Abstract
Supercoiled DNAs varying from 281 to 5302 bp were subjected to shear forces generated by aerosolization or sonication. DNA shearing strongly correlated with length. Typical sized plasmids (⩾3000 bp) degraded rapidly. DNAs 2000–3000 bp persisted ∼10 min. Even in the absence of condensing agents, supercoiled DNA <1200 bp survived nebulization, and increased forces of sonication were necessary to shear it. Circular vectors were considerably more resistant to shearing than linear vectors of the same length. DNA supercoiling afforded additional protection. These results show the potential of shear-resistant Minivector DNAs to overcome one of the major challenges associated with gene therapy delivery.
Keywords: Minivector DNA, Minicircle, hydrodynamic shear, nebulized, aerosolized, lung delivery
Introduction
There are currently many challenges to gene therapy and most involve hurdles with nucleic acid delivery. Although viruses are highly effective and deliver high doses, they can trigger immunological and inflammatory responses and can potentially misregulate endogenous genes. Such responses may result in disease or even death.1, 2, 3, 4 Because of these problems, effort has been expended toward the development of effective non-viral vectors.5 Plasmid DNA vectors are relatively inexpensive, easy to make and store, and have tremendous design capacity. A shortcoming of plasmids is their size (typically >4000 base pairs (bp)), which makes them highly susceptible to shear-induced degradation.6, 7
Another important challenge of gene therapy is targeting the vector to the diseased organ. The lungs are amenable to gene therapy because they are accessible through the nose and mouth. Nebulization to create an aerosol is routinely used to deliver drugs for the treatment of lung diseases. Although toxicity is lessened when plasmid DNA is delivered by aerosol compared with systemic delivery,8, 9 plasmid DNA still cannot be delivered by this method because of extensive degradation.10, 11 DNA degradation during nebulization is thought to mainly occur because of hydrodynamic shear,12 which is the force exerted on the DNA by the rapid flow of the solvent.
DNA degradation by hydrodynamic shear was first studied over 50 years ago by forcing bacteriophage DNA through a hypodermic needle.13 Although several studies since have attempted to determine the mechanisms behind DNA shearing, it is still not fully understood what causes the DNA to degrade.7 Many vehicle systems have been developed to try to protect fragile plasmid vectors (or synthetic small interfering RNA) from delivery-associated shear forces. The cationic agents, polyethylenimine, polylysine, polyamines, and liposomes act to condense and shield the DNA. They protect the DNA from shearing by reducing the hydrodynamic diameter of the molecule.12, 14 However, these vehicles are highly cytotoxic and pro-inflammatory,15, 16 and only partially alleviate DNA shearing.17, 18
For the few cases in which the effect of DNA size has been studied, shear force resistance is inversely correlated with DNA length; 13 000 bp plasmids were more resistant to shearing than 20 000 or 29 000 bp plasmids measured in a rotary disk shearing device6 and 9800 bp plasmids sheared three times more rapidly than 5000 bp plasmids in a jet nebulizer.12 It is reasonable to extrapolate that shorter DNA vectors should be less vulnerable to shearing, but this idea could not be tested previously because it was not possible to generate shorter circular DNA (<2000 bp) in sufficient quantities to do the experiments. Because of the need for a bacterial origin of replication and a selectable marker, thousands of bp are required for the propagation of plasmids.5 Deleting such sequences, through site-specific recombination, results in DNA vectors that are less toxic and less likely to be silenced than plasmids.19, 20 Still thousands of bp in length, however, these ‘Minicircles' remain highly susceptible to shear-induced degradation.
A potential way to make smaller DNA vectors is to ligate linear DNAs in vitro. Depending on sequence, shortening DNA makes it increasingly difficult to bend into a circle because of inherent rigidity.21, 22, 23 Thus, obtaining reasonable quantities of circles less than 1000 bp is difficult; microgram quantities were previously the best achievable yields.24 Manipulating DNA supercoiling through topoisomerase mutations and using site-specific recombination in Escherichia coli, we generate Minivectors as small as ∼250 bp in milligram quantities.25, 26 Nearly devoid of bacterial sequences, Minivectors can be used to express short hairpin RNA (shRNA), microRNA (miRNA) or genes in various cell types, including cells that are refractory to plasmid transfection.26 In this paper, we use Minivectors to determine the role of DNA length, circularity, and supercoiling on shear force resistance. We find that extrapolation from the previous data would not have predicted the behavior of very small, supercoiled circles when subjected to shear forces.
Results
Shearing as a function of DNA length
There are multiple ways to generate hydrodynamic shear: passage through a narrow gauge needle, circulation through high-performance liquid chromatography tubing, nebulization, and sonication.27 We used nebulization because it is both highly reproducible and clinically relevant. In a Collison-like jet nebulizer,28 it is far easier to collect samples from the reservoir, and DNA degradation was identical when sampled from the aerosol or the reservoir (Supplementary Figure 1). For the experiments described below, therefore, we sampled from the reservoir.
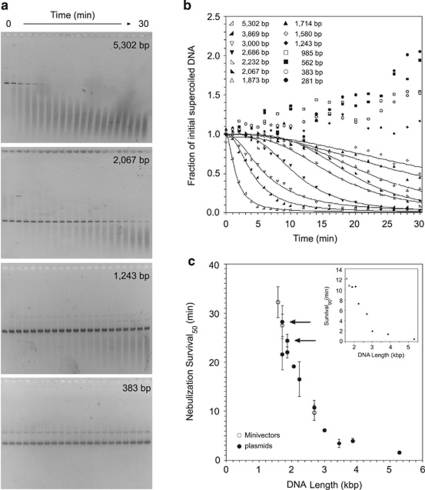
Supercoiled DNAs varying from 281 to 5302 bp (Table 1) were subjected to nebulization and their survival was analyzed by gel electrophoresis (Figure 1). The extent of DNA shearing was quantified by measuring the disappearance of intact DNA vector over time. The ability to withstand shear forces in the nebulizer was, in general, inversely correlated with DNA length. Supercoiled DNA vectors >2600 bp, the size range of DNA vectors used previously for delivery, fragmented completely and rapidly and displayed pseudo-exponential decay (Figure 1b). The sigmoid shape of the decay curves for supercoiled DNA vectors between 1580 and 2232 bp showed an initial resistance to shearing followed by degradation (Figure 1b), a very different behavior from that of the larger vectors. In common, all DNA vectors sheared into fragments that ranged from 200 to 1000 bp (Figure 1a). Supercoiled Minivectors ⩽ 1243 bp slightly concentrated with time in the nebulizer because of evaporation (Figure 1b). To address whether changing DNA concentrations could affect shear force survival, solutions of 1873 bp plasmid ranging from 1 to 10 μg ml−1 were nebulized. The decay kinetics were identical at all concentrations (data not shown).
Table 1. DNA vectors used in this study.
| Length (bp) | DNA vector | Encodesa | Nebulization Survival50 (min) | Sonication Survival50 (min) | Reference or source |
|---|---|---|---|---|---|
| 281 | mv281 | — | >30 | ND | 25 |
| 336 | mv336 | — | ND | 96.13±2.95 | This study |
| 383 | mv-H1-GFPshRNA | shRNA to eGFP | >30 | ND | 26 |
| 562 | Dimer of mv281 | — | >30 | ND | 25 |
| 672 | Dimer of mv336 | — | >30 | 21.86±3.68 | This study |
| 985 | mv-H1-miR31 | Human miR-31 | >30 | ND | This study |
| 999 | mv999 | — | ND | 5.98±1.46 | 25 |
| 1109 | mv-KB4TAL-GLuc | Gaussia luciferase | >30 | ND | This study, Nanolight Technology |
| 1243 | mv-KB4TAL-mCherry | mCherry | >30 | 3.73±0.55 | This study53 |
| 1580 | mv-CMV-GLuc | Gaussia luciferase | 32.3±3.1 | 1.77±0.69 | This study, Nanolight Technology |
| 1711 | pDJC1 | — | 21.6±3.2 28.2±3.4b | ND | This study |
| 1714 | mv-CMV-mCherry | mCherry | 27.5±2.3 | ND | This study53 |
| 1873 | pQR499 | — | 22.0±1.1 24.4±1.3b | 0.85±0.10 | 54 |
| 2067 | pINV2067 | — | 19.1±0.2 | ND | Invitrogen supercoiled DNA ladder |
| 2232 | pAO | — | 16.6±3.5 | ND | 55 |
| 2679 | mv-CMV-Luc2 | Firefly luciferase | 9.6±1.5 | ND | This study, Promega |
| 2686 | pUC18 | — | 10.8±1.4 | ND | Novagen Inc. |
| 3000 | pBLUESCRIPT | — | 6.1±0.1 | ND | Stratagene |
| 3459 | pJB3.5i | attB, attP | 3.4±0.9 | ND | 56 |
| 3869 | pMC339-BbvCI | attB, attP | 4.0±0.5 | 0.17±0.04 | 25 |
| 5302 | pCR2.1-norE | norE | 1.5±0.1 | ND | This study |
Abbreviations: eGFP, enhanced green fluorescent protein; miR, microRNA; ND, not determined; shRNA, short hairpin RNA.
All Minivectors contain attR;25 all plasmids encode bla.
Propagated in LZ54.
Figure 1.
DNA survival during nebulization as a function of DNA length. (a) Representative gels of DNA vectors subjected to nebulization as sampled from the reservoir. (b) Graphical representation of DNA shearing as a function of length (averaged for at least three separate experiments). The curves represent the fit to a sigmoidal function (Supplementary Table 1). Minivectors ⩽1243 bp were not fitted because after 30 min the intact DNA did not decrease. Nebulization survival times were determined for each DNA vector. (c) The time at which 50% of the vector survived (Survival50). Each Survival50 value is the mean from at least three separate experiments. Error bars represent the standard deviation. The arrows denote Survival50 of the two plasmids, pDJC1 and pQR499, that were propagated in LZ54. Minivectors are represented by open circles and plasmids are shown as closed circles. The inset shows Survival90 values, which is the time at which 90% of vectors remained.
To facilitate comparisons among the differently sized DNA vectors, we introduce a term, Survival50, which is the time at which 50% of intact vectors remain. Nebulization Survival50 as a function of DNA vector length is shown (Figure 1c). Comparing the data in this way revealed important and unexpected results. First, Survival50 did not depend monotonically on DNA length, but instead exhibited a threshold-like behavior as defined by the asymptote. Below this threshold, which may reflect a critical hydrodynamic diameter, DNA vectors did not appreciably shear. Second, Minivectors survived better than plasmids of the same length.
There are several differences between plasmids and Minivectors. Plasmids contain origins of replication and genes encoding antibiotic resistance; Minivectors contain attR.25 In addition, different bacterial strains are used for the propagation of plasmids and Minivectors.25, 26 As a consequence, Minivectors have increased supercoiling (σ ∼−0.09 compared with σ ∼−0.075 for plasmids29), which should further decrease their hydrodynamic diameter, and this may explain their increased protection from shear forces. To address the potential effect of increased supercoiling, we propagated a plasmid or a Minivector of the same length in LZ54,30 the bacterial strain used to generate Minivectors. The 1711 bp plasmid, pDJC1, propagated in LZ54 persisted as long as the 1714 bp Minivector (Figure 1c, compare open circle with closed circle, arrow). Plasmid pQR499 (1873 bp) isolated from LZ54 also persisted longer than when it had been propagated in DH5α (Figure 1c, arrow). Increased supercoiling, therefore, and not differences in sequence between plasmids and Minivectors, accounted for the increased shear resistance.
The advantages of reducing vector size were even more pronounced when Survival90, the time at which 90% of vectors remained, was considered (Figure 1c, inset). Below 3000 bp, there was a very steep inverse correlation between DNA length and Survival90 values. For DNA vectors <2000 bp, Survival90 values were longer than 10 min, as a result of the initial resistance to shearing and as seen by the sigmoidal decay. Considering various percentages of DNA vector survival might be useful for calculating the relative benefits and risks of using a particular DNA vector length (Supplementary Table 1).
Effect of DNA circularity and supercoiling on shear force resistance
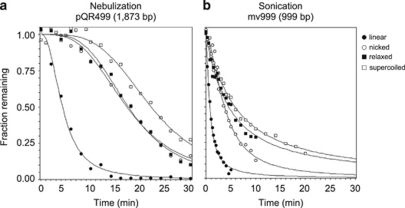
To better understand the contribution of DNA supercoiling to shear force resistance, we compared linear, nicked and relaxed forms of the vectors (Figure 2). Half of the negatively supercoiled 1873 bp plasmid, pQR499, survived 22 min, longer than either the relaxed, open-circular (17.5 min) or nicked (17.2 min) forms of the vector. The nearly identical Survival50 values of the nicked and relaxed plasmids revealed, somewhat unexpectedly, that a single-strand break does not significantly weaken this vector during nebulization. Circularity had a bigger impact on Survival50 than supercoiling did; half of the linear form of the 1873 bp DNA was sheared in ∼4 min. The fragility of linear DNA during nebulization was the same whether the ends were blunt, had 2-base 3′-overhangs or had 4-base 5′-overhangs (data not shown); therefore, the nature of the ends did not affect DNA shearing.
Figure 2.
Effect of DNA topology on DNA survival. (a) How topology of plasmid DNA (1873 bp) influences its survival during nebulization. The fraction of DNA vector of each topology over time (averaged from at least three experiments) is shown. The curves are shown fitted to a sigmodial function. (b) How topology of Minivectors (999 bp) influences its survival during sonication. The fraction of DNA was quantified the same as in (a).
Because shorter vectors completely survived nebulization, we used sonication to determine how supercoiling affected their shearing. Like nebulization, sonication also generates highly reproducible shear forces, but these forces can be much greater and can be generated for longer periods of time. The difference in the shear force magnitude is exemplified by the differences in Survival50 values between the supercoiled 3869 bp plasmid (sonication Survival50=0.17 min and nebulization Survival50=4 min) and the 1873 bp plasmid (sonication Survival50=0.85 min and nebulization Survival50=22 min). Despite the difference in shear force magnitude, the two methods for shearing yielded the same general result—a strong correlation of DNA vector length and shearing (Figure 3). For example, the 3869 bp plasmid was sheared >500 times more rapidly (Survival50=0.17 min) than a vector approximately one-tenth that length, the 336 bp Minivector (Survival50=96 min).
Figure 3.
DNA survival during sonication as a function of DNA length. Because more force was necessary to degrade the smaller Minivectors, sonication was employed. Sonication survival times were determined for each DNA vector in the same manner as for nebulization in Figure 1. The plot shows the time at which 50% of each vector survived (Survival50) versus its length. Each Survival50 value is the mean from at least three separate experiments. Error bars represent the standard deviation.
It was impossible to distinguish Survival50 by sonication for the circular forms of the 1873 bp plasmid, which were all about a minute, but the value for the linear form was only 0.2 min. Although the DNA vectors could be subjected to sonication much longer than they could to nebulization, sonicating DNA for more than 90 min is impractical. Therefore, to determine how DNA supercoiling affected shear force survival required a balance between a Minivector large enough for differences to be detected and small enough for those differences to be separated. A 999 bp Minivector fit these criteria. Like the 1873 bp plasmid sheared by nebulization, this Minivector sheared according to its topology; linear sheared with a sonication Survival50=0.8 min, nicked=3.8 min, relaxed=4.6 min and supercoiled=6 min.
With regard to shear force resistance, circularity is the most important property of a DNA vector for protection. This protection is ∼4- to 6-fold when comparing the nicked/relaxed and linear forms of each vector (Figure 2). Supercoiling provides additional protection from shear forces. The supercoiled 999 and 1873 bp DNA vectors persisted 58 and 28% longer, respectively, than their nicked counterparts (Figure 2).
Discussion
Model for DNA shearing
Early studies of DNA shearing showed that the hydrodynamic shear force exerted on a DNA molecule increases with the square of the hydrodynamic diameter (the length-dependent diameter of the DNA molecule in the fluid stream).12, 31 Our data are consistent with this relationship. Circularity, decreasing length and increasing supercoiling, properties that we find to protect DNA vectors from shearing, should all reduce hydrodynamic diameter. Intuitively, the hydrodynamic diameter of a circular DNA vector should be approximately half the length of the linear form,7 and this idea is supported by experimental data.32 Our findings that the 1873 bp linear DNA sheared as rapidly as the 3869 bp supercoiled plasmid and that the linear 999 bp DNA sheared comparably to a plasmid roughly twice its length is consistent with this concept. In addition, our observation that supercoiling affords additional protection is supported by the increased survival of supercoiled DNA vectors compared with the nicked or relaxed forms of the same length.
Because smaller DNA vectors experience less force, they survive longer, but what finally makes them break? If single-stranded breaks were generated, we would expect to see a transient appearance of nicked intermediate as supercoiled DNA is sheared. Nicked DNA, however, was not observed during shearing of the supercoiled vectors, indicating that shearing generates primarily double-stranded breaks. The absence of any full-length linear degradation products indicates that following the initial double-strand break, additional breaks occur in rapid succession, consistent with the extremely rapid conversion of linear vectors to smaller fragments.
The pronounced lag observed before the smaller DNA vectors degraded indicates that the shear forces in the nebulizer must not initially be harsh enough to break these smaller vectors, and that the continuous re-circulation of the DNA vectors may stretch or contort them into a conformation that is then more susceptible to shearing. There are two proposed competing effects of DNA supercoiling on shear survival: torsional strain, which might make the molecule more susceptible to shearing, and DNA compaction, which should make the molecule more resistant to shearing.7 Although our data clearly show that the beneficial effect of compaction is the dominant effect, torsional stress-induced deformation of DNA33, 34, 35 may contribute to the eventual shearing of the vectors, thereby counteracting the protective effect of supercoiling-mediated DNA compaction. As the reservoir volume decreases, the rate of recirculation increases (Supplementary Figure 2), consequently the frequency of exposure to shear forces increases, and there may be less time for DNA to recover from shear force-induced contortion. The stretched, contorted and weakened DNA is then sheared more readily. This model predicts that DNA base pair steps that are more likely to contort would be the weak points of the molecule that shear. It is possible that the nebulization protocol during treatment of a patient could be altered to decrease this frequency of recycling, perhaps as simply as by maintaining a constant reservoir volume.
Therapeutic consequences of DNA shearing
The most obvious detriment of DNA shearing is a reduction in the amount of intact vector remaining for therapy. Larger DNA vectors completely degrade into small fragments over the course of nebulization. These fragments are presumably unable to elicit a therapeutic effect as the transgene or shRNA together with the promoter are destroyed. It might be tempting to increase the DNA dose to compensate for this loss. Doing so, however, could have negative consequences. First, increasing the amount of DNA vector would also require a commensurate increase in the amount of toxic delivery vehicle. Second, delivery of increased degraded DNA fragments could induce DNA repair and recombination pathways or trigger apoptosis. Third, cells could randomly ligate the fragmented DNA. Although there is the potential of the ligations forming large episomal concatemers that could persist to cause stable long-term transgene expression,36 there is also the possibility that the randomly ligated DNA fragments could join together to generate new, potentially toxic sequences. One way to avoid these problems would be to halt nebulization earlier. For example, DNA vectors <2000 bp, for which Survival90 values were greater than 10 min, could be halted after 10 min. Shortening nebulization treatments would not be necessary for the smaller Minivectors (<1000 bp) because very little shearing occurs, even after 30 min.
DNA vector delivery to the lungs
Identifying DNA vector lengths that survive shear forces has important implications for gene therapy delivery no matter what the delivery route; however, the use of a Collison-like jet nebulizer makes our data particularly germane for the consideration of DNA delivery to the lungs. The lungs are readily accessible by intratracheal, intranasal and aerosol delivery methods, and any of these routes is amenable to DNA vector delivery. Aerosol delivery, in particular, is non-invasive, delivers directly to the affected tissue and may help prevent off-target complications. In addition, aerosol delivery allows DNA to be delivered to the lungs in much higher quantities than systemic administration.37
A number of promising gene therapy targets have been identified for the treatment of pulmonary diseases. Dozens of clinical trials are ongoing that target, for example, cystic fibrosis (http://www.wiley.com/legacy/wileychi/genmed/clinical/). Disappointingly, however, these existing therapies have so far been unsuccessful.38 Asthma as a disease target has high potential for therapeutic RNA interference, including shRNA39 and miRNA,40 but the bottleneck for success for treatment of asthma is the same as for other diseases—delivery of the nucleic acid therapy. The observation that Minivectors can overcome some of the most difficult obstacles associated with delivery renews enthusiasm for gene therapy for the lungs.
Additional therapeutic benefits of small, supercoiled DNA vectors
In a previous study from our group, Minivectors of 383 bp, encoding an shRNA against a pathogenic protein in lymphoma, survived intact in human serum for >48 h, whereas synthetic small interfering RNA and plasmids were degraded within a couple of hours.26 Certain DNA sequences, such as polyA signals and antibiotic resistance genes that are commonly found in DNA vectors, are particularly susceptible to attack by nucleases.41 Some of the biostability of Minivectors in the serum may be a consequence of such sequences being absent from the Minivectors.
Extra DNA on traditional plasmids contain bacterial sequences that induce immunotoxic responses, primarily because of immune responses to CpG motifs42 that are approximately four times more prevalent in bacterial than mammalian DNA.43 In addition to toxicity, bacterial sequences also induce transcriptional silencing of episomal transgenes.44 ‘Minicircles' that lack bacterial sequences exhibit significantly increased transgene expression.20, 44, 45
In addition to increased shear force survival, reducing the length of DNA vectors also improves cell transfection.26, 46, 47 Minivectors of a few hundred bp transfect cells that are refractory to transfection with conventional plasmids.26 The smaller size of Minivectors means more copies per unit mass. This fact, coupled with the very high transfection efficiency,26 suggests that a lower DNA vector dose and, consequently, less toxic transfection vehicle may be used.
Our data show that survival of shear forces associated with nebulization requires DNA vectors to be circular and shorter than 2000 bp. Although it may seem that therapeutically useful Minivectors may express only shorter sequences, such as shRNA, miRNA and small genes, in fact we have not lost the ability to express larger genes. Larger genes can be split into multiple smaller segments that when expressed will reconstitute in vivo to form a functional protein. The concept of splitting genes has been studied for many decades, and one of the first reports involved splitting the gene encoding β-galactosidase.48 Other examples of genes that have been split and reconstituted in vivo include the genes encoding ubiquitin,49 PurN,50 adenylate kinase51 and the yellow fluorescent protein, Venus.52 Because they are nearly devoid of bacterial sequences, almost all of the Minivector is available to encode a useful sequence. Even at 2000 bp, Minivectors can encode a promoter and ∼1500 bp of gene sequence (either an intact small gene or gene fragment), significantly increasing the arsenal of useful therapeutic sequences.
To our knowledge, no previous study has investigated such a broad range of DNA sizes and topologies as reported here. These data will be useful for identifying the factors most important for resisting shear forces involved in gene therapy delivery. Knowing how to mitigate DNA vector degradation will facilitate the development of new, more efficacious gene therapy vectors. In addition, the use of the nebulizer makes our data particularly germane to clinical applications. The ability of Minivectors to resist shear forces associated with gene therapy delivery renews hope of treating human diseases using DNA vectors.
Materials and methods
Chemicals, reagents and equipment
Acrylamide was from EMD Chemicals (Gibbstown, NJ, USA), agarose from ISC BioExpress (Kaysville, UT, USA) and SYBR Gold from Invitrogen (Hercules, CA, USA). Restriction enzymes were from New England Biolabs (Ipswich, MA, USA). All other chemicals were from Fisher Scientific (Waltham, MA, USA). Plasmid Maxi kit was from Qiagen (Valencia, CA, USA) and Amicon Ultra centrifugal filters were from Millipore (Billerica, MA, USA). The Aerotech II jet nebulizer was from CSI-US Inc. (Bedford, MA, USA) and the Aridyne 2000 compressor was from Allied Healthcare Products (St Louis, MO, USA). The 1/8 in probe sonicator (Model 60 Sonic Dismembrator) was from Fisher Scientific. PC Image and Total Lab software were from Fotodyne (Hartland, WI, USA) and Total Lab (Durham, NC, USA), respectively. KaleidaGraph (version 4.1) was from Synergy Software (Reading, PA, USA).
DNA generation and manipulation
DNA vectors used in this study are listed in Table 1. Throughout the text, we refer to these by their length. Following the convention of using ‘p' in front of plasmid names, we designate Minivectors with ‘mv'. Parent plasmids used to generate Minivectors are designated ‘pMV'. pMV-KB4TAL-GLuc and pMV-CMV-GLuc were gifts from Dr Stephen Gottschalk (Baylor College of Medicine, Houston, TX, USA) and pMV-CMV-Luc2 was a gift from Dr David Spencer (Baylor College of Medicine). pMV-H1-miR31, pMV-KB4TAL-mCherry and pMV-CMV-mCherry were gifts from Dr Martin M Matzuk and Dr Zhifeng Yu (Baylor College of Medicine). pQR499 was a gift from Dr John Ward (University College London, London, UK). pDJC1 was constructed by digesting pQR499 with TfiI and AflIII. The recessed ends of the digested pQR499 were filled in with T4 DNA polymerase and subsequently ligated with T4 DNA ligase. Plasmids were generated in Escherichia coli DH5α cells, isolated using a Plasmid Maxi kit as per the manufacturer's instructions, and subsequently desalted and concentrated using Amicon Ultra centrifugal filters.
Minivectors were obtained as follows. Minivector parent plasmids were transformed into E. coli strain LZ54.30 Large-scale λ Int-mediated recombination and Minivector isolation was performed as described.25, 26 To generate nicked DNA vector, nicking endonuclease Nt.BbvCI was used following the manufacturer's protocols. To obtain relaxed DNA, nicked DNA was religated using T4 DNA ligase. Linearization was performed with either BspHI, EcoRV, PvuI or ScaI, depending on which overhang end was desired, as per the manufacturer's protocols. Nicked, relaxed and linear DNAs were extracted with phenol:chloroform:isoamyl alcohol (25:24:1), extracted with chloroform, and precipitated with ethanol. Each DNA topology was resuspended in TE buffer (10 m Tris-HCl, 1 m EDTA, pH 8) and verified using gel electrophoresis before nebulization or sonication.
DNA shearing
For nebulization, 10 ml of DNA at 1 μg ml−1 in TE buffer was added to an Aerotech II jet nebulizer. Air was delivered to the nebulizer at a rate of 10 l min−1 and gauge pressure of 50 p.s.i. by an Aridyne 2000 compressor. For the studies of the effects of DNA length on nebulization survival, 15 μl aliquots were removed from the nebulizer reservoir before and at intervals throughout nebulization up to 30 min, at which point the DNA solution was depleted. Because of the dramatic changes that occurred early, aliquots were taken at 1 min intervals initially and at 2 min intervals after 10 min.
For sonication, 1 ml of DNA at 1 μg ml−1 in TE buffer in a 1.7 ml eppendorf tube was incubated on ice during sonication with a 1/8 in probe sonicator at setting 5, which has an output of 3–7 W (root mean square). For consistency, the probe was inserted halfway down into the solution and the sides of the tube were avoided. 15 μl aliquots were removed before sonication and at the time points indicated.
All DNA shearing experiments were performed a minimum of three separate times. DNA was analyzed by electrophoresis on 1% agarose gels for >1000 bp or 2% agarose gels or 5% acrylamide (29:1 acrylamide:bis-acrylamide) gels for <1000 bp in 40 m Tris-acetate and 2 m EDTA. All gels were submitted to 125 V for 2 h, stained with SYBR Gold for 20 min and visualized using PC Image. Total Lab was utilized to quantify the remaining intact DNA. Data were analyzed and fitted using KaleidaGraph.
Acknowledgments
We thank Alexander Seryshev and Alex J Brewer III for technical assistance, Dr David B Corry and Dr Peter Hu for helpful discussions, and Dr John Ward, Dr Stephen Gottschalk, Dr David Spencer, Dr Martin M Matzuk and Dr Zhifeng Yu for reagents. DJC was funded by Infection and Immunity Training Grant T32 AI55413-05. DES was supported by the Molecular Genetic Technology Program at the University of Texas MD Anderson Cancer Center. BEG was supported by the Clayton Foundation for Research. This work was supported by a grant from the National Institutes of Health RO1A1054830 to LZ. This paper is dedicated to Margie Catanese, Frank Fogg, Rosemary Cmarik, Sissy (Zechiedrich) James, and all who have lost the battle against lung cancer.
Dr Jonathan Fogg and Dr Lynn Zechiedrich are co-inventors on an issued patent, and together with Dr Daniel Catanese and Dr Brian Gilbert, are also co-inventors on another patent application. These patents cover the Minivector technology used in the work reported in this paper.
Footnotes
Supplementary Information accompanies the paper on Gene Therapy website (http://www.nature.com/gt)
Supplementary Material
References
- Somia N, Verma IM. Gene therapy: trials and tribulations. Nat Rev Genet. 2000;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Couzin J, Kaiser J. Gene therapy. As Gelsinger case ends, gene therapy suffers another blow. Science. 2005;307:1028. doi: 10.1126/science.307.5712.1028b. [DOI] [PubMed] [Google Scholar]
- Kaiser J. Gene therapy. Panel urges limits on X-SCID trials. Science. 2005;307:1544–1545. doi: 10.1126/science.307.5715.1544a. [DOI] [PubMed] [Google Scholar]
- Gill DR, Pringle IA, Hyde SC. Progress and prospects: the design and production of plasmid vectors. Gene Therapy. 2009;16:165–171. doi: 10.1038/gt.2008.183. [DOI] [PubMed] [Google Scholar]
- Levy MS, O'Kennedy RD, Ayazi-Shamlou P, Dunnill P. Biochemical engineering approaches to the challenges of producing pure plasmid DNA. Trends Biotechnol. 2000;18:296–305. doi: 10.1016/s0167-7799(00)01446-3. [DOI] [PubMed] [Google Scholar]
- Lengsfeld CS, Anchordoquy TJ. Shear-induced degradation of plasmid DNA. J Pharm Sci. 2002;91:1581–1589. doi: 10.1002/jps.10140. [DOI] [PubMed] [Google Scholar]
- Densmore CL. The re-emergence of aerosol gene delivery: a viable approach to lung cancer therapy. Curr Cancer Drug Targets. 2003;3:275–286. doi: 10.2174/1568009033481886. [DOI] [PubMed] [Google Scholar]
- Di Gioia S, Conese M. Polyethylenimine-mediated gene delivery to the lung and therapeutic applications. Drug Des Dev Ther. 2008;2:163–188. doi: 10.2147/dddt.s2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz LA, Johnson JL, Black M, Cheng SH, Hogan ME, Waldrep JC. Delivery of DNA–cationic liposome complexes by small-particle aerosol. Hum Gene Ther. 1996;7:731–741. doi: 10.1089/hum.1996.7.6-731. [DOI] [PubMed] [Google Scholar]
- Stern M, Sorgi F, Hughes C, Caplen NJ, Browning JE, Middleton PG, et al. The effects of jet nebulisation on cationic liposome-mediated gene transfer in vitro. Gene Therapy. 1998;5:583–593. doi: 10.1038/sj.gt.3300629. [DOI] [PubMed] [Google Scholar]
- Lentz YK, Worden LR, Anchordoquy TJ, Lengsfeld CS. Effect of jet nebulization on DNA: identifying the dominant degradation mechanism and mitigation methods. J Aerosol Sci. 2005;36:973–990. [Google Scholar]
- Davison PF. The effect of hydrodynamic shear on the deoxyribonucleic acid from T(2) and T(4) bacteriophages. Proc Natl Acad Sci USA. 1959;45:1560–1568. doi: 10.1073/pnas.45.11.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belur LR, Podetz-Pedersen K, Frandsen J, McIvor RS. Lung-directed gene therapy in mice using the nonviral Sleeping Beauty transposon system. Nat Protoc. 2007;2:3146–3152. doi: 10.1038/nprot.2007.460. [DOI] [PubMed] [Google Scholar]
- Chollet P, Favrot MC, Hurbin A, Coll JL. Side-effects of a systemic injection of linear polyethylenimine–DNA complexes. J Gene Med. 2002;4:84–91. doi: 10.1002/jgm.237. [DOI] [PubMed] [Google Scholar]
- Wolfert MA, Dash PR, Nazarova O, Oupicky D, Seymour LW, Smart S, et al. Polyelectrolyte vectors for gene delivery: influence of cationic polymer on biophysical properties of complexes formed with DNA. Bioconjug Chem. 1999;10:993–1004. doi: 10.1021/bc990025r. [DOI] [PubMed] [Google Scholar]
- Brunot C, Ponsonnet L, Lagneau C, Farge P, Picart C, Grosgogeat B. Cytotoxicity of polyethyleneimine (PEI), precursor base layer of polyelectrolyte multilayer films. Biomaterials. 2007;28:632–640. doi: 10.1016/j.biomaterials.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Darquet AM, Cameron B, Wils P, Scherman D, Crouzet J. A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Therapy. 1997;4:1341–1349. doi: 10.1038/sj.gt.3300540. [DOI] [PubMed] [Google Scholar]
- Chen ZY, He, CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- Cloutier TE, Widom J. Spontaneous sharp bending of double-stranded DNA. Mol Cell. 2004;14:355–362. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- Horowitz DS, Wang JC. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984;173:75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Shore D, Langowski J, Baldwin RL. DNA flexibility studied by covalent closure of short fragments into circles. Proc Natl Acad Sci USA. 1981;78:4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J, Furrer P, Stasiak A, Dubochet J, Egelman EH, Bates AD. The twist, writhe and overall shape of supercoiled DNA change during counterion-induced transition from a loosely to a tightly interwound superhelix. J Mol Biol. 1994;235:825–847. doi: 10.1006/jmbi.1994.1042. [DOI] [PubMed] [Google Scholar]
- Fogg JM, Kolmakova N, Rees I, Magonov S, Hansma H, Perona JJ, et al. Exploring writhe in supercoiled minicircle DNA. J Phys: Condens Matter. 2006;18:S145–S159. doi: 10.1088/0953-8984/18/14/S01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Fogg JM, Zechiedrich L, Zu Y. Transfection of shRNA-encoding Minivector DNA of a few hundred base pairs to regulate gene expression in lymphoma cells. Gene Therapy. 2011;18:220–224. doi: 10.1038/gt.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW.Protocol 1: generation of a library of randomly overlapping DNA insertsIn: Argentine J (ed.).Molecular Cloning: A Laboratory Manual Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY; 200612.10–12.25. [Google Scholar]
- May KR. The collison nebulizer: description, performance and application. J Aerosol Sci. 1973;4:235–238. [Google Scholar]
- Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, et al. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- Zechiedrich EL, Khodursky AB, Cozzarelli NR. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman RD, Davidson N. Hydrodynamic shear breakage of DNA. Biopolymers. 1972;11:2601–2624. doi: 10.1002/bip.1972.360111217. [DOI] [PubMed] [Google Scholar]
- Lee CS, Davidson N. Flow dichroism of deoxyribonucleic acid solutions. Biopolymers. 1968;6:531–550. doi: 10.1002/bip.1968.360060408. [DOI] [PubMed] [Google Scholar]
- Lankas F, Lavery R, Maddocks JH. Kinking occurs during molecular dynamics simulations of small DNA minicircles. Structure. 2006;14:1527–1534. doi: 10.1016/j.str.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc Natl Acad Sci USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall GL, Zechiedrich L, Pettitt BM. In the absence of writhe, DNA relieves torsional stress with localized, sequence-dependent structural failure to preserve B-form. Nucleic Acids Res. 2009;37:5568–5577. doi: 10.1093/nar/gkp556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Yant SR, He CY, Meuse L, Shen S, Kay MA. Linear DNAs concatemerize in vivo and result in sustained transgene expression in mouse liver. Mol Ther. 2001;3:403–410. doi: 10.1006/mthe.2001.0278. [DOI] [PubMed] [Google Scholar]
- Bennett WD, Brown JS, Zeman KL, Hu SC, Scheuch G, Sommerer K. Targeting delivery of aerosols to different lung regions. J Aerosol Med. 2002;15:179–188. doi: 10.1089/089426802320282301. [DOI] [PubMed] [Google Scholar]
- O'Sullivan BP, Freedman SD. Cystic fibrosis. Lancet. 2009;373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- Kozma N, Halasz M, Polgar B, Poehlmann TG, Markert UR, Palkovics T, et al. Progesterone-induced blocking factor activates STAT6 via binding to a novel IL-4 receptor. J Immunol. 2006;176:819–826. doi: 10.4049/jimmunol.176.2.819. [DOI] [PubMed] [Google Scholar]
- Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010;285:30139–30149. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzoni AR, Ribeiro SC, Monteiro GA, Prazeres DM. The impact of polyadenylation signals on plasmid nuclease-resistance and transgene expression. J Gene Med. 2007;9:392–402. doi: 10.1002/jgm.1031. [DOI] [PubMed] [Google Scholar]
- Bigger BW, Tolmachov O, Collombet JM, Fragkos M, Palaszewski I, Coutelle C. An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J Biol Chem. 2001;276:23018–23027. doi: 10.1074/jbc.M010873200. [DOI] [PubMed] [Google Scholar]
- Swartz MN, Trautner TA, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J Biol Chem. 1962;237:1961–1967. [PubMed] [Google Scholar]
- Chen ZY, He CY, Meuse L, Kay MA. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Therapy. 2004;11:856–864. doi: 10.1038/sj.gt.3302231. [DOI] [PubMed] [Google Scholar]
- Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiss P, Cameron B, Rangara R, Mailhe P, Aguerre-Charriol O, Airiau M, et al. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res. 1999;27:3792–3798. doi: 10.1093/nar/27.19.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Xiang P, Li Q. Investigations of the effect of DNA size in transient transfection assay using dual luciferase system. Anal Biochem. 2005;346:289–294. doi: 10.1016/j.ab.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Ullmann A, Perrin D, Jacob F, Monod J. Identification par complementation in vitro et purification d′un segment peptidique de la β-galactosidase d′Escherichia coli [letter] J Mol Biol. 1965;12:918–923. doi: 10.1016/s0022-2836(65)80338-2. [DOI] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier M, Nixon AE, Shim JH, Benkovic SJ. Combinatorial protein engineering by incremental truncation. Proc Natl Acad Sci USA. 1999;96:3562–3567. doi: 10.1073/pnas.96.7.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PQ, Liu S, Thompson JC, Silberg JJ. Thermostability promotes the cooperative function of split adenylate kinases. Protein Eng Des Sel. 2008;21:303–310. doi: 10.1093/protein/gzn005. [DOI] [PubMed] [Google Scholar]
- Hoff KG, Culler SJ, Nguyen PQ, McGuire RM, Silberg JJ, Smolke CD. In vivo fluorescent detection of Fe–S clusters coordinated by human GRX2. Chem Biol. 2009;16:1299–1308. doi: 10.1016/j.chembiol.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Stuchinskaya T, Mitchenall LA, Schoeffler AJ, Corbett KD, Berger JM, Bates AD, et al. How do type II topoisomerases use ATP hydrolysis to simplify DNA topology beyond equilibrium? Investigating the relaxation reaction of nonsupercoiling type II topoisomerases. J Mol Biol. 2009;385:1397–1408. doi: 10.1016/j.jmb.2008.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HY, Liu LF. DNA looping alters local DNA conformation during transcription. J Mol Biol. 1991;219:615–622. doi: 10.1016/0022-2836(91)90658-s. [DOI] [PubMed] [Google Scholar]
- Bliska JB, Cozzarelli NR. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987;194:205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.