Abstract
Oligomerization of connexins is a critical step in gap junction channel formation. Some members of the connexin family can oligomerize with other members and form functional heteromeric hemichannels [e.g. Cx43 (connexin 43) and Cx45], but others are incompatible (e.g. Cx43 and Cx26). To find connexin domains important for oligomerization, we constructed chimaeras between Cx43 and Cx26 and studied their ability to oligomerize with wild-type Cx43, Cx45 or Cx26. HeLa cells co-expressing Cx43, Cx45 or Cx26 and individual chimaeric constructs were analysed for interactions between the chimaeras and the wild-type connexins using cell biological (subcellular localization by immunofluorescence), functional (intercellular diffusion of microinjected Lucifer yellow) and biochemical (sedimentation velocity through sucrose gradients) assays. All of the chimaeras containing the third transmembrane domain of Cx43 interacted with wild-type Cx43 on the basis of co-localization, dominant-negative inhibition of intercellular communication, and altered sedimentation velocity. The same chimaeras also interacted with co-expressed Cx45. In contrast, immunofluorescence and intracellular diffusion of tracer suggested that other domains influenced oligomerization compatibility when chimaeras were co-expressed with Cx26. Taken together, these results suggest that amino acids in the third transmembrane domain are critical for oligomerization with Cx43 and Cx45. However, motifs in different domains may determine oligomerization compatibility in members of different connexin subfamilies.
Keywords: chimaera, connexin26, connexin43, gap junction, intercellular communication, oligomerization
INTRODUCTION
Gap junction channels connect the cytoplasms of adjacent cells allowing intercellular passage of ions and molecules up to 1 kDa in size between coupled cells. Each gap junction channel is an oligomer composed of twelve protein subunits of connexin. The connexins are polytopic membrane proteins containing four TM (transmembrane) (TM1–TM4), three cytoplasmic [NT (N-terminal), IL (intracellular loop) and CT (C-terminal)], and two extracellular (E1 and E2) domains (reviewed in [1]). Six connexins oligomerize within the secretory pathway to form a hemichannel or connexon. Different connexins oligomerize in different intracellular compartments, for example Cx43 (connexin 43) in the trans-Golgi network [2,3] and Cx32 in the ER (endoplasmic reticulum) [3]. After trafficking to the plasma membrane through the secretory pathway, connexons dock with complementary connexons of the adjacent cell leading to the formation of gap junction channels.
Many cell types express more than one connexin subtype, making possible oligomerization between identical subunits to form homomeric connexons or between different subunits to form heteromeric connexons. Homomeric connexons may form homotypic gap junction channels (when docking takes place between two connexons made of the same connexin), or heterotypic gap junction channels (when docking involves two homomeric connexons made of different connexins). The resulting homotypic, heterotypic and heteromeric gap junction channels have specific and greatly diverse properties (e.g. single channel conductance, permeability, gating and regulation by kinases) [4–9].
We and others have shown that Cx43 forms heteromeric channels with Cx45, Cx40 and Cx37, but not with the ‘β’ connexins, Cx26 and Cx32 [4,10–14]. However, there is limited available information regarding the molecular determinants for connexin compatibility and oligomerization between different connexins. Recently, Maeda et al. [15] published the crystal structure of the gap junction channel formed by human Cx26 at 3.5 Å (1 Å = 0.1 nm) resolution. This structure confirms that the docking between the two adjoining connexons involves both E1 and E2, consistent with previous studies showing that amino acid sequences in E2 determine compatibility of connexins for heterotypic pairing [16]. This structure also indicates that the inter-protomer interactions that stabilize the hexameric connexon are mostly located in the extracellular half of transmembrane helices and in the extracellular loops [15]. However, the domains determining oligomerization compatibility remain unknown. Our previous studies suggest that the CT domain of Cx43 is not needed for oligomerization, since Cx43tr251, a truncated form of Cx43, forms functional homomeric channels [17]. When expressed alone, Cx43tr251 does not form gap junction plaques detectable by immunofluorescence; however, when co-expressed with wild-type Cx43 or Cx45, Cx43tr251 forms gap junction plaques [17]. The experiments presented below were designed to identify connexin domains that determine oligomerization compatibility. To identify critical domains for connexin compatibility, we generated a series of chimaeric constructs in which domains were reciprocally exchanged between two incompatible connexins, Cx43 and Cx26, to determine whether they oligomerize with wild-type connexins.
EXPERIMENTAL
Strategy for generation of chimaeric constructs
Chimaeric constructs were generated by PCR using LA Taq DNA polymerase and primer sequences based on rat Cx43 and Cx26 cDNAs (see Supplementary Table S1 at http://www.BiochemJ.org/bj/435/bj435ppppadd.htm). Chi1–4 (chimaeras 1–4) were generated through a double PCR protocol in which the first PCR product was used as a primer (megaprimer) for the second PCR [18,19]. Chi5 and Chi6 were produced through three successive PCR reactions; the first two PCR reactions used standard oligonucleotide primers with different templates, whereas the third PCR used the product of the first PCR as one primer and the product of the second PCR as template. Details of templates and primers used to generate the different chimaeras are indicated in Supplementary Table S2 (at http://www.BiochemJ.org/bj/435/bj435ppppadd.htm). The chimaeric constructs were all subcloned into pcDNA3.1/Hygro+ (Invitrogen).
Transfections and cell culture
Geneticin-resistant HeLa cells stably transfected with Cx43 or Cx45 were grown as described previously [4,5]. Puromycin-resistant HeLa cells transfected with wild-type Cx26 (HeLaCx26) were provided by Dr Bruce Nicholson (University of Texas Health Science Center, San Antonio, TX, U.S.A.) [9]. Stable co-expression of chimaeras with Cx43 or Cx26 was obtained after transfection of the chimaeric constructs into HeLaCx43 or HeLaCx26 cells respectively, and clonal selection with 100 μg/ml Hygromycin (Calbiochem/EMD). For co-expression with Cx45, HeLaCx45 cells were transiently transfected with the chimaeric constructs.
Indirect immunofluorescence
Cells grown on glass coverslips were fixed in methanol/acetone (1:1) and permeabilized with 1% Triton X-100 prior to incubation with primary antibodies. Single- and double-labelling immunofluorescence was performed using rabbit polyclonal anti-HA (haemagglutinin) antibodies (71–5500; Zymed/Invitrogen) to detect chimaeras, and mouse monoclonal anti-Cx43 (MAB 3068; Chemicon/Millipore), anti-Cx26 (13–8100; Zymed/Invitrogen), anti-Cx45 (MAB 3100; Chemicon/Millipore) or anti-Golgi 58K (Sigma Chemical) antibodies. Cy3-conjugated goat anti-mouse and Cy2-conjugated goat anti-rabbit IgG antibodies were obtained from Jackson ImmunoResearch. Cells were examined using a Zeiss Axiophot II microscope, and images were captured using an AxioCam digital camera.
Immunoblotting
Total cell homogenates were prepared as described previously [4]. Protein from total cell homogenates (100 μg) or of Triton X-100-soluble or -insoluble fractions (50–100 μl) were resolved on SDS/PAGE gels (8% or 10%), and electrotransferred onto Immobilon-P membranes (Millipore). The membranes were subjected to immunoblotting using anti-Cx43, anti-Cx45, anti-Cx26 or anti-HA antibodies as described previously [4,17]. Antibody binding was detected by chemiluminescence (ECL; Amersham) followed by exposure to X-ray film.
Triton X-100 extraction and sedimentation velocity of connexin monomers and oligomers through sucrose density gradients
HeLa cell cultures at 70% confluence were harvested in ice-cold PBS containing 2 mM PMSF and the 100 000 g Triton X-100-soluble fractions were subjected to sedimentation velocity through 10–20% or 5–20% (w/v) linear sucrose gradients as described previously [20]. After centrifugation for 22 h at 100 000 g, 250 μl fractions were collected and analysed by immunoblotting. The monomeric connexin peak was assigned based on the percentage of sucrose at which SDS-solubilized Cx43 or chimaeras sedimented. The intensity of the bands in the different fractions was determined by densitometry.
Assessment of intercellular communication
Cells cultured on coverslips at 80% confluence were impaled with a micropipette filled with 4% Lucifer yellow (Sigma) in 150 mM LiCl. One cell within a cluster was microinjected with a picospritzer (model PLI-188; Nikon) or FemtoJet-injectMan microinjector (Eppendorf) using 0.2- to 0.3-s pulses of 1 to 2 psi (1 psi = 6.9 kPa) for 1 to 2 min. The extent of intercellular transfer was determined by recording the number of adjacent cells containing the tracer excluding the impaled cell after visualization by epifluorescence microscopy [4].
RESULTS
General strategy for production and analysis of Cx26–Cx43 chimaeric connexins
To identify domains that are critical for connexin oligomerization, we generated pairs of chimaeric constructs in which domains were reciprocally exchanged between two incompatible connexins, Cx43 and Cx26 (Figure 1). On the basis of our previous studies demonstrating that the CT domain is not required for Cx43 oligomerization [17], the chimaeras containing the backbone of Cx43 were generated using Cx43tr251 cDNA as template to obtain chimaeras similar in size to Cx26. Each chimaera contained an HA epitope appended to its CT domain to allow its detection and differentiation from wild-type Cx43 which was detected using antibodies directed against the CT domain, which was only present in the full-length protein.
Figure 1. Diagram of the chimaeric constructs used in the present study.
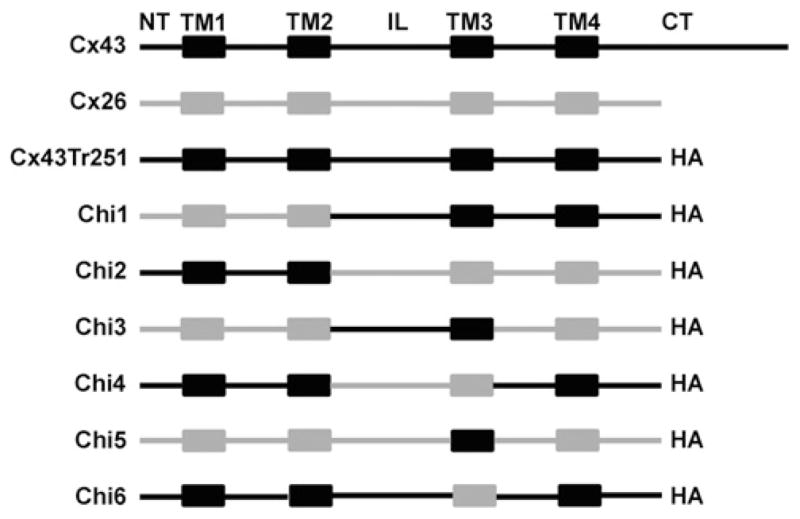
In each chimaera, the different domains are represented in grey if derived from rat Cx26 and in black if derived from rat Cx43. Chi1 contained amino acids 1–96 from Cx26 and 99–251 from Cx43. Chi2 contained amino acids 1–98 from Cx43 and 97–226 from Cx26. Chi3 contained amino acids 1–96 from Cx26, 99–184 from Cx43 and 165–226 from Cx26. Chi4 contained amino acids 1–98 from Cx43, 97–164 from Cx26 and 185–251 from Cx43. Chi5 contained amino acids 1–143 from Cx26, 154–184 from Cx43 and 165–226 from Cx26. Chi6 contained amino acids 1–153 from Cx43, 144–164 from Cx26 and 185–251 from Cx43. All chimaeras contained the HA tag appended at its C-terminus.
Co-expression of Chi1 or Chi2 with Cx43
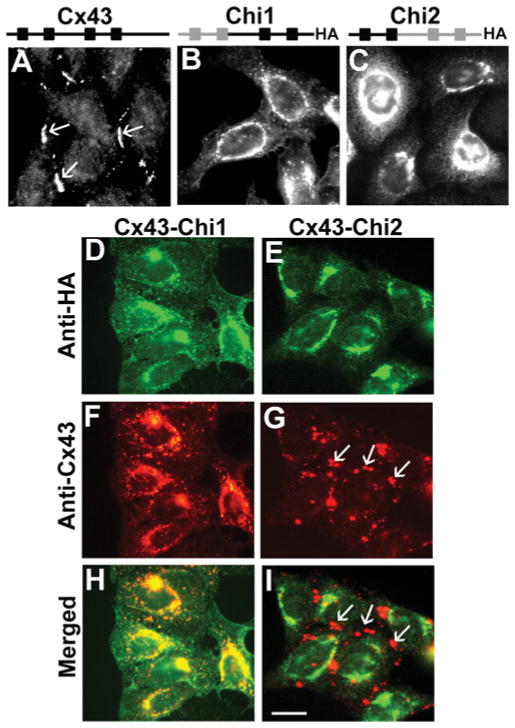
The first set of reciprocal chimaeras, Chi1 and Chi2, contained approximately half of Cx43 and Cx26 each (NT domain to TM2 or IL to CT domain) (Figure 1). The initial screen for interactions with wild-type Cx43 was performed by immunolocalization in transfected HeLa cells. When expressed alone in stably transfected HeLa cells (HeLaCx43), Cx43 localized at appositional membranes (as expected for gap junction plaques) and within the cytoplasm (probably within biosynthetic/secretory compartments) (Figure 2A). When expressed by themselves, Chi1 and Chi2 were only detected within the cytoplasm (Figures 2B and 2C). Double immunofluorescence showed that these chimaeras co-localized with the Golgi 58K protein (results not shown). When co-expressed with Cx43 in stably co-transfected HeLaCx43-Chi1 cells, Chi1 co-localized with Cx43 intracellularly; neither Cx43 nor Chi1 were detected at gap junction plaques (Figures 2D, 2F and 2H). In HeLaCx43-Chi2 cells, Chi2 only localized intracellularly, whereas Cx43 was mainly detected at gap junction plaques with little, if any, chimaera co-localization in an intracellular compartment (Figures 2E, 2G and 2I),. These results suggest that oligomerization of Chi1 with Cx43 retained the wild-type protein within the cytoplasm and that Chi2 did not interact with wild-type Cx43.
Figure 2. Chi1, but not Chi2, co-localizes with Cx43.
Top, schematic representation of constructsshowing Cx43 domains in black and Cx26 domains in grey. Photomicrographs show the immunofluorescence detection of Cx43 (A), Chi1 (B) and Chi2 (C) in stably transfected HeLa cells. Immunofluorescent detection of Chi1 (D) and Chi2 (E) and Cx43 (F and G) in HeLaCx43–Chi1 and HeLaCx43–Chi2 cells respectively. Merged images show co-localization of Cx43 with Chi1 (H) but not with Chi2 (I). Gap junction plaques are indicated by arrows. Scale bar, 15 μm.
Introduction of a second connexin (wild-type or mutant) into a cell can alter intercellular communication. Therefore, we studied intercellular transfer of microinjected Lucifer yellow in the transfected cells. HeLa cells transfected with wild-type Cx43 showed extensive intercellular transfer of Lucifer yellow as expected (Table 1). In HeLaCx43-Chi1 cells, transfer of Lucifer yellow was infrequent, and the number of gap junction tracer-filled neighbours was severely reduced as compared with HeLaCx43 cells (Table 1). In HeLaCx43-Chi2 cells, transfer of Lucifer yellow was observed after all injections, and the number of tracer-filled cells did not differ from that observed in cells expressing Cx43 alone. These results are consistent with a dominant-negative interaction between Chi1 and Cx43 and the absence of a functional interaction between Chi2 and Cx43.
Table 1. Intercellular transfer of Lucifer yellow in HeLa cells expressing wild-type and chimaera connexins.
Individual cells were microinjected with Lucifer yellow to determine the incidence and extent of gap junction coupling. n, number of injections; incidence of coupling, percentage of injections resulting in intercellular dye transfer; tracer-filled neighbours, number of coupled cells (means ± S.D.);
| Connexin–chimaera expression | n | Incidence of coupling (%) | Tracer-filled neighbors |
|---|---|---|---|
| Cx43 | 17 | 100 | 6.3 ± 2.4 |
| Cx43–Chi1 | 32 | 15 | 0.4 ± 1.1* |
| Cx43–Chi2 | 9 | 100 | 5.2 ± 2.3 |
| Cx43–Chi3 | 18 | 5 | 0.0* |
| Cx43–Chi5 | 40 | 0 | 0.0* |
| Cx26 | 30 | 100 | 10.0 ± 3.6 |
| Cx26–Chi1 | 9 | 100 | 13.2 ± 5.7 |
| Cx26–Chi3 | 9 | 55 | 1.7 ± 1.1* |
| Cx26–Chi5 | 30 | 13 | 1.0 ± 0.9* |
P < 0.001 as compared with cells expressing wild-type Cx43 or Cx26.
Interactions between connexins that result in formation of oligomers (or that do not) can be detected by sedimentation through sucrose gradients, because of the differences in sedimentation velocities between connexin monomers and connexins that have oligomerized into connexons [2,3,17,20,21]. Therefore, to examine potential molecular interactions between Cx26/Cx43 chimaeras and Cx43, we examined their sedimentation velocity through sucrose gradients.
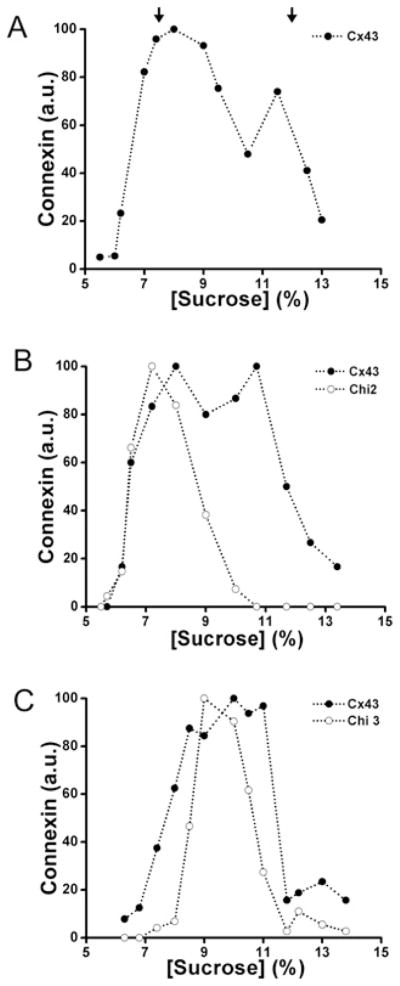
When analysed using a 10–20% sucrose gradient, wild-type Cx43 (solubilized from HeLaCx43 cells) showed a two peak pattern with one peak centred at 11–12% sucrose and a second peak centred at 14–15% sucrose (Figure 3A). Treatment of cell homogenates with SDS (which disrupts connexin oligomers) collapsed immunoreactive Cx43 into a single peak at 11–12% sucrose (Figure 3A). Thus the two peaks observed in the absence of SDS treatment corresponded to monomeric and oligomerized Cx43. In some experiments, we used 5–20% sucrose gradients. In these gradients, Cx43 showed the expected two peaks centred at 7–8% and 12% sucrose corresponding to monomers and hexamers respectively (Figure 4A).
Figure 3. Co-expression of Chi1 with wild-type Cx43 altered the oligomerization of Cx43.
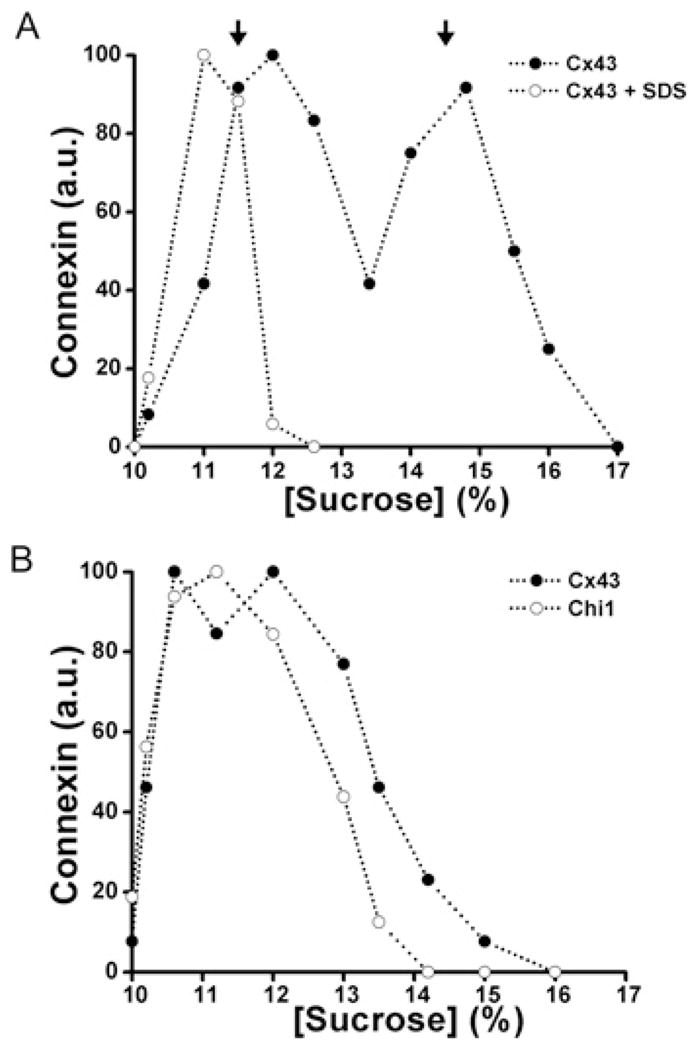
Graphs represent the levels of Cx43 and Chi1 detected by immunoblotting of 10–20 % sucrose gradient fractions of Triton X-100-soluble material from HeLa cells transfected with Cx43 alone (A) or Cx43 and Chi1 (B). The levels of Cx43 from HeLaCx43 cell homogenates subjected to sedimentation velocity through sucrose gradients after treatment with SDS are also represented in (A) (○). Arrows‘ indicate the percentage of sucrose at which Cx43 monomers and hexamers sediment.
Figure 4. Co-expressed Chi3, but not Chi2 co-oligomerizes with Cx43.
Graphs represent the levels of Cx43 and chimaeras detected by immunoblotting of fractions from 5–20 % sucrose gradients after sedimentation velocity of Triton X-100-soluble material from HeLa cells stably transfected with Cx43 alone (A), Cx43 and Chi2 (B) or Cx43 and Chi3 (C). Arrows indicate the percentage of sucrose at which Cx43 monomers and hexamers sediment.
The sedimentation velocities of wild-type Cx43 and chimaeras were determined in stably co-transfected cells. Material solubilized from HeLaCx43–Chi1 cells contained only a single, broad peak of Cx43 at low percentages of sucrose that was closely overlapped by the peak of Chi1 (Figure 3B). These results suggest that Chi1 interacted with Cx43, impairing its oligomerization into hexamers. In contrast, in material from HeLaCx43 cells transfected with the reciprocal chimaera, Chi2, wild-type Cx43 was present in two peaks (similar to those detected in cells expressing Cx43 alone) whereas Chi2 was only present in a single low sucrose percentage peak, the monomer peak (Figure 4B). These results suggest that Chi2 did not interact with or influence the oligomerization of Cx43.
Thus, co-expression of Chi1 with Cx43 affected the localization, function, and sedimentation velocity of the wild-type protein whereas none of these processes were affected by co-expression of Chi2. The simplest explanation for these results is that the Cx43 regions present in Chi1 (IL–TM4) are required for oligomerization with Cx43 whereas those in Chi2 (NT–TM2) are dispensable.
Co-expression of Chi3–Chi6 with Cx43
To define further the region of Cx43 responsible for oligomerization, we generated four additional chimaeras, Chi3 and Chi4 (in which only the IL–TM3 regions were interchanged between Cx26 and Cx43) and Chi5 and Chi6 (in which only TM3 was interchanged) (Figure 1). When expressed alone in transiently transfected HeLa cells, each of these chimaeras localized within the cytoplasm (Figures 5A–5D). The frequency of detection of connexin at appositional membranes was much lower for any of the chimaeras than for the wild-type protein.
Figure 5. Chi3 and Chi5, but not Chi4 and Chi6 co-localize with co-expressed Cx43.
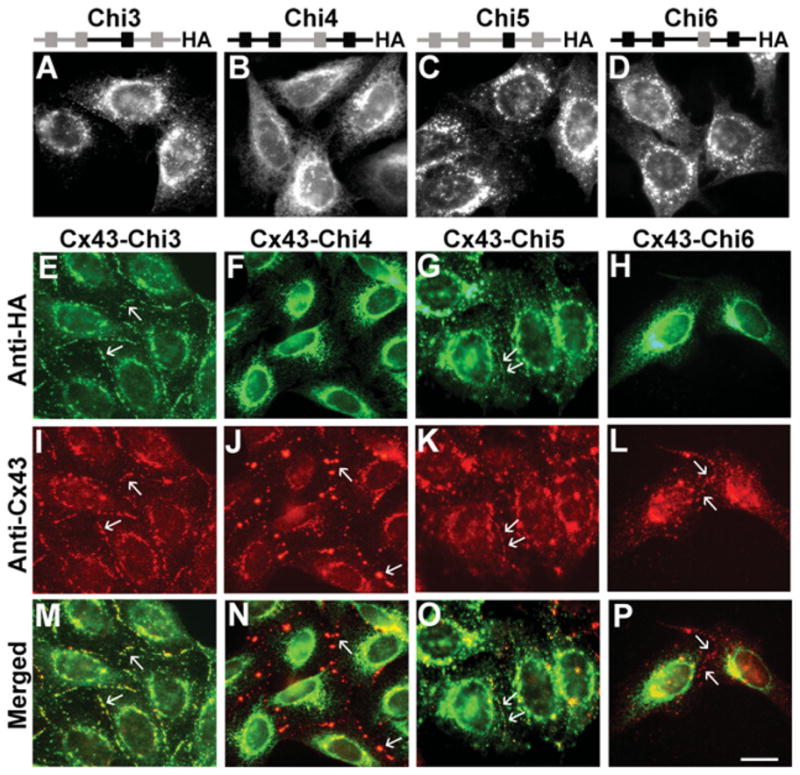
Top panels, schematic representation of expressedconstructs showing Cx43 domains in black and Cx26 domains in grey. Photomicrographs show the immunofluorescence detection of Chi3 (A), Chi4 (B), Chi5 (C) and Chi6 (D) in HeLa cells stably transfected with the respective cDNA construct. (E–P) Immunofluorescent detection of Chi3 (E), Chi4 (F), Chi5 (G), Chi6 (H) and Cx43 (I–L) in HeLaCx43–Chi3, HeLaCx43–Chi4, HeLaCx43–Chi5 and HeLaCx43–Chi6 cells respectively. Merged images show co-localization of Cx43 with Chi3 (M) or Chi5 (O) but not with Chi4 (N) or Chi6 (P). Arrows indicate gap junction plaques. Scale bar, 10 μm.
In co-transfected cells, two chimaeras, Chi3 and Chi5, co-localized with wild-type Cx43 (Figure 5) both intracellularly and at gap junction plaques (Figures 5E, 5I and 5M; and 5G, 5K and 5O). In contrast, Chi4 and Chi6 only localized intracellularly in HeLaCx43–Chi4 (Figures 5F and 5N) and HeLaCx43–Chi6 (Figures 5H and 5P) cells. In these cells, Cx43 was mainly detected at gap junction plaques (Figures 5J and 5L), and there was little, if any, co-localization between Cx43 and Chi4 or Chi6 in the cytoplasm. These results suggest that co-expression of Cx43 with Chi3 or Chi5 ‘rescued’ plaque formation by these chimaeras, a change in distribution that can best be explained by co-oligomerization with wild-type Cx43. The results also suggest that Chi4 and Chi6 did not interact with wild-type Cx43.
The two chimaeras that appeared to interact with Cx43 on the basis of immunofluorescence were further tested to see if they affected Cx43-mediated intercellular communication by measuring intercellular transfer of Lucifer yellow in co-transfected cells. Transfer of Lucifer yellow was infrequent, and the number of gap junction tracer-filled neighbours was substantially reduced in HeLaCx43 cells co-transfected with Chi3 or Chi5 (Table 1).
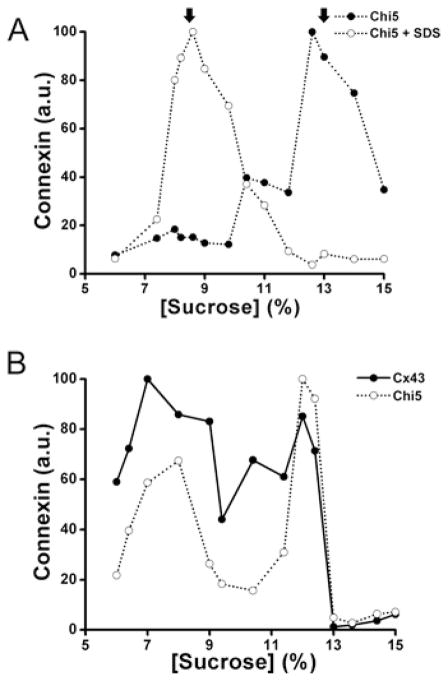
The sedimentation velocity of these chimaeras and co-expressed Cx43 were analysed using Triton X-100-soluble extracts. When samples prepared from HeLaCx43–Chi3 cells were analysed using 5–20% sucrose gradients, wild-type Cx43 and Chi3 co-sedimented in a major peak centred at 9–10% sucrose and a small peak centred at 12–13% sucrose (Figure 4C). These results are consistent with the presence of both Cx43 and Chi3 in oligomers of intermediate size and in hexamers. Fractions isolated from HeLaCx43–Chi4 cells showed two peaks of wild-type Cx43, but only one low sucrose percentage peak of Chi4, suggesting that Chi4 (similar to Chi2) did not oligomerize or interact with Cx43 (results not shown). Gradients prepared from HeLaChi5 showed a main peak centred at ~13% sucrose that collapsed to 8–9% sucrose when the Triton X-100-soluble fraction was pretreated with SDS before loading the gradients (Figure 6A). Gradients prepared from HelaCx43–Chi5 cells exhibited overlapping patterns of sedimentation for Cx43 and Chi5 with two peaks centred at ~8% and at 12% sucrose (Figure 6B). These two peaks contained monomers and oligomers respectively, since we observed only one major peak at ~8% sucrose containing both Cx43 and Chi5 when HeLaCx43–Chi5 Triton X-100-soluble fractions were treated with SDS before loading the gradient (results not shown). These data suggest that Cx43 and Chi5 co-oligomerize and form heteromers in HeLa cells.
Figure 6. Chi5 co-sediments with Cx43.
Graphs represent the levels of Cx43 and chimaeras detected by immunoblotting of 5–20 % sucrose gradient fractions of Triton X-100-soluble material from HeLaChi5 (A) and HeLaCx43–Chi5 (B) cells. When expressed alone (A), Chi5 was detected in a peak of 13–15 % sucrose that probably represented oligomers, since it was reduced to a peak of ~8–10 % sucrose by preincubation with SDS. In contrast, Cx43 and Chi5 were detected together in peaks at ~8 % and 12 % sucrose (B). Arrows indicate the sedimentation peaks of Chi5 oligomers and disrupted oligomers.
The common sequence among the interacting chimaeras is TM3, suggesting the importance of this domain for Cx43 oligomerization.
Cx43-interacting chimaeras also co-localized with wild-type Cx45
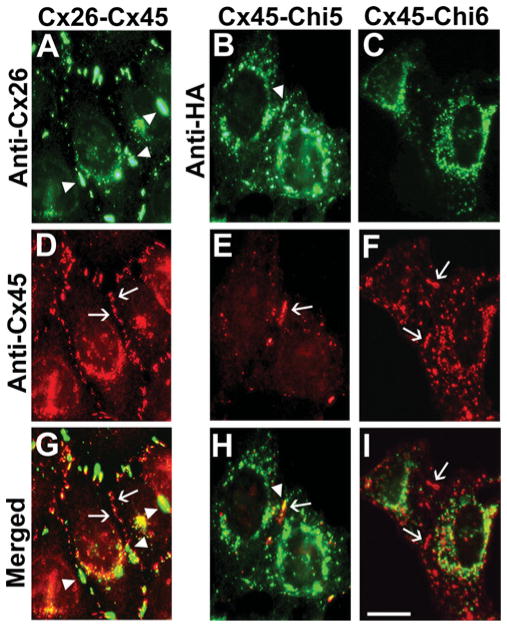
In a previous study, we showed that Cx45 interacts and oligomerizes with Cx43 forming heteromeric channels with unique properties [4]; this implies that Cx43 and Cx45 contain motifs that confer compatibility for hetero-oligomerization. Since it has not previously been reported whether Cx45 can oligomerize with Cx26, we first examined cells co-expressing these two wild-type connexins. When Cx45 was co-expressed with Cx26, both connexins localized in intracellular compartments and at gap junction plaques (Figures 7A, 7D and 7G). The overlap of the staining at appositional membranes was limited, and the sizes and shapes of Cx26- or Cx45-containing plaques appeared very different. When the chimaeras were transiently transfected into HeLaCx45 cells, we observed co-localization of Chi1, Chi3 and Chi5 (but not Chi2, Chi4 or Chi6) with Cx45 in the same compartments (Figure 7 and results not shown). These results suggest that the Cx43 TM3 also contains motifs that determine oligomerization compatibility between Cx43 and Cx45.
Figure 7. Chi5, but not Chi6, co-localizes with Cx45 in co-transfected cells.
Photomicrographs show immunofluorescentlocalization of wild-type Cx26 or Cx45 or chimaeras in transfected HeLa cells. (A, D and G) Co-expressed Cx26 (A) and Cx45 (D) both localized to gap junction plaques at appositional membranes, but there is limited co-localization at these sites (G). Co-transfected Chi5 localizes to gap junction plaques with Cx26 (B, E and H). Co-transfected Chi6 was not found with Cx26 at gap junction plaques (C, F and I). Gap junction plaques are indicated by arrows. Scale bar, 10 μm.
Cx43-interacting chimaeras also co-localized with wild-type Cx26
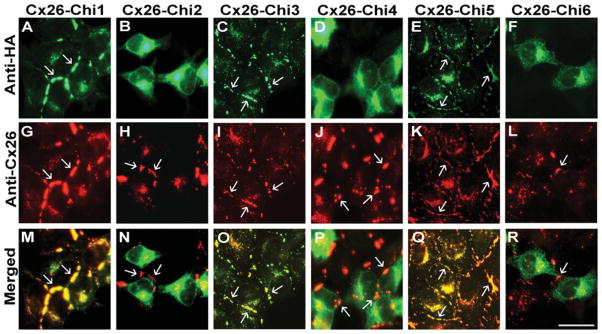
To test for connexin domains important in interactions with Cx26, we performed double label immunofluorescence microscopy in HeLaCx26 cells transfected with the various chimaeras. Immunofluorescence analysis showed co-localization of Cx26 with Chi1 (Figures 8A, 8G and 8M), Chi3 (Figures 8C, 8I and 8O) and Chi5 (Figures 8E, 8K and 8Q) intracellularly and in large gap junction plaques. The shapes and sizes of plaques containing Cx26 or these chimaeras appeared very similar. In contrast, Chi2 (Figures 8B and 8N), Chi4 (Figures 8D and 8P) and Chi6 (Figures 8F and 8R) were only found intracellularly and did not co-localize with Cx26, which formed gap junction plaques in co-expressing cells (Figures 8H and 8N, 8J and 8P, and 8L and 8R). These results imply that Chi1, Chi3 and Chi5 oligomerized with Cx26, whereas Chi2, Chi4 and Chi6 did not. As the TM3 domain in these chimaeras derives from Cx43 and not Cx26, this suggests that TM3 is not the critical region that determines oligomerization compatibility for Cx26.
Figure 8. Wild-type Cx26 co-localizes with Chi1, Chi3 and Chi5, but not Chi2, Chi4 or Chi6.
Photomicrographs show double immunofluorescencelabelling of Cx26 (red) and chimaeras (green) in HeLaCx26 cells co-transfected with Chi1 (A, G and M), Chi2 (B, H and N), Chi3 (C, I and O), Chi4 (D, J and P), Chi5 (E, K and Q) or Chi6 (F, L and R). Co-localization of Cx26 and chimaeras appears yellow in the merged panels (M, O and Q). Gap junction plaques are indicated by arrows. Scale bar, 25 μm.
To test for possible functional effects of chimaera interactions with Cx26, intercellular transfer of Lucifer yellow was tested in HeLa cells co-expressing wild-type Cx26 and the co-localizing chimaeras. HeLaCx26 cells showed extensive intercellular transfer of Lucifer yellow (Table 1). The effects of co-expressed chimaeras on Cx26 varied with different chimaeras. Whereas co-expression of Chi1 had no effect on the incidence or extent of transfer of Lucifer yellow, co-expression of Chi3 or Chi5 significantly reduced the incidence and extent of transfer of Lucifer yellow.
DISCUSSION
Our experiments in the present study of co-expression of Cx26–Cx43 chimaeras with wild-type Cx43 have provided strong evidence that the TM3 of Cx43 is required for oligomerization with Cx43. Several pieces of evidence (summarized in Table 2) support this conclusion. First, double label immunofluorescence showed co-localization of Chi1, Chi3 and Chi5, but not of Chi2, Chi4 or Chi6 with Cx43 in the same subcellular compartments. We found that co-expression of Chi1 led to cytoplasmic retention of Cx43, whereas Chi3 (which formed gap junction plaques very inefficiently when expressed by itself) or Chi5 were ‘rescued’ by wild-type Cx43. Formation of mixed hexamers has previously been invoked to explain rescue of a chimaera/mutant to the plasma membrane by a wild-type connexin or cytoplasmic retention of a wild-type connexin when co-expressed with a mutant [3,11,17,21–23]. Second, Chi1, Chi3 and Chi5 all acted as dominant-negative inhibitors of intercellular transfer of a gap junction tracer when co-expressed with Cx43. Such a significant decrease in gap junction intercellular communication in cells co-expressing connexins has frequently been attributed to formation of heteromeric oligomers. Third, in sedimentation velocity experiments, the distributions of chimaeras or Cx43 in the monomer and oligomer fractions were only altered when chimaeras containing the Cx43 TM3 were co-expressed with wild-type Cx43; Chi2 and Chi4 which contain the Cx26 TM3 did not oligomerize and had no effects on the peak of Cx43 oligomers. The smallest Cx43 domain present in all of the Cx43-interacting chimaeras was TM3.
Table 2.
Overview of evidence from the present study
| Chimaera | Cx26 domains | Cx43 domains | Co-localization with Cx43 | Co-localization with Cx26 | Co-sedimentation with Cx43 | Inhibitory effect on LY transfer of Cx43 | Inhibitory effect on LY transfer of Cx26 |
|---|---|---|---|---|---|---|---|
| 1 | NT–TM2 | IL–CT | Yes (IC) | Yes (P) | Yes | Yes | No |
| 2 | IL–CT | NT–TM2 | No | No | No | No | ND |
| 3 | NT–TM2, E2–CT | IL–TM3 | Yes (P) | Yes (P) | Yes | Yes | Yes |
| 4 | IL–TM3 | NT–TM2, E2–CT | No | No | No | ND | ND |
| 5 | NT–IL, E2–CT | TM3 | Yes (P) | Yes (P) | Yes | Yes | Yes |
| 6 | TM3 | NT–IL, E2–CT | No | No | ND | ND | ND |
IC, intracellular; LY, Lucifer yellow; ND, not determined; P, gap junction plaques.
The importance of sequences in TM3 for determining proper connexin trafficking, the intracellular compartment of oligomerization, and the formation of gap junction plaques has also been inferred from previous studies. Ahmad et al. [24] have reported that a Cx32 chimaera in which TM3 was replaced by a transmembrane domain from CFTR (cystic fibrosis transmembrane conductance regulator) does not oligomerize and is retained in the cytoplasm. Using a chimaeric strategy, Maza et al. [23] have obtained data suggesting that sequences in the TM3 and E2 domains, especially Arg153 and Gln173, regulate Cx43 oligomerization by preventing formation of Cx43 hexamers in the ER. Lagrée et al. [11] found that a Cx43 mutant (Cx43R153W–GFP) containing a tryptophan residue at position 153, matching the corresponding amino acid in Cx32, rescued a transport-deficient Ds-Red-tagged wild-type Cx43 to the plasma membrane but did not rescue wild-type Cx32. The observed effects of our chimaeric constructs on the behaviour of wild-type Cx43 do not seem to depend on the presence of Arg153, because Arg153 was not included as part of the Cx43 TM3 in Chi5, the chimaera in which only TM3 was interchanged between Cx26 and Cx43. This suggests that Arg153 may not be required to determine compatibility for connexin oligomerization.
Similarly to the results obtained in cells co-expressing Cx43, we found that Chi1, Chi3 and Chi5 (but not Chi2, Chi4 and Chi6) co-localized with Cx45. Thus, the sequences that determine oligomerization compatibility of Cx43 with itself and of Cx43 with Cx45 may reside in the same region, TM3. These results are of biological importance for cells in which both Cx43 and Cx45 are expressed (such as bone cells and cardiac myocytes) due to formation of heteromeric channels. Gap junction channels containing both Cx43 and Cx45 differ from the homomeric channels in voltage dependence, single channel conductance, permeability and kinase-dependent regulation [4,25]. Therefore, a dominant mutation of one connexin (e.g. Cx43) might have more deleterious effects in cells that co-express several connexins, because it could also affect the function of another co-expressed oligomerization-compatible connexin (e.g. Cx45).
Since none of the Cx26-interacting chimaeras contained the Cx26 TM3, it appears that this region is not a critical determinant of oligomerization compatibility for Cx26. Rather, since all of the Cx26-interacting chimaeras contained the NT to the TM2 domain of Cx26, it is likely that part of this region is involved in determining Cx26 oligomerization compatibility. In fact, it has been previously suggested that NT amino acids are involved in determining oligomerization compatibility with β-type connexins, since Cx43 mutants co-localize with Cx32 and affect its function when amino acids 12 and 13 in Cx43 are mutated to match those in the analogous positions in Cx32 [11]. However, this conclusion cannot apply to all β-type connexins, since Gemel et al. [26] found that alteration of these residues in Cx43 or Cx40 to match those of Cx26 did not confer compatibility to oligomerize with Cx26.
Interestingly, we found that two of the three Cx26-interacting chimaeras altered the functional properties of Cx26 gap junction channels. Co-expression of Chi3 or Chi5 significantly decreased intercellular transfer of Lucifer yellow through Cx26 channels. These results suggest that oligomerization of Cx26 with Chi3 or Chi5 produces gap junction channels with altered permeability or gating. This may result from a conformational change that partially or completely occludes the channel pore when TM3 (with or without the intracellular loop) of Cx26 is replaced by the homologous segment(s) of Cx43.
The recently published structure of Cx26 [15] indicates that the main pore-lining α-helix is TM1 with some contributions of TM2, whereas TM3 and TM4 face the hydrophobic membrane environment. In this structure, the main inter-subunit interactions are located in the extracellular half of transmembrane helices TM2 and TM4 (including residues Arg75, Thr186 and Glu187) and in the extracellular loops [15]. Since these residues are highly conserved among members of the connexin family, it is unlikely that these domains confer oligomerization compatibility. In the Cx26 structure [15], TM3 contributes to the formation of an aromatic cluster located in the groove between connexin subunits. Perhaps this interaction is critical for connexin oligomerization compatibility of Cx43.
Incompatibility of Cx26 and Cx43 resulting from differences in their molecular determinants of oligomerization may have substantial biological consequences. Cx26 and Cx43 are co-expressed in some cell types in different tissues including skin [27,28], cochlea [29], testes [30] and brain [31,32]. Mutations in Cx26 are associated with sensorineural deafness. Mutations in Cx43 (usually dominant) have been associated with oculodentodigital dysplasia [33], but sensorineural deafness is not a feature of this disease. The simplest explanation for this observation is that these Cx43 mutants retain their oligomerization compatibility, and they do not affect the function of Cx26 in the cochlea. Some dominant-negative Cx26 mutants are associated with syndromic deafness that may result in part from inhibition of co-expressed β-connexins (Cx30, Cx31) [34]. Some disease-associated Cx26 mutants are associated with skin disease and deafness; it has been postulated that skin disease in these patients is due to dominant-negative interference with the function of co-expressed Cx43 [35]. Thus preservation of oligomerization incompatibility in Cx26 mutants may limit the extent of physiological disruption by not affecting Cx43 function.
In conclusion, we have shown that although TM3 is a critical region for homo- and hetero-oligomerization of Cx43 and Cx45, this region is not critical for determining Cx26 oligomerization compatibility, but may modulate Cx26 gap junction channel permeability. Our results suggest that different regions are critical for oligomerization compatibility of connexins belonging to different subfamilies.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health [grant number HL59199 (to E.C.B.)], by FONDECYT [grant number 1090573] and by Anillo de Ciencia y Tecnología ACT-71 (to A.D.M.). The Centro Interdisciplinario de Neurociencias de Valparaíso is a Chilean Millennium Science Institute.
Abbreviations used
- Chi
chimaera
- CT
C-terminal
- Cx
connexin
- E
extracellular loop
- ER
endoplasmic reticulum
- HA
haemagglutinin
- IL
intracellular loop
- NT
N-terminal
- TM domain
transmembrane domain
Footnotes
AUTHOR CONTRIBUTION
Agustín Martínez and Eric Beyer developed the overall experimental strategy. Agustín Martínez prepared the constructs and performed the majority of the experiments. Jaime Maripillan, Rodrigo Acuña and Peter Minogue performed some of the experiments. Viviana Berthoud contributed to experimental design. Agustín Martínez, Viviana Berthoud and Eric Beyer wrote the paper.
References
- 1.Sàez JC, Berthoud VM, Braes MC, Martínez AD, Beyer EC. Plasma membrane channels formed by connexins: their regulation and functions. Physiol Rev. 2003;83:1359–1400. doi: 10.1152/physrev.00007.2003. [DOI] [PubMed] [Google Scholar]
- 2.Musil LS, Goodenough DA. Multisubunit assembly of an integral plasma membrane channel protein, gap junction connexin43, occurs after exit from the ER. Cell. 1993;74:1065–1077. doi: 10.1016/0092-8674(93)90728-9. [DOI] [PubMed] [Google Scholar]
- 3.Das Sarma J, Wang F, Koval M. Targeted gap junction protein constructs reveal connexin-specific differences in oligomerization. J Biol Chem. 2002;277:20911–20918. doi: 10.1074/jbc.M111498200. [DOI] [PubMed] [Google Scholar]
- 4.Martínez AD, Hayrapetyan V, Moreno AP, Beyer EC. Connexin43 and connexin45 form heteromeric gap junction channels in which individual components determine permeability and regulation. Circ Res. 2002;90:1100–1107. doi: 10.1161/01.res.0000019580.64013.31. [DOI] [PubMed] [Google Scholar]
- 5.Elenes S, Martínez AD, Delmar M, Beyer EC, Moreno AP. Heterotypic docking of Cx43 and Cx45 connexons blocks fast voltage gating of Cx43. Biophys J. 2001;81:1406–1418. doi: 10.1016/S0006-3495(01)75796-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrio LC, Suchyna T, Bargiello T, Xu LX, Roginski RS, Bennett MVL, Nicholson BJ. Gap junctions formed by connexins 26 and 32 alone and in combination are differently affected by applied voltage. Proc Natl Acad Sci USA. 1991;88:8410–8414. doi: 10.1073/pnas.88.19.8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukauskas FF, Angele AB, Verselis VK, Bennett MVL. Coupling asymmetry of heterotypic connexin 45/connexin 43-EGFP gap junctions: properties of fast and slow gating mechanisms. Proc Natl Acad Sci USA. 2002;99:7113–7118. doi: 10.1073/pnas.032062099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber PA, Chang HC, Spaeth KE, Nitsche JM, Nicholson BJ. The permeability of gap junction channels to probes of different size is dependent on connexin composition and permeant-pore affinities. Biophys J. 2004;87:958–973. doi: 10.1529/biophysj.103.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao F, Eckert R, Elfgang C, Nitsche JM, Snyder SA, Hülser DF, Willecke K, Nicholson BJ. A quantitative analysis of connexin-specific permeability differences of gap junctions expressed in HeLa transfectants and Xenopus oocytes. J Cell Sci. 1998;111:31–43. doi: 10.1242/jcs.111.1.31. [DOI] [PubMed] [Google Scholar]
- 10.Gemel J, Valiunas V, Brink PR, Beyer EC. Connexin43 and connexin26 form gap junctions, but not heteromeric channels in co-expressing cells. J Cell Sci. 2004;117:2469–2480. doi: 10.1242/jcs.01084. [DOI] [PubMed] [Google Scholar]
- 11.Lagree V, Brunschwig K, Lopez P, Gilula NB, Richard G, Falk MM. Specific amino-acid residues in the N-terminus and TM3 implicated in channel function and oligomerization compatibility of connexin43. J Cell Sci. 2003;116:3189–3201. doi: 10.1242/jcs.00604. [DOI] [PubMed] [Google Scholar]
- 12.Valiunas V, Gemel J, Brink PR, Beyer EC. Gap junction channels formed by coexpressed connexin40 and connexin43. Am J Physiol Heart Circ Physiol. 2001;281:H1675–H1689. doi: 10.1152/ajpheart.2001.281.4.H1675. [DOI] [PubMed] [Google Scholar]
- 13.Falk MM, Buehler LK, Kumar NM, Gilula NB. Cell-free synthesis and assembly of connexins into functional gap junction membrane channels. EMBO J. 1997;16:2703–2716. doi: 10.1093/emboj/16.10.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brink PR, Cronin K, Banach K, Peterson E, Westphale EM, Seul KH, Ramanan SV, Beyer EC. Evidence for heteromeric gap junction channels formed from rat connexin43 and human connexin37. Am J Physiol. 1997;273:C1386–C1396. doi: 10.1152/ajpcell.1997.273.4.C1386. [DOI] [PubMed] [Google Scholar]
- 15.Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- 16.White TW, Bruzzone R, Wolfram S, Paul DL, Goodenough DA. Selective interactions among the multiple connexin proteins expressed in the vertebrate lens: the second extracellular domain is a determinant of compatibility between connexins. J Cell Biol. 1994;125:879–892. doi: 10.1083/jcb.125.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez AD, Hayrapetyan V, Moreno AP, Beyer EC. A carboxyl terminal domain of connexin43 is critical for gap junction plaque formation but not for homo- or hetero-oligomerization. Cell Commun Adhes. 2003;10:323–328. doi: 10.1080/15419060390263092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar G, Sommer SS. The “megaprimer” method of site-directed mutagenesis. Biotechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 19.Pont-Kingdon G. Construction of chimeric molecules by a two-step recombinant PCR method. Biotechniques. 1994;16:1010–1011. [PubMed] [Google Scholar]
- 20.Berthoud VM, Montegna EA, Atal N, Aithal NH, Brink PR, Beyer EC. Heteromeric connexons formed by the lens connexins, connexin43 and connexin56. Eur J Cell Biol. 2001;80:11–19. doi: 10.1078/0171-9335-00132. [DOI] [PubMed] [Google Scholar]
- 21.Das Sarma J, Meyer RA, Wang F, Abraham V, Lo CW, Koval M. Multimeric connexin interactions prior to the trans-Golgi network. J Cell Sci. 2001;114:4013–4024. doi: 10.1242/jcs.114.22.4013. [DOI] [PubMed] [Google Scholar]
- 22.Wang M, Martínez AD, Berthoud VM, Seul KH, Gemel J, Valiunas V, Kumari S, Brink PR, Beyer EC. Connexin43 with a cytoplasmic loop deletion inhibits the function of several connexins. Biochem Biophys Res Commun. 2005;333:1185–1193. doi: 10.1016/j.bbrc.2005.05.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maza J, Das Sarma J, Koval M. Defining a minimal motif required to prevent connexin oligomerization in the endoplasmic reticulum. J Biol Chem. 2005;280:21115–21121. doi: 10.1074/jbc.M412612200. [DOI] [PubMed] [Google Scholar]
- 24.Ahmad S, Martin PE, Evans WH. Assembly of gap junction channels: mechanism, effects of calmodulin antagonists and identification of connexin oligomerization determinants. Eur J Biochem. 2001;268:4544–4552. doi: 10.1046/j.1432-1327.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- 25.Koval M, Geist ST, Westphale EM, Kemendy AE, Civitelli R, Beyer EC, Steinberg TH. Transfected connexin45 alters gap junction permeability in cells expressing endogenous connexin43. J Cell Biol. 1995;130:987–99525. doi: 10.1083/jcb.130.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gemel J, Lin X, Veenstra RD, Beyer EC. N-terminal residues in Cx43 and Cx40 determine physiological properties of gap junction channels, but do not influence heteromeric assembly with each other or with Cx26. J Cell Sci. 2006;119:2258–2268. doi: 10.1242/jcs.02953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Risek B, Klier FG, Gilula NB. Developmental regulation and structural organization of connexins in epidermal gap junctions. Dev Biol. 1994;164:183–196. doi: 10.1006/dbio.1994.1190. [DOI] [PubMed] [Google Scholar]
- 28.Goliger JA, Paul DL. Wounding alters epidermal connexin expression and gap junction-mediated intercellular communication. Mol Biol Cell. 1995;6:1491–1501. doi: 10.1091/mbc.6.11.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikuchi T, Kimura RS, Paul DL, Takasaka T, Adams JC. Gap junction systems in the mammalian cochlea. Brain Res Rev. 2000;32:163–166. doi: 10.1016/s0165-0173(99)00076-4. [DOI] [PubMed] [Google Scholar]
- 30.Brehm R, Marks A, Rey R, Kliesch S, Bergmann M, Steger K. Altered expression of connexins 26 and 43 in Sertoli cells in seminiferous tubules infiltrated with carcinoma-in-situ or seminoma. J Pathol. 2002;197:647–653. doi: 10.1002/path.1140. [DOI] [PubMed] [Google Scholar]
- 31.Nagy JI, Li X, Rempel J, Stelmack G, Patel D, Staines WA, Yasumura T, Rash JE. Connexin26 in adult rodent central nervous system: demonstration at astrocytic gap junctions and colocalization with connexin30 and connexin43. J Comp Neurol. 2001;441:302–323. doi: 10.1002/cne.1414. [DOI] [PubMed] [Google Scholar]
- 32.Spray DC, Moreno AP, Kessler JA, Dermietzel R. Characterization of gap junctions between cultured leptomeningeal cells. Brain Res. 1991;568:1–14. doi: 10.1016/0006-8993(91)91373-9. [DOI] [PubMed] [Google Scholar]
- 33.Paznekas WA, Boyadjiev SA, Shapiro RE, Daniels O, Wollnik B, Keegan CE, Innis JW, Dinulos MB, Christian C, Hannibal MC, Jabs EW. Connexin 43 (GJA1) mutations cause the pleiotropic phenotypes of oculodentodigital dysplasia. Am J Hum Genet. 2003;72:408–418. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martínez AD, Acuña R, Figueroa V, Maripillan J, Nicholson B. Gap junction channels dysfunction in deafness and hearing loss. Antioxid Redox Signal. 2009;11:309–322. doi: 10.1089/ars.2008.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouan F, White TW, Brown N, Taylor AM, Lucke TW, Paul DL, Munro CS, Uitto J, Hodgins MB, Richard G. Trans-dominant inhibition of connexin-43 by mutant connexin-26: implications for dominant connexin disorders affecting epidermal differentiation. J Cell Sci. 2001;114:2105–2113. doi: 10.1242/jcs.114.11.2105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.