Abstract
Major depressive disorder (MDD) is a common complex disorder with a partly genetic etiology. We conducted a genome-wide association study of the MDD2000+ sample (2431 cases, 3673 screened controls and >1 M imputed single-nucleotide polymorphisms (SNPs)). No SNPs achieved genome-wide significance either in the MDD2000+ study, or in meta-analysis with two other studies totaling 5763 cases and 6901 controls. These results imply that common variants of intermediate or large effect do not have main effects in the genetic architecture of MDD. Suggestive but notable results were (a) gene-based tests suggesting roles for adenylate cyclase 3 (ADCY3, 2p23.3) and galanin (GAL, 11q13.3); published functional evidence relates both of these to MDD and serotonergic signaling; (b) support for the bipolar disorder risk variant SNP rs1006737 in CACNA1C (P=0.020, odds ratio=1.10); and (c) lack of support for rs2251219, a SNP identified in a meta-analysis of affective disorder studies (P=0.51). We estimate that sample sizes 1.8- to 2.4-fold greater are needed for association studies of MDD compared with those for schizophrenia to detect variants that explain the same proportion of total variance in liability. Larger study cohorts characterized for genetic and environmental risk factors accumulated prospectively are likely to be needed to dissect more fully the etiology of MDD.
Keywords: major depressive disorder, depression, genome-wide association study, CACNA1C, ADCY3, GAL
Introduction
Major depressive disorder (MDD) is a common and debilitating disorder with pervasive impact on the quality of life for both sufferers and their families. Lifetime prevalence is estimated to be ∼15%,1, 2 and is consistently estimated to be twice as common in women as men.3, 4 MDD is associated with high morbidity, reflected in estimates of burden of disease and years lost in productivity,1, 5 and excess mortality from suicide6 and other causes. Our understanding of the etiology of MDD remains fragmented, despite wide-ranging research, but is the key to effective prevention and treatment.
MDD is familial, with heritability estimated to be 0.37 (95% confidence interval (CI) 0.31–0.42) and both early age of onset and recurrence of depression are associated with higher familial aggregation.7, 8 This implied genetic etiology has motivated studies designed to identify specific genetic variants associated with MDD. Results from genome-wide linkage, reviewed in Boomsma et al.,9 and candidate gene association studies10 have shown little consistency and hopes for new progress have spurred on a generation of genome-wide association studies (GWAS). Five GWAS for MDD have been published to date,11, 12, 13, 14, 15 each using control samples screened negative for MDD. None of these studies has identified variants that achieve genome-wide significance. Taken together, the results of these studies imply that specific genetic variants individually make very small contributions to the etiology of MDD. In this study, we present the largest GWAS for MDD to date, the MDD2000+ study comprising 2431 cases and 3673 screened controls. We compare our results with reports of the other published MDD GWAS and present a formal meta-analysis of our results with the two other largest studies (5763 cases and 6901 controls).
Materials and methods
Overview
The MDD2000+ project comprises a total of 2431 cases with MDD and 3673 screened controls from different sources and genotyped on different platforms (Tables 1 and 2). Samples were provided by the Queensland Institute of Medical Research (QIMR, Australia), The Netherlands Study of Anxiety and Depression (NESDA), The Netherlands Twin Registry (NTR), the University of Edinburgh (UK), and the Molecular Genetics of Schizophrenia study (controls only, US). Genotyping was conducted on different Illumina and Affymetrix platforms and because the overlap in genotyped single-nucleotide polymorphisms (SNPs) is limited, association analysis is based on a set of >1 M imputed SNPs. The number of SNPs for each analysis set (Table 1), represents genotyped SNPs surviving all quality control (QC) criteria that were used for imputation.
Table 1. Analysis set statistics: numbers of samples, numbers of SNPs, ages at interview and MDD onset.
| Analysis set | Platform and approximate number of SNPs genotyped | Post-quality control autosomal SNPs | Post-quality control X chromosome SNPs | Sample source | N | No. of males | No. of females | No of recurrent and early onseta | Age at interview mean±s.d. | Age of onset mean±s.d. |
|---|---|---|---|---|---|---|---|---|---|---|
| I317 | Illumina 317 k | 289 130 | 8722 | QIMR casesb | 84 | 0 | 84 | 31 | 47.5±12.0 | 34.5±12.5 |
| QIMR controlsb | 237 | 0 | 237 | — | 45.4±13.2 | — | ||||
| I370 | Illumina 370 k | 276 135 | 6846 | QIMR casec | 737 | 325 | 412 | 264 | 42.9±9.1 | 27.8±10.4 (N=735) |
| QIMR controlsc | 795 | 443 | 352 | — | 41.0±12.6 (N=794) | — | ||||
| Illumina 610 k | NTR controlsf | 577 | 269 | 308 | — | 49.7±13.4 | — | |||
| I610 | Illumina 610 k | 510 092 | 12 822 | QIMR casesd | 169 | 38 | 131 | 45 | 42.8±10.5 | 31.2±11.5 |
| QIMR controlsd | 428 | 132 | 296 | — | 41.9±12.4 | |||||
| A6.0 | Affymetrix 600 k | 560 631 | 14 447 | QIMR casese | 941 | 298 | 643 | 480 | 42.2±9.9 | 25.4±9.8 (N=940) |
| NESDA/NTR casese | 127 | 45 | 82 | 36 | 41.3±11.7 (N=126) | 28.3±10.5 (N=92) | ||||
| UoE casese | 373 | 151 | 222 | 289 | 31.8±14.5 (N=352) | 23.3±10.9 (N=352) | ||||
| MGS controls | 1636 | 918 | 718 | — | 52.5±17.2 | — | ||||
| ALL | Imputed | 1 251 157 | 37 832f | Cases | 2431 | 857 | 1574 | 1145 | 41.1±11.4 (N=2409) | 26.7±10.8 (N=2372) |
| Controls | 3673 | 1762 | 1911 | — | 48.0±15.8 (N=3613) | — |
Abbreviations: MAF, minor allele frequency; MDD, major depressive disorder; MGS, Molecular Genetics of Schizophrenia; NESDA, The Netherlands Study of Anxiety and Depression; NTR, The Netherlands Twin Registry; QIMR, Queensland Institute of Medical Research; SNP, single-nucleotide polymorphism; UoE, University of Edinburgh.
aEarly onset <31 years.
b,c,d,eSamples with the same superscript had cases and controls genotyped together sometimes in multiple batches.
fX chromosome imputed SNPs with MAF>0.01.
The NTR controls were genotyped on the Illumina Human610-Quad but were included in the I370 analysis set to balance proportions of cases and controls in each set.
Table 2. Descriptive statistics by analysis set and sample source.
| Analysis set | Sample source | Female (%) | Early onset <31 years (%) | Recurrent (%) | Recurrent and early onset (%) | Family history MDD (%) | Recurrent and/or early onset and/or family history (%)a | Education level (% low, middle, high) | Partner status (% with partner) | Regular smoker (%) | Neuroticism score Mean±s.d.b |
|---|---|---|---|---|---|---|---|---|---|---|---|
| I317 | QIMR cases (N=84) | 100 | 46 | 48 | 37 | 25 | 58 | 40,52,9 (N=81) | 73 (N=84) | 20 (N=82) | 0.46±1.02 (N=79) |
| QIMR controls (N=237) | 100 | 0 | — | 38,50,12 (N=228) | 81 (N=237) | 16 (N=230) | −0.58±0.80 (N=224) | ||||
| I370 | QIMR cases (N=737) | 56 | 65 | 43 | 36 | 40 | 76 | 27,39,34 (N=724) | 70 (N=737) | 69 (N=729) | 0.39±1.12 (N=371) |
| QIMR controls (N=795) | 44 | 0 | — | 28,38,34 (N=744) | 81 (N=779) | 59 (N=747) | −0.54±0.91 (N=524) | ||||
| NTR controls (N=577) | 53 | 0 | — | 7,56,37 (N=573) | 92 (N=565) | 17 (N=573) | −0.43 ±0.76 (N=560) | ||||
| I610 | QIMR cases (N=169) | 78 | 55 | 37 | 27 | 22 | 69 | 35,40,25 (N=161) | 66 (N=169) | 47 (N=163) | 0.40±1.01 (N=140) |
| QIMR Controls (N=428) | 69 | 0 | — | 25,51,24 (N=410) | 86 (N=423) | 38 (N=405) | −0.29±1.00 (N=354) | ||||
| A6.0 | QIMR cases (N=941) | 68 | 77 | 66 | 51 | 35 | 95 | 24,39,37 (N=924) | 67 (N=940) | 51 (N=923) | 0.47±1.17 (N=708) |
| NESDA/NTR Cases (N=127) | 65 | 71 | 38 | 28 | 85 (N=106) | 91 | 27,37,35 (N=124) | 67 (N=106) | 43 (N=126) | 0.43±0.86 (N=122) | |
| UoE cases (N=373) | 60 | 78 | 100 | 78 | 46 (N=362) | 100 | NA | NA | NA | NA | |
| MGS controls (N=1636) | 44 | NA | — | NA | NA | NA | NA | ||||
| ALL | Cases (N=2431) | 65 | 71 | 60 | 47 | 39 | 86 | ||||
| Controls (N=3673) | 52 | — |
Abbreviations: MDD, major depressive disorder; MGS, Molecular Genetics of Schizophrenia; NA, not available; NESDA, The Netherlands Study of Anxiety and Depression; NTR, The Netherlands Twin Registry; QIMR, Queensland Institute of Medical Research; UoE, University of Edinburgh.
Family history for QIMR and NTR cases and controls is based on direct psychiatric interview assessment of family members; and was a screening criterion for controls. For NESDA and UoE cases family history is self-report.
For QIMR cases and controls standardized residuals from regression of arcsin-transformed scores on age and sex in a population sample of >18 000.60 For NESDA/NTR cases, standardized scores across based on these cases and GAIN-MDD cases and controls. For NTR controls z-transformed scores based on a population sample of >19 000.
Subject recruitment
QIMR
Study participants were adult twins and their families recruited through the Australian Twin Registry (http://www.twins.org.au). Only unrelated individuals were included in MDD2000+. All participants provided written informed consent under study protocols approved by the QIMR Human Research Ethics Committee. MDD cases were identified through psychiatric questionnaires, either the shortened Composite International Diagnostic Interview16 or the SSAGA-OZ interview instrument (a version of the Semi-Structured Assessment for the Genetics of Alcoholism17 modified for use in Australia), a comprehensive psychiatric interview designed to assess MDD and other psychiatric disorders17 according to DSM-IIIR18 and DSM-IV19 criteria. Structured interviews were administered by trained telephone interviewers, closely supervised by a clinical psychologist. Briefly, from 1988 to 1990, study participants were mailed an extensive health and lifestyle questionnaire, which included the shortened revised Eysenck personality questionnaire.20 Sum scores of 12 item responses in each personality domain resulted in quantitative scores for neuroticism. Between 1992–2000, an unselected subset of these participants were interviewed by telephone using the SSAGA-OZ.21 Over the period 1996–9 sibling pairs that were either concordant or discordant for extreme neuroticism scores (one sibling in the top or bottom decile, the other sibling in the top or bottom quintile) were recruited to complete the Composite International Diagnostic Interview, which provides DSM-IV19 lifetime diagnoses of MDD.22 Finally, some study participants completed the SSAGA-OZ telephone questionnaire in 2003–2007 as part of alcohol and nicotine dependence studies (the nicotine addiction genetics and Inter-related Project Grant studies described in Table 2 of Hansell et al.23). The Inter-related Project Grant/nicotine addiction genetics studies captured 28% of families who had already participated in earlier studies ascertaining those with either (a) large sibship size or containing a proband with either (b) nicotine dependence or (c) alcohol dependence. For this study, all cases met DSM-IV lifetime criteria for MDD. Screening items for mania were not consistent across interviews and screening items for psychosis were not included; the ability to assess accurately these less common criteria is difficult in large-scale community settings. Therefore, it is possible that a small number of individuals with a primary diagnosis of bipolar disorder or schizophrenia are included in the case group. If multiple cases were present within families then one case was selected in the following order of preference: age of onset <31 years, multiple episodes of depression, co-morbid anxiety disorders and high neuroticism score. Unrelated controls were selected as genotyped individuals from families in which no individuals qualified for diagnoses of MDD or anxiety disorders. If multiple controls were available from a family, the individual with the lowest neuroticism score was preferentially selected, otherwise an individual was selected at random.
NESDA and NTR
Additional MDD cases were selected from two parallel studies, NESDA (http://www.nesda.nl) and NTR (http://www.tweelingenregister.org); NTR also provided screened controls. These samples do not overlap with those included in a prior MDD GWAS12 but are drawn from the same parent studies (although a small number of related individuals are included, see meta-analysis section below). Details of the data collection methods are described elsewhere.9 Similar inclusion and exclusion criteria were used to select MDD cases from both the NESDA and NTR studies. Inclusion criteria were a lifetime diagnosis of DSM-IV MDD as determined by the Composite International Diagnostic Interview,16 age 18–65 years, and self-reported western European ancestry. Those not fluent in Dutch or with a primary diagnosis of schizophrenia or schizoaffective disorder, obsessive–compulsive disorder, bipolar disorder or severe substance use or dependence were excluded. All subjects provided written informed consent after study approval by the appropriate Institutional Review Boards.
University of Edinburgh
MDD cases were recruited through in- and out-patient services of psychiatric hospitals in Scotland and were tertiary referrals from primary care. All patients were interviewed by an experienced psychiatrist using the Schedule for Affective Disorders and Schizophrenia-Lifetime,24 supplemented by hospital case note review and information from informants. Final determination of MDD as the primary DSM-IV diagnosis was made by consensus of two psychiatrists. All cases had a lifetime history of recurrent MDD and IQ >70. The study was approved by the Central Office of Research Ethics Committees in Scotland and all subjects gave informed written consent for the collection of DNA samples for use in genetic studies.
Molecular Genetics of Schizophrenia controls
Briefly, random digit dialing was used to achieve a representative sample of individuals from the United States. Participants completed an online questionnaire including the short form Composite International Diagnostic Interview, supplemented by questions about schizophrenia, psychosis and bipolar disorder. Controls were required to never have met criteria for MDD. All subjects provided informed consent. These controls have been used in GWAS for multiple psychiatric disorders including MDD.13, 14
Genotyping and QC
Full details are given in the Supplementary File 1. All analysis sets were put through a common QC pipeline based on that used by the Psychiatric GWAS Consortium.25 SNPs were removed based on the following criteria: minor allele frequency (MAF) <0.01, Hardy–Weinberg equilibrium test P<1 × 10−6 in controls, missingness >0.01 or 0.02, missingness difference between cases and controls of 0.01 or 0.02, difference in frequency with HapMap of 0.07 or 0.15 (more stringent thresholds were applied when cases and controls were not genotyped concurrently, and were chosen to balance SNPs lost versus genomic control inflation factors). Samples were removed with missingness >0.02, if related to other samples or if identified as European ancestry outliers. Imputation was conducted in four analysis sets (I317, I370, I610, A6.0) in batches of ∼300 mixed cases and controls to a common set of SNPs present in HapMap3 CEU/TSI, consisting of 410 haplotypes, using Beagle 3.04.26, 27
Statistical analyses
Association analysis was conducted on 2431 MDD cases and 3673 screened controls using allelic dosages for imputed SNPs passing QC. The test of association was logistic regression, including the first three ancestry principal components (PC) and analysis set as covariates. The INFO score (ratio of observed to expected dosage variance), a measure of the quality of imputation of SNPs was used to interpret results. A second association analysis was conducted restricting cases to those with recurrent early onset MDD (REO, age of MDD onset <31 years with multiple episodes of MDD). Separate analyses were conducted by sex, as different prevalences of MDD between males and females could imply existence of sex-specific genetic risk variants.
Statistical power
Detailed power calculations are provided in the Supplementary File 1. The MDD2000+ sample affords >90% power to detect an associated variant with MAF 0.36 and genotype relative risk (GRR) 1.33 (the median values across all published complex disease/trait GWAS, with P<5 × 10−8).28
Comparison of association results with other studies including meta-analysis
We compared our results with those published for other GWAS of MDD. In particular, we conducted a formal genome-wide meta-analysis for autosomal SNPs from MDD2000+, GAIN–MDD12 and UK15 studies (that is, the three largest MDD GWAS). The GAIN–MDD sample, after removing individuals related to Dutch cases or controls of MDD2000+, comprised 1696 MDD cases and 1634 screened controls and the UK sample comprised 1636 MDD cases and 1594 screened controls. Meta-analysis29 on the logistic regression results from each study, restricting the imputed SNPs to those with INFO scores between 0.8 and 1.1.
Gene-based tests
To determine whether any genes harbored an excess of SNPs with small P-values, we undertook genome-wide gene-based tests which account for both gene length and linkage disequilibrium between SNPs using VEGAS.30 SNPs were allocated to one or more autosomal genes using gene boundaries ±50 kb. We investigated 183 candidate genes for MDD.12, 13, 14, 15, 31
Results
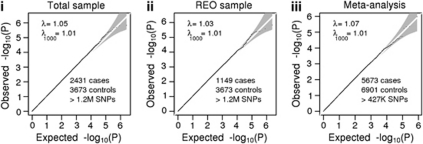
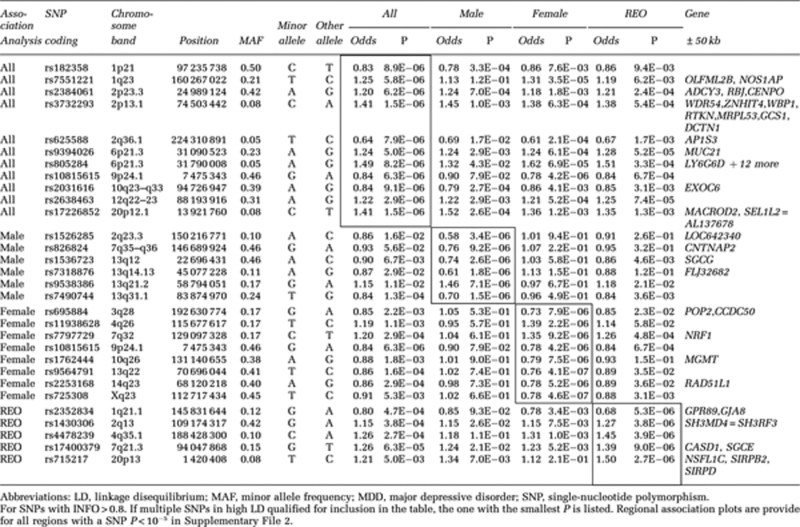
Of 1,251,157 imputed SNPs, 1,079,979 (86%) had MAF>0.01 and INFO>0.8. The quantile–quantile plots of the observed versus expected −log(P) from the four (total, male, female and REO) association analyses are presented in Figure 1 and Supplementary File 1. The genomic control λ (the median χ2 association statistic divided by the median expected under the null) shows no evidence for inflation of the test statistics. For example, for the full analysis, λ=1.05 (and standardized to a sample size of 1000, λ1000=1.02). There were no genome-wide significant results with P<5 × 10−8. The Manhattan plot of association P-values for all subjects is presented in the Supplementary File 1 and Table 3 shows associations with <1 × 10−5 in any analysis. Regional association and forest plots are provided in Supplementary File 2. SNPs associated with P<0.001 in any analysis are provided in Supplementary File 3.
Figure 1.
Quantile–quantile plots for the association analyses of (i) all cases and controls and (ii) recurrent early onset (iii) meta-analysis.
Table 3. Regions containing at least one SNP with a P-value of P<10−5 in any one of the four association analyses—total sample (all), males only, females only and recurrent early onset (REO) MDD ordered by chromosomal position.
Comparison of association results with other studies including meta-analysis
We compared the MDD2000+ results with other published studies. Muglia et al.11 listed 27 SNPs with P<1 × 10−5; 25 SNPs overlapped with our analysis but none had P<0.05 and association with the same allele. Of the top 200 SNPs in one or both of their study samples, ∼90% were analysed in our study but ∼5% had P<0.05, consistent with chance expectations. As the MDD2000+ controls were also used in GenRed13 and STARD,14 we did not make formal comparisons of our results, but we note that ∼5% of their top SNPs (P<1 × 10−5) had P<0.05 in MDD2000+. The SNP rs2251219 identified as a mood disorder risk factor in a GWAS meta-analysis (three bipolar studies plus the GAIN–MDD study)32 but was not associated in MDD2000+ (P=0.51, MAF=0.40, odds ratio (OR)=0.97, 95% CI 0.90–1.05). PCLO was the top finding in the GAIN–MDD study,12 replicated in Australian (QIMR)12 and Dutch33 samples, but we found no evidence for association with PCLO variants (for example, P=0.51 for rs2522833), despite partial overlap (562 cases and 264 controls) in the QIMR samples used here and in the original replication sample (see Supplementary File 1 for details). Meta-analysis results of the three largest MDD GWAS—427362 SNPs in MDD2000+, GAIN–MDD and the UK studies—yielded λ1000=1.01 (Figure 1iii). In all, 19 SNPs gave P<5 × 10−5 compared with ∼21 expected under the null hypothesis (ignoring linkage disequilibrium) and are listed in the Supplementary File 1.
Gene-based tests
MDD2000+ SNPs mapped to 18 454 genes. We list all genes with P<1 × 10−4 and all lie within linkage regions identified in the SLEP34 database of research findings for psychiatric disorders. Gene ontology annotations and names are provided for these genes (Table 4). Three are plausible candidate genes for MDD (GAL, ADCY3, PDK4) and others have a role in ion binding (ZDHHC19, ZNF83). None of the genes listed in Table 4 shows any evidence for association in the gene-based test applied to GAIN–MDD or the UK MDD GWAS results, although association at P<0.001 is retained for GAL in the gene-based test applied to the meta-analysis results.
Table 4. Top ten ranked genes from gene-based test for genes ±50 kb from start and stop positions.
| Rank | Band | Gene ID | No. of SNPs | Length (kb) | Start (bp) of gene | Stop (bp) of gene | Gene test P | SNP with minimum P-value | SNP stats3 | Gene name | Molecular function from GO annotation | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 2p23.3 | ADCY3 | a. b. c. d. | 84 169 29 28 | 100 | 24 895 541 | 24 995 559 | a. b. c. d. | 4.3E−05 6.3E−01 7.3E−01 3.1E−01 | rs2384061 | 6.3E−06 A 0.42 1.20 | Adenylate cyclase 3 | Nucleotide binding; magnesium ion binding; calmodulin binding; ATP binding; calcium- and calmodulin-responsive adenylate cyclase activity |
| 8 | 5q35 | NPM1 | a. b. c. d. | 52 52 16 15 | 23 | 170 747 402 | 170 770 493 | a. b. c. d. | 3.3E−04 6.6E−01 9.6E−01 1.7E−01 | rs11134697 | 2.7E−04 G 0.47 1.16 | Nucleophosmin | Nucleic acid binding; DNA binding; RNA binding; protein binding; rRNA binding; ribosomal large subunit binding; ribosomal small subunit binding |
| 3 | 6q27 | UNC93A1 | a. b. c. d. | 95 147 31 29 | 25 | 167 624 890 | 167 649 491 | a. b. c. d. | 4.4E−05 2.3E−01 6.1E−01 4.9−01 | rs2076008 | 1.3E−04 G 0.26 1.19 | Unc-93 homolog A | — |
| 5 | 6q27 | TTLL21 | a. b. c. d. | 84 131 27 25 | 18 | 167 658 563 | 167 676 167 | a. b. c. d. | 1.1E−04 3.5E−01 5.6E−01 5.2E−01 | rs2076008 | 1.3E−04 G 0.26 1.19 | Tubulin tyrosine ligase-like family, member 2 | Catalytic activity; tubulin–tyrosine ligase activity; ATP binding; ligase activity |
| 7 | 7q21.3 | PDK42 | a. b. c. d. | 77 133 29 28 | 13 | 95 050 744 | 95 063 861 | a. b. c. d. | 2.1E−04 6.1E−01 1.8E−01 3.5E−01 | rs11531570 | 3.4E−05 A 0.41 1.18 | Pyruvate dehydrogenase kinase, isozyme 4 | Two-component sensor activity; nucleotide binding; pyruvate dehydrogenase (acetyl transferring) kinase activity; ATP-binding; transferase activity |
| 10 | 7q21–q22 | ASB42 | a. b. c. d. | 104 182 46 46 | 52 | 94 953 219 | 95 005 007 | a. b. c. d. | 3.6E−04 6.5E−01 4.5E−01 5.6E−01 | rs11531570 | 3.4E−05 A 0.41 1.18 | Ankyrin repeat and SOCS box-containing 4 | — SOCS=suppressor of cytokine signalling |
| 4 | 9q31.1 | TEX10 | a. b. c. d. | 51 90 17 14 | 51 | 102 104 190 | 102 154 995 | a. b. c. d. | 4.7E−05 1.8E−01 8.5E−01 1.1E−01 | rs1930243 | 2.7E−04 A 0.49 1.15 | Testis expressed 10 | Binding |
| 6 | 11q13.3 | GAL | a. b. c. d. | 48 52 11 11 | 7 | 68 208 558 | 68 215 219 | a. b. c. d. | 1.5E−04 2.4E−01 7.8E−01 2.4E−03 | rs2156464 | 2.7E−05 A 0.19 1.24 | Galanin | Neuropeptide hormone activity |
| 9 | 15q22.3 | USP3 | a. b. c. d. | 74 121 23 23 | 87 | 61 583 862 | 61 670 716 | a. b. c. d. | 3.5E−04 9.8E−01 8.5E−01 5.8E−02 | rs7183892 | 4.5E−04 C 0.15 1.20 | Ubiquitin-specific peptidase 3 | Ubiquitin thiolesterase activity; ubiquitin-specific protease activity; peptidase activity; cysteine-type peptidase activity; zinc ion binding; metal ion binding |
| 1 | 20p12.1 | SEL1L2 = AL137678 | a. b. c. d. | 98 209 46 42 | 141 | 13 778 049 | 13 919 262 | a. b. c. d. | 1.1E−05 9.8E−01 7.5E−01 3.9E−01 | rs17226852 | 1.5E−06 C 0.08 1.41 | Sel-1 suppressor of lin-12-like 2 | Binding |
Abbreviations: ATP, adenosine triphosphate; GO, gene ontology; MAF, minor allele frequency; MDD, major depressive disordes; SNP, single-nucleotide polymorphism; rRNA, ribosomal RNA.
a, MDD2000+ b, GAIN-MDD; c, UK GWAS; d, meta-analysis. 1The ±50 kb regions of UNC93A and TTLL2 overlap. 2The ±50 kb regions of PDK4 and ASB4 overlap.
P<1 × 10−2 in bold. Top associated SNPs lie in region ±50 kb from the listed start and stop positions. Regional association plots for ADCY3, GAL and CACNA1C are in Supplementary File 2. SNP stats: P-value, minor allele, MAF, odds ratio.
Of the 183 candidate genes investigated in other GWAS for MDD, we could test for association with 180 (Supplementary File 4). None were associated with P<0.00028 (0.05/180). The six most associated were IL10, OPRM1, HTT, HTR1B, GRIN1 and CACNA1C. Of these, OPRM1 was reported to have P<0.05 in the GAIN–MDD gene-based test and CACNA1C was top ranked in the GenRed13 study; the other studies all used a gene test based on best single associated SNP corrected for gene length. In this study, CACNA1C ranked 808 out of 18 454 (4th percentile). Our top associated SNP in CACNA1C is rs98545 (P=0.0019, OR=0.83, MAF=0.15) which is 387 kb from, and in near linkage equilibrium (r2=0.06) with, rs1006737 (P=0.020, OR=1.10, MAF=0.35), the SNP first identified in genome-wide association studies of bipolar disorder;35 an interaction between these SNPs was not significant (P=0.21). The 3rd ranked of the candidate genes (Huntingtin, HTT) was included in the candidate list because mood disorders are characteristic of presymptomatic carriers of the Huntington's disease trinucleotide repeat polymorphisms.31 HTT ranked in the top 0.7% of all genes; the SNP showing highest individual association within the gene is rs363099 (also known as rs4690074, P=4.7 E−04, OR=0.86, MAF=0.31) located in exon 29, 85 kb from the exon 1 CAG repeat.
Discussion
Study results
The MDD2000+ study is the largest GWAS for MDD reported to date, comprising 2431 cases with MDD and 3673 screened controls. Our analysis was of >1 M SNPs but none achieved genome-wide significance. Indeed, the quantile–quantile plots (Figure 1) show that the distribution of observed associations closely follows that expected under the null hypothesis. These results are consistent with other GWAS of MDD.11, 12, 13, 14, 15 Furthermore, comparison of our results with these other studies including formal meta-analysis of results from MDD2000+ and the next two largest studies12, 15 (a total of 5763 cases and 6901 controls) showed little evidence for replication between studies.
To determine whether any genes harbored an excess of low-associated variants, we undertook gene-based tests which account for both gene length and linkage disequilibrium between SNPs. Our top associated genes were not replicated in the gene-based test when applied to the GAIN–MDD or UK MDD results, although some genes retained low association P-values when applied to the meta-analysis results (Table 4). Despite this there are some suggestive results worthy of note. Among the top ranked genes is GAL, which encodes the neuropeptide galanin, proposed by Weiss et al.36 as having an important role in MDD. Their hypothesis, based on animal models is that GAL released in the ventral tegmentum inhibits the activity of dopaminergic cells resulting in decreased motor activity and anhedonia. GAL is a regulator of brain serotonin and 5-HT1A receptor-mediated transmission,37 agonists of GAL receptors have been proposed as potential drug targets for MDD.38 GAL also has an important role in the hippocampal processing of cognition.39 Our most associated SNP in GAL (rs2156464, P=2.7 × 10−5, OR=1.24, CI=1.12–1.37, MAF=0.19) lies in the same haplotype block as an association reported for panic disorder.40 Another top ranked gene (ADCY3) encodes the enzyme adenylate cyclase 3 that catalyses synthesis of cyclic adenosine monophosphate, the association underpinned by SNP rs2384061 (P=6.3 × 10−6, OR=1.20, CI=1.11–1.29, MAF=0.42, Table 3). ADCY3 is an plausible candidate gene because depressed patients display reduction in platelet ADCY activity.41 Perhaps most interesting is that ADCY and GAL activity are inter-related42 and ADCY3 binds CACNA1C (calcium channel, voltage dependent, L type, α-1C subunit).43 A SNP (rs1006737, intron 3) within the gene CACNA1C is associated with bipolar disorder44 schizophrenia and MDD.45 In this study, CACNA1C ranked in the top 4% of genes and we replicated the association with rs1006737 (P=0.020, OR=1.10, CI=1.01–1.19, MAF=0.34), but the top associated SNP in CACNA1C was rs98545 (P=0.0019, OR=0.83, CI 0.75–0.94, MAF=0.15, intron 29). rs98545 lies 387 kb from rs1006737; they are in near linkage equilibrium (r2=0.06) and in our data their effects are additive in a logistic regression. Neither rs1006737 nor rs98545 were genotyped in the UK MDD or GAIN–MDD studies; imputation of GAIN–MDD genotypes showed weak association with rs1006737 (P=0.070, OR=1.10, CI=0.99–1.21, MAF=0.32) but not with rs98545 (P=0.52). We cannot exclude that the association of rs98545 has occurred by chance in our sample. Regional association plots for ADCY3, GAL and CACNA1C can be found in Supplementary File 2.
Despite the large sample size of MDD2000+, a number of limitations may reduce the realized power for detection of association. First, our study combines data sets genotyped on different primary platforms, and the power of our sample will vary between SNPs depending on the linkage disequilibrium structure between genotyped and imputed SNPs. Second, some of our cases and controls were genotyped separately because of economic constraints. We were fortunate that we had >300 QIMR cases genotyped on both the Affymetrix and Illumina platforms, which we used extensively for QC. Third, when combining cases and controls genotyped as separate sets, it was necessary to impose more stringent QC constraints than for cases and controls genotyped on the same platform. These constraints were successful in removing artefactual differences between the case–control sets, but could also have eliminated true associations. Fourth, in MDD2000+ with the aim of maximizing sample size we have combined the clinical cases from the University of Edinburgh cohort with the community samples from QIMR and NTR. The extent to which this is important depends on the unknown genetic etiology of MDD. Lastly, 26% of both cases and controls contributed by the QIMR sample (which makes the largest contribution in this study) were from families ascertained on the basis of a sibling with nicotine dependence, as reflected by the high proportion of smokers in the QIMR samples. Smoking is associated with depression and if some genes are involved in smoking behavior and MDD, then the power to detect genetic variants for MDD may be reduced.
Implications
Our analysis and meta-analysis represent the largest and most powerful investigation into the genetic architecture of MDD to date. The lack of clear-cut evidence for association allows us to exclude genetic main effects that are common (MAF>0.1) with moderate to strong effect sizes (GRR>1.4). As noted earlier, the MDD2000+ sample alone possessed >90% power to detect the median MAF (0.36) and GRR (1.33) of GWAS associations published to date with P<5 × 10−8. Moreover, as we would expect to detect >20% of variants with OR 1.2 and MAF 0.2–0.7 (Supplementary File 1), and having detected none that are genome-wide significant, we can probably draw an even more stringent boundary on the risk allele architecture under a main effect model. These results are informative for researchers planning association studies of MDD.
These results are unsurprising in the light of results for a range of complex genetic disorders published since this study was conceived in 2007.46 Results from GWAS of psychiatric disorders47 have, on the whole, yielded replicated associations only for low prevalence/high heritability disorders, that is, autism, bipolar disorder and schizophrenia, with tobacco dependence being the exception.48 As a high prevalence/low heritability disorder, the challenge for MDD was always going to be harder. From Yang et al.,49 we estimate that sample sizes 2.4-fold greater are needed for GWAS of MDD (prevalence 0.15) compared with schizophrenia (prevalence 0.007) to identify a variant that explains the same proportion of phenotypic variance to liability of these disorders (Supplementary File 1). This result reflects the smaller mean difference in phenotypic liability between cases and controls for MDD compared with schizophrenia. Hospital-based MDD cohorts may represent a more extreme phenotype, with lower prevalence and higher heritability.50 Using a prevalence for such clinical samples to be 0.06 (the average across sexes)51 still requires a sample size ∼1.8 times greater for a case–control study of MDD compared with one for schizophrenia.
Further, for main effect models, as the heritability on the liability scale (h2) is a function of the number of variants (n), their frequencies (qi) and their effect sizes (ai),
 |
(with total variance of liability of 1), then the lower heritability of MDD (∼0.37, although this may be higher in clinical samples)51 compared to schizophrenia (∼0.81), must be explained by fewer risk alleles, lower risk allele frequencies and/or smaller effect sizes. It seems most plausible that MDD might have smaller effect sizes compared with schizophrenia (except in the unlikely event that the number of loci for MDD is substantially fewer than for schizophrenia). We estimate that sample sizes 4–5 times those needed for schizophrenia studies may be needed for MDD to detect variants that explain an equal proportion of the known genetic variance (Supplementary File 1).
Study designs
It is clear that larger sample sizes are needed to achieve the power required to detect common variants of smaller effect, and that larger sample sizes will be needed for MDD compared with less prevalent but more heritable disorders. The prize is major: any replicated hit provides a priceless window into the etiology of this idiopathic disorder. However, alternative and complementary strategies are possible, particularly those that consider a broader range of genetic models and relax further assumptions of etiological homogeneity.
First, recognizing that effect size is the average effect of a variant across all genetic and environmental backgrounds,52 it may be possible to forgo larger sample sizes to concentrate on more homogenous subsets in which larger effect sizes may be detectable. There has been much debate about the genetic heterogeneity of MDD. Some genetic heterogeneity is consistent with the polygenic model; each affected individual could potentially harbor a different combination of risk variants which could imply a spectrum of phenotypic symptoms, with those presenting similar symptom profiles carrying more similar profiles of risk alleles, consistent with symptom sharing of closely related individuals.53 For this reason, most GWAS for MDD have prioritized genotyping of the less prevalent and more heritable recurrent early onset MDD, but even so genome-wide significant associations have been elusive. Stratification by sex has also not yielded consistent results. Other possibilities certainly exist but phenotypes may be unavailable (for example, accurate delineation of typical versus atypical MDD)54 or prohibitively expensive (for example, magnetic resonance imaging). Use of quantitative scores of severity and reliability may be the best strategy to balance sample size with phenotype definition,55 as MDD symptom profiles are consistent with a quantitative rather than dichotomous liability.56
Second, in addition to achieving a more homogenous genetic background, larger estimated effect sizes may come from selecting more homogenous environmental backgrounds. The low heritability of liability for MDD implies an important role for environmental risk factors. Although genotype × environment interaction cannot explain the so-called ‘missing heritability',52 it can contribute to small effect sizes. Although genotype × environment studies are conceptually attractive, the lessons learned from the most studied genotype × environment hypothesis for MDD (5HTTLPR and stressful life event) are sobering.57 However, this broad conclusion has been challenged58, 59 with Uher and McGuffin58 arguing that replication has been achieved when stressful life event were recorded objectively, temporally and before the onset to MDD. More mileage might be gained by considering MDD in the context of more clearly defined exposures, such as pregnancy, in which women with perinatal and post-partum MDD have been exposed to a similar event.
Whichever way we look at it, and whether risk variants are common or rare, it seems that the challenge for MDD will be much harder than for the less prevalent more heritable psychiatric disorders. Larger samples are required whether we attempt to identify associated variants with small effect across average backgrounds or attempt to enhance detectable effects sizes by selection of homogeneity of genetic or environmental background. In the long-term, a greater understanding of the etiology of MDD will require large prospective, longitudinal, uniformly and broadly phenotyped and genotyped cohorts that allow the joint dissection of the genetic and environmental factors underlying MDD.
Acknowledgments
We thank the twins and their families registered at the Australian Twin Registry for their participation in the many studies that have contributed to this research. We thank Dixie Statham (sample collection); Leanne Wallace, Anthony Caracella and staff of the Molecular Epidemiology Laboratory (DNA processing); David Smyth, Harry Beeby, and Daniel Park (IT support). Funding was provided by the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496675, 496739, 552485, 552498, 613608), the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254), and the US National Institutes of Health (NIH grants AA07535, AA10248, AA13320, AA13321, AA13326, AA14041, MH66206, DA12854, DA019951). A portion of the genotyping on which this study was based (Illumina 370K scans on 4300 individuals) was carried out at the Center for Inherited Disease Research, Baltimore through an access award to our late colleague Dr Richard Todd (Psychiatry, Washington University School of Medicine, St Louis). Statistical analyses were partly conducted at the Genetic Cluster Computer (http://www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003). SEM, SM and GWM are supported by the National Health and Medical Research Council Fellowship Scheme. NRW and DRN are supported by the Australian Research Council Future Fellowship Scheme.
Funding support was provided by the Netherlands Scientific Organization (904-61-090, 904-61-193, 480-04-004, 400-05-717, Centre for Medical Systems Biology (NWO Genomics), the Neuroscience Campus Amsterdam and the EMGO+ institute; the European Union (EU/WLRT-2001-01254), ZonMW (Geestkracht program, 10-000-1002), NIMH (RO1 MH059160) and matching funds from participating institutes in NESDA and NTR. The NTR controls were genotyped in the Genomics platform (certified service provider (CSPro(R)) for Illumina) at the LIFE and BRAIN Center Bonn (funded by NWO-SPI 56-464-1419).
We would like to record our appreciation for much practical support from our late colleague Walter J Muir, Professor of Developmental Psychiatry at the University of Edinburgh and especially for his many stimulating ideas for genetic studies in depression. We thank Margaret Van Beck for sample collection and Lee Murphy, Wellcome Trust Clinical Research Facility, Western General Hospital, Crewe Road South, Edinburgh, for sample management. The collection of the Edinburgh cohort was supported by grants from The Wellcome Trust, London and the Chief Scientist Office of the Scottish Government.
Additional funding was provided by National Institute of Mental Health Schizophrenia (NIMH) grant MH080403 to EJGCvdO and PFS. Control subjects from the NIMH Genetics Initiative, data and biomaterials are being collected by the ‘Molecular Genetics of Schizophrenia II' collaboration. The investigators and coinvestigators are: ENH/Northwestern University, Evanston, IL, MH059571, Pablo V Gejman, MD (Collaboration Coordinator; PI), Alan R Sanders, MD; Emory University School of Medicine, Atlanta, GA, MH59587, Farooq Amin, MD (PI); Louisiana State University Health Sciences Center; New Orleans, Louisiana, MH067257, Nancy Buccola APRN, BC, MSN (PI); University of California-Irvine, Irvine, CA, MH60870, William Byerley, MD (PI); Washington University, St Louis, MO, U01, MH060879, C Robert Cloninger, MD (PI); University of Iowa, Iowa, IA, MH59566, Raymond Crowe, MD (PI), Donald Black, MD; University of Colorado, Denver, CO, MH059565, Robert Freedman, MD (PI); University of Pennsylvania, Philadelphia, PA, MH061675, Douglas Levinson, MD (PI); University of Queensland, Queensland, Australia, MH059588, Bryan Mowry, MD (PI); Mt. Sinai School of Medicine, New York, NY, MH59586, Jeremy Silverman, PhD (PI).
Author contributions
Funding: PFS, EJCGvdO, PAFM, MLP, DIB, NGM, GWM, ACH, DHB, BWJHP, IBH
QIMR sample: Phenotype: MLP, PAFM, DS, ACH, NGM; Biobank: GWM, AKH
NTR sample: DIB, JJH, GW, CM, EJCG
NESDA sample: BWJHP, JHS
University of Edinburgh sample: DHRB, DMM, KAMcG, AWM
UK GWAS: CML, PMcG
Analysis Set QC: DRN, SDG, NRW, BPMcE, SEM
Joint analysis set QC, imputation, association analysis and regional plots of MDD2000+: SR
Post-association analysis analyses: NRW, SDG, EMB, JZL, SM
Manuscript preparation: NRW, PFS and all other authors
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Australian Bureau of Statistics 4326.0—National Survey of Mental Health and Wellbeing: Summary of Results, 2007 http://wwwabsgovau/AUSSTATS/abs@nsf/mf/43260 .
- Kessler RC, Berglund P, Demler O, Jin R, Walters EE. Lifetime prevalence and age-of-onset distributions' of DSM-IV disorders in the national comorbidity survey replication. Arch Gen Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- Weissman MM, Bland R, Joyce PR, Newman S, Wells JE, Wittchen HU. Sex-differences in rates of depression—cross-national perspectives. J Affect Disord. 1993;29:77–84. doi: 10.1016/0165-0327(93)90025-f. [DOI] [PubMed] [Google Scholar]
- Wilhelm K, Mitchell P, Slade T, Brownhill S, Andrews G. Prevalence and correlates of DSM-IV major depression in an Australian national survey. J Affect Disord. 2003;75:155–162. doi: 10.1016/s0165-0327(02)00040-x. [DOI] [PubMed] [Google Scholar]
- Craddock N, Forty L. Genetics of affective (mood) disorders. Eur J Hum Genet. 2006;14:660–668. doi: 10.1038/sj.ejhg.5201549. [DOI] [PubMed] [Google Scholar]
- Fairweather-Schmidt AK, Anstey KJ, Mackinnon AJ. Is suicidality distinguishable from depression? Evidence from a community-based sample. Aust N Z J Psychiatry. 2009;43:208–215. doi: 10.1080/00048670802653331. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Gatz M, Gardner CO, Pedersen NL. Age at onset and familial risk for major depression in a Swedish national twin sample. Psychol Med. 2005;35:1573–1579. doi: 10.1017/S0033291705005714. [DOI] [PubMed] [Google Scholar]
- Boomsma DI, Willemsen G, Sullivan PF, Heutink P, Meijer P, Sondervan D, et al. Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet. 2008;16:335–342. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- Lopez-Leon S, Janssens A, Ladd A, Del-Favero J, Claes SJ, Oostra BA, et al. Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry. 2008;13:772–785. doi: 10.1038/sj.mp.4002088. [DOI] [PubMed] [Google Scholar]
- Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ, et al. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, de Geus EJC, Willemsen G, James MR, Smit JH, Zandbelt T, et al. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, et al. Genome-wide association study of recurrent early-onset major depressive disorder Mol Psychiatry2 February 2010; e-pub ahead of print. [DOI] [PMC free article] [PubMed]
- Shyn SI, Shi J, Kraft JB, Potash JB, Knowles JA, Weissman MM, et al. Novel loci for major depression identified by genome-wide association study of sequenced treatment alternatives to relieve depression and meta-analysis of three studies Mol Psychiatry29 December 2009; e-pub ahead of print. [DOI] [PMC free article] [PubMed]
- Lewis CM, Ng MY, Bulter AW, Cohen-Woods S, Uher R, Pirlo K, et al. Genome-wide association study of major depression in the UK population. Am J Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- World Health Organisation . Composite International Diagnostic Interview Version 2.1. World Health Organization: Geneva; 1997. [Google Scholar]
- Bucholz KK, Cloninger CR, Dinwiddie DH, Hesselbrock VM, Nurnberger JL, Reich T, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report of the reliability of the SSAGA. J Stud Alcoholism. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders, revised 3rd edn. American Psychiatric Association: Washington, DC; 1987. [Google Scholar]
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders, revised 4th edn. American Psychiatric Association: Washington DC; 1994. [Google Scholar]
- Eysenck SBG, Eysenck HJ, Barrett P. A revised version of the pyschoticism scale. Pers Individ Dif. 1985;6:21–30. [Google Scholar]
- Bierut LJ, Heath AC, Bucholz KK, Dinwiddie SH, Madden PA, Statham DJ, et al. Major depressive disorder in a community-based twin sample: are there different genetic and environmental contributions for men and women. Arch Gen Psychiatry. 1999;56:557–563. doi: 10.1001/archpsyc.56.6.557. [DOI] [PubMed] [Google Scholar]
- Kirk KM, Birley AJ, Statham DJ, Haddon B, Lake RI, Andrews JG, et al. Anxiety and depression in twin and sib pairs extremely discordant and concordant for neuroticism: prodromus to a linkage study. Twin Res. 2000;3:299–309. doi: 10.1375/136905200320565274. [DOI] [PubMed] [Google Scholar]
- Hansell NK, Agrawal A, Whitfield JB, Morley KI, Zhu G, Lind PA, et al. Long-term stability and heritability of telephone interview measures of alcohol consumption and dependence. Twin Res Hum Genet. 2008;11:287–305. doi: 10.1375/twin.11.3.287. [DOI] [PubMed] [Google Scholar]
- Endicott J, Spitzer RL. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry. 1978;35:837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- The Psychiatric GWAS Consortium Genome-wide association studies: history, rationale and prospects for psychiatric disorders. Am J Psychiatry. 2009;166:540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH, Smoller JW, Mysore J, Sun M, Gillis T, Purcell S, et al. Prevalence of incompletely Penetrant Huntington's disease alleles among individuals with major depressive disorder. Am J Psychiatry. 2010;167:574–579. doi: 10.1176/appi.ajp.2009.09070973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD, et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat Genet. 2010;42:128–131. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hek K, Mulder CL, Luijendijk HJ, van Duijn CM, Hofman A, Uitterlinden AG, et al. The PCLO gene and depressive disorders: replication in a population-based study. Hum Mol Genet. 2010;19:731–734. doi: 10.1093/hmg/ddp529. [DOI] [PubMed] [Google Scholar]
- Konneker T, Barnes T, Furberg H, Losh M, Bulik CM, Sullivan PF. Rapid publication—a searchable database of genetic evidence for psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:671–675. doi: 10.1002/ajmg.b.30802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MAR, Perlis RH, Chambert K, et al. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JM, Bonsall RW, Demetrikopoulos MK, Emery MS, West CH. Galanin: a significant role in depression. Ann N Y Acad Sci. 1998;863:364–382. doi: 10.1111/j.1749-6632.1998.tb10707.x. [DOI] [PubMed] [Google Scholar]
- Ogren SO, Schott PA, Kehr J, Yoshitake T, Misane I, Mannstrom P, et al. Modulation of acetylcholine and serotonin transmission by galanin. Relationship to spatial and aversive learning. Ann N Y Acad Sci. 1998;863:342–363. doi: 10.1111/j.1749-6632.1998.tb10706.x. [DOI] [PubMed] [Google Scholar]
- Ogren SO, Kuteeva E, Hokfelt T, Kehr J. Galanin receptor antagonists—a potential novel pharmacological treatment for mood disorders. CNS Drugs. 2006;20:633–654. doi: 10.2165/00023210-200620080-00003. [DOI] [PubMed] [Google Scholar]
- Ogren SO, Kuteeva E, Elvander-Tottie E, Hokfelt T. Neuropeptides in learning and memory processes with focus on galanin. Eur J Pharmacol. 2010;626:9–17. doi: 10.1016/j.ejphar.2009.09.070. [DOI] [PubMed] [Google Scholar]
- Unschuld PG, Ising M, Erhardt A, Lucae S, Kohli M, Kloiber S, et al. Polymorphisms in the galanin gene are associated with symptomseverity in female patients suffering from panic disorder. J Affect Disord. 2008;105:177–184. doi: 10.1016/j.jad.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Hines LM, Tabakoff B, Trait WISS. Platelet adenylyl cyclase activity: a biological marker for major depression and recent drug use. Biol Psychiatry. 2005;58:955–962. doi: 10.1016/j.biopsych.2005.05.040. [DOI] [PubMed] [Google Scholar]
- Karelson E, Langel U. Galaninergic signalling and adenylate cyclase. Neuropeptides. 1998;32:197–210. doi: 10.1016/s0143-4179(98)90038-5. [DOI] [PubMed] [Google Scholar]
- Ingenuity knowledge base http://www.ingenuity.com 2010
- Ferreira MAR, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry. 2010;15:1016–1022. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock NJ, Owen MJ. Genetics of psychosis; insights from views across the genome. Hum Genet. 2009;126:3–12. doi: 10.1007/s00439-009-0703-0. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PAF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wray NR, Visscher PM. Comparing apples and oranges: equating the power of case-control and quantitative trait association studies. Genet Epidemiol. 2010;34:254–257. doi: 10.1002/gepi.20456. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Cohen S, Knight J. Homing in on depression genes. Am J Psychiatry. 2007;164:195–197. doi: 10.1176/ajp.2007.164.2.195. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Katz R, Watkins S, Rutherford J. A hospital-based twin register of the heritability of DSM-IV unipolar depression. Arch Gen Psychiatry. 1996;53:129–136. doi: 10.1001/archpsyc.1996.01830020047006. [DOI] [PubMed] [Google Scholar]
- Falconer D, Mackay T.Introduction to Quantitative Genetics4th edn.Longman: England; 1996. 464pp. [Google Scholar]
- Korszun A, Moskvina V, Brewster S, Craddock N, Ferrero F, Gill M, et al. Familiality of symptom dimensions in depression. Arch Gen Psychiatry. 2004;61:468–474. doi: 10.1001/archpsyc.61.5.468. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kessler RC, Kendler KS. Latent class analysis of lifetime depressive symptoms in the National Comorbidity Survey. Am J Psychiatry. 1998;155:1398–1406. doi: 10.1176/ajp.155.10.1398. [DOI] [PubMed] [Google Scholar]
- Hettema JM, Neale MC, Myers JM, Prescott CA, Kendler KS. A population-based twin study of the relationship between neuroticism and internalizing disorders. Am J Psychiatry. 2006;163:857–864. doi: 10.1176/ajp.2006.163.5.857. [DOI] [PubMed] [Google Scholar]
- Rice JP, Rochberg N, Endicott J, Lavori PW, Miller C. Stability of psychiatric diagnoses—an application to the affective disorders. Arch Gen Psychiatry. 1992;49:824–830. doi: 10.1001/archpsyc.1992.01820100068012. [DOI] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression a meta-analysis. JAMA. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uher R, McGuffin P. The moderation by the serotonin transporter gene of environmental adversity in the etiology of depression: 2009 update. Mole Psychiatry. 2010;15:18–22. doi: 10.1038/mp.2009.123. [DOI] [PubMed] [Google Scholar]
- Rutter M, Thapar A, Pickles A. Gene-environment interactions biologically valid pathway or artifact. Arch Gen Psychiatry. 2009;66:1287–1289. doi: 10.1001/archgenpsychiatry.2009.167. [DOI] [PubMed] [Google Scholar]
- Wray NR, Birley AJ, Sullivan PF, Visscher PM, Martin NG. Genetic and phenotypic stability of measures of neuroticism over 22 years. Twin Res Hum Genet. 2007;10:695–702. doi: 10.1375/twin.10.5.695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.