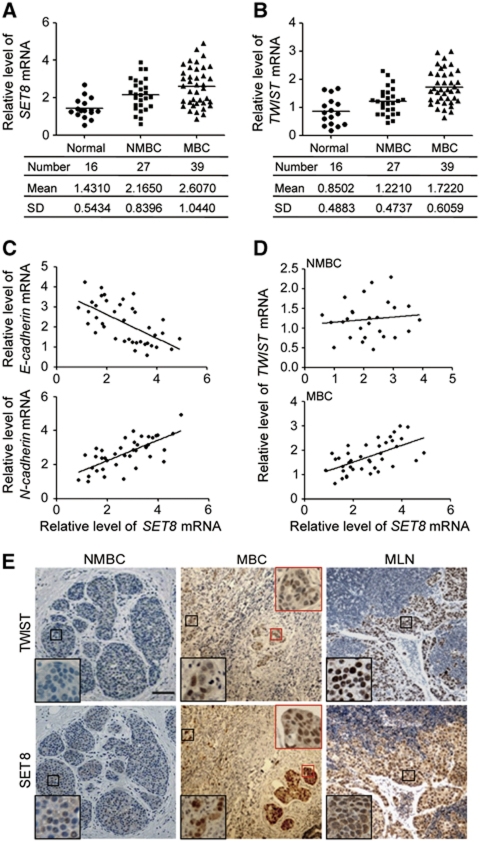
Next, to further support the role of SET8 in EMT and cell invasion, we collected breast samples of breast normal tissue (Normal), non-metastatic breast cancer (NMBC), and metastatic breast cancer (MBC) from breast cancer patients. The expression of SET8 mRNA was analysed by qPCR. We found that SET8 expression is upregulated in MBC (Figure 7A), and there appeared to be a progressive increase in SET8 mRNA levels from normal to metastatic samples, suggesting that SET8 is positively correlated with metastatic capacity. In addition, TWIST mRNA level is also positively correlated with the metastatic potential of breast cancer (Figure 7B), in accordance with a previous report (Yang et al, 2004). E-cadherin and N-cadherin mRNA levels in metastatic tumour samples were also analysed and were plotted against the levels of SET8 mRNA (Figure 7C). Statistical analysis of E-cadherin and SET8 expression found a Pearson's correlation coefficient of −0.6246 (P<0.0001) and a Spearman's correlation coefficient of −0.6358 (P<0.0001). Similar analysis of the expression patterns of N-cadherin and SET8 showed a Pearson's correlation coefficient of 0.7032 (P<0.0001) and a Spearman's correlation coefficient of 0.6698 (P<0.0001), indicating a strong negative correlation between expression of E-cadherin and SET8 and a positive correlation between that of N-cadherin and SET8 in MBCs. A positive correlation between SET8 and TWIST expression was also noted in MBCs (Pearson's correlation coefficient of 0.5936 (P<0.0001) and Spearman's correlation coefficient of 0.5800 (P<0.0001)) but not in NMBCs (Figure 7D). Furthermore, immunohistochemical (IHC) analyses of the sample sections of NMBCs, MBCs, and metastatic lymph nodes (MLNs) revealed that SET8 and TWIST are highly expressed in MBC (i.e., vascular tumour embolism) and MLN, compared with their expression in NMBC (Figure 7E), supporting the function interdependence between SET8 and TWIST in breast cancer metastasis.
Figure 7.
Correlation of SET8 with the metastasis of human breast cancers. The level of SET8 (A) or TWIST (B) mRNA is positively correlated with breast cancer metastasis. Total RNAs from the sample of breast normal tissue (Normal), non-metastatic breast cancer (NMBC), and metastatic breast cancer (MBC) were extracted and the expression of SET8 and TWIST mRNA was measured by qPCR with GAPDH as the reference gene. (C) The level of SET8 mRNA is negatively correlated with E-cadherin mRNA and positively correlated with N-cadherin mRNA in MBCs. The level of SET8, E-cadherin, and N-cadherin mRNAs was measured by qPCR and the relative level of SET8 mRNA was plotted against that of E-cadherin or N-cadherin. (D) The level of SET8 mRNA is positively correlated with that of TWIST mRNA in MBC samples. Total RNAs from NMBC and MBC samples were extracted, and the expression of SET8 and TWIST was measured by qPCR. The relative level of SET8 mRNA was plotted against that of TWIST. (E) Immunohistochemical analyses of SET8 or TWIST expression in NMBC, MBC (the black square represents invasive cancer cells and the red square indicates cancer embolus in circulation), and MLN samples. Representative images are shown. Scale bar, 50 μm.
Discussion
Recent evidence indicates that dysregulation of histone methylation (writing), interpretation (reading), and removal (erasing) is associated with oncogenesis (Chi et al, 2010). Histone methylation miswriting by MLL rearrangement (Krivtsov and Armstrong, 2007) or EZH2 deregulation (Morin et al, 2010) leads to aberrant gene expression and consequent tumourigenesis, and misreading of histone methylation marks such as H3K4me3 contributes to cellular transformation and tumourigenesis (Wang et al, 2009a). Additionally, histone lysine demethylases, such as LSD1 (Wang et al, 2009b) and the jumonji family (Yamane et al, 2007; Wang et al, 2009b), were implicated in several types of tumours. It has been reported that the H4K20 monomethyltransferase SET8 is required for S-phase progression (Jorgensen et al, 2007; Tardat et al, 2007), is engaged in transcriptional regulation (Congdon et al, 2010; Li et al, 2011b), genome replication and stability (Houston et al, 2008; Oda et al, 2009; Tardat et al, 2010), and modulates the proapoptotic and cell-cycle arrest functions of the tumour suppressor p53 (Shi et al, 2007), suggesting that SET8 might be implicated in pathological processes such as tumourigenesis. Our current observations that SET8 promotes the EMT and invasion of breast cancer cells and that the SET8 expression level is positively correlated with metastatic potential in breast carcinoma suggest that SET8 is another histone methylation regulator that might be implicated in the development and progression of breast cancer.
EMT is the initial step of tumour metastasis and is essential for tumour cells to overcome multiple barriers formed by the normal tissues that they encounter en route. TWIST has been defined as a chief modulator in epithelial–mesenchymal conversion and tumour metastasis (Yang et al, 2004; Mironchik et al, 2005; Ansieau et al, 2008; Shiota et al, 2008) and has been associated with advanced tumour stages and poor prognosis in several types of cancer (Yang et al, 2004; Kwok et al, 2005; Mironchik et al, 2005; Horikawa et al, 2007; Hasselblatt et al, 2009; Fu et al, 2011). In this study, we found that SET8 and TWIST are physically associated and functionally interplay to promote EMT. In cells stably expressing SET8, we observed a morphologic transition from the epithelial to fibroblastic-like shape, which represents the phenotypic hallmark of EMT (Yang et al, 2004; Mironchik et al, 2005), implying that SET8 is involved in EMT. To further determine the molecular connection between SET8 and TWIST in EMT promotion, we found that SET8 and TWIST are functionally interdependent in triggering the alterations in the expression of EMT markers, establishing an essential role of SET8 in modulating the TWIST-activated EMT and metastasis of breast cancer.
TWIST has also been characterized as both a repressor and an activator of transcription on distinct target genes (Yang et al, 2004; Mironchik et al, 2005; Alexander et al, 2006; Cheng et al, 2007). The molecular mechanisms underlying this dual mode of transcriptional regulation are not fully understood. Previous reports showed that TWIST transcription is regulated by a variety of factors such as PPARδ (Pan et al, 2009), STAT3 (Cheng et al, 2008), and HIF1α (Yang et al, 2008). Moreover, researches indicate that SET-domain methyltransferase superfamily like SET7/9 and SET8 can, in addition to histone H3 or H4, methylate non-histone proteins (Shi et al, 2007; Subramanian et al, 2008). In light of the report that SET7/9 can methylate oestrogen receptor α at lysine 302 (K302) and further regulate its target genes recruitment and transactivation function (Subramanian et al, 2008) and that SET8 is able to monomethylate p53 at lysine 382 (p53K382me1) and inhibit its proapoptotic and cell-cycle arrest functions (Shi et al, 2007), we postulated that SET8 might methylate TWIST and modulate its transcription activity. However, no SET8-directed methylation of TWIST was detected. We, therefore, turned our attention to the conventional gene regulation model in which TWIST require/recruit SET8 to its target promoters to regulate its gene transcription, and found that SET8 is recruited by TWIST on N-cadherin promoter and its H4K20 monomethylation activity contributes to the N-cadherin activation. On the other hand, we revealed that TWIST functions as a transcriptional repressor and cooperates with SET8 to repress E-cadherin expression, during which SET8 acts as a corepressor by catalysing H4K20 to H4K20me1 on the E-cadherin promoter. These results are consistent with previous researches that TWIST acts as a transcriptional repressor on some targets including E-cadherin (Yang et al, 2004) and with the original observation that SET8 and H4K20me1 are associated with repressed chromatin and involved in a gene repression in vivo (Nishioka et al, 2002; Trojer et al, 2007). In addition, it was recently reported that TWIST interacts with several components of the Mi2/NuRD repressive complex during EMT process (Fu et al, 2011), which is confirmed by our FPLC result showing the Mi2/NuRD proteins is in the same complex with TWIST. It appears that SET8 and the Mi2/NuRD complex may collaborate in fine-tuning the transcriptional activity of TWIST through their chromatin remodelling activities. This dual function of SET8-mediated H4K20me1 in both transcription repression and activation has been well documented (Nishioka et al, 2002; Talasz et al, 2005; Trojer et al, 2007; Kalakonda et al, 2008; Li et al, 2011b). Therefore, it is possible that the dual mode of TWIST in transcription regulation during EMT might be conferred by SET8.
Our study indicates that SET8 is a cofactor for TWIST and may exert its transcriptional activation and repression through its mediated H4K20 monomethylation. However, how SET8-mediated H4K20me1 acts as a repressive mark on E-cadherin and works as an activation mark on N-cadherin is still not known. It is conceivable that different readers of H4K20me1 lead to diverse biological effects of H4K20me1. The reading of the H4K20me1 mark by different effectors, in turn, may be determined by gene-specific DNA context and other transcription cofactors.
In view of the documentation that β1 integrin is engaged in the nuclear/cytoplasmic translocation of TWIST and modulate TWIST-directed N-cadherin transcriptional activation (Alexander et al, 2006), we employed the whole cell extracts for MS analysis to uncover novel functions of TWIST in both cytoplasm and nucleus. Several nuclear/cytoplasmic proteins were co-purified with TWIST, such as RELA, PPP2CA, HES-6, BRAP, and SET8. However, the components of the Mi2/NuRD/MTA2 complex was not contained in the TWIST-co-precipitated proteins, which is inconsistent with the results of a recent report (Fu et al, 2011) and our FPLC results. The discrepancy of the results between MS and FPLC may be due to the technical limitation of MS and a low abundance of the HDAC repressor complex in our assay.
In conclusion, we demonstrated that SET8 promotes EMT and enhances the invasive capacity of breast cancer cells via functional interdependence with TWIST and through dual chromatin remodelling activity. We showed that TWIST and SET8 cooperate to exert their dual transcriptional regulatory function via repression of E-cadherin expression and activation of N-cadherin expression. We demonstrated that SET8 facilitates intravasation and lung metastases of tumour cells in the mouse model of human breast cancer metastasis. Significantly, we found that in breast carcinoma samples that SET8 expression is positively correlated with N-cadherin and TWIST expression and metastasis and negatively correlated with E-cadherin expression. Together, our experiments revealed a role for SET8 in promoting EMT and invasive potential of breast cancer cells, suggesting that SET8 might be a potential therapeutic target for EMT and metastasis of breast cancer.
Materials and methods
Antibodies and reagents
Antibodies used were anti-FLAG, anti-Tubulin, anti-TWIST (rabbit), anti-Fibronectin, anti-Vimentin, anti-HDAC1 (Sigma); anti-MTA2 (Upstate Biotechnology); anti-Mi2 (Santa Cruz Biotechnology); anti-SET8 (Rabbit, Cell Signaling Technology); anti-MYC (MBL); anti-E-cadherin, anti-α-catenin, anti-β-catenin, anti-γ-catenin, anti-N-cadherin (BD Bioscience); anti-H4K20me1, anti-H4K20me2, anti-H4K20me3, anti-H4, anti-TWIST (Mouse) and anti-SET8 (Mouse) (Abcam). Protein A/G Sepharose CL-4B beads were from Amersham Biosciences, and protease inhibitor cocktail was from Roche Applied Science. The siRNA sequences used were control siRNA: 5′-UUCUCCGAACGUGUCACGU-3′; SET8 siRNA: #1, 5′-GGAAGAGAACUCAGUUACA-3′; #2, 5′-CAAAUGCUCUGGAAUGCGU-3′; TWIST siRNA: 5′-GGACAAGCUGAGCAAGAUUCA-3′. siRNA oligonucleotides were transfected into cells using RNAiMAX (Invitrogen) with the final concentration at 20 nM.
Preparation of cell nuclear extracts
MCF-7 cells were cultured in DMEM medium containing 10% fetal calf serum (FCS) and nuclear extracts were prepared essentially as described (Dalton et al, 1997; Smirnova et al, 2000). Briefly, MCF-7 cells were washed twice with cold PBS, scraped, and collected by centrifugation at 1500 g for 5 min. The cell pellet was resuspended in hypotonic buffer (10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, and 0.2 mM phenylmethyl-sulphonyl fluoride), and immediately centrifuged at 1500 g for 5 min. Cells were then resuspended in two times the original packed cell volume of hypotonic buffer, allowed to swell on ice for 10 min, and homogenized with 10 strokes of a Dounce homogenizer (B pestle). Nuclei were collected by centrifugation at 3300 g for 15 min at 4°C. The nuclei were resuspended in lysis buffer (50 mM Tris–Cl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.25% sodium deoxycholate and protease inhibitor mixture), incubated for 20 min on ice on a platform rocking at 150 r.p.m. The supernatant (nuclear fraction) was collected by centrifugation at 14 000 g for 30 min at 4°C. Protein concentration was determined using BCA Pierce protein assay kit (Thermo Scientific) and 6 mg of nuclear proteins was used for FPLC assay.
Fast protein liquid chromatography
MCF-7 cell nuclear extracts were prepared and dialysed against buffer D (20 mM HEPES (pH 8.0), 10% glycerol, 0.1 mM EDTA, 300 mM NaCl) (Applygen Technologies Inc.). Approximately 6 mg nuclear protein was concentrated to 0.5 ml using a Millipore Ultrafree centrifugal filter apparatus (10 kDa nominal molecular mass limit), and then applied to an 850 × 20 mm Superose 6 size exclusion column (Amersham Biosciences) that was equilibrated with buffer D containing 1 mM dithiothreitol and calibrated with protein standards (blue dextran, 2000 kDa; thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; bovine serum albumin, 67 kDa; and RNase A, 13.7 kDa, Amersham Biosciences). The column was eluted at a flow rate of 0.5 ml/min and fractions were separately collected.
ChIP, ChIP-ReIP and qChIP
ChIP and qChIP experiments were performed according to the procedure described previously (Zhang et al, 2004, 2006; Wu et al, 2005; Li et al, 2009; Shi et al, 2011). ChIP-ReIP was done essentially the same as the primary ChIP. Bead elutes from the first IP were incubated with 10 mM DTT at 37°C for 30 min and diluted 1:50 in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl (pH 8.1)) followed by ReIP with the secondary antibodies. For common ChIP and ChIP/ReIP assays, the final target DNA sequence was amplified and resolved on standard agarose DNA gels; for the qChIP, the target sequences were measured by quantitative real-time PCR. Primers used were common ChIP/ReIP primers: E-cadherin: 5′-AGGGTCACCGCGTCTATG-3′ (forward) and 5′-CTTCCGCAAGCTCACAGG-3′ (reverse), N-cadherin: 5′-CCTCATTCTTTGACCTCCTG-3′ (forward) and 5-TCTGTAAATAAGACGACCCAAT-3′ (reverse); and qChIP primers: E-cadherin: 5′-GCAGGTCCCATAACCCACCTA-3′ (forward) and 5′-CATAGACGCGGTGACCCTCTA-3′ (reverse), N-cadherin: 5′-TGCCAGTCACTTGCTAACAAAAG-3′ (forward) and 5′-GTGTGCGCTGGGAGAATAAAG-3′ (reverse).
Construction of E-cadherin or N-cadherin reporter plasmid and luciferase assays
For E-cadherin reporter construction, the sequence of E-cadherin promoter and partial first exon (−736 to +64 bp) was obtained by PCR using primers 5′-AAATTTGAGCTCGCCTGGGCAAGACAGAGC-3′ (forward) and 5′-GAATTTAGATCTGAGCGGGCTGGAGTCTGA-3′ (reverse); the N-cadherin reporter was generated as described previously (Alexander et al, 2006). The first intron (+746 to +3156 bp) from the human N-cadherin gene was obtained by PCR using primers 5′-AAATTTGAGCTCGGCTCTAGGGGCTGGATT-3′ (forward) and 5′-GGTTGGAGATCTTGTTGTTCGGGCGTGTAA-3′ (reverse). These PCR products were digested and ligated into the BglII-SacI sites of pGL3 basic vector (Promega) to generate pGL3-E-cadherin or pGL3-N-cadherin luciferase reporter construct. MDA-MB-231 cells in 48-well plates were transfected with E-cadherin or N-cadherin promoter luciferase reporter, Renilla plasmid, and indicated expression constructs, using LTX-Plus (Invitrogen). The amount of DNA in each transfection was kept constant by addition of empty vector. Forty-eight hours after transfection, the firefly and Renilla luciferases were assayed according to the manufacturer's protocol (Promega), and the firefly luciferase activity was normalized to that of Renilla luciferase. Each experiment was repeated in triplicate.
Transwell invasion assay
The transwell invasion assay was performed using the transwell chamber (Chemicon Incorporation) with a Matrigel-coated filter. MDA-MB-231 cells were cultured in Leibovitz's L-15 medium with 10% FCS at 37°C without CO2, and overexpression or/and knockdown of SET8 or/and TWIST was performed via transfection of corresponding vectors and/or siRNA duplexes. Forty-eight hour later, cells were deprived in serum-free Leibovitz's L-15 medium. After 18 h of deprivation, cells were harvested, washed three times in PBS and resuspended in serum-free culture medium. Afterwards, 1 × 105 of these cells in 300 μl of serum-free media were plated onto the upper chamber of the transwell. The upper chamber was then transferred to a well containing 500 μl of media supplemented with 10% FCS and incubated for 24 h. Cells may actively migrate from the upper to the lower side of the filter due to FCS as attractant. Cells on the upside were removed using cotton swabs, and the invasive cells on the lower side were fixed, stained with 0.1% crystal violet solution, and counted using light microscope. The experiment was repeated three times.
Retroviral production and infection
The generation of the pBabe-TWIST or pBabe--SET8-integrated retroviruses was conducted according to a protocol described by Weinberg's lab (Addgene: production of retroviruses using FuGENE-6). Briefly, the human expression plasmid of pBABE-3 × FLAG–TWIST or pBABE-3 × FLAG–SET8 was generated by subcloning the 3 × FLAG–TWIST or pBABE-3 × FLAG–SET8 fragment into the pBABE-Puro vector using following primers: pBABE-3 × FLAG–TWIST: 5′-GGCGGTGGATCCATGGACTACAAAGACCATGACGGTG-3′ (forward) and 5′-AAATTTCAATTGCTAGTGGGACGCGGACATGGACCA-3′ (reverse); pBABE-3 × FLAG–SET8: 5′-GGCGGTGGATCCATGGACTACAAAGACCATGACGG-3 (forward) and 5′-AAATTCAATTGTTAATGCTTCAGCCACGGGTGGGCT-3 (reverse). The retroviral plasmid vectors pBABE-3 × FLAG–TWIST/-SET8, pVSV-G, and pGag-Pol were co-transfected into the packaging cell line 293T. Viral supernatants were collected 48 h later, clarified by filtration, and concentrated by ultracentrifugation. The concentrated virus was used to infect 5 × 105 cells (20–30% confluent) in a 60-mm dish with 8 μg ml−1 polybrene. Infected cells were selected by 2 μg ml−1 puromycin (Merck).
Lentiviral production and infection
Assembling of RNAi lentivirus system using pLL3.7 and other LentiLox vectors was carried out according to a protocol described online (http://web.mit.edu/jacks-lab/protocols/pll37.htm). In brief, siRNA sequences targeting human TWIST or SET8 were designed and cloned into the pLL3.7 shuttle vector. The recombinant construct as well as three assisted vectors, VSVG, pMDL g/p RRE, and RSV-REV, were then transiently transfected into 293T cells. Viral supernatants were collected, filtrated, concentrated, and quantified. The concentrated virus was used to infect 5 × 105 cells in a 60-mm dish with 8 μg ml−1 polybrene.
In vivo metastasis
The MDA-MB-231-Luc-D3H2LN cell line (MDA-MB-231 cell line engineered to stably express firefly luciferase) (Xenogen Corporation) was infected with retroviruses or/and lentiviruses carrying empty vector, TWIST and/or SET8 expression construct, control shRNA, TWIST or SET8 shRNA. These cells were inoculated into the left abdominal mammary fat pad (2 × 106 cells) of 6-week-old immunocompromised female BALB/c mice (Charles River, Beijing, China). Lung metastasis bioluminescence imaging was conducted after 6 weeks of initial implantation, in which mice were anesthetized and given 150 μg g−1 of D-luciferin in PBS by i.p. injection. Fifteen minutes after injection, bioluminescence was imaged with a charge-coupled device camera (IVIS; Xenogen). Bioluminescence from relative optical intensity was defined manually, and data were expressed as photon flux (photons s−1 cm−2 Steradian−1) and were normalized to background photon flux which was defined from a relative optical intensity drawn over a mouse that was not given an injection of luciferin. Metastatic cells in circulation were assessed by real-time RT–PCR analysis of the relative level of human GAPDH expression to murine β2-microglobulin in blood samples. Animal handling and procedures were approved by the Peking University Health Science Center Institutional Animal Care and Use Committee.
Tissue specimens and analysis
Breast normal tissues, NMBC, MBC, and MLNs were procured from surgical specimens from patients with breast cancer for which complete information on clinical tumour size and metastatic status was available. For mRNA extraction, samples were frozen in liquid nitrogen immediately after surgical removal. Samples from NMBC, MBC, and MLN were fixed in 10% neutral-buffered formalin overnight, and then processed, paraffin embedded, sectioned, and stained with haematoxylin and eosin according to the standard protocol. For IHC analysis, 3 μm sample sections were incubated with primary antibodies indicated overnight at 4°C in a humidified chamber, followed by incubation with the HRP-conjugated secondary antibodies for 2 h. Staining is completed by 5–10 min incubation with 3.3′-diaminobenzidine (DAB) substrate, which results in a brown-coloured precipitate at the antigen site. All human tissue was collected using protocols approved by the Ethics Committee of the Peking University Health Science Center. For detailed Materials and methods, see Supplementary data.
Supplementary Material
Acknowledgments
This work was supported by grants (30830032 and 30921062 to YS; 30671076 and 30771104 to HZ) from the National Natural Science Foundation of China, by grants (973 Program: 2011CB504204 to YS and HZ) from the Ministry of Science and Technology of China, and by grants (KC1001, SMFA09A11, and 2010SY5403002 to HZ) from State Key Laboratory of Space Medicine Fundamentals and Application of China.
Author contributions: FY conceived and performed the experiments, analysed data, and wrote the manuscript. LS, QL, and XH performed experiments and analysed data. LL analysed data. HZ conceived and supervised the experiments, analysed data, and wrote the manuscript. YS supervised the experiments and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A (2010) CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 40: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, Ayala GE, Peterson LE, Ittmann M, Spencer DM (2007) Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell 12: 559–571 [DOI] [PubMed] [Google Scholar]
- Alexander NR, Tran NL, Rekapally H, Summers CE, Glackin C, Heimark RL (2006) N-cadherin gene expression in prostate carcinoma is modulated by integrin-dependent nuclear translocation of Twist1. Cancer Res 66: 3365–3369 [DOI] [PubMed] [Google Scholar]
- Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S, Maestro R, Voeltzel T, Selmi A, Valsesia-Wittmann S, Caron de Fromentel C, Puisieux A (2008) Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 14: 79–89 [DOI] [PubMed] [Google Scholar]
- Castanon I, Von Stetina S, Kass J, Baylies MK (2001) Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development 128: 3145–3159 [DOI] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L (2010) CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell 40: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH (2007) Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res 67: 1979–1987 [DOI] [PubMed] [Google Scholar]
- Cheng GZ, Zhang WZ, Sun M, Wang Q, Coppola D, Mansour M, Xu LM, Costanzo C, Cheng JQ, Wang LH (2008) Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J Biol Chem 283: 14665–14673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG (2010) Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon LM, Houston SI, Veerappan CS, Spektor TM, Rice JC (2010) PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. J Cell Biochem 110: 609–619 [DOI] [PubMed] [Google Scholar]
- Dalton TP, Bittel D, Andrews GK (1997) Reversible activation of mouse metal response element-binding transcription factor 1 DNA binding involves zinc interaction with the zinc finger domain. Mol Cell Biol 17: 2781–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y (2002) Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol 12: 1086–1099 [DOI] [PubMed] [Google Scholar]
- Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J (2011) The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res 21: 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselblatt M, Mertsch S, Koos B, Riesmeier B, Stegemann H, Jeibmann A, Tomm M, Schmitz N, Wrede B, Wolff JE, Zheng W, Paulus W (2009) TWIST-1 is overexpressed in neoplastic choroid plexus epithelial cells and promotes proliferation and invasion. Cancer Res 69: 2219–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa T, Yang J, Kondo S, Yoshizaki T, Joab I, Furukawa M, Pagano JS (2007) Twist and epithelial-mesenchymal transition are induced by the EBV oncoprotein latent membrane protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res 67: 1970–1978 [DOI] [PubMed] [Google Scholar]
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC (2008) Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem 283: 19478–19488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS (2007) The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179: 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JA, Pollard JW (2009) Microenvironmental regulation of metastasis. Nat Rev Cancer 9: 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, Rice JC, Nimer SD (2008) Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 27: 4293–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CA (2009) Parallel progression of primary tumours and metastases. Nat Rev Cancer 9: 302–312 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA (2007) MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 7: 823–833 [DOI] [PubMed] [Google Scholar]
- Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y (2009) Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell 15: 195–206 [DOI] [PubMed] [Google Scholar]
- Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, Chua CW, Chan KW, Chan FL, Glackin C, Wong YC, Wang X (2005) Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res 65: 5153–5162 [DOI] [PubMed] [Google Scholar]
- Li R, Zhang H, Yu W, Chen Y, Gui B, Liang J, Wang Y, Sun L, Yang X, Zhang Y, Shi L, Li Y, Shang Y (2009) ZIP: a novel transcription repressor, represses EGFR oncogene and suppresses breast carcinogenesis. EMBO J 28: 2763–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun L, Zhang Y, Wang D, Wang F, Liang J, Gui B, Shang Y (2011a) The histone modifications governing TFF1 transcription mediated by estrogen receptor. J Biol Chem 286: 13925–13936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Nie F, Wang S, Li L (2011b) Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci USA 108: 3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Han HD, Mangala LS, Ali-Fehmi R, Newton CS, Ozbun L, Armaiz-Pena GN, Hu W, Stone RL, Munkarah A, Ravoori MK, Shahzad MM, Lee JW, Mora E, Langley RR, Carroll AR, Matsuo K, Spannuth WA, Schmandt R, Jennings NB et al. (2010) Regulation of tumor angiogenesis by EZH2. Cancer Cell 18: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, Kutok JL, Hartwell K, Richardson AL, Weinberg RA (2007) Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA 104: 10069–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG (2006) Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 6: 38–51 [DOI] [PubMed] [Google Scholar]
- Mironchik Y, Winnard PT Jr, Vesuna F, Kato Y, Wildes F, Pathak AP, Kominsky S, Artemov D, Bhujwalla Z, Van Diest P, Burger H, Glackin C, Raman V (2005) Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer. Cancer Res 65: 10801–10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R et al. (2010) Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, Massague J (2009) Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 9: 274–284 [DOI] [PubMed] [Google Scholar]
- Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA (2009) MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer 9: 293–302 [DOI] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D (2002) PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9: 1201–1213 [DOI] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D (2009) Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 29: 2278–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oft M, Heider KH, Beug H (1998) TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol 8: 1243–1252 [DOI] [PubMed] [Google Scholar]
- Pan D, Fujimoto M, Lopes A, Wang YX (2009) Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell 137: 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA (2009) Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9: 265–273 [DOI] [PubMed] [Google Scholar]
- Psaila B, Lyden D (2009) The metastatic niche: adapting the foreign soil. Nat Rev Cancer 9: 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky DC, Bissell MJ (2007) NF-kappaB links oestrogen receptor signalling and EMT. Nat Cell Biol 9: 361–363 [DOI] [PubMed] [Google Scholar]
- Rayasam GV, Wendling O, Angrand PO, Mark M, Niederreither K, Song L, Lerouge T, Hager GL, Chambon P, Losson R (2003) NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J 22: 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, Hotfilder M, Lowel D, von Luettichau I, Mossbrugger I, Quintanilla-Martinez L, Kovar H, Staege MS, Muller-Tidow C, Burdach S (2009) EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci USA 106: 5324–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Sun L, Li Q, Liang J, Yu W, Yi X, Yang X, Li Y, Han X, Zhang Y, Xuan C, Yao Z, Shang Y (2011) Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci USA 108: 7541–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O (2007) Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 27: 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota M, Izumi H, Onitsuka T, Miyamoto N, Kashiwagi E, Kidani A, Yokomizo A, Naito S, Kohno K (2008) Twist promotes tumor cell growth through YB-1 expression. Cancer Res 68: 98–105 [DOI] [PubMed] [Google Scholar]
- Smirnova IV, Bittel DC, Ravindra R, Jiang H, Andrews GK (2000) Zinc and cadmium can promote rapid nuclear translocation of metal response element-binding transcription factor-1. J Biol Chem 275: 9377–9384 [DOI] [PubMed] [Google Scholar]
- Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM (2008) Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell 30: 336–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talasz H, Lindner HH, Sarg B, Helliger W (2005) Histone H4-lysine 20 monomethylation is increased in promoter and coding regions of active genes and correlates with hyperacetylation. J Biol Chem 280: 38814–38822 [DOI] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E (2010) The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E (2007) PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol 179: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW, Hollier BG, Ram PT, Lander ES, Rosen JM, Weinberg RA, Mani SA (2010) Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA 107: 15449–15454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ III, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, Wang YH, Reinberg D (2007) L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129: 915–928 [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I, Kitakami J, Ihara S, Hashimoto Y, Hamakubo T, Kodama T, Aburatani H, Sakai J (2009) The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 29: 3544–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD (2009a) Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459: 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, Zhang Y, Li Q, Yi X, Shang Y (2009b) LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138: 660–672 [DOI] [PubMed] [Google Scholar]
- Wu H, Chen Y, Liang J, Shi B, Wu G, Zhang Y, Wang D, Li R, Yi X, Zhang H, Sun L, Shang Y (2005) Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature 438: 981–987 [DOI] [PubMed] [Google Scholar]
- Wu S, Wang W, Kong X, Congdon LM, Yokomori K, Kirschner MW, Rice JC (2010) Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev 24: 2531–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y (2007) PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 25: 801–812 [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117: 927–939 [DOI] [PubMed] [Google Scholar]
- Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ (2008) Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol 10: 295–305 [DOI] [PubMed] [Google Scholar]
- Zhang H, Sun L, Liang J, Yu W, Zhang Y, Wang Y, Chen Y, Li R, Sun X, Shang Y (2006) The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription. EMBO J 25: 4223–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yi X, Sun X, Yin N, Shi B, Wu H, Wang D, Wu G, Shang Y (2004) Differential gene regulation by the SRC family of coactivators. Genes Dev 18: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.