ABSTRACT
Vibrio fischeri, the bacterial symbiont of the Hawaiian bobtail squid, Euprymna scolopes, uses quorum sensing to control genes involved in bioluminescence, host colonization, and other biological processes. Previous work has shown that AinS/R-directed quorum sensing also regulates the expression of rpoQ (VF_A1015), a gene annotated as an RpoS-like sigma factor. In this study, we demonstrate using phylogenetics that RpoQ is related to, but distinct from, the stationary-phase sigma factor RpoS. Overexpression of rpoQ results in elevated chitinase activity but decreased motility and luminescence, three activities associated with symbiosis. The reduction in bacterial luminescence associated with the overexpression of rpoQ occurs both in culture and within the light-emitting organ of the squid host. This suppression of bioluminescence is due to the repression of the luxICDABEG promoter. Our results highlight RpoQ as a novel regulatory component, embedded in the quorum-signaling network that controls several biological processes in V. fischeri.
IMPORTANCE
Quorum signaling is a widely occurring phenomenon that functions in diverse bacterial taxa. It is most often found associated with species that interact with animal or plant hosts, either as mutualists or pathogens, and controls the expression of genes critical to tissue colonization. We present the discovery of rpoQ, which encodes a new regulatory component in the quorum-signaling pathway of Vibrio fischeri. RpoQ is a novel protein in the RpoS family of stationary-phase sigma factors. Unlike many other regulatory proteins involved in the quorum-signaling pathways of the Vibrionaceae, the distribution of RpoQ appears to be restricted to only two closely related species. The role of this regulator is to enhance some quorum-signaling outputs (chitinase activity) while suppressing others (luminescence). We propose that RpoQ may be a recently evolved or acquired component in V. fischeri that provides this organism with an additional level of regulation to modulate its existing quorum-signaling pathway.
Introduction
The marine bacterium Vibrio fischeri can exist either as a free-living organism or in symbiosis with certain bioluminescent animals, including the Hawaiian bobtail squid, Euprymna scolopes (1). Like other bacteria, V. fischeri must be able to specifically adapt to different or changing environments. Two widely distributed and functionally distinct regulatory mechanisms by which bacteria react transcriptionally to changes in their environment include quorum-sensing-mediated signaling to alter transcription factor activity (2) and alternative sigma factors to modify the core transcription machinery (3).
Quorum signaling enables many species of bacteria to coordinate gene expression according to the presence of molecules called autoinducers. V. fischeri encodes three quorum-sensing systems, each of which depends on a specific type of autoinducer (Fig. 1). Together, these quorum-sensing systems regulate the expression of genes involved in luminescence and colonization (2, 4–8). The LuxI-LuxR system directly controls the genes responsible for light production. Briefly, at low cell density, LuxI synthesizes low levels of the autoinducer N-3-oxo-hexanoyl homoserine lactone (3-oxo-C6). As the culture reaches high cell density, 3-oxo-C6 accumulates to a threshold level and binds to the transcriptional regulator LuxR. The LuxR/3-oxo-C6 complex directly activates transcription of the luxICDABEG operon (Fig. 1), which is responsible for the bacterium’s ability to produce luciferase and bioluminescence (9, 10).
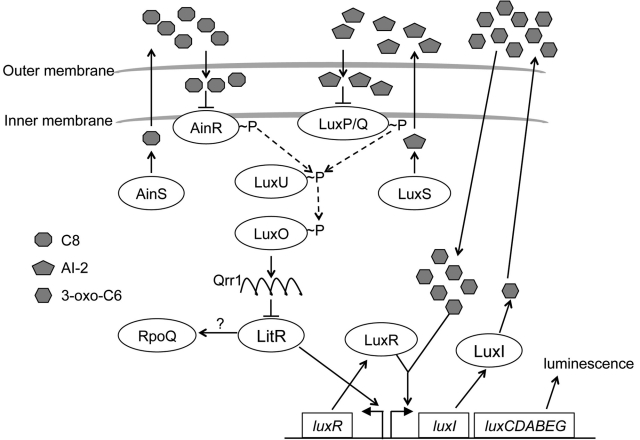
FIG 1 .
Model of quorum-sensing systems in V. fischeri. When the levels of octanoyl homoserine lactone (C8; octagons) and AI-2 (pentagons) are low, AinR and the LuxP/Q complex phosphorylate LuxU. Phosphorylated LuxU, in turn, phosphorylates LuxO, which activates the expression of the small RNA (sRNA) Qrr1. Qrr1 destabilizes the mRNA encoding LitR. Conversely, when the level of C8 and AI-2 autoinducers is high, the signaling cascade is inactivated, resulting in stable LitR production. LitR transcriptionally activates luxR, which encodes the transcriptional regulator LuxR. In the presence of the accumulating autoinducer 3-oxo-hexanoyl homoserine lactone (3-oxo-C6; hexagons), LuxR activates several genes, including the luxCDABEG locus, which is responsible for luciferase production and activity. The dashed arrows indicate phosphoryl group transfers. The question mark indicates an unknown number of steps.
Upstream of LuxI/R within this regulatory network are two other quorum-sensing systems, AinS/R and, to a lesser extent, LuxS/PQ, which each signal through the LuxU-LuxO phosphorelay (5). At low cell density, these two systems are silent, resulting in the phosphorylation of the phosphotransferase LuxU, which in turn phosphorylates the response regulator LuxO. Phosphorylated LuxO activates the expression of a noncoding RNA, qrr1, which destabilizes the mRNA of the master regulator LitR in an Hfq-dependent manner (11). As cell density increases, so does signaling by AinS/R and LuxS/PQ. As a result, LuxO is dephosphorylated, litR mRNA is stabilized and translated, and LitR positively regulates the expression of luxR (12). At even higher cell density, when 3-oxo-C6 accumulates and binds LuxR, luxICDABEG is expressed (6), resulting in light production (Fig. 1).
In addition to luminescence and squid colonization, quorum sensing regulates other biological processes in V. fischeri. Signaling via AinS regulates the acetate switch by inducing acs, which encodes acetyl coenzyme A (acetyl-CoA) synthetase, leading to a removal of previously excreted acetate and thus to a decrease in medium acidification (13). AinS, as well as LitR, was shown to also regulate motility (6). In particular, disruption of either ainS or litR results in hypermotile cells.
Alternative sigma factors provide a different mechanism for transcriptional control through the replacement of the primary sigma factor, RpoD (14). RpoS is a particularly well-characterized example of the σ70 family of sigma factors, which controls cellular responses to stress conditions, including the presence of reactive oxygen species, starvation, DNA damage, extreme temperature, ethanol, and hyperosmolarity (15–17). In some bacteria, RpoS is required for virulence as well as quorum-sensing regulation (18–22). In V. fischeri, RpoS regulates catalase activity but is not involved in the regulation of quorum-regulated genes like acs (13, 23). The sigma factor σ54, encoded by rpoN, can control nitrogen metabolism, motility, bioluminescence, and host colonization in many bacteria (24–27). Similarly, RpoN contributes to the regulation of these activities and to biofilm formation in V. fischeri (28).
Recently, in addition to an authentic RpoS factor, RpoS-like sigma factors have been identified in several bacteria, such as Vibrio alginolyticus ZJ-51 and 12G01, Vibrio splendidus LGP32, Vibrio sp. MED222, and Vibrio campbellii BAA_1116. There are no studies about any of these RpoS-like sigma factors except for RpoX in V. alginolyticus ZJ-51 (29). While RpoS and RpoX are both involved in biofilm formation and stress response, they have different functions. In this study, we focus on VF_A1015, which is annotated as encoding an RpoS-like sigma factor and is positively regulated by ainS (6). This gene is located on the second chromosome of V. fischeri and is linked to other genes predicted to be involved in signaling. Because of its apparent regulation by quorum signaling, we have renamed VF_A1015 as rpoQ (30); here we present evidence of both its position and role within the V. fischeri quorum-signaling pathways.
RESULTS
rpoQ encodes a novel sigma factor-like protein regulated by quorum sensing.
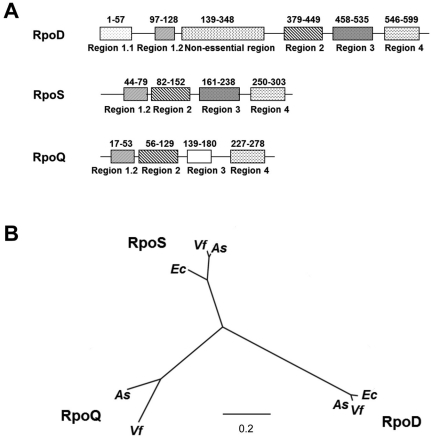
VF_A1015, here designated rpoQ, was originally annotated as encoding an RpoS-like sigma factor (6). This annotation arose because the amino acid sequence of RpoQ is 45% identical, and is of similar length, to that of RpoS. In contrast to rpoS, which is present within all fully-sequenced members of the Vibrionaceae, we were able to identify rpoQ homologues within the genomes of only two species: V. fischeri and Aliivibrio (Vibrio) salmonicida (VSAL_II0319). Examination using the Pfam database revealed that RpoQ contains four conserved domains: (i) σ70 region 1.2, (ii) σ70 region 2, (iii) σ70 region 3, and (iv) σ70 region 4, each of which is significant except σ70 region 3 (Fig. 2A). In contrast, RpoS and RpoD of V. fischeri each have an identifiable σ70 region 3 in the predicted location. Region 3 is involved in binding to the core RNA polymerase and recognition of the extended −10 promoter. Only regions 2 and 4 are highly conserved and play important roles in binding the core RNA polymerase and in recognition of the −10 and −35 promoters, respectively (31). Phylogenetic analysis of the amino acid sequences spanning from region 2 to region 4 of the RpoQ, RpoS, and RpoD homologues among two species of the Vibrionaceae (V. fischeri and A. salmonicida) and Escherichia coli indicates that RpoQ is a protein that is distinct from RpoS (Fig. 2B).
FIG 2 .
Structure and phylogeny of Rpo proteins. (A) Domain comparison between the V. fischeri RpoD, RpoS, and RpoQ proteins. Numbers above the conserved domains indicate amino acid positions. (B) Neighbor-joining phylogenetic analysis of RpoQ. Sequences spanning region 2 through region 4 were used to generate the phylogenetic tree with the PHYLIP software program. The tree was bootstrapped with 1,000 replicates, and all nodes were supported 100%. Abbreviations used are as follows: Vf, Vibrio fischeri; As, Aliivibrio salmonicida; Ec, Escherichia coli. The scale bar represents genetic distances in substitutions per residue.
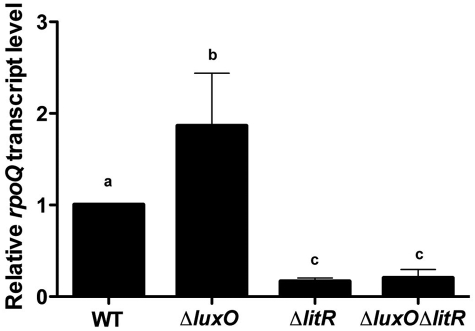
Previous results from a global transcriptional analysis (6) suggested that expression of rpoQ is controlled by the response regulator LuxO, which is involved in quorum sensing (Fig. 1). To validate and extend this analysis, we used quantitative reverse transcriptase PCR (qRT-PCR) to measure the expression of rpoQ in different mutant backgrounds of V. fischeri. Consistent with the previous study, the level of rpoQ expression is elevated about 2-fold in a ΔluxO mutant when compared to that of a wild-type strain (Fig. 3). In contrast to the ΔluxO mutant, both a ΔlitR single mutant and a ΔluxO ΔlitR double mutant exhibited 4-fold-lower levels of rpoQ expression than the wild type (Fig. 3), demonstrating that the regulation of rpoQ by LuxO operates through LitR.
FIG 3 .
qRT-PCR analysis of rpoQ expression in V. fischeri strains MJM1100 (wild type [WT]), TIM305 (ΔluxO), TIM358 (ΔlitR), and TIM355 (ΔluxOΔlitR). The strains were grown in LBS medium and harvested at an OD600 of 0.5. The transcript levels of rpoD were used as a normalizing control. Data are relative to wild-type levels, set at 1.0. Graphical and error bars indicate the averages and standard deviations of data from three independent experiments, respectively. Shared letters above the bars indicate no statistically significant difference (P > 0.05), whereas different letters indicate that there is a significant difference (P < 0.01) in rpoQ transcript levels between those strains (analysis of variance [ANOVA] and Tukey’s honestly significant difference [HSD] test).
RpoQ regulates gene expression in V. fischeri.
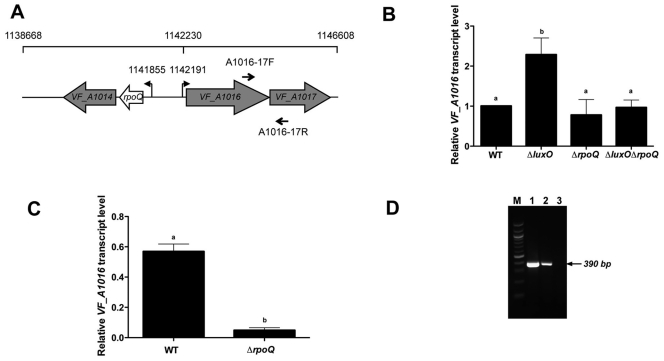
To determine whether RpoQ regulates expression of other genes in V. fischeri, we generated a ΔrpoQ in-frame deletion mutant that avoids potential polar effects on the downstream gene VF_A1014 (Fig. 4A). Previous work suggested that, like rpoQ, the genes VF_A1016 and VF_A1017, which are predicted to encode a histidine kinase and a response regulator, respectively, are also regulated by the LuxO (6). Due to the genetic linkage between this operon and rpoQ (they are divergently described, with a 449-nucleotide intergenic region) (Fig. 4A), we explored the possibility that the LuxO-dependent regulation of VF_A1016 and VF_A1017 requires RpoQ activity. Consistent with the previous report (6), we found that the expression level of VF_A1016 was elevated more than 2-fold in a ΔluxO mutant over that in the wild-type strain (Fig. 4B). In contrast, VF_A1016 expression levels were similar to wild-type levels in a ΔrpoQ mutant and remained low in the ΔluxO ΔrpoQ double mutant. These data indicate that RpoQ is required for LuxO-dependent changes in VF_A1016. Furthermore, when rpoQ was integrated into the Tn7 site of the chromosome of a ΔluxO ΔrpoQ double mutant (strain CA6), the transcription level of VF_A1016 was partially restored relative to that for the ΔluxO single mutant (see Fig. S1 in the supplemental material). Based on the finding that VF_A1016 was regulated by quorum signaling, we were led to test the possibility that transcription of VF_A1016 is reduced in the ΔrpoQ mutant at a higher cell density. As predicted, the transcription level of VF_A1016 is higher (12-fold) in the wild-type strain than in the ΔrpoQ mutant (Fig. 4C). In addition, RT-PCR analysis indicated that VF_A1016 and VF_A1017 are cotranscribed (Fig. 4D). Together, these results suggest that the LuxO signaling system transcriptionally controls RpoQ, which in turn activates transcription of a two-component system. Using 5′ rapid amplification of cDNA ends (RACE), we mapped the transcriptional start sites of rpoQ and VF_A1016 (Fig. 4A). Neither start site was detected in the ΔrpoQ mutant, supporting the idea that transcription of rpoQ and VF_A1016 requires RpoQ (data not shown).
FIG 4 .
LuxO and RpoQ regulate transcription of the VF_A1016-VF_A1017 operon. (A) Schematic representation of the rpoQ locus. Open reading frames (ORFs) are indicated as the four block arrows. VF_A1014 is predicted to encode a GGDEF-EAL-domain-containing protein of unknown function. VF_A1016 and VF_A1017 are predicted to encode a two-component histidine kinase sensor and response regulator, respectively. Numbers on the scale bar indicate the location (in bp from the origin) along the second chromosome of V. fischeri. Transcriptional start sites of rpoQ and VF_A1016 determined by 5′-RACE are shown upstream of corresponding genes. Primer sites used in RT-PCR analysis shown in panel D are indicated by arrows. (B) qRT-PCR analysis of VF_A1016 expression in V. fischeri strains MJM1100 (WT), TIM305 (ΔluxO), CA1 (ΔrpoQ), and CA4 (ΔluxOΔrpoQ). The strains were grown in LBS medium and harvested at an OD600 of 0.5. The transcript levels of rpoD were used to normalize VF_A1016 levels. Data are relative to wild-type levels, set at 1.0. Graphical and error bars indicate the averages and standard deviations of data from three independent experiments, respectively. Shared letters above the bars indicate no statistically significant difference (P > 0.05), whereas different letters indicate that there is a significant difference (P < 0.05) in VF_A1016 transcript levels between those strains (ANOVA and Tukey’s HSD test). (C) qRT-PCR analysis of VF_A1016 expression in V. fischeri strains MJM1100 (WT) and CA1 (ΔrpoQ) at a high cell density. The strains were grown in LBS medium and harvested at an OD600 of 3.0, which is 6 times denser than the cultures used for panel B. The transcript levels of rpoD were used to normalize VF_A1016 levels. Graphical and error bars indicate the averages and standard deviations, respectively, of data from three independent experiments. Different letters indicate that there is a significant difference (P < 0.001) in VF_A1016 transcript levels between the WT and the ΔrpoQ mutant (Student’s t test). (D) RT-PCR analysis of VF_A1016-VF_A1017 transcripts. Templates used are genomic DNA (lane 1), cDNA (lane 2), and RNA (lane 3). The location of the primer pair (A1016-17F/A1016-17R) is shown in panel A. Lane M contains molecular size standards (size range, in bp, from top to bottom: 1,517, 1,200, 1,000, 900, 800, 700, 600, 500/517, 400, 300, 200, and 100).
RpoQ represses luminescence in vivo and in vitro.
Because quorum signaling controls both early and late colonization genes (4, 6, 8, 12), we asked whether RpoQ regulates factors involved in light-organ symbiosis. Juvenile squid colonized by the ΔrpoQ mutant exhibited normal symbiont population levels and luminescence over the first 48 h postcolonization (data not shown), during which the symbionts have expressed both early and late quorum-sensing responses (6). Furthermore, cocolonization experiments using the ΔrpoQ mutant and a wild-type strain did not reveal a competition defect in the absence of RpoQ (data not shown). Together, these results suggest that the initial steps of bacterial colonization of the host, involving known early and late quorum-regulated colonization factors, do not require RpoQ.
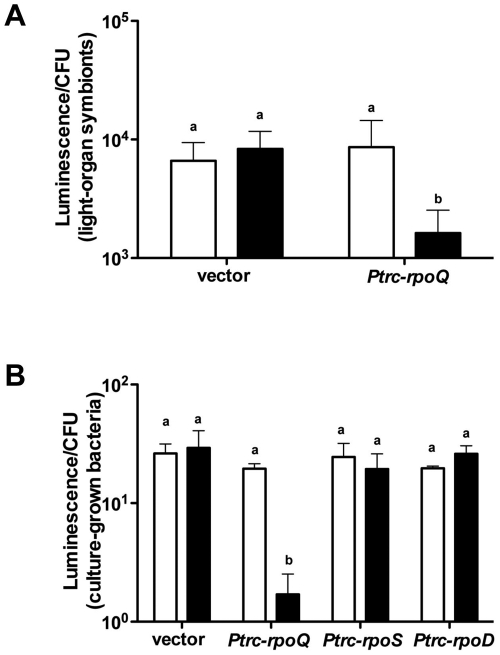
As an alternative approach to examine RpoQ function during colonization, we constructed a plasmid (pXCD10) that conditionally (i.e., in the presence of isopropyl-β-d-thiogalactopyranoside [IPTG]) overexpresses RpoQ, by cloning rpoQ downstream of the trc promoter (32). Such approaches, with caveats for possible artifacts, have been used successfully to manipulate target genes of other extracytoplasmic function (ECF) sigma factors (33–37). Juvenile squid colonized with wild-type V. fischeri harboring pXDC10 were placed in seawater, either with or without IPTG. The level of luminescence of symbionts overexpressing RpoQ was significantly lower than (i.e., <20% of) that of wild-type cells harboring the control plasmid, pTM214 (Fig. 5A). In contrast, there were no significant differences in luminescence per CFU between animals colonized with wild-type cells harboring pTM214 (vector control) with or without IPTG or wild-type cells harboring pXDC10 without IPTG; in addition, the CFU levels were similar among all animals (data not shown), indicating that overexpressing RpoQ did not affect the ability of V. fischeri to colonize the host.
FIG 5 .
The impact of RpoQ on luminescence. (A) Squid luminescence (per CFU) of wild-type V. fischeri cells harboring either pTM214 (vector) or pXDC10 (Ptrc-rpoQ) in the presence (black bars) or absence (white bars) of 1 mM IPTG. Graphical and error bars indicate the averages and standard deviations. One representative experiment performed in triplicate is analyzed. (B) Culture luminescence per CFU of wild-type MJM1100 harboring pTM214 (vector), pXDC10 (Ptrc-rpoQ), pXDC35 (Ptrc-rpoS), and pXDC36 (Ptrc-rpoD), with (black bars) or without (white bars) IPTG addition. Graphical and error bars indicate, respectively, the averages and standard deviations, respectively, of data from three independent experiments. Shared letters above the bars indicate no statistically significant difference (P > 0.05), whereas different letters indicate a significant difference (P < 0.001 in panel A and P < 0.05 in panel B) between the luminescence levels (ANOVA and Tukey’s HSD test).
Consistent with the results described above, the luminescence emitted by cells in culture was reduced more than 10-fold when rpoQ was overexpressed (Fig. 5B). In contrast, overexpression of two other sigma factors, rpoS and rpoD, did not repress luminescence. Taken together, these results provide evidence that the cells are not disturbed by these overexpression studies and show that RpoQ represses luminescence both in vivo and in vitro, as well as further demonstrating a functional distinction between RpoQ and RpoS.
RpoQ represses the luxICDABEG promoter.
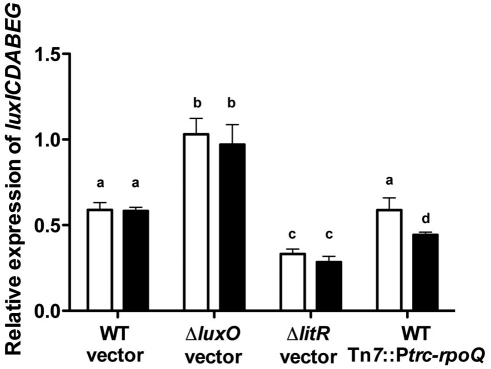
Because overexpression of RpoQ leads to repressed luminescence, we hypothesized that RpoQ affects transcription of the lux operon. To facilitate experiments that test this idea, we integrated the IPTG-inducible rpoQ gene into the chromosome at the Tn7 site, resulting in strain TIM366. Overexpression of this single copy of rpoQ by IPTG addition also repressed luminescence but to only 31% of the extent seen when rpoQ is induced from the multicopy plasmid pXDC10. A two-color fluorescent reporter of luxICDABEG promoter activity (11) was used to determine whether RpoQ affects transcription of this promoter. The reporter plasmid (pTM280) was constructed by cloning the luxICDABEG promoter upstream of gfp, which encodes green fluorescent protein (GFP). This plasmid also constitutively expresses the red fluorescent protein mCherry. The luxICDABEG transcription level is quantitatively measured by the GFP/mCherry fluorescence ratio; this normalization accounts for potential variations in either growth rates or copy number of the reporter plasmid in different strains. We found that there was a significant difference in the level of luxICDABEG transcription in strain TIM366 when rpoQ was induced (Fig. 6). As predicted, luxICDABEG transcription was higher in a ΔluxO mutant and lower in a ΔlitR mutant than that in the wild-type strain, independent of supplemented IPTG. In contrast, overexpression of rpoQ had no effect on luxR promoter activity (data not shown), and overexpression of VF_A1016 did not affect luminescence (see Fig. S2 in the supplemental material).
FIG 6 .
Transcriptional activity of luxICDABEG. Relative expression levels of luxICDABEG (GFP/mCherry) for strains TIM313 (WT vector), TIM315 (ΔluxO vector), CA19 (ΔlitR vector), and TIM366 (WT Tn7::Ptrc-rpoQ) either with (black bars) or without (white bars) the addition of 1 mM IPTG. Each strain carried the two-color fluorescent reporter plasmid pTM280 (PluxI-gfp PtetA-mCherry). Graphical and error bars indicate the averages and standard deviations of data from three independent experiments, respectively. Shared letters above the bars indicate no statistically significant difference (P > 0.05), whereas different letters indicate a significant difference (P < 0.05) between luxICDABEG transcript levels (ANOVA and Tukey’s HSD test).
We next asked whether the repression of the luxICDABEG promoter by RpoQ functioned through (i) LitR, the transcriptional regulator known to affect the activity of the luxICDABEG promoter, (ii) VF_A1014, which is the gene downstream of rpoQ that encodes a GGDEF- and EAL-domain-containing protein, (iii) a nearby gene encoding the response regulator VF_A1017, or (iv) ArcA, the recently discovered negative regulator of luminescence (38). We moved the inducible rpoQ-bearing plasmid pXDC10 into the ΔlitR (TIM358), ΔVF_A1014 (CA8), VF_A1017 (KV1612), and ΔarcA (AMJ2) mutants and measured their effect on the level of culture luminescence. Overexpression of RpoQ repressed luminescence in all of the strains tested (data not shown), suggesting that the repression of luminescence by RpoQ is independent of transcriptional regulation by LitR, VF_A1014, VF_A1017, or ArcA.
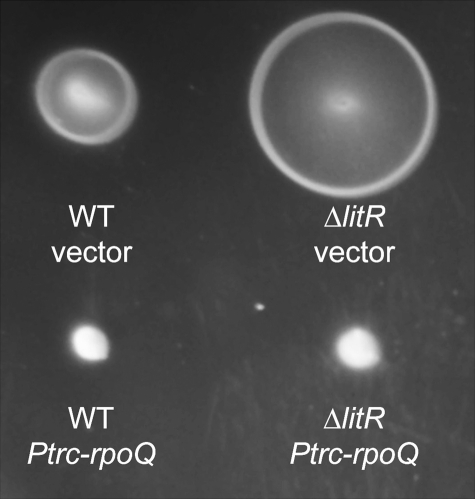
Overexpression of RpoQ decreases motility.
To investigate whether RpoQ is involved in the regulation of other quorum-signaling-dependent phenotypes besides luminescence, we examined the impact of overexpressing RpoQ on V. fischeri flagellar motility. The ΔlitR mutant displayed hypermotile phenotypes, as previously reported (6). In contrast, when RpoQ was overexpressed in either the wild type or the ΔlitR mutant, both were essentially nonmotile (Fig. 7), suggesting that quorum signaling regulates motility, at least in part, through an RpoQ mechanism downstream of LitR. Interestingly, overexpression of VF_A1016 had a small positive effect on wild-type motility (see Fig. S3 in the supplemental material).
FIG 7 .
The motility of wild-type V. fischeri cells harboring either pTM214 (WT vector) or pXDC10 (WT Ptrc-rpoQ), compared to strain TIM358 harboring either pTM214 (ΔlitR vector) or pXDC10 (ΔlitR Ptrc-rpoQ). Relative rates of motility were determined in minimal medium containing 1 mM IPTG and solidified with 0.25% agar. One representative experiment of three is shown. Growth rates of the strains were comparable in the minimal medium (data not shown).
RpoQ elevates chitinase activity.
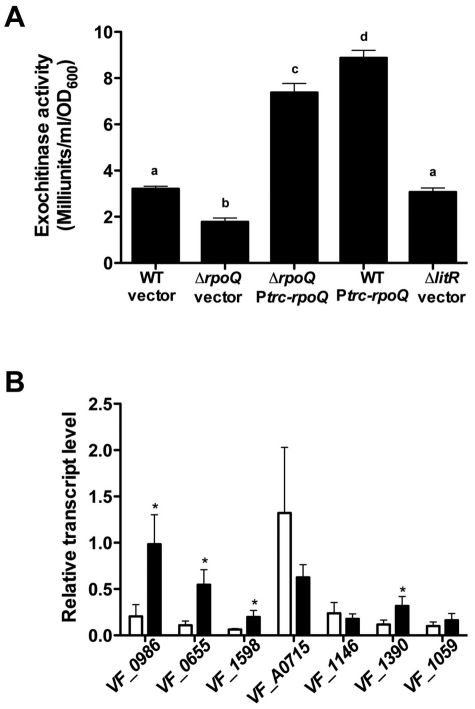
Quorum signaling negatively regulates chitinase activity in Vibrio harveyi; however, in Chromobacterium violaceum it leads to its activation (39, 40). To test whether and how RpoQ might control chitinase activity in V. fischeri, we examined the spent culture supernatants of the wild type and the ΔrpoQ mutant carrying the vector plasmid (pTM214) and the inducible rpoQ allele (pXDC10), as well as the ΔlitR mutant harboring the vector plasmid (pTM214). The supernatant of a culture of the ΔrpoQ mutant showed chitinase activity that was significantly lower (about 50%) than that of the wild type (Fig. 8A). In contrast, the supernatants of both the wild type and the ΔrpoQ mutant overexpressing RpoQ showed chitinase activity that was 2.3- to 2.8-fold-higher than that of the wild type carrying the vector. These results indicate that an increased level of RpoQ elevates chitinase activity in V. fischeri. Interestingly, there was no difference in chitinase activity between the wild-type strain and a ΔlitR mutant (Fig. 8A), suggesting that the basal levels of RpoQ present even in a ΔlitR mutant are sufficient to result in normal chitinase activity (Fig. 9). Furthermore, the supernatant of the wild type overexpressing VF_A1016 also showed chitinase activity that was 2.4-fold higher than that of the wild type carrying the vector (see Fig. S4 in the supplemental material), supporting the possibility that RpoQ may elevate chitinase activity through VF_A1016.
FIG 8 .
Regulation of chitinase genes by RpoQ. (A) Exochitinase activity of wild-type V. fischeri cells harboring either pTM214 (WT vector) or pXDC10 (WT Ptrc-rpoQ), strain CA1 harboring either pTM214 (ΔrpoQ vector) or pXDC10 (ΔrpoQ Ptrc-rpoQ), and TIM358 harboring pTM214 (ΔlitR vector). Secreted chitinase activity was determined in the cell-free supernatant. Graphical and error bars indicate, respectively, the averages and standard deviations of data from three independent experiments. Shared letters above the bars indicate no statistically significant difference (P > 0.05), whereas different letters indicate a significant difference (P < 0.05) in exochitinase activity between those strains (ANOVA and Tukey’s HSD test). (B) Wild-type cells carrying the empty vector plasmid (pTM214; white bars) or the inducible rpoQ allele (pXDC10; black bars) were grown in LBS medium supplemented with 1 mM IPTG and harvested at an OD600 of 1.4 to 1.5. VF_0986, chitodextrinase; VF_0655, endochitinase; VF_1598, exochitinase; VF_A0715, chitodextrinase precursor; VF_1146, chitodextrinase precursor; VF_1390, chitinase; VF_1059, chitinase. Graphical and error bars indicate, respectively, the averages and standard deviations of data from four independent experiments. The asterisks indicate a significant difference in transcription levels between each set of paired strains.
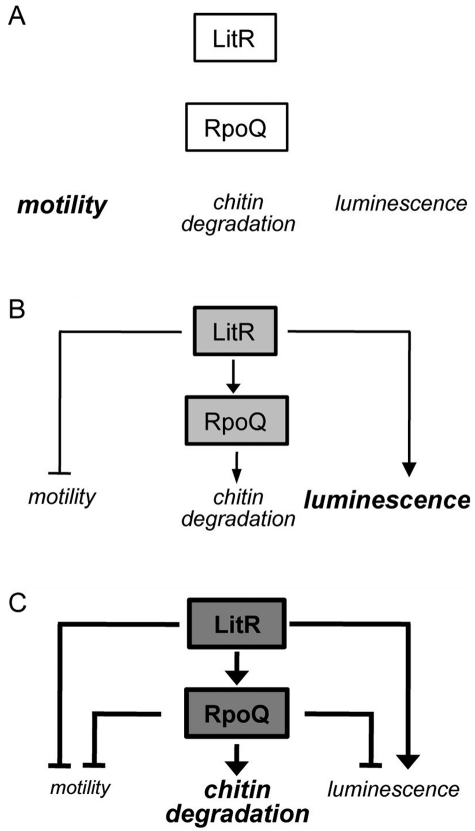
FIG 9 .
A model of RpoQ regulation of quorum-sensing-dependent phenotypes in V. fischeri. (A) At low cell density (no quorum signaling), LitR has little effect on basal levels of motility (high) and luminescence (low). (B) At moderate cell density, LitR signaling represses motility and induces luminescence as previously described (5); LitR also activates transcription of RpoQ. However, while this level of RpoQ remains insufficient to affect motility and luminescence, it does lead to increased chitinase activity. (C) At high cell density, LitR signaling increases enough to induce RpoQ to a higher level, leading to a strong repression of both motility and luminescence, as well as an induction of chitinase activity. Lines with arrowheads indicate positive regulation, while those with a bar at the end indicate negative regulation; the lines may represent a pathway with several steps of regulation.
Previous studies predicted that there are 7 chitinase genes encoded by V. fischeri ES114: VF_0655, VF_0986, VF_1059, VF_1146, VF_1390, VF_1598, and VF_A0715 (41). To test which of these chitinase genes is regulated by RpoQ, qRT-PCR was carried out using the wild-type strain carrying either the empty vector plasmid (pTM214) or the inducible rpoQ allele (pXDC10) in the present of IPTG. The transcription levels of four of the genes (VF_0986, VF_0655, VF_1598, and VF_1390) were significant higher when RpoQ was overexpressed (Fig. 8B). In contrast, the transcription levels of VF_A0715, VF_1146, and VF_1059 were independent of RpoQ. These data reveal that RpoQ activates the transcription of some chitinase genes, resulting in elevated levels of secreted exochitinase activity.
DISCUSSION
In this study, we have shown that RpoQ is a novel component that functions downstream of the core LuxO-LitR quorum-signaling system of V. fischeri (Fig. 1). The domain structure and phylogenic similarities between RpoQ and RpoS (Fig. 2) suggest that the former could have been evolutionarily derived from the latter, perhaps after the divergence between the clade containing V. fischeri and A. salmonicida and the rest of the Vibrionaceae (42). Interestingly, rpoQ and its A. salmonicida homologue, VSAL_II0319, are found on the smaller, second chromosomes of V. fischeri and A. salmonicida, respectively, where many paralogs and nonessential genes are also located (43). Such a position would be consistent with our hypothesis that rpoQ is a recently duplicated gene and diverged in the Vibrionaceae (44, 45).
RpoS has been shown to participate in bacterial quorum sensing; for instance, in Vibrio anguillarum, RpoS induces expression of the LitR homologue, VanT, in a manner independent of the LuxO homologue, VanO (21). Because of the role of VanT in inducing pigment and metalloprotease production, these authors suggest that V. anguillarum RpoS works with the quorum-signaling system to regulate survival and stress responses. Similarly, in Vibrio cholerae, RpoS can upregulate the expression of its LitR homologue, HapR (19, 46). However, in this species, HapR in turn induces the expression of RpoS, which reinforces the stress response (47). In contrast, RpoS does not affect quorum-sensing regulation in V. harveyi (15). Yet another pattern is seen within Pseudomonas aeruginosa, in which quorum sensing regulates the transcription of rpoS. RpoS, in turn, induces expression of lasR and rhlR, which encode homologues of the V. fischeri activator LuxR (20). In addition, RpoS is involved in the repression of rhlI, which encodes one of the autoinducer synthases in P. aeruginosa (21). Thus, in P. aeruginosa, whose quorum-signaling pathway has no LuxU-LuxO phosphorelay nor LitR master regulator homologs, the impact of RpoS functions instead through the LuxR and LuxI homologs. This mode of regulation contrasts with that of V. fischeri RpoQ, which has no effect on luxR expression. As described above, other than in V. fischeri, we can identify an rpoQ homolog only within the genome of the closely related A. salmonicida. Because homologs of other quorum-sensing regulators can also be found in this species (11, 48), we hypothesize that RpoQ is a component of quorum signaling in A. salmonicida as well.
Numerous studies have shown that quorum sensing controls functions important for host interaction, such as luminescence, motility, and chitin utilization (4, 6, 8, 40). Our results revealed that RpoQ is involved in regulation of each of those functions (Fig. 9). The lux regulon is responsible for the production of bioluminescence and has served as a model for the genetic regulation underlying quorum sensing (10, 49, 50). Several genetic elements (e.g., those encoding ArcA and LexA) have been reported to negatively affect luminescence by repressing the expression of the luxICDABEG operon (38, 51, 52). Our finding that overexpression of RpoQ leads to repressed luminescence suggests that RpoQ is also a source of negative regulation in the circuit controlling lux expression.
In addition to luminescence, the quorum-sensing system also regulates motility in several different bacteria (53). In V. fischeri, the AinS-LitR pathway has been shown to be involved in the repression of motility (6, 12). Similarly, overexpression of RpoQ resulted in a significant motility defect (Fig. 7), suggesting that RpoQ may regulate the disappearance of flagellation at the extremely high cell density found within the light organ (54), thereby not only conserving nutrients and energy but also allowing the cells to pack more tightly.
Recently, transcriptional evidence has suggested that the V. fischeri population in the mature squid light-organ symbiosis ferments chitin at night, resulting in the production of formic and acetic acids (55). This hypothesis has been supported by the discovery that host blood cells carry particulate chitin to the tissue surrounding the symbionts (56). Essentially all Vibrio species encode numerous chitinases (41), and the elevated chitinase activity associated with high RpoQ levels may allow V. fischeri to more effectively use host-derived chitin as a nutrient.
How might LitR and RpoQ work together to regulate these three quorum-sensing-dependent phenotypes? At low LitR levels (e.g., in uninduced cells or the ΔlitR mutant), cells produce low luminescence and are highly motile (Fig. 9A). However, low LitR has no effect on chitinase activity. As quorum signaling leads to higher levels of LitR, its increased activity results in higher luminescence and lower motility (Fig. 9B). Meanwhile, at this early stage of induction, LitR activates transcription of rpoQ. However, while inducing chitinase activity, RpoQ levels remain insufficient to have an effect on motility and luminescence. At very high LitR levels, RpoQ is highly induced and begins to have a dominant effect on other downstream genes, reversing luminescence output and increasing the loss of motility, as well as further activating chitinase activity (Fig. 9C). This model is also supported by the observation that luminescence (see Fig. S2 in the supplemental material) and motility (see Fig. S3) but not chitinase activity (see Fig. S4) are directly regulated by LitR.
At this point in our analyses, we do not know whether RpoQ regulates using a sigma-like activity. Nevertheless, the results of a domain comparison suggest that RpoQ is a group 2 sigma factor (Fig. 2A), which generally play a role in upregulating transcription initiation at target promoters (31). Therefore, while it is possible that RpoQ directly binds to the promoter regions of the luxICDABEG operon or motility-related genes, thereby repressing transcription, we favor the hypothesis that RpoQ upregulates transcription of an unknown factor(s) that then represses transcription of the target genes identified here.
In summary, the novel V. fischeri sigma-like factor RpoQ is similar to but distinct from the alternative sigma factor, RpoS, and is regulated by the quorum-sensing system. RpoQ is involved in the regulation of several quorum-sensing-dependent phenotypes at high cell density (Fig. 9), suggesting that it acts as a global regulator of symbiotic maintenance in the host, enhancing some of those phenotypes while attenuating others.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in LB medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, and 1% [wt/vol] NaCl). Liquid cultures of V. fischeri were grown at 28°C with aeration either in LBS medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 2% [wt/vol] NaCl, 50 mM Tris-HCl [pH 7.5]) or in SWT medium (57). When necessary, antibiotics were used at the following concentrations: 25 µg ml−1 chloramphenicol (Cam) and 50 µg ml−1 kanamycin (Kan) for E. coli or 2.5 µg ml−1 Cam and 5 µg ml−1 erythromycin (Erm) for V. fischeri. Stock solutions of the N-3-oxo-hexanoyl homoserine lactone autoinducer (3-oxo-C6) were made in dimethyl sulfoxide.
TABLE 1 .
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference(s) |
|---|---|---|
| Strain | ||
| MJM1100 | Sequenced wild-type V. fischeri ES114 | 30, 43 |
| TIM305 | ES114 ΔluxO | 11 |
| TIM355 | ES114 ΔluxO ΔlitR | 11 |
| TIM358 | ES114 ΔlitR | 11 |
| TIM313 | ES114 Tn7::pEVS107 (erm) | 11 |
| TIM315 | ES114 ΔluxO Tn7::pEVS107 (erm) | 11 |
| TIM366 | ES114 Tn7::pTM319 (Ptrc-rpoQ) | This study |
| CA1 | ES114 ΔrpoQ | This study |
| CA4 | ES114 ΔluxO ΔrpoQ | This study |
| CA6 | ES114 ΔluxO ΔrpoQ Tn7::rpoQ | This study |
| CA8 | ES114 ΔVFA1014 | This study |
| CA19 | ES114 ΔlitR Tn7::pEVS107 (erm) | This study |
| KV1612 | ES114 VFA1017::kan | 66 |
| AMJ2 | ES114 ΔarcA | 38 |
| Plasmid | ||
| pEVS79 | pBC SK(+) oriT cat | 62 |
| pEVS104 | R6KoriRP4 oriT trb tra kan | 62 |
| pEVS107 | R6Kori oriT mini-Tn7 mob erm kan | 64 |
| pVSV105 | R6Kori ori(pES213) RP4 oriT cat | 67 |
| pUX-BF13 | R6Kori tns bla | 65 |
| pSVS112 | pEVS79 ΔrpoQ | This study |
| pTM214 | pVSV105 Ptrc-mCherry | 63 |
| pTM280 | pVSV105 PluxI-gfp + PtetA-mCherry | 11 |
| pTM319 | pEVS107 lacIqPtrc-rpoQ | This study |
| pXDC1 | pEVS79 ΔVFA1014 | This study |
| pXDC10 | pTM214 Ptrc-rpoQ ΔmCherry | This study |
| pXDC23 | pTM214 Ptrc-VF_A1016 ΔmCherry | This study |
| pXDC35 | pTM214 Ptrc-rpoS ΔmCherry | This study |
| pXDC36 | pTM214 Ptrc-rpoD ΔmCherry | This study |
Bioinformatics analysis.
Homologues of V. fischeri RpoQ (VF_A1015) were identified in A. salmonicida strain LFI1238 (VSAL_II0319) by BLAST analysis (58). Homologues of V. fischeri RpoD (VF_2254) and RpoS (VF_2067) were identified in E. coli strain MG1655 and A. salmonicida strain LFI1238 by BLAST. The conserved domains of these sigma factors were found using the Pfam database (59). Amino acid sequences were aligned using the ClustalX 2.0.11 software program (60) and used to generate a neighbor-joining tree using the PHYLIP program (61).
Plasmid and strain construction.
All the restriction enzymes used were from NEB (New England Biolabs, Ipswich, MA). V. fischeri mutants CA1 (ΔrpoQ), CA4 (ΔluxO ΔrpoQ), and CA8 (ΔVF_A1014) were constructed using allelic exchange based on pEVS79 (62). To construct the plasmid pSVS112, which contains the ΔrpoQ allele as an in-frame deletion of the sequence encoding amino acids 10 to 288, the homologous regions upstream and downstream of rpoQ were amplified by PCR using the primer pairs rpoQF1/rpoQR1 and rpoQF2/rpoQR2 (Table 2), digested by XhoI/SalI and SalI/SpeI, and ligated into the Cam resistance-encoding vector pEVS79 (62), which had been digested with XhoI/SpeI. To construct the plasmid pXDC1, which contains the ΔVF_A1014 allele as an in-frame deletion of the sequence encoding amino acids 34 to 421, the homologous regions upstream and downstream of VF_A1014 were amplified by PCR using the primer pairs VF_A1014F1/VF_A1014R1 and VF_A1014F2/VF_A1014R2 (Table 2), digested by XhoI/EcoRI and EcoRI/BamHI, and ligated into pEVS79, which had been digested with XhoI/BamHI. Each of these allelic-exchange vectors was then moved into V. fischeri by triparental mating using the conjugal helper plasmid pEVS104 (62). Conjugants were selected on LBS agar medium containing Cam, after which the appearance of a second recombination event was screened for on LBS agar medium containing no antibiotic. Colonies were purified, and their sequence was confirmed by PCR using the primers rpoQF3/rpoQR3 and rpoQF4/rpoQR4 or VF_A1014F3/VF_A1014R3 and VF_A1014F4/VF_A1014R4. Verification that the expected mutations were fixed in the chromosome was obtained by sequencing.
TABLE 2 .
Primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| A1016-17F | GAGATCCGGAAAGTAGAAAAAAACC |
| A1016-17R | GAAGTCGATGAAAAGTAATGCGTAT |
| CrpoDF | GAATACCGCCGTGGTTACAAA |
| CrpoDR | TTGACGCGAGATACGATTCAGT |
| CrpoQF | TCCTTGCTGAAACATCCCC |
| CrpoQR | AAGTTAGATATGAGAGTCTGCGAAG |
| CVF_A1016F | CACGAAATAAGAACACCACTCAATG |
| CVF_A1016R | TATCGTTAATCAAAACAAGCAAGGTT |
| RpoD-eF | GGGGTACCATGGATCAAAATCCGCAGTCACAGC |
| RpoD-eR | GCGTCGACTTATTCGTCTAAGAAGCTACGTAAT |
| RpoQ-eF | CGAGCTCGCTAGGTACAAGGATATGTTATGG |
| RpoQ-eR | GCGATATCTCAACCCAGTGCTATTTCTAAATCC |
| RpoS-eF | GGGGTACCATGAGTAAAAGCAATGCAGTAACTA |
| RpoS-eR | TTAGTATTCTTCAACTGCAAATAAT |
| rpoQF1 | CCCTCGAGAACGCCAGACATACCAATAATACC |
| rpoQR1 | GCGTCGACTCTCATATCTAACTTTGCATAAGC |
| rpoQF2 | GCGTCGACTTAGAAATAGCACTGGGTTGATAT |
| rpoQR2 | GGACTAGTATTTCGCACTTTTCATACCCTTTA |
| rpoQF3 | ACAGCACCAAGATCTAAGGCACGA |
| rpoQR3 | CACTTCACGTACATTGCTTGGCTG |
| rpoQF4 | GTATTGCCGCTTGTTAGTCT |
| rpoQR4 | ACAACCTCAACATATCCATGACA |
| RNA-linker | AUAUGCGCGAAUUCCUGUAGAACGAACACUAGAAGAAA |
| RACE-adapter | GCGCGAATTCCTGTAGA |
| rpoQ-RACE | CTGATAGCGATTTATTTCCTTTAA |
| rpoQ-RACE-nested | GTTCTTCGCAGACTCTCATATCTA |
| VF_A1016-RACE | GAGAGATAAGAATTAAGGGAACAA |
| VF_A1016-RACE-nested | GACCAAAACTCTTGATTTGATAGA |
| VF_A1014F1 | CGCTCGAGACGGTATTGCCGCTTGTTAGTCTTT |
| VF_A1014R1 | CGGAATTCATTATTCTTAACAAGATCATA |
| VF_A1014F2 | CGGAATTCGGTACAGGGTACTCCTCTTTAT |
| VF_A1014R2 | CGGGATCCAATTAGCCTGTAATCGTTGGCTTT |
| VF_A1014F3 | GTTATTTAATGTGGCGGCCTATGAC |
| VF_A1014R3 | AACGCTTTCGCGACTCTAACAAGCA |
| VF_A1014F4 | TGGATTTAGAAATAGCACTGGGTTG |
| VF_A1014R4 | ATGTGCAAGATAAATCGTCGATGCC |
Nucleotides in bold represent restriction enzyme sites added to the 5′ region of the primer.
To construct the plasmid pXDC10, which is an rpoQ overexpression vector, rpoQ was amplified by PCR using the primer pair RpoQ-eF/RpoQ-eR (Table 2), digested with SacI/EcoRV, and ligated into the vector pTM214 (58) after digestion with SacI/XmnI. To construct the plasmid pXDC35, which is an rpoS overexpression vector, rpoS was amplified by PCR using primer pair RpoS-eF/RpoS-eR (Table 2), digested with KpnI, and ligated into the vector pTM214 (63) after digestion with KpnI/XmnI. To construct the plasmid pXDC36, which is an rpoD overexpression vector, rpoD was amplified by PCR using primer pair RpoD-eF/RpoD-eR (Table 2), digested by KpnI/SalI, and ligated into the vector pTM214 (63). All constructs were verified by sequencing and then conjugated into the wild-type strain MJM1100 using pEVS104 (62). Conjugates were selected on LBS agar medium containing Cam.
To generate the plasmid pTM319, pXDC10 was digested by NotI and made blunt by T4 DNA polymerase. The resulting product was cut by SpeI and ligated into pEVS107 (64) after digestion with KpnI, making the end blunt with T4 DNA polymerase and cutting by SpeI. Integration of pTM319 into the Tn7 site of V. fischeri was performed using the helper plasmids pEVS104 (62) and pUX-BF13 (65). Conjugants were selected on LBS agar medium containing Erm.
Quantitative qRT-PCR measurements.
Total RNA from V. fischeri cultures was isolated using a QuickExtract RNA extraction kit and DNase I (Epicentre Biotechnologies, Madison, WI). RNA served as the template for cDNA synthesis with random hexamer primers and avian myeloblastosis virus (AMV) reverse transcriptase (Promega, Madison, WI). Quantitative reverse transcriptase PCR (qRT-PCR) measurements were performed using an iCycler thermal cycler (Bio-Rad Laboratories, Hercules, CA), as described previously (11). Amplification was performed at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. The primer sequences used (CrpoDF/CrpoDR, CrpoQF/CrpoQR, and CVF_A1016F/CVF_A1016R) are listed in Table 2. The CT values of the genes are between 20 and 30.
RT-PCR analyses.
Total RNA from V. fischeri cultures was isolated as described above, and RT-PCR was performed using the Access RT-PCR system (Promega, Madison, WI). Wild-type genomic DNA was used as the positive control, while an RNA sample with no reverse transcriptase added was the negative control. Primer pairs A1016-17F/A1016-17R (Table 2) were used to amplify the intergenic regions between VF_A1016 and VF_A1017.
5′ RACE.
Six micrograms of total RNA was extracted from ΔluxO (TIM305) and ΔluxO ΔrpoQ (CA4) cultures grown to an optical density at 600 nm (OD600) of 0.9 and subjected to dephosphorylation by tobacco acid pyrophosphatase (TAP) (Epicentre Biotechnologies, Madison, WI) for 30 min at 37°C. The RNA oligoRNA linker was ligated to total RNA using T4 RNA ligase (New England Biolabs, Inc., Ipswich, MA) according to the manufacturer’s instructions. cDNA was synthesized using AMV RT (Promega Corp., Madison, WI) according to the manufacturer’s instructions with the rpoQ-specific primer rpoQ-RACE and the VF_A1016-specific primer VF_A1016-RACE, respectively. PCR amplification was performed using the nested primers RACE-adapter and either rpoQ-RACE-nested or VF_A1016-RACE-nested. The single band that was present within the reaction containing TAP but absent from the TAP-minus control reaction was subcloned using the Topo TA cloning kit (Invitrogen) and sequenced as recommended by the manufacturer.
Culture luminescence assays.
Luminescence assays were performed using cultures of V. fischeri as described previously (11) with the following modification. Overnight cultures were diluted 1:100 in LBS medium either with or without 1 mM IPTG and grown at 28°C for 2 h. Cultures were then diluted 1:10 in medium containing 120 nM of the autoinducer 3-oxo-C6. After 3 h, luminescence and CFU were measured and reported in relative luminescence units (RLU), where 1 RLU corresponds to 1.9 × 104 quanta s−1.
Squid colonization and luminescence experiments.
Overnight cultures of V. fischeri were diluted 1:100 in fresh LBS medium and grown aerobically at 28°C to an OD600 of 1.0. Cultures were diluted to between 2,000 and 5,000 CFU/ml in filter-sterilized artificial seawater (Instant Ocean; IO) (Spectrum Brands Inc., Atlanta, GA) containing newly hatched squid. At 24 h postinoculation, sets of animals were placed in fresh IO either with or without 1 mM IPTG. At 48 h postinoculation, the luminescence of each animal was measured using a TD 20/20 luminometer (Turner Design, Sunnyvale, CA). To measure the RLU produced by symbiotic bacteria, animals were homogenized and the homogenate was measured for luminescence; the homogenate was also serially diluted and spread on LBS agar medium to determine CFU levels.
Fluorescence promoter-reporter assay.
V. fischeri cells were grown as described in the culture luminescence assay. Cultures were cooled quickly using an ice-slurry mix before 1-ml samples were harvested by centrifugation at 4°C for 2 min at 8,000 × g. Cell pellets were resuspended in 1 ml of defined seawater minimal medium (50 mM MgSO4, 10 mM CaCl2, 300 mM NaCl, 10 mM KCl, 0.01 mM FeSO4, 0.33 mM K2HPO4, 18.5 mM NH4Cl, 50 mM Tris-HCl [pH 7.5]) without a carbon source. Samples of 100 µl (each) in volume were placed in a microtiter plate well, and the OD600, as well as the levels of GFP and mCherry fluorescence, were measured using a Tecan Genios Pro plate reader (Tecan Group, Männedorf, Switzerland). A 485-nm excitation and 535-nm emission filter set was used for GFP measurements, and a 535-nm excitation and 612-nm emission filter set was used for mCherry measurements. A control nonfluorescent strain of V. fischeri was used to subtract any autofluorescence.
Motility assay.
Strains of V. fischeri were grown in SWT medium to an OD600 of approximately 0.3. A 3-µl culture sample was then stabbed into a defined seawater minimal medium (described above) supplemented with 0.5 g N-acetylglucosamine per liter and containing 0.25% agar. After 10 h, the diameters of the rings of migrating cells were measured.
Chitinase activity.
Exochitinase activity was measured using the chromogenic artificial substrate 4-nitrophenyl N,N′-diacetyl-β-d-chitobioside (Sigma-Aldrich Corp., St. Louis, MO). Briefly, V. fischeri strains were grown in LBS medium supplemented with 1 mM IPTG overnight at 28°C. The OD600 of the cultures was determined, and cell-free supernatants were collected by centrifugation. The substrate was then added to the supernatants and, after incubation for 30 min, the OD420 was determined and used to estimate the specific exochitinase activity (mU/ml/OD600) according to the manufacturer’s instructions.
SUPPLEMENTAL MATERIAL
qRT-PCR analysis of VF_A1016 expression in V. fischeri ΔluxO Tn7::erm, ΔluxO ΔrpoQ Tn7::erm, and ΔluxO ΔrpoQ Tn7::rpoQ strains. The strains were grown in LBS medium and harvested at an OD600 of 0.7 to 0.8. The transcript levels of rpoD were used to normalize VF_A1016 levels. Graphical and error bars indicate, respectively, the averages and standard deviations of data from three independent experiments. Shared letters above the bars indicate no statistically significant difference (P > 0.05), whereas different letters indicate a significant difference (P < 0.05) in VF_A1016 transcript levels between those strains (ANOVA and Tukey’s HSD test). Download Figure S1, TIF file, 0.1 MB.
Culture luminescence per CFU of (i) wild-type MJM1100 harboring either pTM214 (vector) or the inducible VF_A1016 allele (Ptrc-VF_A1016) and (ii) ΔlitR and ΔlitR ΔrpoQ strains harboring either pTM214 (vector) or inducible litR (Ptrc-litR). The cultures were grown either with (black bars) or without (white bars) IPTG addition. Graphical and error bars indicate, respectively, the averages and standard deviations of data from three independent experiments. Download Figure S2, TIF file, 0.1 MB.
The motility of (i) wild-type V. fischeri cells harboring either pTM214 (WT vector) or the inducible VF_A1016 allele (WT Ptrc-VF_A1016) and (ii) ΔlitR and ΔlitR ΔrpoQ cells harboring either pTM214 (vector) or the inducible litR allele (Ptrc-litR). Relative rates of motility were determined in minimal medium containing 1 mM IPTG and solidified with 0.25% agar. One representative experiment of three is shown. Growth rates of the strains were comparable in the minimal medium (data not shown). Download Figure S3, TIF file, 0.4 MB.
The exochitinase activity of (i) wild-type V. fischeri cells harboring either pTM214 (WT vector) or the inducible VF_A1016 allele (WT Ptrc-VF_A1016), and (ii) ΔlitR and ΔlitR ΔrpoQ cells harboring either pTM214 (vector) or the inducible litR allele (Ptrc-litR). Secreted chitinase activity was determined in the cell-free supernatant. Graphical and error bars indicate, respectively, the averages and standard deviations of data from three independent experiments. Shared letters above the bars indicate no statistically significant difference (P > 0.05), whereas different letters indicate a significant difference (P < 0.05) in exochitinase activity between those strains (ANOVA and Tukey’s HSD test). Download Figure S4, TIF file, 0.1 MB.
ACKNOWLEDGMENTS
We thank the members of the Ruby and M. McFall-Ngai laboratories for valuable advice and discussion and the manuscript’s reviewers for their insightful comments.
This work was supported by NIH grant RR12294 to E.G.R. and M. McFall-Ngai, by NSF grant IOS-0817232 to M. McFall-Ngai and E.G.R., by a China Scholarship Council’s State Scholarship Fund Award to X.C., and by fellowships 5F32GM084620 and 1K99GM097032 from the NIGMS to T.M.
Footnotes
Citation Cao X, et al. 2012. The novel sigma factor-like regulator RpoQ controls luminescence, chitinase activity, and motility in Vibrio fischeri. mBio 3(1):e00285-11. doi:10.1128/mBio.00285-11.
REFERENCES
- 1. Lee KH, Ruby EG. 1994. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60:1565–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Milton DL. 2006. Quorum sensing in Vibrios: complexity for diversification. Int. J. Med. Microbiol. 296:61–71 [DOI] [PubMed] [Google Scholar]
- 3. Kazmierczak MJ, Wiedmann M, Boor KJ. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bose JL, Rosenberg CS, Stabb EV. 2008. Effects of LuxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch. Microbiol. 190:169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lupp C, Ruby EG. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 186:3873–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lupp C, Ruby EG. 2005. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187:3620–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lupp C, Urbanowski M, Greenberg EP, Ruby EG. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319–331 [DOI] [PubMed] [Google Scholar]
- 8. Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meighen EA. 1994. Genetics of bacterial bioluminescence. Annu. Rev. Genet. 28:117–139 [DOI] [PubMed] [Google Scholar]
- 10. Sitnikov DM, Schineller JB, Baldwin TO. 1995. Transcriptional regulation of bioluminescence genes from Vibrio fischeri. Mol. Microbiol. 17:801–812 [DOI] [PubMed] [Google Scholar]
- 11. Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. 2010. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol. Microbiol. 77:1556–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131–143 [DOI] [PubMed] [Google Scholar]
- 13. Studer SV, Mandel MJ, Ruby EG. 2008. AinS quorum sensing regulates the Vibrio fischeri acetate switch. J. Bacteriol. 190:5915–5923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Osterberg S, Del Peso-Santos T, Shingler V. 2011. Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 65:37–55 [DOI] [PubMed] [Google Scholar]
- 15. Lin YH, Miyamoto C, Meighen EA. 2002. Cloning, sequencing, and functional studies of the rpoS gene from Vibrio harveyi. Biochem. Biophys. Res. Commun. 293:456–462 [DOI] [PubMed] [Google Scholar]
- 16. Merrikh H, Ferrazzoli AE, Bougdour A, Olivier-Mason A, Lovett ST. 2009. A DNA damage response in Escherichia coli involving the alternative sigma factor, RpoS. Proc. Natl. Acad. Sci. U. S. A. 106:611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tian Y, et al. 2008. Role of RpoS in stress survival, synthesis of extracellular autoinducer 2, and virulence in Vibrio alginolyticus. Arch. Microbiol. 190:585–594 [DOI] [PubMed] [Google Scholar]
- 18. Dong T, Schellhorn HE. 2010. Role of RpoS in virulence of pathogens. Infect. Immun. 78:887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nielsen AT, et al. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS. Pathog. 2:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schuster M, Hawkins AC, Harwood CS, Greenberg EP. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973–985 [DOI] [PubMed] [Google Scholar]
- 21. Weber B, Croxatto A, Chen C, Milton DL. 2008. RpoS induces expression of the Vibrio anguillarum quorum-sensing regulator VanT. Microbiology 154:767–780 [DOI] [PubMed] [Google Scholar]
- 22. Whiteley M, Parsek MR, Greenberg EP. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Visick KL, Ruby EG. 1998. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and approach to stationary phase. J. Bacteriol. 180:2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klose KE, Mekalanos JJ. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501–520 [DOI] [PubMed] [Google Scholar]
- 25. Lilley BN, Bassler BL. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940–954 [DOI] [PubMed] [Google Scholar]
- 26. Reitzer L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57:155–176 [DOI] [PubMed] [Google Scholar]
- 27. Stewart BJ, McCarter LL. 2003. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185:4508–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolfe AJ, Millikan DS, Campbell JM, Visick KL. 2004. Vibrio fischeri sigma54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 70:2520–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao JJ, Chen C, Zhang LP, Hu CQ. 2009. Cloning, identification, and characterization of the rpoS-like sigma factor rpoX from Vibrio alginolyticus. J. Biomed. Biotechnol. 2009:126986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mandel MJ, Stabb EV, Ruby EG. 2008. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics 9:138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paget MS, Helmann JD. 2003. The sigma70 family of sigma factors. Genome Biol. 4:203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Amann E, Ochs B, Abel KJ. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315 [DOI] [PubMed] [Google Scholar]
- 33. Beare PA, For RJ, Martin LW, Lamont IL. 2003. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 47:195–207 [DOI] [PubMed] [Google Scholar]
- 34. Koster M, van Klompenburg W, Bitter W, Leong J, Weisbeek P. 1994. Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J. 13:2805–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Llamas MA, et al. 2008. Characterization of five novel Pseudomonas aeruginosa cell-surface signalling systems. Mol. Microbiol. 67:458–472 [DOI] [PubMed] [Google Scholar]
- 36. Llamas MA, et al. 2006. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J. Bacteriol. 188:1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Llamas MA, et al. 2009. A novel extracytoplasmic function (ECF) sigma factor regulates virulence in Pseudomonas aeruginosa. PLoS Pathog. 5:e1000572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bose JL, et al. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65:538–553 [DOI] [PubMed] [Google Scholar]
- 39. Chernin LS, et al. 1998. Chitinolytic activity in Chromobacterium violaceum: substrate analysis and regulation by quorum sensing. J. Bacteriol. 180:4435–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Defoirdt T, Ruwandeepika HAD, Karunasagar I, Boon N, Bossier P. 2010. Quorum sensing negatively regulates chitinase in Vibrio harveyi. Environ. Microbiol. Rep. 2:44–49 [DOI] [PubMed] [Google Scholar]
- 41. Hunt DE, Gevers D, Vahora NM, Polz MF. 2008. Conservation of the chitin-utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol. 74:44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Urbanczyk H, Ast JC, Higgins MJ, Carson J, Dunlap PV. 2007. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb.nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int. J. Syst. Evol. Microbiol. 57:2823–2829 [DOI] [PubMed] [Google Scholar]
- 43. Ruby EG, et al. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. U. S. A. 102:3004–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bavishi A, Abhishek A, Lin L, Choudhary M. 2010. Complex prokaryotic genome structure: rapid evolution of chromosome. Genome 53:675–687 [DOI] [PubMed] [Google Scholar]
- 45. Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. 2010. Why genes evolve faster on secondary chromosomes in bacteria. PLoS Comput. Biol. 6:e1000732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497–515 [DOI] [PubMed] [Google Scholar]
- 47. Joelsson A, Kan B, Zhu J. 2007. Quorum sensing enhances the stress response in Vibrio cholerae. Appl. Environ. Microbiol. 73:3742–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hjerde E, et al. 2008. The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics 9:616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stevens AM, Dolan KM, Greenberg EP. 1994. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. U. S. A. 91:12619–12623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stevens AM, Greenberg EP. 1997. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J. Bacteriol. 179:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Devine JH, Shadel GS, Baldwin TO. 1989. Identification of the operator of the lux regulon from Vibrio fischeri ATCC 7744. Proc. Natl. Acad. Sci. U. S. A. 86:5688–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lyell NL, Dunn AK, Bose JL, Stabb EV. 2010. Bright mutants of Vibrio fischeri ES114 reveal conditions and regulators that control bioluminescence and expression of the lux operon. J. Bacteriol. 74:7059–7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Daniels R, Vanderleyden J, Michiels J. 2004. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 28:261–289 [DOI] [PubMed] [Google Scholar]
- 54. Ruby EG, Asato LM. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159:160–167 [DOI] [PubMed] [Google Scholar]
- 55. Wier AM, et al. 2010. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc. Natl. Acad. Sci. U. S. A. 107:2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Heath-Heckman EA, McFall-Ngai MJ. 2011. The occurrence of chitin in the hemocytes of invertebrates. Zoology 114:191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 61. Felsenstein J. 1989. PHYLIP-phylogeny inference package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 62. Stabb EV, Ruby EG. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413–426 [DOI] [PubMed] [Google Scholar]
- 63. Miyashiro T, et al. 2011. The N-acetyl-d-glucosamine repressor NagC of Vibrio fischeri facilitates colonization of Euprymna scolopes. Mol. Microbiol. 82:894–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McCann J, Stabb EV, Millikan DS, Ruby EG. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 69:5928–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bao Y, Lies DP, Fu H, Roberts GP. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167–168 [DOI] [PubMed] [Google Scholar]
- 66. Hussa EA, O’Shea TM, Darnell CL, Ruby EG, Visick KL. 2007. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J. Bacteriol. 189:5825–5838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qRT-PCR analysis of VF_A1016 expression in V. fischeri ΔluxO Tn7::erm, ΔluxO ΔrpoQ Tn7::erm, and ΔluxO ΔrpoQ Tn7::rpoQ strains. The strains were grown in LBS medium and harvested at an OD600 of 0.7 to 0.8. The transcript levels of rpoD were used to normalize VF_A1016 levels. Graphical and error bars indicate, respectively, the averages and standard deviations of data from three independent experiments. Shared letters above the bars indicate no statistically significant difference (P > 0.05), whereas different letters indicate a significant difference (P < 0.05) in VF_A1016 transcript levels between those strains (ANOVA and Tukey’s HSD test). Download Figure S1, TIF file, 0.1 MB.
Culture luminescence per CFU of (i) wild-type MJM1100 harboring either pTM214 (vector) or the inducible VF_A1016 allele (Ptrc-VF_A1016) and (ii) ΔlitR and ΔlitR ΔrpoQ strains harboring either pTM214 (vector) or inducible litR (Ptrc-litR). The cultures were grown either with (black bars) or without (white bars) IPTG addition. Graphical and error bars indicate, respectively, the averages and standard deviations of data from three independent experiments. Download Figure S2, TIF file, 0.1 MB.
The motility of (i) wild-type V. fischeri cells harboring either pTM214 (WT vector) or the inducible VF_A1016 allele (WT Ptrc-VF_A1016) and (ii) ΔlitR and ΔlitR ΔrpoQ cells harboring either pTM214 (vector) or the inducible litR allele (Ptrc-litR). Relative rates of motility were determined in minimal medium containing 1 mM IPTG and solidified with 0.25% agar. One representative experiment of three is shown. Growth rates of the strains were comparable in the minimal medium (data not shown). Download Figure S3, TIF file, 0.4 MB.
The exochitinase activity of (i) wild-type V. fischeri cells harboring either pTM214 (WT vector) or the inducible VF_A1016 allele (WT Ptrc-VF_A1016), and (ii) ΔlitR and ΔlitR ΔrpoQ cells harboring either pTM214 (vector) or the inducible litR allele (Ptrc-litR). Secreted chitinase activity was determined in the cell-free supernatant. Graphical and error bars indicate, respectively, the averages and standard deviations of data from three independent experiments. Shared letters above the bars indicate no statistically significant difference (P > 0.05), whereas different letters indicate a significant difference (P < 0.05) in exochitinase activity between those strains (ANOVA and Tukey’s HSD test). Download Figure S4, TIF file, 0.1 MB.