Abstract
Biodegradable poly(ester amine) (PEA)-based and poly(amido amine) (PAA)-based nanoparticles were developed for efficient in vitro siRNA delivery to human umbilical vein endothelial cells (HUVECs). They were screened, characterized, and compared with traditionally studied DNA-containing particles. Several of the polymeric nanoparticles tested were found to be effective for delivering functional siRNA to green fluorescent protein (GFP) + HUVECs, achieving 60%–75% GFP knockdown while maintaining high viability. While PEAs have been used previously to form polyplexes or nanoparticles for DNA delivery, highly effective siRNA delivery in hard-to-transfect human cell types has not been previously reported. PEAs and linear nondendrimeric PAAs were also found to be effective for DNA delivery to HUVECs using GFP-encoding plasmid DNA (up to 50%–60% transfection efficiency). PEAs and PAAs can be separated into groups that form polymeric nanoparticles effective for siRNA delivery, for DNA delivery, or for both.
Keywords: nanoparticles, siRNA, DNA, gene delivery, human umbilical vein endothelial cells
Introduction
Delivery of siRNA and DNA is an attractive option for a variety of applications, from basic science research to potential clinical use. The ability to either induce or reduce expression of specific genes in vitro may lend itself to cell differentiation or guidance for regenerative medicine.1,2 Aberrant expression of genes has also been found to play an important role in many diseases.3,4 The ability to mimic these pathological states could be useful for basic studies in a laboratory setting, while the ability to reverse those states has potential for clinical use in curing or ameliorating specific diseases5–10 or for promoting tissue repair.11
Human umbilical vein endothelial cells (HUVECs) are a particularly suitable model cell in which to study these effects. HUVECs and related cells have high relevance in the fields of tissue engineering,12,13 cancer therapy,14,15 and other conditions with altered angiogenesis.16,17 Importantly, upregulation of proangiogenic genes may be crucial for tissue engineering to allow nutrient and gas transfer throughout a construct.18 The opposite effect would be necessary for the treatment of diseases like cancer19 or macular degeneration,20 in which excess angiogenesis or neovascularization is a cause of pathology.21 The ability to deliver either siRNA or DNA to these and other cell types would allow the flexibility to alter gene expression in either case. While siRNA and DNA molecules are chemically similar and share the ability to complex with polymeric nanoparticles via electrostatic interactions, their mechanisms of action and intracellular targets are different; siRNA acts primarily in the cytoplasm while DNA must reach the nucleus,22 providing distinct challenges in the delivery of each via transfection agents.
Many attempts to improve delivery of nucleic acids have relied on viruses which, while efficient, are limited to small cargo sizes and are much more likely to cause insertional mutagenesis, an immune response, or severe systemic toxicity. 23 Attempts to improve siRNA delivery technologies include lipid-based formulations24 and emulsion particles using mixtures of polymers or biomaterials.25,26 Still, efficiency must be improved before translating this technology to the clinic. Natural and synthetic cationic polymers have been previously studied because they are potentially safer and easier to manufacture,27,28 but nondegradable polymers like poly(ethyleneimine) suffer from high toxicity,29 which correlates with high molecular weight.30 Although polymeric gene delivery has been traditionally less efficient than virus-mediated gene transfer,31 members of one class of polymers, ie, poly(ester amine)s, have been found to be very effective in DNA delivery to a number of cell types while causing less toxicity than leading commercially available transfection reagents,32–34 likely due to the ability of polymers to degrade under physiological conditions and their theoretically unlimited variation in structure and properties, providing the flexibility to choose a polymer structure based on the particular purpose for which it is best suited.
Here, we expand upon the methods and chemical moieties that have in the past been used in gene delivery35 to identify poly(ester amine)s (PEAs) and poly(amido amine)s (PAAs) from a chemically diverse library that could complex well with either siRNA, plasmid DNA, or both. In particular, PEAs in this form have not been previously shown to enable effective siRNA delivery. With new polymer structures and formulation conditions, we show here that they are not only effective but also superior to the leading commercially available reagent, Lipofectamine™ 2000. After formation and characterization, the polymer-nucleic acid nanoparticles were used to deliver bioactive siRNA and DNA to HUVECs, and their effectiveness in both applications was compared. The goals of this study were to determine if small changes to the polymer structure could tune the efficacy of a polymeric nanoparticle for nucleic acid delivery and to determine any relationships between physicochemical properties and transfection efficiency profiles.
Materials and methods
Materials
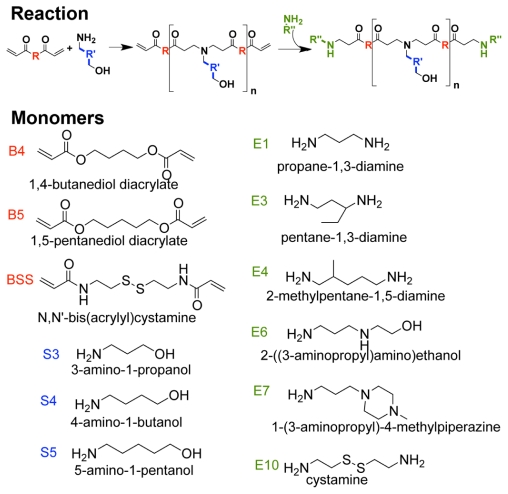
Monomers for polymer synthesis (Figure 1) were purchased from Alfa Aesar (Ward Hill, MA) [1,4-butanediol diacrylate (B4), N,N′ - bis (acryloyl) cystamine (BSS), 3-amino-1-propanol (S3), 4-amino-1-butanol (S4), 5-amino-1-pentanol (S5), 1-(3-aminopropyl)-4- methylpiperazine (E7), cystamine dihydrochloride (E10)], Fluka (Milwaukee, WI) [2-(3-aminopropylamino)ethanol (E6)], Monomer-Polymer and Dajac Labs (Feasterville, PA) [1,5-pentanediol diacrylate (B5)], Sigma Aldrich (St Louis, MO) [1,3-diaminopropane (E1)], and TCI America (Chicago, IL) [1,3-diaminopentane (E3), 2-methyl-1,5-diaminopentane (E4)]. HUVECs and endothelial growth medium-2 (EGM-2) were purchased from Lonza (Walkersville, MD) and used as recommended. Plasmid pDsRed-Max-N1 DNA (Addgene plasmid 21718,36 Cambridge, MA) was amplified by Aldevron (Fargo, ND), and siRNA against eGFP with 5′-CAAGCUGACCCUGAAGUUCTT (sense) and 3′-GAACUUCAGGGUCAGCUUGCC (antisense) (Silencer® positive control) and a scrambled siRNA sequence with 5′-AGUACUGCUUACGAUACGGTT (sense) and 3′-CCGUAUCGUAAGCAGUACUTT (antisense) (Silencer® negative control 1) were designed by and purchased from Ambion Inc (Carlsbad, CA). Lipofectamine 2000 and Opti- MEM I were from Invitrogen (Carlsbad, CA) and used according to the manufacturer’s instructions. All other materials used were reagent grade.
Figure 1.
Monomer structures used to synthesize polymers. Backbone (B) monomers were polymerized with sidechain (S) monomers. The B-S base polymers were then endcapped with small molecules (E).
Polymer synthesis
The specific monomers used in this study were chosen in order to sample varied parts of a large chemical space while maintaining structural similarities between the backbone, sidechain, and endgroup monomers utilized. PEAs were synthesized as previously reported.34,37,38 Briefly, one diacrylate backbone monomer (B) was mixed with one amine-containing sidechain (S) at 1.2:1 or 1.05:1 ratio and stirred overnight at 90°C. Acrylate-terminated B-S base polymers resulted from Michael addition of amines to the acrylate groups. Each base polymer was dissolved in dimethyl sulfoxide (DMSO) at 167 mg/mL; 480 μL of base polymer solution was mixed with 320 μL of a solution of one endcap monomer (E) in DMSO at 0.5 M, and the mixture was shaken for 24 hours at room temperature. Endcapped polymers were stored at 4°C as 100 mg/mL solutions in DMSO. For PAA synthesis, backbone monomer BSS was mixed with a sidechain (S) monomer at 200 mg/mL in DMSO, stirred overnight at 90°C, and endcapped as described above. The polymers are described henceforth by their constituent monomers, eg, the copolymer of B4 and S5 (B4-S5) can be endcapped with E6, then abbreviated as B4-S5-E6, or 456.
Polymer degradation
To ensure that the polymers used in this study were degradable under mild and physiological conditions, polymers were dissolved in 1× phosphate-buffered solution at 20 mg/mL. The polymer solutions were agitated using a shaker at 37°C. At various time points, 20 mL were removed, the pH was measured, and the samples were snapfrozen and lyophilized. Three sample replicates were analyzed at each time point. The samples were reconstituted in butylated hydroxytoluene-stabilized tetrahydrofuran with 5% DMSO and 1% piperidine, filtered through a 0.2 μm filter, and measured with gel permeation chromatography (Waters, Milford, MA).
Nanoparticle preparation
A 0.18 μM (2.7 μg/mL) solution of siRNA in sodium acetate buffer (25 mM, pH 5) or 60 μg/mL plasmid DsRed-Max DNA in sodium acetate was prepared. Polymer was diluted in sodium acetate at various weight/weight (w/w) ratios, then added at a 1:1 volume ratio to the nucleic acid solution. The complexes were mixed by pipetting and incubated at room temperature for 10 minutes to allow complexation and were used immediately.
Nanoparticle characterization
After 10 minutes, the suspension of complexed nanoparticles was diluted in 1× phosphate-buffered solution to a final concentration between 108 and 109 particles/mL and measured by nanoparticle tracking analysis using a NanoSight LM10. Three samples were prepared and measured for each condition. When using normalized concentration values, samples were normalized to the amount (mass) of nucleic acid rather than to the number of particles in order to keep the amount of the bioactive agent constant when comparing groups. Particles were also imaged by transmission electron microscopy (TEM) using a Phillips/FEI BioTwin CM120. For TEM, nanoparticle suspensions without stain were adsorbed onto carbon-coated copper grids, which were allowed to dry overnight before imaging. For zeta potential analysis, nanoparticles were prepared as described above, then diluted in 1× phosphate-buffered solution. The diluted particles were measured by dynamic light scattering using a Nano Series zetasizer (Malvern, Worcestershire, UK) at a final concentration of 5 μg/mL.
Gel retardation assays were carried out by adding polymer of varying concentrations in sodium acetate buffer to a constant concentration of DNA or siRNA in sodium acetate, similar to the normal particle preparation protocols described above. After 10 minutes of incubation, a solution of 30% glycerol in water was added in a 1:5 volumetric ratio as a loading buffer. Bromophenol blue or other dyes were not added, because they were found to interfere with binding. Samples were loaded into 1% agarose gel with 1 μg/mL ethidium bromide at 125 ng DNA or siRNA per well. Samples were run for 15 and 30 minutes for siRNA and DNA, respectively, under 100 V, then visualized using ultraviolet exposure.
Transfection of HUVECs
HUVECs were stably transduced using a lentiviral pPT-eGFP plasmid vector (cytomegalovirus promoter). For siRNA transfections, the cells were seeded at 13,000 cells/cm2 in 96-well plates in complete EGM-2 and allowed to adhere overnight. Then, siRNA against eGFP or a scrambled control sequence was diluted in sodium acetate buffer (25 mM, pH 5). Polymers were diluted in sodium acetate buffer and combined with each of the GFP (green fluorescent protein)-siRNA and the control scrRNA at 60, 100, or 150 w/w. After ten minutes, nanoparticle complexes were added directly to the cells in medium at a final volume ratio of 1:5 nanoparticles to medium and a final siRNA concentration of 60 nM per well. As a positive control, Lipofectamine 2000 complexes were prepared in Opti-MEM I according to the manufacturer’s instructions and were added to the wells at the same final siRNA concentration as the experimental polymer groups. After 4 hours of incubation, the particles were aspirated and the medium replaced with fresh EGM-2.
Viability was assessed 24 hours after transfection using an MTS assay (CellTiter AqueousONE, Promega, Madison, WI) according to the manufacturer’s protocol. On each day following transfection, the amount of GFP present in each well was measured with a fluorescence plate reader (Synergy 2, Biotek, Seattle, WA). After three days, cells were imaged with a Zeiss Observer A.1, then trypsinized, resuspended in phosphate-buffered solution with 2% fetal bovine serum, and measured with an Accuri C6 flow cytometer equipped with an Intellicyt high-throughput loader and reader. The geometric mean of the eGFP signal measured with emission at 530/30 nm was calculated for untreated cells, cells treated with GFP-siRNA, and cells treated with scrRNA. Nonspecific autofluorescence of each sample at 530/30 nm emission was subtracted using GFP-positive HUVECs of the same lot and passage number that underwent the same treatment with polymer-siRNA nanoparticles. The geometric mean GFP signal per cell for each polymer-siRNA sample was normalized to the GFP signal per cell in polymer-scrRNA control samples. FlowJo was used for flow cytometry analysis.
For DNA transfections, HUVECs were seeded at 26,000 cells/cm2 in 96-well plates in complete EGM-2 and allowed to adhere overnight. DNA polymer nanoparticles were prepared as previously described. Briefly, plasmid DsRed-Max and polymer were each separately diluted in 25 mM sodium acetate, then mixed at a 1:1 volume ratio for a polymer-to-DNA w/w ratio of either 30, 60, or 90. After ten minutes, nanoparticles were added directly to the cells in medium, at a final volume ratio of 1:5 nanoparticles to medium and a final DNA concentration of 5 μg/mL per well. Viability was assessed 24 hours after transfection using an MTS assay. After 48 hours, the cells were trypsinized and resuspended in phosphate-buffered solution with 2% fetal bovine serum. Transfection efficiency was assessed using flow cytometry, with DsRed emission in the 580/40 nm channel.
Statistics
Comparisons between GFP-siRNA and scrRNA within the same polymer were done using the Student’s t-test. Comparisons across multiple groups of siRNA or DNA treatment conditions were done using one-way analysis of variance with a post hoc Dunnett’s test to assess significance compared with the Lipofectamine 2000-positive control group. Significance is shown as *P < 0.05, **P < 0.01, or ***P < 0.001 unless otherwise stated. All graphs show mean ± standard error of the mean unless otherwise stated. All the transfection experiments (DNA or siRNA) were performed in quadruplicate and all the sizing/ zeta potential experiments (nanoparticle tracking analysis or dynamic light scattering) were performed in triplicate.
Results and discussion
Polymer characterization
Both types of polymers used in this study are biodegradable, which is essential to release nucleic acid intracellularly and prevent toxicity; PEAs degrade by hydrolysis and PAAs by bioreduction in the reducing cytosolic compartment. Polymer structures selected for this study were among those previously used for DNA delivery,39 as well as some additional structures used in preliminary optimization studies for siRNA delivery to human cells.
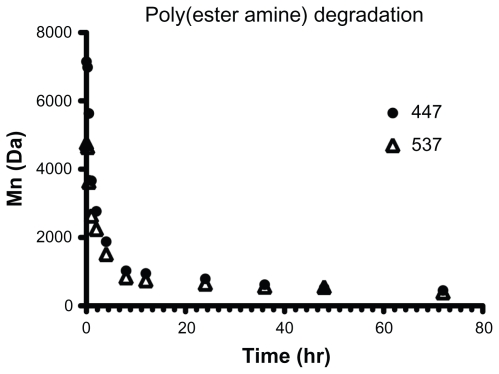
The polymers tested were shown to degrade under mild and physiologically relevant conditions by using representative PEAs 447 and 537, with an initial number-weighted molecular weight (Mn) of 7152 Da and 4759 Da, respectively. By gel permeation chromatography analysis, both polymers degraded fully, defined as having an Mn less than twice the molecular weight of one backbone repeat unit within 24 hours in an aqueous buffer solution (Figure 2). After 72 hours, the Mn of 447 and 537 was 450 ± 12 Da and 390 ± 47 Da, respectively.
Figure 2.
Polymers 447 and 537 degraded readily in an aqueous buffer solution when incubated with agitation at 37°C.
Note: Error bars represent the standard error of the mean of three samples.
Characterization of DNA and siRNA nanoparticles
The ability of PEAs and PAAs to condense nucleic acids into particles was measured by nanoparticle tracking analysis, whereby the size of each individual particle in a sample is measured based on its rate of diffusion. This is in contrast with dynamic light scattering, which, while having certain advantages, is often poorly suited for analysis of polydispersed distributions. Because nanoparticle tracking analysis directly calculates number-weighted size measurements, size distributions should be less biased toward small numbers of large particles or aggregates. The validity of the results was determined by TEM.
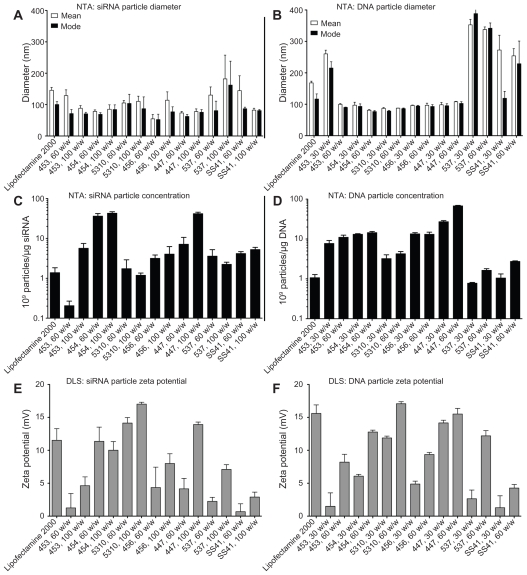
The mean± standard error of the mean is shown for the number-weighted mean and mode particle hydrodynamic diameter for both DNA and siRNA particles (Figure 3A and B). Particle concentration, normalized to the amount (μg) of siRNA or DNA in each sample, is shown in Figure 3C and D and the zeta potential in Figure 3E and F.
Figure 3.
The mean and mode diameter of measured particles is shown in (A) siRNA and (B) DNA. The concentration of particles formed under these conditions is shown in (C) siRNA and (D) DNA. The zeta potential in 1× phosphate-buffered solution is shown in (E) siRNA and (F) DNA.
Abbreviations: NTA, nanoparticle tracking analysis; DLS, dynamic light scattering.
Given that the cationic polymers form nanoparticles with nucleic acids via electrostatic interactions, it is not surprising that similar trends are seen between siRNA and DNA particles; those polymers that can condense DNA efficiently into particles that remain stable in a solution of salt buffer also tend to be effective in forming siRNA particles. Most of the particles fall within the range of 50–150 nm in diameter, which has been shown to be a range in which particles are easily taken up by cells for nucleic acid delivery.40 However, we hypothesized that there would be significant differences between some of the DNA-containing and siRNA-containing particles due to the diversity in polymer structures and nanoparticle formulation ratios that we have used. Our results indicate this to be the case. Some polymers that complex well with DNA at 30 w/w (eg, 453) form very few particles with siRNA at low or moderate amounts of polymer (<90 w/w). The small size and low concentration of particles formed between siRNA and 453 at 60 w/w indicates that free polymer remains in solution and, being amphiphilic, may simply dissolve in water rather than participate in particle formation, as it does at higher concentrations. While this polymer forms many particles with DNA at low w/w, it also has a larger mean and mode particle size in those conditions, which may indicate instability of the nanoparticle complexes. Therefore, it may be possible to correlate cationic polymer efficiency in DNA complexation with its efficiency in siRNA complexation.
Small changes in the chemical structure of a polymer, such as the use of different endgroups, sometimes caused a drastic change in the properties of the resulting particles. For example, starting with the same base polymer B5-S3, modification with the E10 endgroup formed particles that were small in size (about 80 nm for both DNA and siRNA), whereas modification of B5-S3 with the E7 endgroup formed larger particles (about 300 nm for DNA and about 200 nm for siRNA). Similarly, with the B4-S5 base polymer, even the single-carbon difference between the endgroups E3 and E4 caused a difference in size. Surface charge or zeta potential similarly showed that small changes in structure could affect particle properties (Figure 3E and F). As expected, all the particle formulations studied here had a positive surface charge due to the cationic polymers, ranging from 1–20 mV when suspended in phosphate-buffered solution. The zeta potentials of particles formed with siRNA were generally similar to that of particles formed with DNA. Although only a relatively small number of structures were explored in this study, these results suggest that further work on a larger group of polymers could be useful to understand better the relationship between the chemical structure and properties of the resulting particles. Of particular interest is the potential to correlate the particle-forming capability of a polymer with its transfection efficiency.
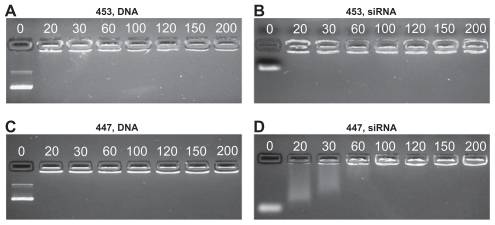
In general, siRNA required a higher polymer-to-nucleic acid w/w ratio for effective electrostatic complexation than plasmid DNA. This may be due to the high multivalency/ avidity of plasmid DNA, which has over 200-fold more negatively charged phosphate groups per molecule than siRNA. Differences in particle formation efficiency were also observed using gel electrophoresis. DNA movement through the gel was completely stopped even at the lowest w/w tested (20 w/w), while some polymers, like 447, needed to be in higher concentration for siRNA, which is not fully retarded until 60 w/w 447 is added (Figure 4).
Figure 4.
Poly(ester amine) 453 fully retards both DNA and siRNA at 20 w/w or lower, (A and B) 447 fully retards DNA, but up to 60 w/w is needed for siRNA retardation (C and D).
Note: Numbers in each image refer to the polymer w/w ratio to DNA or siRNA.
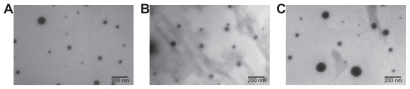
TEM was used qualitatively to visualize the nanoparticles (Figure 5). The particles appeared approximately spherical. Crucially, the trends in size and distribution found using nanoparticle tracking analysis were also seen in TEM micrographs, verifying that this method gave accurate size distributions. It is expected that TEM measurements would be slightly smaller, as nanoparticle tracking analysis measures the hydrodynamic diameter while TEM shows dry particles; however, trends should remain similar regardless of the measurement modality. For example, nanoparticles of siRNA complexed with PEA 5310 at 60 w/w had a mean diameter of 105 ± 13 nm, measured by nanoparticle tracking analysis, while DNA nanoparticles with PEAs 453 and 537 at 60 w/w had mean diameters of 100 ± 5 nm and 338 ± 15 nm, respectively. TEM images showed the same trends, with mean diameters of 61 ± 10, 93 ± 16, and 190 ± 31 nm for siRNA-5310, DNA-453, and DNA-537, respectively.
Figure 5.
Transmission electron micrographs show particles composed of siRNA-5310 (A), DNA-453 (B), and DNA-537 (C).
Note: Scale bar in each 200 nm.
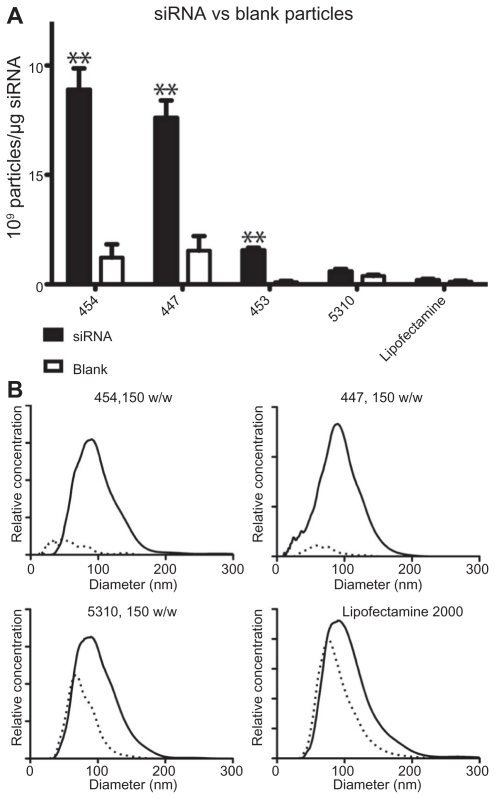
To verify that the trends in Figure 3 were not due simply to the inherent tendency of the polymers to aggregate in aqueous medium, sizing studies were repeated on a subset of the polymers using the same procedure and concentrations as for siRNA particles above, but without any nucleic acid (polymer only). Most of the tested polymers that appeared to complex well with nucleic acid based on concentration and size distribution, such as 454 and 447, were also found to form several-fold more particles when complexed with siRNA than they did on their own (Figure 6A). Interestingly, some polymers, like 5310, and Lipofectamine 2000 showed no statistically significant difference in the number of particles formed either alone or when complexed with siRNA. However, Figure 6B does show a slight shift in the peak particle diameter between 5310 or Lipofectamine alone and in complex with siRNA, which may indicate that those particles formed with siRNA are different from those formed simply via self-assembly of amphiphilic molecules in aqueous medium with no siRNA.
Figure 6.
Blank nanoparticles composed of polymer only or siRNA-polymer nanoparticles at 150 w/w were compared by concentration (A) and size distribution (B).
Notes: In (A), **P < 0.01 comparing siRNA and blank particles. In (B), solid line represents siRNA-polymer nanoparticles; dotted line represents polymer only.
siRNA transfection of HUVECs
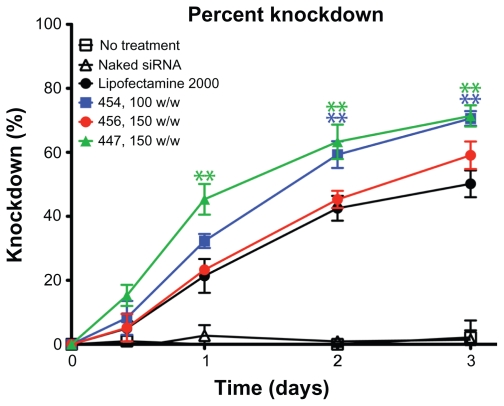
HUVECs stably expressing eGFP were transfected with siRNA-polymer nanoparticles. The measured GFP signal over time was compared with that of cells treated with scrRNA-polymer nanoparticles, because preliminary work had shown that this was an accurate measure of the amount of GFP in each well (see Figure S1). Selected polymers, as well as positive and negative controls, are shown in Figure 7. Background fluorescence was measured from GFP cells in medium and was subtracted from all other readings. Percent knockdown was calculated by normalizing GFP fluorescence from the GFP-siRNA-treated cells to the scrRNA-treated cells. In groups that received either no treatment, treatment with only siRNA, or treatment with polymer complexed with scrRNA, the GFP signal increased over time as the HUVECs proliferated. In groups treated with GFP-siRNA complexed with certain polymers (453, 454, 456, 447, and Lipofectamine 2000), the GFP signal decreased compared with the controls. The difference between cells treated with GFP-siRNA and scrRNA was statistically significant (P < 0.05) for the polymer formulations shown in Figure 7.
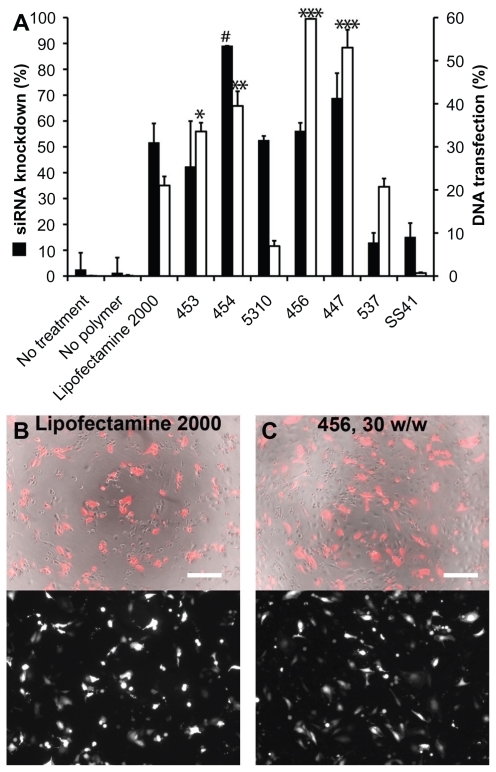
Figure 7.
The percent knockdown of green fluorescent protein from cells treated with green fluorescent protein-siRNA, normalized to scrRNA.
Notes: **P < 0.01 superior knockdown compared with Lipofectamine™ 2000. Error bars represent the standard error of the mean.
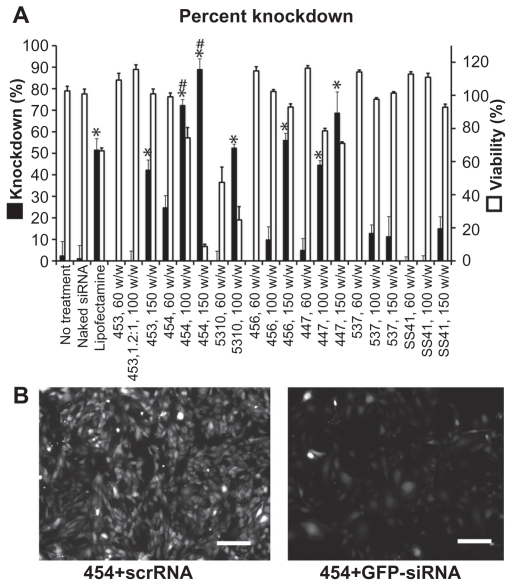
The decrease in average GFP signal per cell three days after transfection is shown in Figure 8. Cell counts and viability were not statistically different between the GFP-siRNA and scrRNA groups. However, flow cytometry analysis showed an increase in the 530 nm emission per cell in some of the groups treated with the scrRNA-polymer control. This increase was statistically significant compared with the untreated groups (P < 0.05) for polymers 454 (150 w/w), 5310 (100 w/w), 447 (100 and 150 w/w), and SS41 (150 w/w). Because it was accompanied by a corresponding increase in emission at 580 nm, this was considered to be increased autofluorescence due to treatment with certain polymers. This increased autofluorescence was not correlated with cell cytotoxicity (>90% viability for SS41 at 150 w/w and approximately 80% viability for 447 at 100 w/w). For analysis, the reported knockdown was calculated by comparing GFP-siRNA groups with scrRNA groups using the same polymer. Importantly, there were some polymer formulations that showed efficient knockdown, high viability, and no significant change in autofluorescence due to treatment. Polymers 453, 454, 5310, 456, and 447 and Lipofectamine all showed statistically significant (one-way analysis of variance, post hoc Dunnett’s test, P < 0.05) knockdown compared with untreated controls. The best formulation of 454 (100 w/w) showed 72.3% ± 0.6% knockdown of the GFP signal, which was superior to knockdown by the positive control, Lipofectamine 2000 (51.6% ± 5.2%), with comparable or better viability. While knockdown is even greater when the concentration of 454 is increased, this is accompanied by high toxicity as well, showing that the formulation must be optimized for safe as well as effective delivery.
Figure 8.
Flow cytometry (A) and fluorescence microscopy with polymer 454 at 100 w/w, exposure time 100 msec on the green fluorescent protein channel (B) both show significant green fluorescent protein knockdown after treatment with green fluorescent protein-siRNA compared with scrRNA.
Notes: *P < 0.05 significant knockdown compared with no treatment control; #P < 0.05 significant knockdown compared with Lipofectamine™ 2000 positive control. Scale bar in (B) 200 μm.
DNA transfection
Experiments done with siRNA-polymer nanoparticles were repeated with plasmid DsRed-Max DNA by slightly modifying the particle fabrication method described earlier. Optimal polymer-to-DNA ratios were found to lie between 30 and 90 w/w (preliminary data not shown), and DsRed expression was measured using flow cytometry two days after transfection. Figure 9 shows the siRNA knockdown efficiency (up to 72.3% ± 0.6%) and DNA transfection efficiency (up to 59.7% ± 2.0%) for all tested polymers at the optimal w/w ratio found for each polymer. As with the nanoparticle sizing study, many of the polymers that are efficient for delivery of one type of nucleic acid also work in delivery of the other, although the optimal polymers for delivery of each type of nucleic acid were found to differ slightly.
Figure 9.
For each polymer, the best siRNA knockdown and best DNA transfection efficiency from the optimal formulation of polymer-nucleic acid are plotted together in (A).
Notes: #P < 0.05 superior siRNA delivery compared with Lipofectamine™ 2000; *P < 0.05, **P < 0.01, ***P < 0.001 superior DNA delivery compared with Lipofectamine 2000. DNA transfection efficiency after 24 hours with Lipofectamine 2000 (B) and 456, 30 w/w (C) are also shown. For (B) and (C) top, phase contrast + DsRed merged; bottom, DsRed only; scale bar 200 μm.
Fluorescence imaging suggested relatively high transfection using Lipofectamine 2000 after 24 hours, as seen in Figure 9B. However, there was also high toxicity, as evident in the image, which likely explains the low measurement of transfection efficiency from flow cytometry. On the other hand, many of the PEAs used caused significantly higher transfection after 48 hours with much lower nonspecific toxicity.
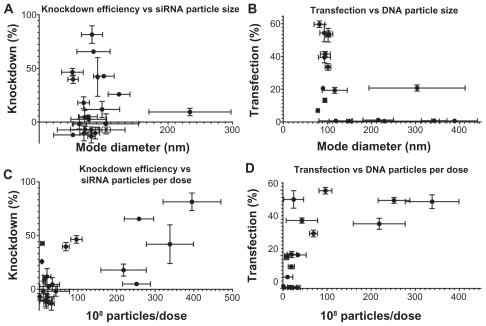
Interestingly, the polymers that formed the best nanoparticles with a mean and mode diameter of 50–150 nm and high concentration of formed particles also tended to be among those that were very efficient in nucleic acid delivery. As seen in Figure 10A and B, there is a narrow range of sizes within which nanoparticles are effective. The one polymer in both the siRNA and DNA groups that did not fit the trend was 537 at 100 w/w and 90 w/w, respectively, and showed moderate efficacy despite its large size. However, both nanoparticle tracking analysis and TEM showed that 537 forms a polydispersed population of nanoparticles with siRNA and DNA, suggesting that a subpopulation of smaller particles may be able to interact with and transfect cells, while larger particles are less effective and therefore cause the lower overall efficiency seen using the polymer. These graphs suggest that an optimal size may be necessary but not sufficient for effective transfection.
Figure 10.
Moderate correlations can be seen between the physical properties of the nanoparticles and their effectiveness in transfecting cells. (A) and (B) show a narrow size range in which particles are effective, with polymer 537 as an exception; (C) and (D) show a trend of increasing efficacy with increasing number of particles formed.
Some of the differences in delivery efficacy may be explained by differences in particle concentration. While all experimental conditions have the same dose of nucleic acid, the number of individual particles that self-assemble with this constant amount of nucleic acid varies with polymer structure. The particle concentration may be indicative of the efficiency with which polymers are able to complex their nucleic acid cargo. Figure 10C and D show that there is a small positive correlation between the number of nanoparticles per dose and siRNA knockdown efficiency (r2 = 0.4678) or DNA transfection efficiency (r2 = 0.4344). Interestingly, this trend is more easily apparent in the siRNA particles, since some DNA particles were able to achieve moderate to high transfection at a relatively low number of particles per dose. This suggests that, with the polymers tested, fewer particles may be needed for effective DNA delivery than for siRNA delivery.
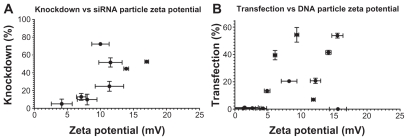
Similarly, dynamic light scattering was used to measure the zeta potential of polymer-nucleic acid complexes and showed some slight trends as well. The increasing transfection efficacy with zeta potential is visible for siRNA nanoparticles with r2 = 0.4905; while DNA transfection generally increases with zeta potential, the correlation is less clear with a correlation coefficient of only r2 = 0.2427 (Figure 11). Transfection efficiency is expected to be dependent on multiple factors and all of these physicochemical properties must be taken into account when designing effective polymers for nucleic acid delivery. These studies reveal the complexity of PEA and PAA nanoparticle systems wherein there is not a single parameter that defines transfection efficiency. Here, we show quantitatively the correlation of particle size, zeta potential, and concentration on transfection efficacy over a range of different polymer types, as well as demonstrating the likely effect of polymer structure.
Figure 11.
Slight correlations can be seen between the surface charge of the nanoparticles and their effectiveness in transfecting cells with siRNA (left) or DNA (right).
It is also interesting to note the differences in trends observed for DNA delivery compared with siRNA delivery. For example, with the B4-S5 base polymer, the E4 endgroup nanoparticles have smaller size compared with the E3 endgroup nanoparticles, and the smaller 454 nanoparticles are more effective for siRNA delivery than 453, although their DNA delivery is comparable. Similarly, the base polymer B5-S3 formed different particles depending on its endgroup, which was reflected in the degree of success of transfection. The relatively large nanoparticles formed with the 537 polymer are more effective for DNA delivery while having only a moderate effect when using siRNA.
However, comparison with another B5-S3 polymer, 5310, makes it clear that several factors in combination are likely to dictate the success of transfection. The smaller particles formed with 5310 were expected in part to be due to the positive charge of the E10 endgroup and the relatively high positive zeta potential of 5310. While these particles were moderately effective for siRNA delivery, with up to 52.5% ± 1.7% knockdown in the best formulation, it was one of the least efficient polymers for DNA delivery (7.0% ± 1.2% of cells transfected). The opposite is true of 456, which was the most effective for DNA delivery (59.7% ± 2.0%) and only moderately effective for siRNA delivery, even though the particles it formed were similar in concentration, size distribution, and surface charge compared with other less effective polymers. The biochemical properties of these polymers, including their endgroups and degradation mechanisms, must also play an important role in determining nanoparticle efficacy. It is important to note that the intracellular targets and effects are not the same for siRNA and DNA delivery; the latter must enter the nucleus intact and be able to be transcribed for protein production, while the former acts primarily in the cytoplasm. The initial barriers are expected to be similar in these two cases, with nucleic acid-polymer particles entering the cell via endocytosis or macropinocytosis and then escaping the endosomal compartment to be trafficked through the cytoplasm. In addition to the differences in particle formation capacity shown in this study, it is possible that certain PEAs or PAAs are better suited for intracellular steps downstream from initial internalization and endosomal escape. For example, it is possible that the cytosolic mechanism of action of siRNA makes quick polymer degradation or nucleic acid release more crucial than in DNA delivery, where slower degradation could protect the plasmid for a longer time as it is trafficked towards the nucleus. In particular, based on our observations in this study, we hypothesize that E10- terminated polymers, a newly developed structure, may have improved efficacy for siRNA compared with DNA due to the presence of disulfide bonds that can enable quicker release of nucleic acid cargo to the cytoplasm.
Formulation parameters were found to be extremely important in transitioning from DNA to siRNA delivery. Although more polymer per nucleic acid residue was needed in these studies for siRNA delivery than for DNA delivery, because the molecular weight of siRNA is so much smaller than that used for the DNA transfections, the total mass of nucleic acid, and therefore the total amount of polymer added to cells in siRNA transfections, was still less than that used for DNA. In the literature, PEA-siRNA nanoparticles were found to be unable to transfect HeLa cells without the use of gold nanoparticles as a scaffold,41 and other groups using similar systems report less knockdown than that described here, or used higher siRNA doses of up to 125 nM, usually in the absence of serum in their transfection media.42,43 It is likely that the higher weight ratios used in the present study (100–150 w/w) as well as the ability to screen several distinct molecular entities enabled our enhanced efficacy, which highlights the importance of different fabrication conditions for nanoparticle self-assembly to encapsulate DNA versus siRNA.
It is encouraging that some formulations, such as 454 at 100 w/w and 456 at 150 w/w, are able to cause significant knockdown while maintaining 80% viability or greater, reflective of the mild nature of these reagents compared with other commonly studied materials for siRNA delivery. For example, many efforts toward efficient siRNA delivery have used lipid-based formulations, such as the Lipofectamine 2000 used here as a point of comparison. However, these may be highly cytotoxic44 and exhibit low colloidal stability.45 A cationic polymer commonly studied for nucleic acid delivery, polyethyleneimine, also shows high toxicity and often must be chemically modified to ameliorate this before use,46 as well as requiring high doses for effective knockdown.47 Other studies using nonlipid formulations, like exosomes48 or gold nanoparticle immobilization,49 have also used much higher in vitro siRNA concentrations of up to 200 nM to achieve similar knockdown to the 60 nM reported here, while also using easier-to-transfect cell types like CHO-K1 or HeLa cells. It has also been speculated that surface adsorption or conjugation methods, while fairly effective, may have lower efficiency because of the reduced availability of immobilized siRNA.50 Some siRNA delivery systems require added heat or chemicals during synthesis45,51 that can be potentially destructive to siRNA. In contrast, the method described here allows for quick, simple complexation under mild conditions.
Conclusion
Here, we report for the first time that synthetic PEA-siRNA self-assembled nanoparticles are effective for siRNA delivery and show significant knockdown of HUVECs at relatively low doses of siRNA. We also report end-modified PAAs capable of more modest intracellular nucleic acid delivery. Experiments with nanoparticles containing siRNA resulted in up to 60%–75% knockdown in HUVECs after 3 days. Maximum DNA delivery with these polymeric nanoparticles caused transgene expression in 59.7% ± 2.0% of HUVECs after 2 days. Importantly, different parameters may be most crucial for each type of nucleic acid cargo. A high nanoparticle formation efficiency and a size distribution centered near 100 nm appeared necessary but not sufficient for successful transfection. In general, higher weight ratios of polymer to nucleic acid (100–150 w/w) were required for siRNAcontaining nanoparticles versus nanoparticles formed with plasmid DNA (30–90 w/w).
While there were some polymers that were similarly effective in delivering both types of nucleic acids, other polymers delivered one type of nucleic acid well but were fairly ineffective in delivering the other. This raises the possibility that some of these cationic polymers may be able to deliver multiple DNA and siRNA molecules together simultaneously within the same nanoparticles. In another strategy, specialized polymers optimized for the delivery of only one type of nucleic acid may be more efficient for a certain application. While these results were shown in HUVECs, a difficult-to-transfect human primary cell type with many potential applications in tissue engineering and drug delivery, it is expected that these nanoparticles would be effective for delivery to other cell types as well.
Supporting figure
GFP+ HUVECs were seeded in 96-well plates at known densities and allowed to settle and adhere for 1 hr before measuring fluorescence with a microplate reader. Linear regression showed a positive, linear correlation following the relation RFU = 33073*(#cells)+6599 with correlation coefficient r2 = 0.985. The gray, shaded region shows the 95% confidence interval of the regression line.
Note: Error bars represent standard deviation of multiple replicates at each cell density.
Abbreviations: GFP, green fluorescent protein; HUVECs, human umbilical vein endothelial cells; RFU, relative fluorescence units.
Acknowledgments
This work was supported in part by the Johns Hopkins University Institute for NanoBioTechnology, Maryland Technology Development Corporation and Maryland Stem Cell Research Fund (2009-MSCRFE-0098-00), and the National Institutes of Health (R21CA152473). Preliminary studies were funded by the Provost’s Undergraduate Research Award at Johns Hopkins. SYT thanks the National Science Foundation for fellowship support. GFP-positive HUVECs were transduced by Brian Goh using a viral vector provided by Dr David Yue. The authors thank Joel C Sunshine for synthesizing some of the polymers used in this study, Ron B Shmueli for assistance with HUVEC culture and transfection, and Corey J Bishop and the Johns Hopkins School of Medicine Microscope Facility for assistance with TEM.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jang JH, Houchin TL, Shea LD. Gene delivery from polymer scaffolds for tissue engineering. Expert Rev Med Devices. 2004;1(1):127–138. doi: 10.1586/17434440.1.1.127. [DOI] [PubMed] [Google Scholar]
- 2.Bonadio J. Tissue engineering via local gene delivery: update and future prospects for enhancing the technology. Adv Drug Deliv Rev. 2000;44(2–3):185–194. doi: 10.1016/s0169-409x(00)00094-6. [DOI] [PubMed] [Google Scholar]
- 3.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: current status and prospective. Int J Cancer. 2007;120(5):953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 4.Yadav S, van Vlerken LE, Little SR, Amiji MM. Evaluations of combination MDR-1 gene silencing and paclitaxel administration in biodegradable polymeric nanoparticle formulations to overcome multidrug resistance in cancer cells. Cancer Chemother Pharmacol. 2009;63(4):711–722. doi: 10.1007/s00280-008-0790-y. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen DN, Green JJ, Chan JM, Langer R, Anderson DG. Polymeric materials for gene delivery and DNA vaccination. Adv Mater. 2009;21(8):847–867. doi: 10.1002/adma.200801478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pringle IA, Hyde SC, Gill DR. Non-viral vectors in cystic fibrosis gene therapy: recent developments and future prospects. Expert Opin Biol Ther. 2009;9(8):991–1003. doi: 10.1517/14712590903055029. [DOI] [PubMed] [Google Scholar]
- 7.Ptasznik A, Nakata Y, Kalota A, Emerson SG, Gewirtz AM. Short interfering RNA (siRNA) targeting the Lyn kinase induces apoptosis in primary, and drug-resistant, BCR-ABL1(+) leukemia cells. Nat Med. 2004;10(11):1187–1189. doi: 10.1038/nm1127. [DOI] [PubMed] [Google Scholar]
- 8.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. Local and systemic delivery of VEGF siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J Control Release. 2008;129(2):107–116. doi: 10.1016/j.jconrel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Howard KA, Paludan SR, Behlke MA, Besenbacher F, Deleuran B, Kjems J. Chitosan/siRNA nanoparticle-mediated TNF-alpha knockdown in peritoneal macrophages for anti-inflammatory treatment in a murine arthritis model. Mol Ther. 2009;17(1):162–168. doi: 10.1038/mt.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karvinen H, Yla-Herttuala S. New aspects in vascular gene therapy. Curr Opin Pharmacol. 2010;10(2):208–211. doi: 10.1016/j.coph.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Mooney DJ, Vandenburgh H. Cell delivery mechanisms for tissue repair. Cell Stem Cell. 2008;2(3):205–213. doi: 10.1016/j.stem.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Kannan RY, Salacinski HJ, Sales K, Butler P, Seifalian AM. The roles of tissue engineering and vascularisation in the development of micro-vascular networks: a review. Biomaterials. 2005;26(14):1857–1875. doi: 10.1016/j.biomaterials.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Rouwkema J, de Boer J, Van Blitterswijk CA. Endothelial cells assemble into a 3-dimensional prevascular network in a bone tissue engineering construct. Tissue Eng. 2006;12(9):2685–2693. doi: 10.1089/ten.2006.12.2685. [DOI] [PubMed] [Google Scholar]
- 14.Folkins C, Shaked Y, Man S, et al. Glioma tumor stem-like cells promote tumor angiogenesis and vasculogenesis via vascular endothelial growth factor and stromal-derived factor 1. Cancer Res. 2009;69(18):7243–7251. doi: 10.1158/0008-5472.CAN-09-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rak J, Al-Nedawi K, Meehan B, Kerbel RS, Allison AC. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci U S A. 2009;106(10):3794–3799. doi: 10.1073/pnas.0804543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouis D, Hospers GA, Meijer C, Molema G, Mulder NH. Endothelium in vitro: a review of human vascular endothelial cell lines for blood vessel-related research. Angiogenesis. 2001;4(2):91–102. doi: 10.1023/a:1012259529167. [DOI] [PubMed] [Google Scholar]
- 17.Ohno-Matsui K, Morita I, Tombran-Tink J, et al. Novel mechanism for age-related macular degeneration: An equilibrium shift between the angiogenesis factors VEGF and PEDF. J Cell Physiol. 2001;189(3):323–333. doi: 10.1002/jcp.10026. [DOI] [PubMed] [Google Scholar]
- 18.Laschke MW, Harder Y, Amon M, et al. Angiogenesis in tissue engineering: Breathing life into constructed tissue substitutes. Tissue Eng. 2006;12(8):2093–2104. doi: 10.1089/ten.2006.12.2093. [DOI] [PubMed] [Google Scholar]
- 19.Liotta LA, Steeg PS, Stetler-Stevenson WG. Cancer metastasis and angiogenesis: an imbalance of positive and negative regulation. Cell. 1991;64(2):327–336. doi: 10.1016/0092-8674(91)90642-c. [DOI] [PubMed] [Google Scholar]
- 20.Witmer AN, Vrensen GF, Van Noorden CJ, Schlingemann RO. Vascular endothelial growth factors and angiogenesis in eye disease. Prog Retin Eye Res. 2003;22(1):1–29. doi: 10.1016/s1350-9462(02)00043-5. [DOI] [PubMed] [Google Scholar]
- 21.Bhise NS, Shmueli RB, Sunshine JC, Tzeng SY, Green JJ. Drug delivery strategies for therapeutic angiogenesis and antiangiogenesis. Expert Opin Drug Deliv. 2011;8(4):485–504. doi: 10.1517/17425247.2011.558082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh N, Agrawal A, Leung AK, Sharp PA, Bhatia SN. Effect of nanoparticle conjugation on gene silencing by RNA interference. J Am Chem Soc. 2010;132(24):8241–8243. doi: 10.1021/ja102132e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 24.Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61(10):850–862. doi: 10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 25.Woodrow KA, Cu Y, Booth CJ, Saucier-Sawyer JK, Wood MJ, Saltzman WM. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat Mater. 2009;8(6):526–533. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen J, Schubert S, Cohen J, Mynar J, Wich P, Frechet J. Acid-degradable cationic dextran particles for the delivery of siRNA therapeutics. Bioconjug Chem. 2011;22(6):1056–1065. doi: 10.1021/bc100542r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffen-burger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 2005;12(13):1023–1032. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 28.Kim SW, Park TG, Jeong JH. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev. 2006;58(4):467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Florea BI, Meaney C, Junginger HE, Borchard G. Transfection efficiency and toxicity of polyethylenimine in differentiated Calu-3 and nondifferentiated COS-1 cell cultures. AAPS Pharm Sci. 2002;4(3):E12. doi: 10.1208/ps040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kissel T, Kunath K, von Harpe A, et al. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release. 2003;89(1):113–125. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]
- 31.Putnam D. Polymers for gene delivery across length scales. Nat Mater. 2006;5(6):439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 32.Bhise NS, Gray RS, Sunshine JC, Htet S, Ewald AJ, Green JJ. The relationship between terminal functionalization and molecular weight of a gene delivery polymer and transfection efficacy in mammary epithelial 2-D cultures and 3-D organotypic cultures. Biomaterials. 2010;31(31):8088–8096. doi: 10.1016/j.biomaterials.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunshine J, Green JJ, Mahon K, et al. Small molecule end group of linear polymer determine cell-type gene delivery efficacy. Adv Mater. 2009;21(48):4947–4951. doi: 10.1002/adma.200901718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41(6):749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Biodegradable polymeric vectors for gene delivery to human endothelial cells. Bioconjug Chem. 2006;17(5):1162–1169. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- 36.Strack RL, Strongin DE, Bhattacharyya D, et al. A noncytotoxic DsRed variant for whole-cell labeling. Nat Methods. 2008;5(11):955–957. doi: 10.1038/nmeth.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sunshine JC, Akanda MI, Li D, Kozielski KL, Green JJ. Effects of base polymer hydrophobicity and end-group modification on polymeric gene delivery. Biomacromolecules. 2011;12(10):3592–3600. doi: 10.1021/bm200807s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Non-viral gene delivery nanoparticles based on poly(beta-amino esters) for treatment of glioblastoma. Biomaterials. 2011;32(23):5402–5410. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green JJ, Zugates GT, Tedford NC, et al. Combinatorial modification of degradable polymers enables transfection of human cells comparable to adenovirus. Adv Mater. 2007;19(19):2836–2842. [Google Scholar]
- 40.Midoux P, Breuzard G, Gomez JP, Pichon C. Polymer-based gene delivery: a current review on the uptake and intracellular trafficking of polyplexes. Curr Gene Ther. 2008;8(5):335–352. doi: 10.2174/156652308786071014. [DOI] [PubMed] [Google Scholar]
- 41.Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Letters. 2009;9(6):2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jere D, Xu CX, Arote R, Yun CH, Cho MH, Cho CS. Poly(beta-amino ester) as a carrier for si/shRNA delivery in lung cancer cells. Biomaterials. 2008;29(16):2535–2547. doi: 10.1016/j.biomaterials.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 43.Vandenbroucke RE, De Geest BG, Bonne S, et al. Prolonged gene silencing in hepatoma cells and primary hepatocytes after small interfering RNA delivery with biodegradable poly(beta-amino esters) J Gene Med. 2008;10(7):783–794. doi: 10.1002/jgm.1202. [DOI] [PubMed] [Google Scholar]
- 44.Spagnou S, Miller AD, Keller M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry. 2004;43(42):13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- 45.Adair JH, Parette MP, Altinoglu EI, Kester M. Nanoparticulate alternatives for drug delivery. ACS Nano. 2010;4(9):4967–4970. doi: 10.1021/nn102324e. [DOI] [PubMed] [Google Scholar]
- 46.Sutton D, Kim SJ, Shuai XT, et al. Efficient suppression of secretory clusterin levels by polymer-siRNA nanocomplexes enhances ionizing radiation lethality in human MCF-7 breast cancer cells in vitro. Int J Nanomed. 2006;1(2):155–162. doi: 10.2147/nano.2006.1.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breunig M, Hozsa C, Lungwitz U, et al. Mechanistic investigation of poly(ethylene imine)-based siRNA delivery: disulfide bonds boost intracellular release of the cargo. J Control Release. 2008;130(1):57–63. doi: 10.1016/j.jconrel.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 48.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 49.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc. 2009;131(6):2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elbakry A, Zaky A, Liebl R, Rachel R, Goepferich A, Breunig M. Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano Lett. 2009;9(5):2059–2064. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 51.Qi L, Gao X. Quantum dot-amphipol nanocomplex for intracellular delivery and real-time imaging of siRNA. ACS Nano. 2008;2(7):1403–1410. doi: 10.1021/nn800280r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GFP+ HUVECs were seeded in 96-well plates at known densities and allowed to settle and adhere for 1 hr before measuring fluorescence with a microplate reader. Linear regression showed a positive, linear correlation following the relation RFU = 33073*(#cells)+6599 with correlation coefficient r2 = 0.985. The gray, shaded region shows the 95% confidence interval of the regression line.
Note: Error bars represent standard deviation of multiple replicates at each cell density.
Abbreviations: GFP, green fluorescent protein; HUVECs, human umbilical vein endothelial cells; RFU, relative fluorescence units.