Abstract
Inhibitors of apoptosis proteins (IAPs) are a highly conserved class of multifunctional proteins. Rac1 is a well-studied Rho GTPase that controls numerous basic cellular processes. While the regulation of nucleotide binding to Rac1 is well understood, the molecular mechanisms controlling Rac1 degradation are not known. Here, we demonstrate X-linked IAP (XIAP) and cellular IAP1 (c-IAP1) directly bind to Rac1 in a nucleotide-independent manner to promote its polyubiquitination at Lys147 and proteasomal degradation. These IAPs are also required for degradation of Rac1 upon CNF1 toxin treatment or RhoGDI depletion. Consistently, downregulation of XIAP or c-IAP1 by various strategies led to an increase in Rac1 protein levels in primary and tumour cells, leading to an elongated morphology and enhanced cell migration. Further, XIAP counteracts Rac1-dependent cellular polarization in the developing zebrafish hindbrain and promotes the delamination of neurons from the normal tissue architecture. These observations unveil an evolutionarily conserved role of IAPs in controlling Rac1 stability thereby regulating the plasticity of cell migration and morphogenesis.
Keywords: apoptosis, IAP, Rac1
Introduction
Inhibitors of apoptosis proteins (IAPs) are the only known endogenous inhibitors of caspases as they directly bind to partially processed caspases and prevent their activation (Salvesen and Duckett, 2002). IAPs are characterized by the presence of a Baculoviral IAP repeat (BIR) domain, a highly conserved protein–protein interaction motif (Srinivasula and Ashwell, 2008). Eight mammalian IAPs are known (X-linked IAP (XIAP), cellular IAP1 (c-IAP1), c-IAP2, ML-IAP, Survivin, BRUCE, ILP2 and NAIP) and five of these (XIAP, c-IAP1, c-IAP2, ILP2 and ML-IAP) also possess a RING domain with E3 ubiquitin ligase activity (Vaux and Silke, 2005). Some IAPs (c-IAPs, XIAP, ILP2 and DIAP2) also possess an evolutionarily conserved ubiquitin-associated domain (UBA), which exhibits ubiquitin-binding properties (Blankenship et al, 2008; Gyrd-Hansen et al, 2008; Rajalingam and Dikic, 2009). IAPs have been directly implicated in lymphoproliferative and neoplastic disorders. XIAP mutations were identified in patients with X-linked lymphoproliferative syndrome (XLP), c-IAP2 is associated with mucosa-associated lymphoid tissue (MALT) lymphoma and overexpression of IAPs in tumours is correlated with poor prognosis (Srinivasula and Ashwell, 2008). Further, c-IAP1 has been recently recognized as an oncogene, as amplification of c-IAP1 promoted murine hepatocellular carcinoma in cooperation with c-Myc (Zender et al, 2006; Xu et al, 2007).
One of the current strategies of tumour therapeutics is to specifically downregulate IAPs so that the tumour cells can be sensitized to conventional chemotherapy (Gyrd-Hansen and Meier, 2010). During apoptosis, permeabilization of the mitochondrial outer membrane leads to the release of natural IAP antagonists Smac (Second mitochondrial activator of caspases)/DIABLO (direct IAP binding protein with low pI) and Omi (also called HtrA2), which directly bind to IAPs via a highly conserved N-terminal four residue (AVPI in Smac and AVPS in Omi) IAP binding motif (IBM) (Verhagen et al, 2000; Vaux and Silke, 2003). To this end, several IAP antagonist compounds (IACs) mimicking the N-terminus (AVPI) of the natural IAP antagonist Smac have been developed and some of them are already in clinical trials (Gyrd-Hansen and Meier, 2010). IACs promote degradation of c-IAPs and cell death in a cell type-dependent manner (Varfolomeev et al, 2007; Vince et al, 2007).
Apart from the strong association of IAPs with pathological disorders, the physiological role of IAPs is not well understood. In Drosophila, IAPs are important cell death regulators as loss-of-function mutations in diap1 gene cause spontaneous cell death (Goyal et al, 2000; Lisi et al, 2000). Gene knockout studies in mice revealed that c-IAP1, c-IAP2 and XIAP are dispensable for normal development and survival (Srinivasula and Ashwell, 2008). The absence of overt phenotypes in IAP-deficient mice was initially interpreted to indicate functional redundancy among the IAPs. Recent studies revealed that IAPs also play a crucial role in modulating NF-κB, MAPK signalling, proliferation and migration (Dogan et al, 2008; Gyrd-Hansen et al, 2008; Gyrd-Hansen and Meier, 2010; Liu et al, 2011; Lopez et al, 2011). In this report, we unveil a novel role for IAPs in controlling the protein stability of Rho GTPase, Rac1.
Rho GTPases are a distinct group of the Ras family of small GTPases characterized by the presence of a Rho-specific insert domain located between the fifth β-strand and the fourth α-helix of the GTPase (Vega and Ridley, 2008). Rac1, initially discovered as Ras-related C3 botulinum toxin substrate 1, is ubiquitously expressed and has been shown to play a crucial role in control of the actin cytoskeleton, cell migration, axonal guidance, wound healing and tissue repair, production of superoxide and cellular transformation (Heasman and Ridley, 2008). The Rac family of Rho GTPases comprises Rac1, Rac2, Rac3 and RhoG. The major differences between the family members are found only in the C-terminal sequence. The activity of Rho GTPases is primarily controlled by GEF and GAP proteins and they cycle between the GTP- and GDP-bound forms (Heasman and Ridley, 2008). Apart from nucleotide binding, Rho GTPases can also be modulated by ubiquitination and degradation (Nethe and Hordijk, 2010). While the regulation of nucleotide binding to Rac1 is well understood, the precise molecular mechanisms controlling Rac1 degradation are not known. A very recent study revealed that Sumoylation of Rac1 by PIAS3 is required for maintenance of Rac1–GTP levels and to sustain cell migration (Castillo-Lluva et al, 2010). Smurf1, an HECT domain containing E3 ligase has been shown to mediate polyubiquitination and degradation of RhoA (Wang et al, 2003). Degradation of Rho GTPases was first identified during host–pathogen interactions (Doye et al, 2002; Lerm et al, 2002).
Depending on the cellular background, Rac1 could promote or inhibit tumour invasion and metastasis (Malliri and Collard, 2003; Vega and Ridley, 2008). The cross talk between the Rho GTPases, especially between Rac1 and RhoA controls the plasticity of tumour cell motility as well as epithelial–mesenchymal transition (EMT) in several tumour types (Friedl and Wolf, 2003). While Rho-ROCK signalling plays a more crucial role in an amoeboid mode of migration, high levels of active Rac1 and an elongated morphology represent a mesenchymal mode of migration (Sahai and Marshall, 2003; Sanz-Moreno et al, 2008). Depending on the extracellular cues, tumour cells switch from one mode to the other (Sahai and Marshall, 2003; Wolf et al, 2003; Sanz-Moreno et al, 2008).
Here, we demonstrate that silencing of IAPs led to Rac1 stabilization, elongated morphology and enhanced migration. In addition, we find that XIAP and c-IAP1 function as the E3 ubiquitin ligases of Rac1 directly conjugating polyubiquitin to Lys147 thereby directing Rac1 for its degradation through proteasomes. Further, Rac1 has recently been shown to regulate rhombic lip-derived neuronal differentiation in the developing cerebellum (Tahirovic et al, 2010). Consistent with these findings, Rac1 inactivation specifically in rhombic lip cells of the developing zebrafish cerebellum results in polarity defects and delamination of the progenitor cells, resulting in isolated rounded neural cells or even the accumulation of large ectopic cellular clusters in the cerebrospinal fluid of the hindbrain ventricle. This cell behaviour is phenocopied by enhancing endogenous XIAP expression in the rhombic lip in a Rac1-dependent manner, consistent with IAP-mediated Rac1 degradation. Thus, fine-tuned regulation of Rac1 activity by XIAP is important to ensure the integrity of neural proliferation regions crucial for proper brain development and plasticity of cell migration.
Results
Loss of IAPs led to an elongated morphology in normal and tumour cells
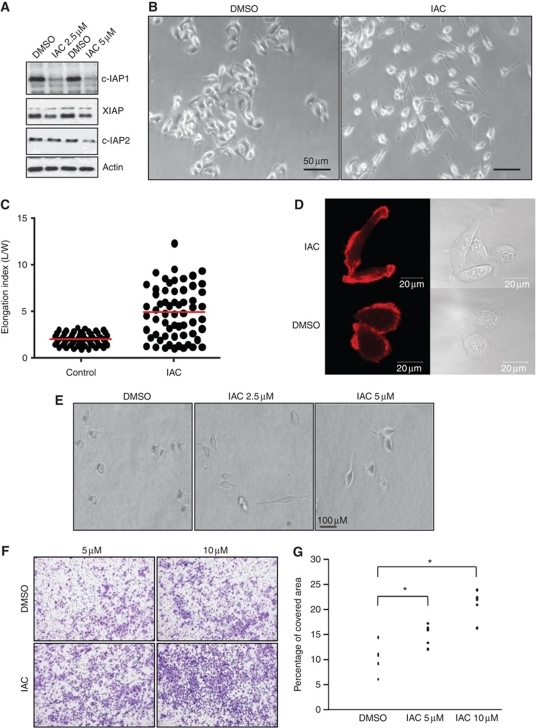
As IAPs are highly expressed in tumours, one of the current strategies of tumour therapeutics is to specifically downregulate IAPs so that the tumour cells can be sensitized to conventional chemotherapy. To this end, a large number of Smac-based IAP antagonists have been developed (Chen and Huerta, 2009; Gyrd-Hansen and Meier, 2010). One class of Smac mimetic compounds (BV6, referred to here as IAC) has been shown to trigger the autoubiquitination of c-IAPs in a time- and concentration-dependent manner leading to TNFα-mediated apoptosis in some cell types (Varfolomeev et al, 2007; Varfolomeev and Vucic, 2008). We have recently demonstrated that depletion of X- or c-IAPs led to stabilization of C-RAF and cell migration in a panel of tumour cell lines (Dogan et al, 2008). To check if treatment with IAC recapitulates these effects, we treated various cancer cell lines with sublethal doses of IAC. Lower doses of IAC triggered a complete loss of c-IAP1 and a partial loss of c-IAP2 and XIAP at 16 h post treatment in HeLa cells (Figure 1A). Interestingly, IAC-treated HeLa cells exhibited highly elongated morphology when seeded onto gelatin or collagen matrices with F-actin-rich protrusions (Figure 1B–D). IAC-treated HeLa cells also exhibited elongated morphology when embedded on a thick layer of collagen (Figure 1E). Transwell migration experiments revealed that IAC-treated cells exhibited enhanced directional cell migration (Figure 1F and G). These results confirmed that IAPs regulate the plasticity of tumour cell motility.
Figure 1.
Depletion of IAPs with IAC induces elongated morphology and migration. (A) HeLa cells were treated with two concentrations of IAC for 16 h and the depletion of various IAPs was monitored by immunoblots. (B) Phase-contrast images of HeLa cells grown on plastic dishes coated with gelatin. The cells were seeded in the presence of DMSO or IAC (5 μM) for 15 h. (C) Elongation index of cells shown in (B). (D) HeLa cells were transfected with LifeAct to stain polymerized actin in live cells. The cells were seeded onto collagen-coated plates and the morphological changes were observed by time-lapse microscopy. (E) Morphological changes of HeLa cells embedded in thick collagen matrices 24 h after IAC treatment. (F) HeLa cells were allowed to migrate in a transwell migration chamber after IAC treatment. The migrated cells were then fixed and stained with crystal violet. Data from one representative experiment are shown. (G) The quantifications of transwell migration experiments from three independent experiments are shown (*P<0.05).
Loss of IAPs led to an increase in Rac1 levels
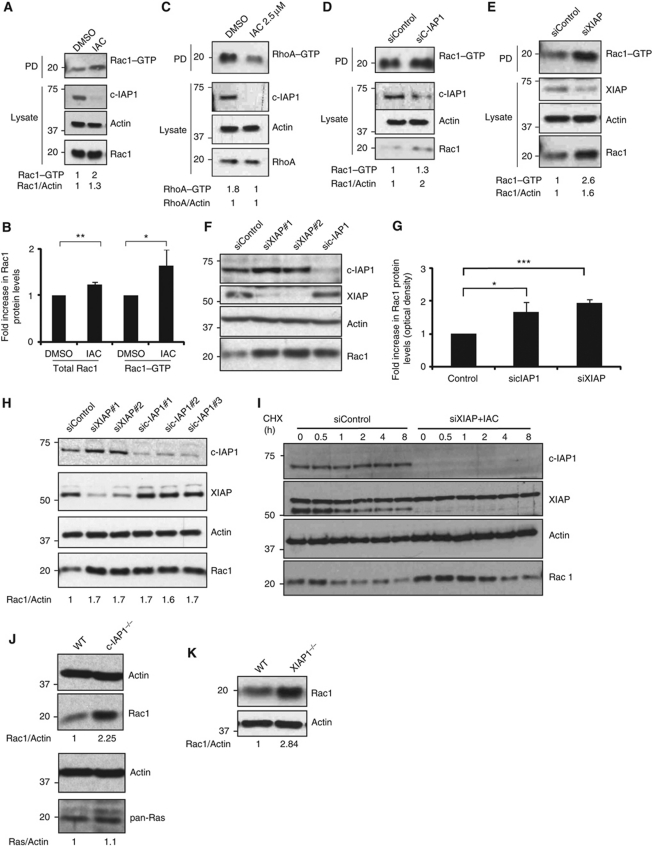
As loss of IAPs can stabilize C-RAF kinase (Dogan et al, 2008), we checked for C-RAF/ERK1/2 activation in cells after challenging them with sublethal concentrations of IAC. As expected, an increase in C-RAF levels with a concordant increase in phosphorylated active forms of C-RAF and ERK1/2 is evident in IAC-treated HeLa and Sbcl2 cells (Supplementary Figure S1A and B). We then examined if RAF/MAPK activation contributes to the elongated morphology in IAC-treated cells. Interestingly, activation of the C-RAF/MAPK pathway is required for IAC-mediated directional migration but not for elongated morphology (Supplementary Figure S1C–E). A mesenchymal mode of migration is controlled by activation of Rac1 or inactivation of RhoA (Sahai and Marshall, 2003). To test this possibility, we performed active Rac1 and RhoA pull-down assays in IAC-treated cells. Consistent with the elongated morphology, we detected relatively high levels of GTP-bound active Rac1 in IAC-treated cells (Figure 2A and B). Interestingly, we detected a modest but reproducible increase in total Rac1 levels in IAC-treated cells (Figure 2A and B). As expected, the amount of GTP-bound RhoA was reduced in IAC-treated cells with no alterations in total RhoA levels (Figure 2C). As IAC treatment can lead to downregulation of c-IAPs and XIAP to various extents, we tested for the role of individual IAPs in regulating Rac1 levels. Intriguingly, an increase in total as well as active Rac1 levels was detected when the cells were transfected with c-IAP1 and XIAP siRNAs (Figure 2D and E). As total Rac1 levels were found to be increased in IAP-depleted cells, we tested the possibility of IAPs directly influencing Rac1 protein stability. As expected, we detected an increase in the protein (∼2-fold) but not in the Rac1 mRNA levels in IAC-treated HeLa cells (Supplementary Figure S2A). Silencing of XIAP or c-IAP1 or both led to a significant increase in total Rac1 protein levels in HeLa cells (Figure 2F and G; Supplementary Figure S2B). This effect is not confined to HeLa cells as an increase in Rac1 protein was readily detected in 293T cells transfected with more than one set of siRNA targeted against XIAP or c-IAP1 (Figure 2H). Identical results were obtained in other tumour and normal cell lines when IAPs are depleted with IAC or siRNAs (Supplementary Figure S2C–J). We have then tested if loss of IAPs led to an increased half-life of Rac1 protein. Combined depletion of XIAP and c-IAP1 enhanced the half-life of endogenous Rac1 protein in HeLa cells (Figure 2I).
Figure 2.
IAPs regulate the stability of Rac1. (A) HeLa cells were treated either with IAC (2.5 μM) or with DMSO for 16 h and the active GTP-bound form of active Rac1 or RhoA (C) was precipitated as mentioned in Materials and methods. (B) Quantification of total and active Rac1 levels (by densitometry as described in Materials and methods) in control and IAC-treated cells is shown. Data from three independent experiments are shown. Error bars represent ±s.d. of the mean. The depletion of c-IAP1 and the total levels of Rac1 and RhoA were monitored in the total lysates. (D) HeLa cells were transfected with c-IAP1 or (E) XIAP siRNAs for 48 h and the amount of GTP-bound Rac1 was precipitated. The levels of total Rac1 were monitored in the cell lysates by immunoblots. (F) HeLa cells were transfected with various siRNAs for 48 h. Total cell lysates were western blotted for monitoring the protein levels of various IAPs and Rac1. (G) The increase in Rac1 levels was quantified by densitometry. The data from three independent experiments are shown (*P<0.05, **P<0.01, ***P<0.005). Error bars represent ±s.d. of the mean. (H) 293T cells were transfected with two sets of XIAP and three sets of c-IAP1 siRNAs. Rac1 levels were monitored by western blots. Quantification of Rac1 increase is provided. (I) Combined loss of XIAP and c-IAP1 enhances the protein half-life of Rac1. HeLa cells transfected with XIAP siRNAs were treated with IAC and cycloheximide was added to cells to monitor the protein half-life of Rac1. (J) Western blot analyses showing protein levels of Rac1 and Ras in wild-type (WT) and c-IAP1−/− or (K) WT and XIAP−/− MEFs. The signalling intensities of Rac1 and Ras bands were quantified by densitometry and normalized to actin levels. Figure source data can be found in Supplementary data.
To further confirm these observations, we checked mouse embryonic fibroblasts (MEFs) derived from mice deficient in various IAPs. Consistently, high levels of Rac1 protein were detected in c-IAP1- and XIAP-deficient MEFs in their early passages (Figure 2J and K). The effect on Rac1 is specific as the related small GTPases Ras or RhoA are not upregulated in c-IAP1-deficient or IAC-treated cells (Figure 2C and J). These results confirmed that IAPs control the protein stability of Rac1 in primary and tumour cells.
Rac1 is required for IAC-induced elongated morphology and cell migration
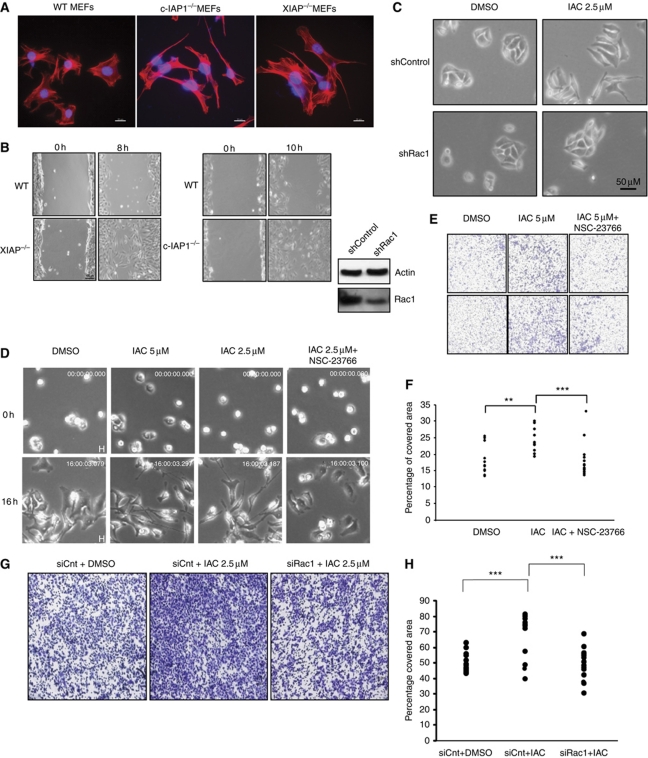
We then examined if Rac1 is required for the enhanced migration and elongated morphology observed in IAP-depleted cells. Consistent with the increase in Rac1 levels, XIAP- and c-IAP1-deficient MEFs exhibited Rac1-dependent elongated morphology and enhanced cell migration (Figure 3A and B; Supplementary Figure S3). Interestingly, c-IAP1-deficient MEFs exhibited a more elongated morphology than XIAP-deficient MEFs. We then tested if the phenotypes we observed in tumour cell lines are dependent on Rac1 levels. Short hairpin RNA-mediated depletion of Rac1 prevented the IAC-induced elongated morphology (Figure 3C). As expected, pretreatment of cells with the Rac1 inhibitor NSC-23766 strongly reduced the elongated morphology and the rate of migration in IAC-treated cells (Figure 3D; Supplementary Figure S4; Supplementary Movies S1,S2,S3,S4). Further, treatment of HeLa cells with Rac1 inhibitor or Rac1 siRNAs also prevented the enhanced migration of IAC-treated cells in transwell migration chambers (Figure 3E–H). These results revealed that depletion of IAPs led to activation and upregulation of Rac1 and inactivation of RhoA, which in turn drove a mesenchymal mode of migration.
Figure 3.
Rac1 is required for IAC-mediated elongated morphology and cell migration. (A) Representative images from wild-type, c-IAP1−/− and XIAP−/− MEFs grown on glass and stained with Phalloidin-TRITC (red). (B) Phase-contrast images at 0 and 8 or 10 h from a representative wound healing experiment performed with wild-type and XIAP−/− or c-IAP1−/− MEFs. (C) Stable cell lines expressing Rac1 or control shRNAs were treated with IAC and the morphological changes were monitored by phase-contrast microscopy. The representative fields from each condition as well as western blot showing the knockdown efficiency of Rac1 shRNA are shown. (D) HeLa cells seeded onto gelatin were treated with DMSO, IAC or IAC+NSC-23766 (50 μM) and observed under a microscope at various time intervals. Cropped snap shots from Supplementary movies are shown. (E) HeLa cells were pretreated with Rac1 inhibitor NSC-23766 and the influence of IAC in mediating transwell migration was monitored. (F) The transwell migration experiments shown in (E) were quantified by computer-assisted algorithms as mentioned in Materials and methods. The data from three independent experiments are shown (**P<0.01, ***P<0.005). (G) HeLa cells were pretreated with control or Rac1 siRNAs and the influence of IAC in mediating transwell migration was monitored. (H) The transwell migration experiments shown in (G) were quantified by computer-assisted algorithms as mentioned in Materials and methods. The data from three independent experiments are shown (**P<0.01, ***P<0.005).
IAPs directly bind to Rac1
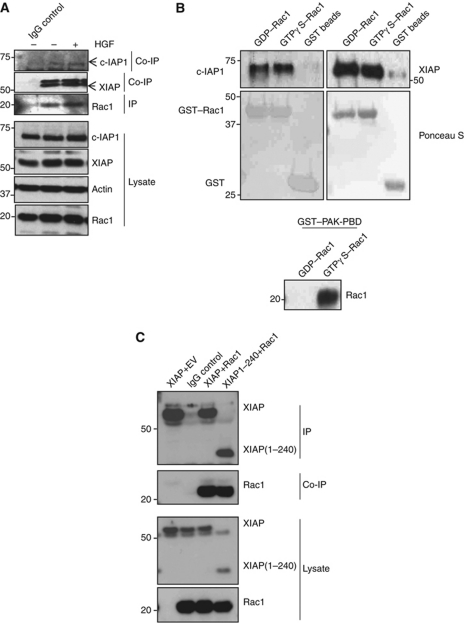
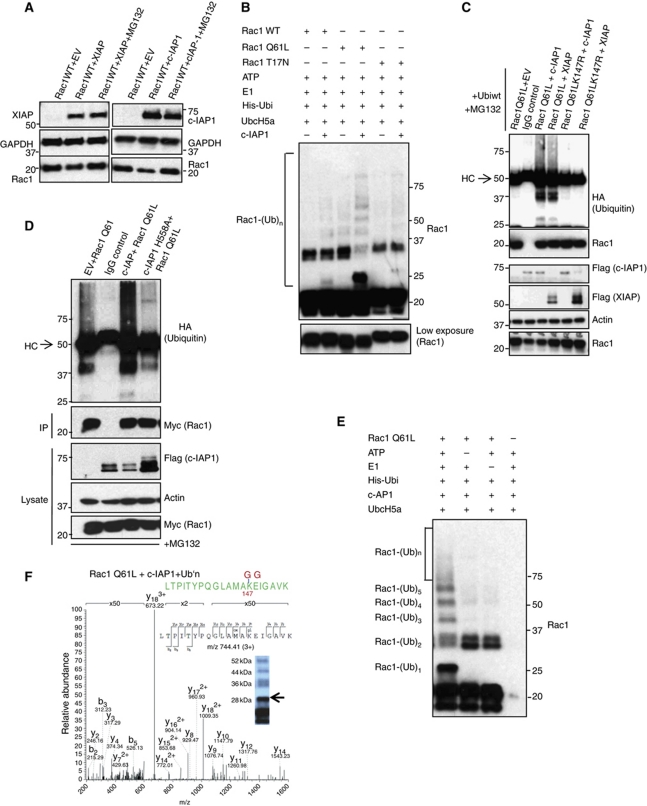
We then explored the molecular mechanisms behind IAP-mediated Rac1 stability. Previous studies with Drosophila DIAP1 revealed a direct interaction between Rac1 and DIAP1 (Geisbrecht and Montell, 2004). As IAPs are highly conserved, we examined if human IAPs can also bind to Rac1. IAP–Rac1 interaction could be detected at endogenous levels, as XIAP and c-IAP1 readily co-precipitated with Rac1 (Figure 4A). In-vitro binding experiments with purified proteins revealed a direct interaction between XIAP or c-IAP1 and Rac1 in a nucleotide-independent manner (Figure 4B). In contrast, GST–PAK1-PBD bound only to Rac1 loaded with GTPγS. These results are consistent with observations made with DIAP1, suggesting that the interaction between IAPs and Rac1 is evolutionarily conserved. Studies with various mutants of XIAP revealed that RING, UBA and BIR3 domains of XIAP are dispensable for interaction with Rac1 (Figure 4C). As Rac isoforms differ only in their C-terminus, we checked if XIAP could interact with Rac2 and Rac3. In-vitro pull-down experiments with purified proteins revealed a direct interaction between XIAP and Rac2 or Rac3 (Supplementary Figure S5A). These results confirmed the direct interaction between IAPs and Rac proteins.
Figure 4.
IAPs can bind to Rac1 in vitro and in vivo. (A) Endogenous interaction between Rac1 and IAPs. HeLa cells grown in 10 cm dishes were stimulated with HGF (100 ng, 15 h) and lysed with appropriate buffer. Endogenous Rac1 was then immunoprecipitated and the immunocomplexes were resolved in an SDS–PAGE and western blotted for co-precipitating XIAP and c-IAP1. (B) In-vitro binding assays with purified proteins showing XIAP and c-IAP1 binding to GST–Rac1 in a GTP/GDP-independent manner. The GST–Rac1 is pretreated with GTPγS and GDP to analyse the dependency of nucleotides in binding with IAPs. The nucleotide loading was confirmed by PAK1-PBD pull-down experiments, which revealed the GTP-specific binding. (C) The interaction between Myc-tagged XIAP or XIAP (1–240) and HA–Rac1 is tested after expressing them in 293T cells. The unspecific binding of antibodies in immunoprecipitation experiments was always checked with respective isotype controls. Figure source data can be found in Supplementary data.
XIAP and c-IAP-1 are the direct E3 ubiquitin ligases of Rac1
To test if IAPs degrade Rac1 in a proteasome-dependent manner, we co-expressed IAP and Rac1 in the presence of proteasome inhibitors. Indeed, inhibition of proteasomes with two different inhibitors prevented IAP-mediated degradation of Rac1 (Figure 5A; Supplementary Figure S5B). Previous studies have revealed that activated Rac1 is predisposed to ubiquitin-dependent degradation and lysine 147 was identified as a potential ubiquitin acceptor for Rac1 (Doye et al, 2002; Lerm et al, 2002; Visvikis et al, 2008). As expected, activated Rac1 was more susceptible to XIAP-mediated degradation (Supplementary Figure S5C). To test if IAPs are the direct E3 ligases of Rac1, we checked whether Rac1 can be directly ubiquitinated by c-IAP1, a strong E3 ligase. Rac1Q61L was ubiquitinated more efficiently in vitro by c-IAP1 in the presence of Ubc5Ha, one of the designated E2s for c-IAP1 (Figure 5B). These results suggest that the activated form of Rac1 is a better ubiquitination substrate, which explains why the Rac1Q61L mutant is more susceptible for degradation. Further, in-vivo ubiquitination experiments in 293T cells revealed that co-expression of XIAP or c-IAP1 promoted the polyubiquitination of activated Rac1, which could be rescued by mutation of lysine 147 (Figure 5C). In line with these, overexpression of XIAP or c-IAP1 led to downregulation of co-expressed Rac1 and not Rac2 or Rac1Q61LK147 or Rac1b (Supplementary Figure S5D–F). Interestingly, expression of wild type but not c-IAP1H588A enhanced the polyubiquitination of activated Rac1 in vivo (Figure 5D; Supplementary Figure S6A). Consistently, the RING mutant of c-IAP1 strongly reduced the decay rate of Rac1 (Supplementary Figure S6B). To further confirm these observations, we performed mass spectrometric analysis, which revealed that c-IAP1 and XIAP can conjugate ubiquitin directly to lysine 147 of activated Rac1 (Figure 5E and F; Supplementary Figure S6C and D). Interestingly, c-IAP1 ubiquitinated RacQ61L more efficiently than XIAP with Ubc5Ha as an E2 enzyme (Supplementary Figure S6C). These results revealed that XIAP and c-IAP-1 are the direct E3 ubiquitin ligases of Rac1.
Figure 5.
Binding of IAPs to Rac1 promotes Rac1 polyubiquitination and degradation. (A) The rescue in the levels of Rac1 under the influence of proteasomal inhibitors MG132 upon IAP overexpression is monitored by western blotting. (B) c-IAP1 directly ubiquitinates Rac1. Purified recombinant Rac1 proteins with various mutations were subjected to in-vitro ubiquitination with c-IAP1 as described in Materials and methods. The conjugation of ubiquitin to Rac1 was monitored by immunoblots. The lower exposure of Rac1 was shown below. (C) XIAP and c-IAP1 promoted the polyubiquitination of activated Rac1. 293T cells were transfected with Myc–Rac1Q61L or Myc–Rac1Q61LK147R in combination with Flag–IAPs and HA–ubiquitin constructs. Rac1Q61L was immunoprecipitated by using Myc antibody and the immunocomplexes were western blotted for HA (ubiquitin), Rac1 and Flag for IAPs. The cells were treated with MG132 for 6 h before lysing. (D) Ubiquitination of Myc–Rac1Q61L by c-IAP1 and c-IAPH588A in 293T cells is analysed as mentioned in (C). (E) c-IAP1 directly ubiquitinates Rac1 at lysine 147. Recombinant Rac1Q61L was subjected to an in-vitro ubiquitination reaction as mentioned in Materials and methods with c-IAP1. The modification of Rac1 protein was monitored by immunoblots. (F) Samples from in-vitro ubiquitination reaction of Rac1 with c-IAP1 (E) were subjected to mass spectrometric analysis after digestion with trypsin. MS/MS spectrum of the Rac1 peptide carrying Gly-Gly modification at position K147 is shown. The mass of the triply charged precursor ion at 744.41 was measured with a mass deviation of 0.08 p.p.m. Arrow indicates the gel band where the peptide was detected. Figure source data can be found in Supplementary data.
IAPs are required for CNF1 toxin- and RhoGDI depletion-mediated Rac1 degradation
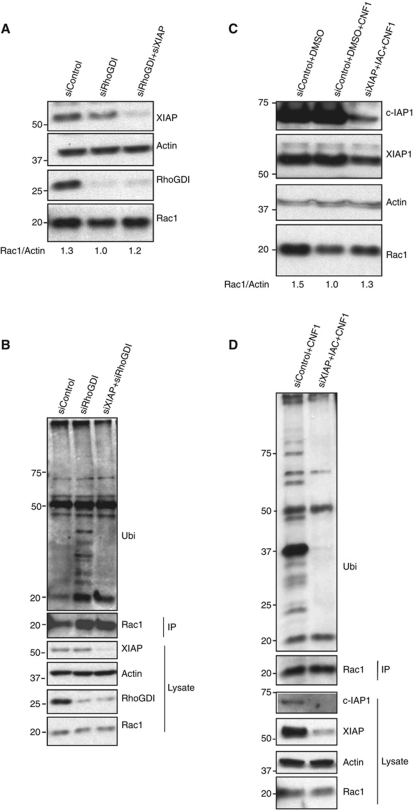
We then set out to exploit a system where Rac1 ubiquitination and proteasomal degradation can be accomplished at endogenous levels. Recent studies revealed an evolutionarily conserved role of RhoGDI1 in regulating the homeostasis of Rho GTPases. Keith Burridge and colleagues demonstrated that upon depletion of RhoGDI1 by siRNAs, several Rho GTPases including Rac1 are dramatically degraded in a proteasome-dependent manner (Boulter et al, 2010). We tested if IAPs are required for degradation of Rac1 under these settings. Indeed, combined depletion of XIAP and RhoGDI1 rescued Rac1 protein levels in these cells (Figure 6A). Consistent with these results, RhoGDI depletion promoted the ubiquitination of endogenous Rac1, which could be fully rescued by the co-depletion of XIAP (Figure 6B). Further, we employed purified CNF1 toxin from enteropathogenic E. coli to target Rac1 for degradation (Lerm et al, 2002). Treatment of 293T cells with 200 ng of CNF1 toxin led to a reduction in Rac1 levels at 16 h post treatment. As expected, loss of XIAP and c-IAP1 resulted in a rescue of CNF1-mediated polyubiquitination and proteasomal degradation of endogenous Rac1 (Figure 6C and D). These results reaffirmed the crucial in-vivo role of IAPs in controlling the stability of Rac1.
Figure 6.
IAPs are required for polyubiquitination and proteasomal degradation of endogenous Rac1. (A) HeLa cells were transfected with control, XIAP and RhoGDI siRNAs. The levels of various proteins were monitored by immunoblots. (B) HeLa cells were transfected with control, RhoGDI or RhoGDI and XIAP siRNAs. Endogenous Rac1 was immunoprecipitated and the conjugation of polyubiquitin chains to Rac1 was monitored by immunoblots. The cells were treated with MG132 for 6 h before lysing to preserve ubiquitinated Rac1. (C) IAPs are required for CNF1-mediated Rac1 degradation. 293T cells transfected with siRNAs and/or treated with IAC are further treated with 200 ng/ml of CNF1 toxin for 6 h. The levels of Rac1 and IAPs are monitored. (D) 293T cells transfected with control or XIAP siRNAs+IAC are treated with CNF1 toxin as mentioned earlier. Endogenous Rac1 was immunoprecipitated and conjugation of ubiquitin is monitored. The cells were treated with MG132 for 6 h before lysing. Figure source data can be found in Supplementary data.
XIAP overexpression in zebrafish rhombic lip cells phenocopies defects caused by Rac1 inactivation
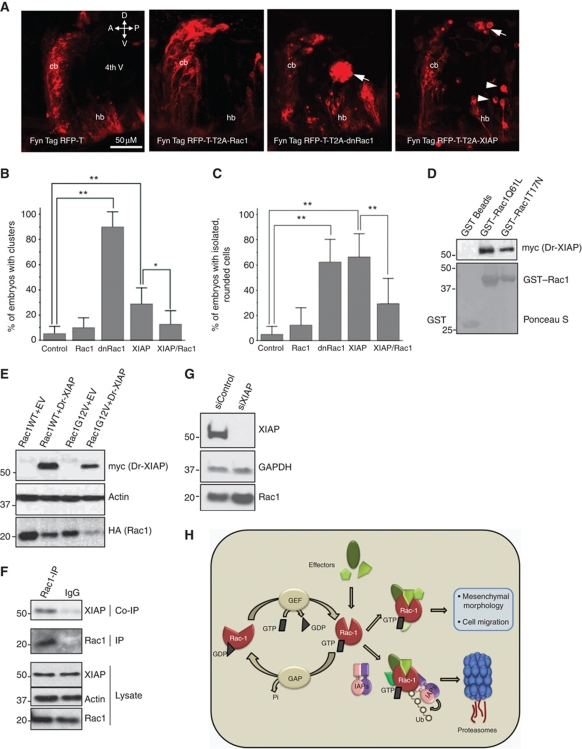
To reveal genetic interactions between Rac1 and XIAP, we have resorted to characterize the cellular integrity in a prominent proliferation zone of the developing cerebellum, the upper rhombic lip (URL). Recent in-vivo time-lapse studies in zebrafish embryos revealed the importance of neuronal progenitor polarization in this germinal zone. These cells initiate highly directional collective migration over long distances to finally settle in the hindbrain tegmentum and later in the granule cell layer of the cerebellum. Cerebellar rhombic lip-derived cells give rise to neurons of several tegmental hindbrain nuclei and granule neurons of the cerebellar cortex, respectively. Thus, this proliferation zone is of major relevance to vertebrate development (Koster and Fraser, 2001; Rieger et al, 2009; Volkmann et al, 2010). Furthermore, neural cells of this germinal zone give rise to the most common childhood brain tumour (medulloblastoma), which disseminates through delamination into the cerebrospinal fluid of the ventricle. As Rac1 is also implicated in regulating cerebellar development in vertebrates, we set out to test the potential role of XIAP in this morphogenetic process (Tahirovic et al, 2010). We turned to the stable transgenic zebrafish strain Tg(atoh1a:Gal4TA4)hzm2 which expresses the Gal4 variant KalTA4 under control of an atonal1a regulatory element and can be used to generate conditional expression of transgenes in rhombic lip cells in a mosaic manner (Distel et al, 2010). A Gal4-dependent expression construct driving a dominant-negative Rac1 (T17N) variant together with the membrane-targeted fluorescent protein FynTagRFP-T was injected into one-cell stage Tg(atoh1a:Gal4TA4)hzm2 embryos and fluorescent cells were analysed at 48 h.p.f. In around 90% of Rac1T17N expressing embryos (n=62; six experiments), clusters of fluorescent rhombic lip cells were observed in the hindbrain ventricle likely due to cell polarity defects, something rarely observed in control embryos expressing FynTagRFP-T (5±6%) (Figure 7A and B). Strikingly, an increase in endogenous zebrafish XIAP (Dr-XIAP) expression in rhombic lip cells by use of an analogous Gal4-dependent Dr-XIAP/TagRFP-T expression construct resulted in similar ectopic cluster formation in the hindbrain ventricle in 29±13% of expressing embryos (Figure 7A and B). The fact that fewer of the Dr-XIAP-expressing embryos extruded clusters of cells into the ventricle as compared with those expressing Rac1T17N is not surprising considering that Dr-XIAP-mediated Rac1-depletion requires several hours and that residual Rac1 activity is sufficient to ensure proper cerebellar development, as heterozygous Rac1-deficient mice display no obvious cerebellar malformation (Tahirovic et al, 2010). However, many of the Dr-XIAP-expressing embryos did show progenitor cells with rounded morphology similar to cells within the clusters and those cells were often ectopically located within the ventricle (Figure 7A, white arrowheads). This precursor state of ectopic cluster formation was observed in about 65% of embryos expressing either Rac1 or Dr-XIAP, compared with 5% of unaffected controls (Figure 7C). Notably, the cells extruded from the tissue, whether as clusters or individually, were not simply dying cells, as the majority of the abnormal Dr-XIAP and dnRac1-expressing cells were not labelled by acridine orange (Supplementary Figure S7A). To further test whether the phenotype observed in Dr-XIAP-overexpressing progenitor cells is dependent on Rac1, we performed rescue experiments for which we generated additional double and triple transgene expression constructs to co-express membrane-targeted FynTagRFP and Rac1 or FynTagRFP/Dr-XIAP/Rac1 in a Gal4-dependent manner. These experiments revealed that overexpression of Rac1 alone does not result in obvious aberrations of rhombic lip and cerebellum development (Figure 7A). As expected, simultaneous elevation of Rac1 levels to Dr-XIAP expression was able to rescue the delamination phenotype and significantly reduced the number of both Dr-XIAP-induced ectopic cell clusters and rounded cells in the cerebrospinal fluid of the zebrafish hindbrain ventricle, indicating a genetic interaction of Dr-XIAP and Rac1 in vivo (Figure 7B and C).
Figure 7.
Expression of dnRac1 or XIAP in the hindbrain rhombic lip causes cells to delaminate into the ventricle. (A) Lateral view of the cerebellum and anterior hindbrain of 48 h.p.f. Tg(atoh1a:Gal4TA4)hzm2 embryos expressing FynTagRFP-T, FynTagRFP-T/Rac1, FynTagRFP-T/dnRac1 or FynTagRFP-T/XIAP. The cells were found either as isolated cells (arrowheads) or as clusters (arrows) in the cerebrospinal fluid of the ventricle. Abbreviations: 4th V; 4th ventricle; A, anterior; cb, cerebellum; D, dorsal; hb, hindbrain; P, posterior; V, ventral. (B) Graph showing the percentage of embryos in each condition with one or more abnormal TagRFP-T-expressing cell cluster. (C) Graph showing the percentage of embryos with one or more isolated, rounded TagRFP-T-expressing cells. For both graphs, n=6 independent experiments, one-way ANOVA and Tukey’s post hoc test. **P<0.01, *P<0.05. Error bars represent ±s.d. (D) Zebrafish XIAP binds to Rac1. Dr-XIAP produced in rabbit reticulolysates was subjected to GST pull-down experiments with either GST or GST–RacQ61L or with GST–RacT17N. (E) Dr-XIAP and Rac1 constructs were expressed in 293T cells and the degradation of Rac1 was monitored in the immunoblots. (F) Rac1 was immunoprecipitated from immortalized cerebellar granule neurons from mice and the co-precipitation of XIAP was monitored by immunoblots. (G) mCGN cells were transfected with XIAP siRNA and the increase in total Rac1 levels was monitored by immunoblots. (H) Proposed model for IAP-mediated degradation of Rac1. GEFs and GAPs control the binding of GTP/GDP to Rac1. XIAP–c-IAP1 complex directly binds to Rac1 and promote the conjugation of polyubiquitin chains to Lysine147 of activated Rac1 in vivo. Loss of IAPs stabilizes Rac1 protein leading to elongated/mesenchymal mode of migration in normal and tumour cells. Figure source data can be found in Supplementary data.
To confirm that Dr-XIAP can regulate Rac1 directly, we tested for a Dr-XIAP–Rac1 interaction. Indeed, zebrafish XIAP strongly binds to Rac1 in a nucleotide-independent manner (Figure 7D). Further, expression of Dr-XIAP, but not GFP, mRNA led to a decrease in the level of total Rac1 protein in zebrafish embryos (Supplementary Figure S7B). In line with these observations, expression of Dr-XIAP in 293T cells led to degradation of human Rac1 (Figure 7E).
To further test if XIAP and Rac1 can interact at endogenous levels in URL cells, we obtained immortalized cerebellar granule neurons (CGNs) from postnatal day 4 (P4) cerebella of mice. Intriguingly, we could nicely co-precipitate XIAP with Rac1 in these cells (Figure 7F). Further, siRNA-mediated depletion of XIAP led to an increase in endogenous Rac1 levels in these cells (Figure 7G). These results reveal an evolutionarily conserved role of IAPs in regulating Rac1 degradation. As XIAP is highly expressed in medulloblastomas, it is tempting to propose that augmented XIAP expression in rhombic lip cells may predispose these cells to tumour formation.
Discussion
The work reported here demonstrates a direct role for IAPs in controlling the stability of Rac1, tissue integrity and plasticity of cell migration. Despite evidence that Rho GTPases are regulated by ubiquitination and degradation, the E3 ubiquitin ligase(s) responsible for Rac1 ubiquitination and degradation are not known (Nethe and Hordijk, 2010). Our studies reveal the molecular mechanisms behind Rac1 degradation, a crucial mode of Rac1 regulation apart from GEFs, GAPs and GDIs. Both XIAP and c-IAP1 can conjugate polyubiquitin chains directly to lysine 147 of activated Rac1 both in vitro and in vivo and target them for proteasomal degradation. Consistent with these biochemical data, depletion of physiological levels of these IAPs by RNAi-mediated knockdown or treatment of cells with IACs or genetic deletion led to an increase in the protein levels of endogenous Rac1 leading to elongated cell morphology and enhanced cell migration. In these lines, loss of IAPs also prevented polyubiquitination of endogenous Rac1 when the cells are treated with CNF1 toxin or RhoGDI depletion, which promotes proteasomal degradation (Figure 6). The role of IAPs in controlling Rac1 functions is possibly evolutionarily conserved, as overexpression of DIAP1 has been previously demonstrated to prevent RacN17-mediated border cell migration defects during Drosophila development though the mechanisms are not known (Geisbrecht and Montell, 2004). Consistent with the observations made with DIAP1, we detect that human and zebrafish IAPs can also bind to Rac1 in a nucleotide-independent manner. Loading of Rac1 with either GDP or GTP fails to impair the interaction of Rac1 with XIAP or c-IAP1 in vitro, suggesting that IAPs are possibly targeting the C-terminus of Rac1-like SET or β-pix through their BIR domains.
Our data reveal that the polyubiquitination of Rac1 is dependent on the RING domain of c-IAP1. However, XIAP is still needed as loss of either XIAP or c-IAP1 or both has led to a similar increase in Rac1 protein levels, indicating functional non-redundancy among these IAPs in controlling Rac1 stability. Though XIAP and c-IAP1 can directly conjugate ubiquitin chains to Rac1 in the presence of Ubc5Ha in vitro, a XIAP–c-IAP1 complex might function as a more efficient E3 ligase in vivo to recruit the same or different E2s to ubiquitinate Rac1 in cells (Figure 7H). IAPs have been shown to form homomers and heteromers (IAPosomes) and RING-mediated heteromerization between XIAP and c-IAP1 has been demonstrated before (Silke et al, 2005; Rajalingam et al, 2006). Recent studies with RIP2, another substrate of c-IAPs also revealed a non-redundant role for these IAPs in controlling RIP2 ubiquitination supporting an emerging theme of IAP–IAP heteromers functioning in the ubiquitination of their substrates (Bertrand et al, 2009; Mace et al, 2010). As several kinds of ubiquitin chains can be synthesized by IAPs, it would be interesting to characterize the kind of ubiquitin chains synthesized by IAPs on Rac1 and the E2 enzymes exploited for the same. As IAPs can auto- and cross-ubiquitinate themselves, it would indeed be a challenging task to decipher the intricate machinery controlling the conjugation of specific chains and selection of substrates to regulate various cellular processes (Vaux and Silke, 2005).
XIAP–c-IAP1-mediated ubiquitination of Rac1 might be spatially regulated by binding to other proteins like Caveolin and RhoGDI (Nethe and Hordijk, 2010; Figure 7H). Further, XIAP and c-IAP1 could target only a proportion of Rac1 in a localized manner to regulate migration. RhoGDI depletion primarily leads to the degradation of cytosolic Rac1 (Boulter et al, 2010) and XIAP is required for this process (Figure 6A and B). This may also contribute to the not so robust increase in Rac1 levels upon IAP depletion in some cell types. Previous studies have revealed that the polybasic region (PBR), nuclear localization, isoprenylation and binding to other proteins like Karyopherin alpha2 are required for the degradation of Rac1 (Lanning et al, 2004; Pop et al, 2004; Sandrock et al, 2010). Along these lines, it is interesting to indicate that c-IAP1 was shown to be localized to the nuclear compartment in many cell types (Samuel et al, 2005; Cartier et al, 2011). Clearly, the spatiotemporal dynamics of the Rac1–c-IAP1 interaction need further elucidation.
As Rac1 controls numerous cellular processes, it is indeed surprising that mice deficient in individual IAPs lack any major phenotype(s), which indicates the possible presence of redundant E3 ligase(s) for Rac1. RhoA, for instance, has been shown to be ubiquitinated by Smurf1 as well as by Cullin-RING ubiquitin ligases, and Rac1 has been recognised as a chaperone client (Wang et al, 2003; Chen et al, 2009; Boulter et al, 2010). We propose that in the absence of IAPs (XIAP and/or c-IAP1), the increase in Rac1 protein levels may be sensed and cleared off by the more general ‘protein quality control’ system to maintain homeostasis. Further, IAPs cross-regulate each other and it is well known that loss of one IAP leads to an increase in the protein levels of other IAPs. In this connection, it is interesting to point out that the robust increase in Rac1 levels observed in XIAP-deficient MEFs is reduced in later passages possibly due to the upregulation of c-IAP1 or by the activation of chaperone machinery. Thus, we expect serious developmental defects in XIAP—c-IAP-1 double-deficient mice.
Our studies in zebrafish cerebella development, however, reveal a role of XIAP in regulating progenitor cell polarity as augmenting XIAP expression leads to delamination of these cells that can be rescued by Rac1 co-expression. Further, neural cells of this germinal zone primarily express Rac1 isoform and give rise to the most common childhood brain tumour (medulloblastoma), which disseminates through delamination into the cerebrospinal fluid of the ventricle—a resemblance to the phenotype observed by our mosaic manipulation of Rac1 function. It is interesting to note that cell lines derived from medulloblastomas such as DAOY or ONS-76 express high levels of XIAP and our observations suggest that augmenting XIAP expression may predispose these cells to tumour formation.
IAP-depleted tumour cells exhibited an elongated morphology and mesenchymal mode of movement with protruding lamellipodia in a Rac1-dependent manner. These effects are evident even in cells with mixed morphologies, like WM852 cells, as treatment of these cells with IAC led to further enhancement in their elongated morphology (data not shown). As IAPs control Rac1 and C-RAF protein levels, one major concern is that treatment with IAP antagonists may unexpectedly promote tumour cell migration and metastasis (especially when the tumour cells failed to undergo apoptosis). However, this possibility needs to be carefully verified as Rac1 signalling influences sequential stages of tumour cell invasion (Malliri and Collard, 2003). Further, a mesenchymal mode of migration limits lung colonization of tumour cells as Rac1 signalling can lead to promotion or inhibition of tumour dissemination depending on the cell/tissue background (Malliri and Collard, 2003; Sanz-Moreno et al, 2008). In these lines, Lopez et al (2011) observed reduced migration upon c-IAP1 depletion, which could be attributed to the difference in the cell types employed and siRNA-mediated depletion of XIAP in MCF-7 cells has led to an increase in Rac1 levels (Supplementary Figure S2H). Further, loss of IAPs can also influence Rho signalling by stabilizing C-RAF kinase (Ehrenreiter et al, 2005). IAPs are recognised as direct metastasis genes and the Survivin–XIAP heteromer has been shown to promote liver metastases of cells derived from breast and prostate adenocarcinomas (Mehrotra et al, 2010). In another study, loss of XIAP has actually led to an aggressive form of disease in an immune-competent mouse prostate cancer model (Hwang et al, 2008). Our current observations are very much relevant to these phenotypes as IAP-mediated Rac1 regulation might contribute to the aetiology of Rho-ROCK-mediated tumour cell invasion and metastases.
Our studies also have direct implications in IAP-based tumour therapeutics and assist in selection of patients for treatment with this class of novel drugs. As IAPs influence pro-survival pathways, a more thorough study of long-term responses of various tumours to sublethal doses of IAC is clearly warranted. In conclusion, our results reveal an evolutionarily conserved side of IAPs in regulating Rac1, a Rho GTPase that influences several pathways of fundamental significance to numerous cellular processes. As Rac1 also plays an important role in tissue regeneration (Dipersio, 2007), we propose that IAP antagonists might be pursued for treating tissue injury.
Materials and methods
Transfection of siRNAs and plasmids
In order to silence XIAP, c-IAP1 and Rac1 expression by RNA interference, ∼75 000 cells/well were seeded in a 12-well plate at least 20 h prior to transfection. siRNAs directed against various genes and scrambled control siRNA as negative control were transfected using the Hiperfect (Qiagen) or DharmaFECT®Duo (Dharmacon) or Lipofectamine RNAimax (Invitrogen) transfection kits. At 24 h post transfection, cells were trypsinized and one half of the cells was seeded out on glass coverslips in a 12-well plate for immunofluorescence analysis if needed, while the other half was seeded for western blot analysis. The cells were normally lysed at 48 h post transfection. Unless otherwise mentioned, we have transfected siRNAs at a final concentration of 60 nM. The following siRNAs were employed in this study:
XIAP siRNA-1: Validated siRNA, Catalogue No: SI00299446 (Qiagen);
XIAP3′UTR siRNA: Target Sequence: CTGACTGATCTAATTGTATTA (Qiagen);
c-IAP1 siRNA-1: J-004390-12-0010 (Dharmacon on target plus siRNA);
c-IAP1 siRNA-2: Target sequence: CTAGGAGACAGTCCTATTCAA (Qiagen); c-IAP1 siRNA-3: J-004390-13-0010 (Dharmacon on target plus siRNA);
Rac1 mouse siRNA: Thermo Scientific Cat #J-041170-06;
RhoGDI1 siRNA: 5′-UCAAUCUUGACGCCUUUCCTT-3′ (Sigma-Genosys);
Control siRNA: Catalogue No: D-001210-01-20 (Dharmacon);
Rac1 siGENOME SMARTpool siRNA: Catalogue No: M-003560-06-0005 (Dharmacon);
Rac1 shRNA: (TRCN0000004872; Sigma-Aldrich) CCGGCCCTACTGTCTTTGACAATTACTCGAGTAATTGTCAAAGACAGTAGGGTTTTT.
Lentiviral particles carrying control and Rac1 shRNAs were obtained from Sigma and the stable cell lines were produced following the manufacturer's instructions.
For the co-knockdown experiments, the cells were transfected with siRNAs at a final concentration of 60 nM each.
Detection of Rac1/RhoA activity
HeLa cells were treated with various concentrations of IAC for 15 h, or transfected with various siRNAs for 48 h. The GTP-bound Rac1 or RhoA was precipitated using EZ-Detect™ Rac1/RhoA Activation Kit (PIERCE) following the manufacturer's instructions.
Transwell migration experiments
HeLa cells were transfected with siRNAs as described above. HeLa cells (2 × 105) or MEFs either transfected with siRNAs for 36 h or cells treated with various concentrations of IAC were first treated with 10 μM of U0126 for 2 h if needed, and then seeded onto 8 μm transwell migration chambers (Corning, #3422) and the membranes were precoated with 0.1% Collagen type I derived from Calf skin (Sigma, #C8919). The cells were stimulated with serum-free RPMI, or RPMI with 10% FCS, added to the lower chamber. The cells were left to migrate for 12 h or overnight. Cells on the upper part of the membrane were scraped using a cotton swab and the migrated cells were fixed in 3.7% (v/v) paraformaldehyde and stained with 0.4% Crystal Violet in 10% ethanol. The experiment was performed in triplicates for all conditions described. From every transwell, several images were taken under a phase-contrast microscope at × 10 magnification and two broad fields were considered for quantification. Analyses were performed using two special algorithms as described before (Dogan et al, 2008). The results of the analysis of the individual photos are depicted as dots in the various graphs. Student's t-test was performed to check for the significance (*P<0.05, **P<0.01, ***P<0.005).
Zebrafish injection and microscopy
Zebrafish were maintained as described (Westerfield, 1995). Zebrafish embryos were injected with expression plasmids (25 ng/μl each together with 25 ng/μl mRNA encoding Tol1 transposase, 1.5 nl) at the one-cell stage. Raised embryos were screened for expression at 48 h.p.f., dechorionated, fixed overnight in 4% paraformaldehyde/PBS, and subsequently rinsed and stored in PBS at 4°C. For image recording, embryos were embedded in 1.2% ultra low melting point agarose and confocal Z-stacks were recorded with a Zeiss LSM510 confocal microscope. For acridine orange staining, live transgene-expressing embryos were soaked at 48 h.p.f. for 1 h in acridine orange (Invitrogen) diluted 1:1000 in 30% Danieau's, 0.15 mM PTU, 0.01% Tricaine, washed 3 × 1 min in media without dye, and embedded and imaged as described above.
Ubiquitination experiments
For detecting the ubiquitination of Rac1 in vivo, 293T cells were transfected with myc–Rac1Q61L in combination with HA-Ubi wt, with or without XIAP, c-IAP1 and c-IAP1H588A, using Polyethylenimine (Polysciences) reagent at a final concentration 1 μg/ml for 48 h. The cells were lysed on the plate with lysis buffer containing: 50 mM Tris–HCl pH 7.5, 250 mM NaCl, 10% glycerol, 1% Triton-X, 0.01% 2-mercaptoethanol, 1 mM sodium orthovanadate, 1.5 mM MgCl2, 25 mM sodium fluoride, 0.01% leupeptin, 0.01% aprotinin, 2 mM pepstatin and 1 mM phenylmethylsulphonylfluoride (PMSF) and were incubated on rotator for 30 min at 4°C. Lysates were cleared by centrifugation for 15 min at 14 000 r.p.m. Supernatants were incubated with appropriate antibodies for 15 h. The antigen–antibody complexes were precipitated by sepharose coupled protein A/G beads (Roche). The beads were washed with the lysis buffer, and bound proteins were analysed by SDS–PAGE and immunoblotting. To avoid the loss of ubiquitinated proteins, the cells were treated with 10 μM of proteasome inhibitor MG-132 (Calbiochem) for 5–6 h before lysis.
His–ubiquitin pull-down assay was performed according to previously published protocol (Song et al, 2010). Briefly, 293T cells were co-transfected with pRK5 myc–Rac1Q61L, pCS2 His–ubiquitin, with or without CMV Flag–c-IAP1 in 10 cm dishes. Cells were treated with MG132 for 6 h before pull down. Forty-eight hours post transfection, cells were lysed in Buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM Imidazole, pH 8.0) and sonicated. His–ubiquitin pull-down was carried out by using 50 μl of equilibrated Ni-NTA agarose per sample for 3 h at room temperature. Finally, beads were washed under denaturating conditions and conjugated proteins eluted in 50 μl of 2 × Laemmli/Imidazole and boiled. Eluates were analysed by western blot analyses using anti-c-myc antibodies.
In-vitro ubiquitylation of Rac1 was performed with Rac1Q61L purified from bacteria. Ubiquitylation reaction was performed in the presence of Ubiquitylation buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2, 1 mM DTT), 100 nM c-IAP1 (R&D Systems), 100 nM E1 (Boston Biochem), 150 μM UbcH5a (Boston Biochem), 107 μM His–Ubiquitin (Boston Biochem), 50 mM EDTA, 1 × Mg-ATP (Enzo Life Sciences), 1 U inorganic pyrophosphatase (Fluka) and 50 mM DTT. The reaction was incubated at 37°C for 1 h. For western blot analyses, the reaction was stopped using Laemmli buffer and Rac1 was visualized using Rac1 antibody (C11, Santa Cruz).
Cell culture matrices
Cells were also cultured on dishes or plates coated with 0.1% (1 mg/ml) Collagen (Sigma, #C8919) or 0.1% of Gelatin (Sigma, #G2500) or with 1.6–2.4 mg/ml collagen (Advanced BioMatrix, #5005) for thin and thick layers of collagen matrices to study 2D effects of IACs. For testing the effects of IACs on 3D matrices, HeLa cells were mixed with collagen (Millipore, #ECM 675) and then added onto cell culture plates according to the manufacturer's instructions. The cells embedded in the matrix were imaged for 3D effects.
Supplementary Material
Acknowledgments
We would like to thank Genentech–Roche for kindly providing BV6-IAC compound. We thank Sabine Haeder and Kristina Wagner for excellent technical assistance; Domagoj Vucic, Erez Raz, Richard Marais, Reza Ahmadian, Harald Wajant, Michael Rape and Metello Innocenti for various reagents and cell lines; David Vaux, Diep Chau and John Silke for kindly providing various IAP-deficient MEFs; and P Friedl and E Sahai for advise on collagen matrices. We thank Ivan Dikic for the critical reading of the manuscript and for his support. This work is supported by an Emmy Noether programme grant RA1739/1-1 to KR from the DFG and by funds from the Helmholtz Association and the DFG (KO1949/3-1) to RWK.
Author contributions TKO and TD performed most of the experiments and analysed interpreted data. JM, RPS and CLA performed additional experiments. RKW, JCH and KN conceived-performed zebrafish experiments and analysed interpreted data. CK, DMH, GS, GSH and MR provided valuable reagents/materials and contributed to microscopy/data analysis. AC and BM performed mass spectrometric analysis. KR conceived-designed the project, analysed interpreted data and wrote the paper with inputs from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M (2009) Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity 30: 789–801 [DOI] [PubMed] [Google Scholar]
- Blankenship JW, Varfolomeev E, Goncharov T, Fedorova AV, Kirkpatrick DS, Izrael-Tomasevic A, Phu L, Arnott D, Aghajan M, Zobel K, Bazan JF, Fairbrother WJ, Deshayes K, Vucic D (2008) Ubiquitin binding modulates IAP antagonist stimulated proteasomal degradation of c IAP1 and c IAP2. Biochem J 417: 1–3 [DOI] [PubMed] [Google Scholar]
- Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K (2010) Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat Cell Biol 12: 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier J, Berthelet J, Marivin A, Gemble S, Edmond V, Plenchette S, Lagrange B, Hammann A, Dupoux A, Delva L, Eymin B, Solary E, Dubrez L (2011) Cellular inhibitor of apoptosis protein-1 (CIAP1) can regulate E2F1-mediated control of cyclin transcription. J Biol Chem 29: 26406–26417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Lluva S, Tatham MH, Jones RC, Jaffray EG, Edmondson RD, Hay RT, Malliri A (2010) SUMOylation of the GTPase Rac1 is required for optimal cell migration. Nat Cell Biol 12: 1078–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DJ, Huerta S (2009) Smac mimetics as new cancer therapeutics. Anti-Cancer Drugs 20: 646–658 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F (2009) Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell 35: 841–855 [DOI] [PubMed] [Google Scholar]
- Dipersio CM (2007) Double duty for Rac1 in epidermal wound healing. Sci STKE 2007: e33. [DOI] [PubMed] [Google Scholar]
- Distel M, Hocking JC, Volkmann K, Koster RW (2010) The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J Cell Biol 191: 875–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan T, Harms GS, Hekman M, Karreman C, Oberoi TK, Alnemri ES, Rapp UR, Rajalingam K (2008) X-linked and cellular IAPs modulate the stability of C-RAF kinase and cell motility. Nat Cell Biol 10: 1447–1455 [DOI] [PubMed] [Google Scholar]
- Doye A, Mettouchi A, Bossis G, Clement R, Buisson-Touati C, Flatau G, Gagnoux L, Piechaczyk M, Boquet P, Lemichez E (2002) CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell 111: 553–564 [DOI] [PubMed] [Google Scholar]
- Ehrenreiter K, Piazzolla D, Velamoor V, Sobczak I, Small JV, Takeda J, Leung T, Baccarini M (2005) Raf-1 regulates Rho signaling and cell migration. J Cell Biol 168: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Wolf K (2003) Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 3: 362–374 [DOI] [PubMed] [Google Scholar]
- Geisbrecht ER, Montell DJ (2004) A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell 118: 111–125 [DOI] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H (2000) Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J 19: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Darding M, Miasari M, Santoro MM, Zender L, Xue W, Tenev T, da Fonseca PC, Zvelebil M, Bujnicki JM, Lowe S, Silke J, Meier P (2008) IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-kappaB as well as cell survival and oncogenesis. Nat Cell Biol 10: 1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Meier P (2010) IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer 10: 561–574 [DOI] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ (2008) Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 9: 690–701 [DOI] [PubMed] [Google Scholar]
- Hwang C, Oetjen KA, Kosoff D, Wojno KJ, Albertelli MA, Dunn RL, Robins DM, Cooney KA, Duckett CS (2008) X-linked inhibitor of apoptosis deficiency in the TRAMP mouse prostate cancer model. Cell Death Differ 15: 831–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster RW, Fraser SE (2001) Direct imaging of in vivo neuronal migration in the developing cerebellum. Curr Biol 11: 1858–1863 [DOI] [PubMed] [Google Scholar]
- Lanning CC, Daddona JL, Ruiz-Velasco R, Shafer SH, Williams CL (2004) The Rac1 C-terminal polybasic region regulates the nuclear localization and protein degradation of Rac1. J Biol Chem 279: 44197–44210 [DOI] [PubMed] [Google Scholar]
- Lerm M, Pop M, Fritz G, Aktories K, Schmidt G (2002) Proteasomal degradation of cytotoxic necrotizing factor 1-activated rac. Infect Immun 70: 4053–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisi S, Mazzon I, White K (2000) Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics 154: 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang D, Luo W, Yu Y, Yu J, Li J, Zhang X, Zhang B, Chen J, Wu XR, Rosas-Acosta G, Huang C (2011) X-linked inhibitor of apoptosis protein (XIAP) mediates cancer cell motility via Rho GDP dissociation inhibitor (RhoGDI)-dependent regulation of the cytoskeleton. J Biol Chem 286: 15630–15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J, John SW, Tenev T, Rautureau GJ, Hinds MG, Francalanci F, Wilson R, Broemer M, Santoro MM, Day CL, Meier P (2011) CARD-mediated autoinhibition of cIAP1's E3 ligase activity suppresses cell proliferation and migration. Mol Cell 42: 569–583 [DOI] [PubMed] [Google Scholar]
- Mace PD, Shirley S, Day CL (2010) Assembling the building blocks: structure and function of inhibitor of apoptosis proteins. Cell Death Differ 17: 46–53 [DOI] [PubMed] [Google Scholar]
- Malliri A, Collard JG (2003) Role of Rho-family proteins in cell adhesion and cancer. Curr Opin Cell Biol 15: 583–589 [DOI] [PubMed] [Google Scholar]
- Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri DC (2010) IAP regulation of metastasis. Cancer Cell 17: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nethe M, Hordijk PL (2010) The role of ubiquitylation and degradation in RhoGTPase signalling. J Cell Sci 123: 4011–4018 [DOI] [PubMed] [Google Scholar]
- Pop M, Aktories K, Schmidt G (2004) Isotype-specific degradation of Rac activated by the cytotoxic necrotizing factor 1. J Biol Chem 279: 35840–35848 [DOI] [PubMed] [Google Scholar]
- Rajalingam K, Dikic I (2009) Inhibitors of apoptosis catch ubiquitin. Biochem J 417: e1–e3 [DOI] [PubMed] [Google Scholar]
- Rajalingam K, Sharma M, Paland N, Hurwitz R, Thieck O, Oswald M, Machuy N, Rudel T (2006) IAP-IAP complexes required for apoptosis resistance of C. trachomatis-infected cells. PLoS Pathog 2: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger S, Senghaas N, Walch A, Köster RW (2009) Cadherin-2 controls directional chain migration of cerebellar granule neurons. PLoS Biol 7: e1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Marshall CJ (2003) Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol 5: 711–719 [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS (2002) IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol 3: 401–410 [DOI] [PubMed] [Google Scholar]
- Samuel T, Okada K, Hyer M, Welsh K, Zapata JM, Reed JC (2005) cIAP1 Localizes to the nuclear compartment and modulates the cell cycle. Cancer Res 65: 210–218 [PubMed] [Google Scholar]
- Sandrock K, Bielek H, Schradi K, Schmidt G, Klugbauer N (2010) The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic 11: 198–209 [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ (2008) Rac activation and inactivation control plasticity of tumor cell movement. Cell 135: 510–523 [DOI] [PubMed] [Google Scholar]
- Silke J, Kratina T, Chu D, Ekert PG, Day CL, Pakusch M, Huang DC, Vaux DL (2005) Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc Natl Acad Sci USA 102: 16182–16187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M (2010) The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev 24: 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula SM, Ashwell JD (2008) IAPs: what's in a name? Mol Cell 30: 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S, Hellal F, Neukirchen D, Hindges R, Garvalov BK, Flynn KC, Stradal TE, Chrostek-Grashoff A, Brakebusch C, Bradke F (2010) Rac1 regulates neuronal polarization through the WAVE complex. J Neurosci 30: 6930–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D (2007) IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131: 669–681 [DOI] [PubMed] [Google Scholar]
- Varfolomeev E, Vucic D (2008) (Un)expected roles of c-IAPs in apoptotic and NFkappaB signaling pathways. Cell Cycle 7: 1511–1521 [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J (2003) Mammalian mitochondrial IAP binding proteins. Biochem Biophy Res Comm 304: 499–504 [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J (2005) IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol 6: 287–297 [DOI] [PubMed] [Google Scholar]
- Vega FM, Ridley AJ (2008) Rho GTPases in cancer cell biology. FEBS Lett 582: 2093–2101 [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL (2000) Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102: 43–53 [DOI] [PubMed] [Google Scholar]
- Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, Brink R, Leverkus M, Tergaonkar V, Schneider P, Callus BA, Koentgen F, Vaux DL, Silke J (2007) IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 131: 682–693 [DOI] [PubMed] [Google Scholar]
- Visvikis O, Lores P, Boyer L, Chardin P, Lemichez E, Gacon G (2008) Activated Rac1, but not the tumorigenic variant Rac1b, is ubiquitinated on Lys 147 through a JNK-regulated process. FEBS J 275: 386–396 [DOI] [PubMed] [Google Scholar]
- Volkmann K, Chen YY, Harris MP, Wullimann MF, Köster RW (2010) The zebrafish cerebellar upper rhombic lip generates tegmental hindbrain nuclei by long-distance migration in an evolutionary conserved manner. J Comp Neurol 518: 2794–2817 [DOI] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL (2003) Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302: 1775–1779 [DOI] [PubMed] [Google Scholar]
- Westerfield M (1995) The Zebrafish Book. Eugene, OR: University of Oregon
- Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P (2003) Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol 160: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhu J, Hu X, Zhu H, Kim HT, LaBaer J, Goldberg A, Yuan J (2007) c-IAP1 cooperates with Myc by acting as a ubiquitin ligase for Mad1. Mol Cell 28: 914–922 [DOI] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, Mu D, Lucito R, Powers S, Lowe SW (2006) Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125: 1253–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.