Abstract
The endosomal sorting complexes required for transport (ESCRT) guides transmembrane proteins to domains that bud away from the cytoplasm. The ESCRT machinery consists of four complexes. ESCRT complexes 0–II are important for cargo recognition and concentration via ubiquitin binding. Most of the membrane bending function is mediated by the large multimeric ESCRT-III complex and associated proteins. Here we present the first in vivo proteome analysis of a member of the ESCRT-III complex which is unique to the plant kingdom. We show with LC–MS/MS, yeast-two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) that coimmunoprecipitated proteins from Arabidopsisthaliana roots expressing a functional GFP-tagged VACUOLAR PROTEIN SORTING 2.2 (AtVPS2.2) protein are members of the ESCRT-III complex and associated proteins. Therefore we propose that at least in plants the large ESCRT-III membrane scaffolding complex consists of a mixture of SNF7, VPS2 and the associated VPS46 and VPS60 proteins. Apart from transmembrane proteins, numerous membrane-associated but also nuclear and extracellular proteins have been identified, indicating that AtVPS2.2 might be involved in processes beyond the classical ESCRT role. This study is the first in vivo proteome analysis with a tagged ESCRT-III component demonstrating the feasibility of this approach and provides numerous starting points for the investigation of the biological process in which AtVPS2.2 is involved.
Keywords: ESCRT-III, AtVPS2.2, GFP-tagged, coimmunioprecipitation, LC−MS/MS, yeast two-hybrid, bimolecular fluorescence complementation, in vivo, dynamins
Short abstract
The endosomal sorting complexes required for transport (ESCRT) guides transmembrane proteins to domains that bud away from the cytoplasm. Most of the membrane bending function is mediated by the ESCRT-III complex. Proteomic analysis was used to identify novel ESCRT-III interactors of Arabidopsis thaliana seedlings expressing a functional GFP-tagged VACUOLAR PROTEIN SORTING2.2 as bait. Some intractors were confirmed by yeast-two-hybrid and bimolecular fluorescence complementation and others will be the target for future investigations.
Introduction
Eukaryotic cells comprise a highly dynamic vesicular trafficking system that selects and delivers proteins and lipids to different cellular locations. On the crossroad between endocytosis, secretion and transport to the vacuoles or lysosomes are sorting endosomes and multivesicular bodies (MVBs).1,2 Membrane proteins dedicated for either lysosomal/vacuolar degradation and functions or exosomal release are sequestered into MVB intraluminal vesicles (ILV).3,4 MVBs fuse with lysosomes/vacuoles to deliver ILVs and their contents. In the secretory or recycling pathway, MVBs fuse with the plasma membrane and release the ILVs into the extracellular milieu.5,6 In recent studies, MVBs are suggested to promote recycling of the RNA-induced silencing complex (RISC) and/or their competence in loading small RNAs.7,8
Both the biogenesis of MVBs and the sorting of cargoes into the ILVs depend on the evolutionary conserved endosomal sorting complex required for transport (ESCRT) system. The ESCRT machinery consists of at least four complexes (ESCRT-0 to -III) and associated proteins. ESCRT-0, -I, -II components are involved in cargo selection via binding to their ubiquitin modifications and recruit ESCRT-III. ESCRT-III mediates the biogenesis of MVBs by exerting membrane bending, scission and fusion. At the plasma membrane, ESCRT-III has topological similar functions such as the budding of enveloped viruses and the abscission of the plasma membrane during cytokinesis.9,10 Some of the newest addition to ESCRT-III functions is the control of autophagosome fusions in yeast, worm, fly and mammalian cells (reviewed in refs (9, 11, 12)), of vacuolar integrity13 and of peroxisomal invaginations.14 ESCRT-III together with its associated proteins are the most ancient and conserved components of the ESCRT-complexes. In Archaea, which have no endomembrane system, ESCRT-III components polymerize between segregating nucleotides during cell division15 and have also been found in secreted vesicles.16
Most components of the ESCRT machinery have been identified in genetic screens affecting vacuolar protein sorting and therefore are named VPS (VACUOLAR PROTEIN SORTING) in yeast. The human orthologs are termed after their physicochemical properties and localization as CHARGED MULTIVESICULAR BODY PROTEINS or CHROMATIN MODIFYING PROTEIN (CHMPs).17 The four core components of the ESCRT-III complex, VPS20/CHMP6, SNF7 (SUCROSE NON FERMENTING7)/CHMP4, VPS2/CHMP2, VPS24/CHMP3 and the associated proteins VPS46/CHMP1, VPS60/CHMP5 are small soluble, highly charged proteins that share a common tertiary structure consisting of six α-helices.18−20 Despite this structural similarity, the proteins are not functionally redundant and operate in a defined order.21−23 While the initiation of ESCRT-III spirals on membranes needs at least two copies of VPS20/CHMP6, the heteropolymeric spiral consists mostly of SNF7/CHMP4 molecules and is capped by few molecules of VPS2/CHMP2 and VPS24/CHMP3.24 The C-terminal fragment of all ESCRT-III components is able to activate the AAA ATPase, VPS4/SUPPRESSOR OF K+ TRANSPORT GROWTH DEFECT (SKD1) which disassembles the heteropolymers.25,26 The assembly of the ESCRT-III spiral on the cytoplasmic side of membranes and on deformed, tubular and dome-like membranes is congruent with ESCRT-III’s role in membrane budding away from the cytoplasm. These budding events are important during the biogenesis of interluminal vesicles of MVBs, and at the plasma membrane for the release of enveloped viruses, exosomes in animal cells and membrane vesicles in the Archaea.
We have shown that all of the ESCRT-III components and their associated proteins are present in plants.27 Recent genetic studies reported that mutants of AtVPS4/AtSKD1 and AtCHMP2A/AtVPS2.1 are lethal28 and a dominant negative version interferes with vacuolar protein trafficking and maintenance once expressed in Arabidopsis thaliana trichomes.13 Furthermore double mutants of ESCRT-III associated AtCHMP1B/AtVPS46.1 are severely compromised in embryo development.29
Only few ESCRT-III interactors have been identified to date. Apart from the recent finding in Arabidopsis that the deubiquitinating enzyme AMSH3 interacts specifically with the classical AtVPS2.1 protein30 all other studies have been carried out in yeast or human cells.31 Therefore we took advantage of the functional GFP-tagged AtVPS2.2 protein as bait to immunoprecipitate and identify in vivo interactors with mass-spectrometry. Among the 89 proteins recognized by this proteomic approach, 35 were considered as specific interactors of AtVPS2.2. Consistent with AtVPS2.2 as putative component of the ESCRT-III complex AtSNF7.1 and AtVPS2.1 were found in the pulled down fraction. However novel interactions with ESCRT-III associated proteins AtCHMP1A/AtVPS46.1, AtCHMP1B/AtVPS46.2 and AtVPS60.1 were identified and confirmed by yeast two hybrid (Y2H) and bifluorescence complementation (BiFC) analyses.
Comparison of AtVPS2.2 interactors with 45 published proteomic analyses of Arabidopsis thaliana and related species such as Brassica napus and Brassica oleracea revealed that approximately one-third has also been identified in vacuolar and extracellular protein extracts. The largest overlap with the published proteomic studies is for 20 proteins that have been isolated in membrane fractions. About half of them are annotated as plasma membrane associated proteins and include the water channel protein AtPIP1B, the H+ ATPase 1 AtAHA1, six dynamin-related proteins and phospholipase D alpha 1 (PLDα1). Similar to ESCRT-III components, dynamins deform membranes in such a way that fusion and fission can occur.32−36 However, the classical mechanochemical property and topology of dynamins is the opposite supporting budding into the cytoplasm.37 We propose a model integrating this conjectural difference of ESCRT-III and dynamins.
The conclusion that the role of AtVPS2.2 may lie beyond the membrane deformation and fission function is supported by the large fraction of proteins annotated in and isolated from nuclei and the localization of AtVPS2.2-GFP in the nucleus. Interestingly early findings in mammalian cells report the association of CHMP1 with the transcriptional repressor polycomb-like protein (Pcl) on condensed chromatin.17
This study is the first proteomic analysis with an ESCRT-III component pointing to its role in the nucleus and as component of subdomains of membranes enriched with dynamins and PLDα1.
Material and Methods
Plant Material, Growth Conditions and Transformation
Wild-type accession Columbia (Col) was obtained from the Arabidopsis Stock Centers (Arabidopsis Biological Resource Center, ABRC, Columbus, USA and European Arabidopsis Stock Center, NASC, Nottingham, UK). Provenance of the vps2–3/hya-3 was described in.38 Plant growth conditions on sterile nutrient agar medium were described previously.39 Plant transformation was done according to the floral dip protocol of Clough and Bent into wild-type (Col) and mutant vps2.2–3 background.40 The MM2d cell suspension culture was obtained from Aventis CropScience and maintained after the protocol of Menges and Murray.41 For protoplast transformation, we principally followed the protocol of Sheen42 with 50 mL of a five day old MM2D culture.
Cloning of AtVPS2.2-GFP and Functional Complementation
Genomic DNA was isolated from seedlings by a modified Cetyltrimethylammoniumbromid (CTAB) method.43 A fragment of 3.3 kb from the At5g44560 gene including 2141 bp coding, 644 bp promoter, 124 bp 5′, 175 bp 3′ untranslated and 214 bp 3′ noncoding sequences, was amplified from genomic DNA with primers 5g44560–5Bam and 5g44560–3Bam and cloned into the BamHI site of the pPZP211 plasmid. The final construct (pPZP-AtVPS2.2) was verified by sequencing and transformed into Agrobacterium tumefaciens (GV3101) by electroporation. For cloning of AtVPS2.2-GFP, a 2807 bp subfragment of pPZP211-AtVPS2.2 was PCR amplified using primer 5g44560_GFP_SacI_pm and 5g44560_GFP_NcoI_R, and subcloned into pCR4-TOPO for sequence verification. Using SacI/NcoI the insert was transferred to the binary GFP-containing vector pGreenII0029–35S-GFP-RL replacing the 35S promoter. The final construct AtVPS2.2-GFP was transformed into Agrobacterium tumefaciens strain GV3101pSoup by electroporation and used to transform wild-type (WS, Col) and hya/vps2.2–2, hya/vps2.2–3 plants. Transformants were selected on MS medium containing kanamycin (100 μg mL–1). The functionality of transgenic lines was determined with root growth measurements on seven day qold seedlings.
Microscopic Analyses
Protoplasts from the BiFC assays, roots of Col;AtVPS2.2-GFP and the F1 of a cross between Col;AtVPS2.2-GFP and the trans-Golgi network (TGN) marker VHAa1-mRFP were observed under the stereomicroscope (DC500, Leica, Germany) and with the confocal laser scanning microscope (CLSM) (TCS-SP2 and SP5 II, Leica, Germany). Whole seedlings were mounted either in water or low melting agarose (LM; 1/2 MS, 1% sucrose) on slides with petroleum jelly borders to create a chamber. To visualize the nuclei 5 days old seedlings were fixed in 3.7% formaldehyde in 1× PBT (130 mM NaCl, 10 mM phosphate puffer pH 7.0, 0.1% Tween 20) for 3 h, washed extensively with 40 mL 1× PBT, stained with 1 μg mL–1 4′,6-diamidino-2-phenylindole (DAPI) for 3 min, washed again extensively with 40 mL tap water and mounted. Excitation wavelengths were 405 nm for DAPI, 488 nm (argon laser) for GFP/YFP and 561 nm for mRFP. Emissions were detected between 425 and 480 nm for DAPI, 500–535 nm for GFP, 518–548 nm for YFP and 570–651 nm for mRFP. Images were taken with a long distance 63× water-immersion objective. Pictures were prepared for publication with the Leica LAS AF Lite, ImageJ and Paint Shop Pro 5 software.
Plant Extraction and Western Blot
Roots and shoots of approximately 50 16-days old seedlings expressing AtVPS2.2-GFP and a control GFP-construct were harvested lysed with 150 μL cracking buffer (60 mM Tris pH 6.8, 1% 2-mercaptoethanol, 1% SDS; 10% glycerol, 0.01% bromophenol blue) and denatured for 10 min at 95 °C. Fifteen μL of these extracts were loaded on a 12% SDS-PAGE and blotted overnight at 4 °C in 1× TB (50 mM Tris/HCl pH 8.3, 50 mM boric acid) with continuous 30 V on a PVDV membrane (Roth, Germany). The membrane was blocked with 5% low fat milk powder in 1× TBST (20 mM Tris.HCl, pH 7.5 and 137 mM NaCl, 0.1% Tween 20) for 1 h at room temperature. The AtVPS2.2-GFP fusion protein was detected in 1× TBST with anti-GFP (Roche, 1:1000) as primary, antimouse IgG-HRP (horseradish peroxidase) as secondary antibody (Amersham, 1:10000) and by chemiluminescence using Roti-Lumin as substrate (Roth, Germany).
Immunoprecipitations
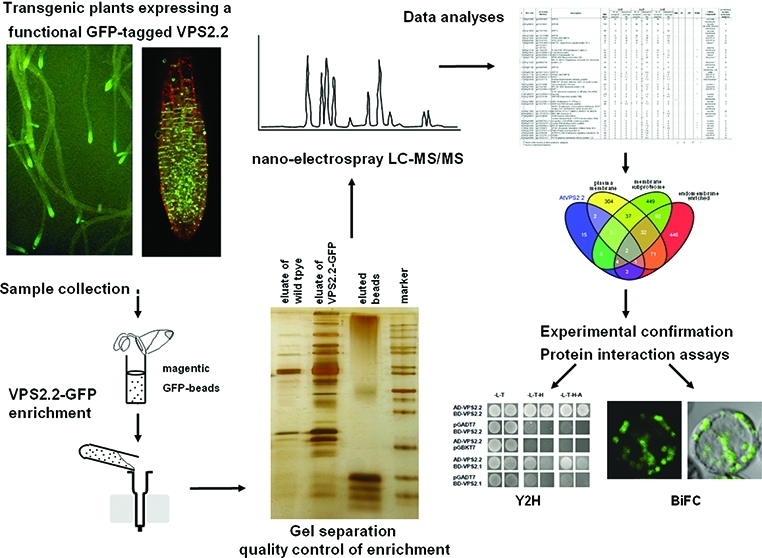
For the pull down assays the μMACS epitope tag protein isolation kit was used (Miltenyi Biotec, Bergisch Gladbach, Germany). Roughly 300 mg roots of 10–14 days old seedlings expressing AtVPS2.2-GFP, wildtype (Columbia) or 35S::GFP were harvested, lysed with 1.5 mL lysis buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris HCl pH 8.0) supplemented with 50 μL Pefablock (0.2 M, Roth, Germany) and incubated on ice for 30 min. After a second addition of 50 μL Pefablock (0.2 M) and 5 μL DTT (1 M) the solution was cleared from cell debris by centrifugation (10 min at 13.000 rpm at 4 °C). 50 μL anti-GFP μMACS MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) were added to the supernatant, mixed by rotating and incubated for 50 min on ice. Samples were loaded on μ Columns and washed five times with 200 μL wash buffer 1 and the last time with 100 μL wash buffer 2 (Miltenyi Biotec, Bergisch Gladbach, Germany). The proteins were eluted with 55 μL elution buffer (Miltenyi Biotec, Bergisch Gladbach, Germany) and 15–20 μL were loaded on a 12% PAGE. Proteins were fixed in 50% MeOH and 5% HAc for 20 min and after washing in distilled water overnight, visualized with a LC–MS/MS compatible silver staining protocol.44 The gel was sensitized 1 min in 1.2 mM Na2S2O3, washed two times 1 min in distilled water and stained for 20 min in 8.8 mM AgNO3 and 0.25% formaldehyde. After two 1 min washes with distilled water, the protein bands were developed with 188 mM Na2CO3 and 0.04% formaldehyd. Staining was stopped with 5% HAc and stored at 4 °C in 1% HAc.
Enzymatic Digest, LC–MS/MS and Data Analysis
Silver stained samples were used for the nanoelectrospray LC–MS/MS investigations. Selected gel bands were cut for the investigation of the first pull down; in case of the second and third pull downs, whole lanes of the “sample” and “control” were excised and cut into slices. The bands were washed several times with high quality water (Maxima, Elga, U.K.) to remove salts and detergents from the gel. Proteins were reduced by dithiothreitol (DTT - Roche, Germany) and alkylated by iodoacetamide (Sigma-Aldrich, Germany) without extraction. Trypsin (Roche Diagnostics, Germany - recombinant; proteomics grade) was used as protease, the digest was carried out overnight at 37 °C and was stopped by adding 10% formic acid (Merck, Germany) in water to a final concentration in the aliquot of approximately 1%. The HPLC used was an UltiMate system (Dionex Corporation, Sunnyvale, CA) equipped with a PepMap C18 purification column (300 μm × 5 mm) and a 75 μm x 150 mm analytical column of the same material. 0.1% TFA (Pierce Biotechnology Inc., Rockford, IL) was used on the Switchos module for the binding of the peptides and a linear gradient of acetonitrile (Chromasolv, Sigma-Aldrich, Germany), and 0.1% formic acid in water was used for the elution. The gradient was (mobile phase A: 5% acetonitrile/0.1% formic acid in water; mobile phase B: 80% acetonitrile/0.1% formic acid in water): 0% B for 8 min, 50% B in 60 min, 95% B in 1 min, 100% B for 5 min, 0% B in 1 min, 0% B for 20 min. LC–MS/MS analyses were carried out with an LTQ (Thermo Fisher Scientific Inc., Fremont, CA) linear ion trap mass spectrometer. The data acquisition software was XCalibur 2.0.7. (Thermo Fisher Scientfic Inc., Fremont, CA). The nanospray source of Proxeon (Odense, Denmark) was used with the distal coated silica capillaries of New Objective (Woburn, MA). The electrospray voltage was set to 1500 V. Peptide spectra were recorded over the range of m/z 450–1600, MS/MS spectra were recorded in information dependent data acquisition, the default charge state was set to 3; the mass range for MS/MS measurements was calculated according to the masses of the parent ions. One full spectrum was recorded followed by 4 MS/MS spectra for the most intense ions, automatic gain control was applied and the collision energy was set to the arbitrary value of 35. Helium was used as collision gas. The instrument was operated in data dependent modus; fragmented ions were set onto an exclusion list for 20 s.
MS/MS spectra were interpreted by Mascot 2.2 (Matrix Science Ltd., London, U.K.) and Scaffold 1.0 or Scaffold 2.0 (Proteome Software Inc., Portland, OR). Peptide tolerance was set to ±2 Da, MS/MS tolerance was set to ±0.8 Da. Carbamidomethylcysteine was set as static, oxidation of methionine residues as variable modification. Number of allowed missed cleavages was set to 2, only fully tryptic peptides were accepted. Raw files acquired from the same lane were merged into one file for the interpretation. Peptide identifications were accepted if they exceeded specific database search engine thresholds. Mascot identifications required at least ion scores greater than both the associated identity scores and 20. X! Tandem identifications required at least −Log (Expect Scores) scores of greater than 2.0. Protein identifications were accepted if they contained at least 2 identified peptides. The database used for Mascot search was the nr protein database of NIH (NCBI Resources, NIH, Bethesda, MD), taxonomy was Arabidopsis. The data of the first two pull down assays were analyzed with the nr database version of 16th Jan 2007, the MS/MS data of the third pull down was searched against the nr database version from 3rd Feb. 2008.
Yeast Two Hybrid (Y2H) Analyses
Total RNA of roots or cell suspension cultures were used for cDNA synthesis according to Karsai et al.45 cDNAs were amplified with primers (Supplemental Table S2, Supporting Information) suited for in frame cloning into the Y2H vectors pGADT7 and pGBKT7, inserted into pCR4-TOPO (Invitrogen, Carlsbad, CA) for sequencing and transferred to the two Y2H vectors. For interaction studies the yeast strain pJ69–4a was cotransformed with the AD and BD vectors by the PEG/LiCl heat shock method46 and selected on synthetic drop out medium (SD, Clontech, Mountain View, CA) lacking Leu and Trp at 29 °C. Protein–protein interactions were scored by spotting a liquid SD culture of the positive cotransformants on selective media lacking Trp, Leu and His (-L/-T/-H). The strength of interactions was determined by replating the cotransformants on SD/-T/-L/-H medium with increasing concentrations of the histidine analog 3-amino-1,2,4-triazol (3, 8, 15, and 25 mM At, SIGMA-Aldrich, Germany).
Construction of BiFC Fusion Proteins
Full length cDNAs of SNF7.1 (At4g29160), VPS2.1 (At2g06530), VPS2.2 (At5g44560), VPS46.1 (At1g17730) and VPS60.1 (At3g10640) were PCR amplified with primers suitable for the BD In-Fusion Universal PCR Cloning System (BD BIOSIENCES, France) and recombined into the vectors pUC-SPYNE and pUC-SPYCE.47 All constructs were confirmed by sequencing. Empty vector combinations were cotransformed as negative controls. Interactions were scored from three independent transformations with the CLSM.
Classification of AtVPS2.2-GFP-interacting Proteins
The proteins were classified according to their molecular function as determined on the basis of the available literature and of the gene ontology database in TAIR (www.arabidopsis.org/tools/bulk/go/index.jsp). The percentage of proteins in each category was calculated by normalizing the number of proteins in each group to the total number of different proteins identified from the screen. Signal sites for the secretory pathway (SP), GPI-anchoring (GPI) (http://gpi.unibe.ch/ and http://mendel.imp.ac.at/sat/gpi/gpi_server.html), palmitoylation (http://csspalm.biocuckoo.org/online.php), myristoylation (N-Myr) (http://plantsp.genomics.purdue.edu/plantsp/html/myrist.html and http://mendel.imp.ac.at/myristate/SUPLpredictor.htm) and prenylation (http://mendel.imp.ac.at/sat/PrePS/index.html) were investigated. Moreover, searches were done for transmembrane domains (TMD) (http://www.plantenergy.uwa.edu.au/ and http://aramemnon.botanik.uni-koeln.de/index.ep). For comparison with other proteomic analyses the Plant Proteome Database (http://ppdb.tc.cornell.edu/) and the VENNY tool48 (http://bioinfogp.cnb.csic.es/tools/venny/index.html) were used.
Results and Discussion
The GFP-tagged AtVPS2.2 is Functional
To reveal if and how the ESCRT-III complex is cooperating with other proteins including potential MVB targets we used transgenic Arabidopsis seedlings expressing a C-terminally GFP-tagged AtVPS2.2 (AtVPS2.2-GFP) under the endogenous promoter as bait for immunoprecipitation studies. Even though it has been shown that a C-terminal fusion to the human SNF7/CHMP4A was able to be integrated into ESCRT-III filaments,49 an overexpressed hSNF7.1–GFP fusion inhibits MVB maturation and viral budding50 and the hSNF7.1-GFP filaments had a tighter appearance.49 While GFP dimerization was excluded to cause later effect the C-terminal fusion enhanced the interaction.51
Therefore the choice for the proteomic approach with an ESCRT-III component was based on the possibility to prove the functionality of the construct. While mutants of AtVPS2.2 exhibit a strong root growth phenotype38,52 mutants of AtVPS2.1 are lethal and the knock out of AtVPS2.3 has a week root growth defect.30 Moreover VPS2 is one of the most conserved ESCRT-III components with up to three copies in the genomes of Metazoa, Fungi, Amoebozoa, Viridiplantae, Phodophyta, Apicomplexa, Ciliates, Stramenopiles, Discicristata and Metamonada.53 VPS2 is the only ESCRT-III component which has been identified so far by a proteomic approach in plants.54
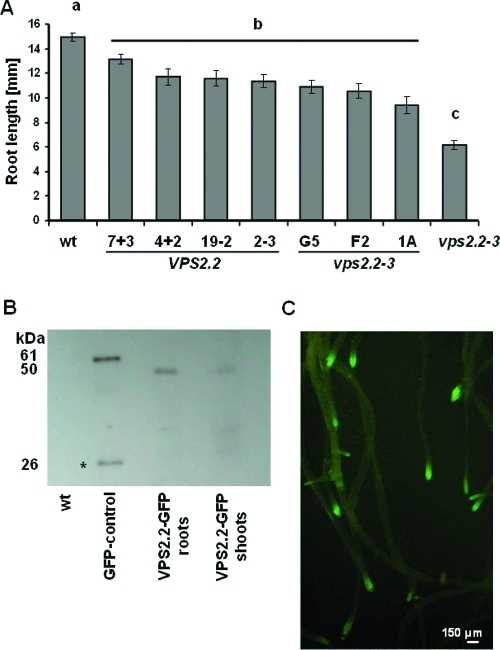
To evaluate the functionality of the AtVPS2.2-GFP construct we first investigated its ability to complement the vps2.2–3 root growth phenotype. Of seven independent transformants with vps2.2–3 background three restored root growth to the level of the AtVPS2.2-GFP expressing wildtype plants (Figure 1A). The expression level and stability of the fusion construct in transgenic seedlings was quantified by Western blots (Figure 1B). Finally, the microscopic analyses show that the AtVPS2.2-GFP is visible primarily in punctuated/vesicular structures of root meristems (Figure 1C, Figure 5B–D,F).
Figure 1.
AtVPS2.2-GFP complements vps2.2–3 phenotypes and is predominantly expressed in root meristems. (A) Wildtype and vps2.2–3 mutants expressing AtVPS2.2-GFP develop significantly longer roots than untransformed vps2.2–3, substantiating the functionality of the construct (pab < 4.4 × 10–5 for all comparison to vps2.2–3). Yet all transformants develop shorter roots than untransformed wildtype seedlings (pbc < 0.004). Root lengths were determined of at least 20 seedlings 7 days after germination on MS medium supplemented with 4.5% sucrose. The significance was calculated by students t-tests. (B) Stability and prevalent root expression of the AtVPS2.2-GFP fusion protein and the specificity of the GFP-antibody was determined by Western blot analyses loading 75 μg total proteins of wildtype root (wt), of seedlings expressing an unstable GFP-fusion protein (GFP-control), roots and shoot extracts of wildtype seedlings expressing AtVPS2.2-GFP. While in the unstable GFP-control extract a degradation product is detectable (star, 26 kD) the AtVPS2.2-GFP fusions (50 kD) are stable. (C) Roots of 14 days old seedlings expressing AtVPS2.2-GFP. AtVPS2.2-GFP is predominantly expressed in root meristems.
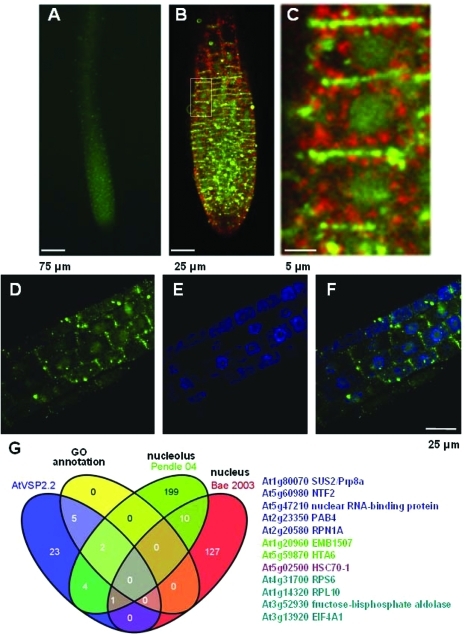
Figure 5.
AtVPS2.2-GFP localizes to distinct region of the plasma membrane, in cytoplasmic vesicles separated for the trans-Golgi network (TGN) marker VHAa1 and in nuclei of root meristems. (A) Root tips of seedlings expressing AtVPS2.2-GFP, (B) root meristems of seedlings expressing AtVPS2.2-GFP and the TNG marker VHAa1-mRFP, (C) close up of the marked area in B demonstrating that AtVPS2.2-GFP localizes strongly to the transverse cell borders, to vesicles of different sizes in the cytoplasm and diffusely in the nucleus. (D) Root area at the upper end of the meristem with diffuse nuclear AtVPS2.2-GFP localization. (E) Same root area as in D but labeled with the fluorescent DNA stain, DAPI. (F) Overlay of D and E. (G) Venn diagram of the overlap between AtVPS2.2-GFP interactors and proteins that have been isolated from and annotated to the nucleus. The colored AGI codes of the proteins correspond to the overlapping sectors. The first authors of the publications used for comparison are also color coded.
Immunopreciptation with AtVPS2.2-GFP Identified New Interactions of ESCRT-III Associated Proteins
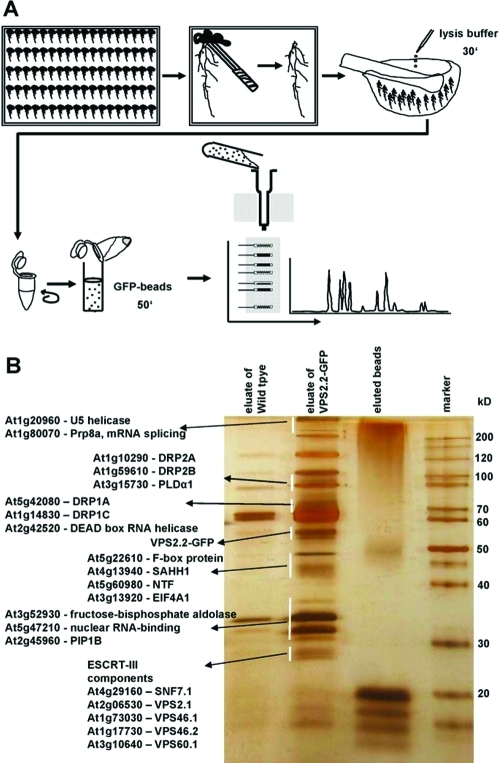
Based on the three criteria supporting the functionality of AtVPS2.2-GFP, root extracts of transgenic seedlings were used for three independent IPs followed by LC–MS/MS analysis (Figure 2). To increase the stringency of the analyses root extracts of wildtype and GFP overexpressing seedlings were included as negative controls. In total 89 proteins were isolated (Supplemental Table S1, Supporting Information). Interactors were defined as specific if they were (i), represented by at least two peptides, (ii) were not pulled down with the negative controls or (iii) the peptide coverage was more than 2-fold enriched in comparison to the negative controls. With these stringent criteria 35 interaction partners of AtVPS2.2 were sorted out (Table 1).
Figure 2.
Strategy to purify and identify AtVPS2.2-GFP interacting proteins. (A) Roots were separated from seedlings, cushed in liquid nitrogen, lysed with cracking buffer on ice and centrifuged to separate cell debris. Anti-GFP μMACS MicroBeads were added to the supernatant, mixed by rotating and after incubation on ice applied onto μ Columns. The eluted proteins were separated on a denaturing 12% (w/v) PAGE and visualized with a LC–MS/MS compatible silver staining protocol. Lanes were excised into up to 29 fractions and subjected to nanoelectrospray LC–MS/MS sequencing. (B) Example of a silver stained PAGE showing the eluates from extracts of control roots (wildtype), transgenes expressing AtVPS2.2-GFP and the eluted beads. Indicated are proteins which have been specifically enriched by this method.
Table 1. AtVPS2.2-GFP Interacting Proteins Identified with the Pull down Screena.
| first IP |
second IP |
third IP |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | AGI code | acession number | description | MW (kD) | nr. of assigned spectra | sequence coverage (%) | nr. of assigned sectra | sequence coverage (%) | nr. of assigned spectra | sequence coverage (%) | TMD | SP | GPI | N-Myr | cellular component | occurrence in other proteomic analyses |
| 1 | At5g42080 | gi|30693985 | DRP1A | 68 | 22 | 29 | 27 | 28 | 28 | 33 | – | – | – | ° − | cell plate | 7 |
| 2 | At1g59610 | gi|15218837 | DRP2B | 100 | 0 | 0 | 59 | 34 | 16 | 13 | – | – | – | – | plasma membrane | 5 |
| 3 | At1g14830 | gi|21537304 | DRP1C | 69 | 12 | 21 | 19 | 27 | 15 | 20 | – | – | – | – | cell plate, plasma membrane, vacuole | 4 |
| 4 | At1g10290 | gi|110737889 | DRP2A | 99 | 0 | 0 | 26 | 16 | 7 | 6.8 | – | – | – | – | plasma membrane, vacuole | 6 |
| 5 | At1g73030 | gi|18410249 | VPS46.2/AtCHMP1A | 23 | 14 | 30 | 11 | 26 | 3 | 13 | – | – | – | – | ESCRT-III | 0 |
| 6 | At5g44560 | gi|15241525 | VPS2.2/HYA | 24 | 5 | 19 | 16 | 31 | 6 | 20 | – | – | – | – | ESCRT-III | 0 |
| 7 | At5g02500 | gi|15241849 (+1) | HSC70–1 (heat shock cognate protein 70–1) | 71 | 0 | 0 | 9 | 11 | 10 | 12 | – | – | – | – | cytosol * | 22 |
| 8 | At3g15730 | gi|15232671, gi|1297302, gi|11994345 | PLDALPHA1 (Phospholipase D alpha 1) | 92 | 0 | 0 | 4 | 3.8 | 10 | 12 | – | – | – | ° − | plasma membrane | 8 |
| 9 | At5g22610 | gi|22326994 (+1) | F-box family protein | 54 | 0 | 0 | 0 | 0 | 11 | 2 | – | – | – | – | unknown | 0 |
| 10 | At4g05320 | gi|28436485 (+35) | UBQ10 (Polyubiquitin 10) | 28 | 0 | 0 | 0 | 0 | 8 | 14 | – | – | – | – | cytosol | 2 |
| 11 | At4g31700 | gi|15236042 | RPS6 (40S Ribosomal protein S6) | 28 | 0 | 0 | 4 | 20 | 3 | 11 | – | – | – | ° − | ribosome * | 9 |
| 12 | At1g14320 | gi|30683726 | RPL10, SAC52 (Suppressor of Acaulis 52), ribosomal protein L10 | 25 | 0 | 0 | 7 | 24 | 0 | 0 | – | – | – | – | ribosome * | 9 |
| 13 | At3g61760 | gi|30695480 | DRP1B | 68 | 0 | 0 | 6 | 7 | 0 | 0 | – | – | – | – | cell plate, plasma membrane, vacuole | 0 |
| 14 | At3g60190 | gi|18411520 (+1) | DRP1E | 70 | 0 | 0 | 4 | 6.4 | 2 | 3.5 | – | – | – | ° − | plasma membrane, cell plate | 7 |
| 15 | At1g17730 | gi|15220819 (+1) | VPS46.1/AtCHMP1B | 23 | 4 | 17 | 2 | 12 | 0 | 0 | – | – | – | – | ESCRT-III | 0 |
| 16 | At2g06530 | gi|18396352 (+11) | VPS2.1 | 25 | 3 | 7.1 | 0 | 0 | 2 | 7.1 | – | – | – | – | ESCRT-III | 1 |
| 17 | At3g52930 | gi|15231715 | fructose-bisphosphate aldolase, putative | 39 | 0 | 0 | 3 | 7.3 | 2 | 7.3 | – | – | – | ° − | mitochondrion * | 22 |
| 18 | At1g20960 | gi|15218086 | EMB1507 (Embryo defective 1507) U5 small nuclear ribonucleoprotein helicase | 247 | 0 | 0 | 4 | 2.3 | 0 | 0 | – | – | – | ° − | nucleus * | 2 |
| 19 | At5g27850 | gi|15241061 | RPL18C (60S ribosomal protein L18) | 21 | 0 | 0 | 4 | 12 | 0 | 0 | – | – | – | – | ribosome | 5 |
| 20 | At4g29160 | gi|15233464 (+2) | SNF7.1 | 24 | 0 | 0 | 0 | 0 | 4 | 16 | – | – | – | – | ESCRT-III | 0 |
| 21 | At1g80070 | gi|15220049 (+1) | SUS2 (abnormal suspensor 2), AtPrp8a, Pre-mRNA Splicing | 278 | 0 | 0 | 3 | 1.4 | 0 | 0 | – | – | – | ° − | nucleus | 2 |
| 22 | At1g16030 | gi|15219109 | HSP70B (Heat shock protein 70B) | 71 | 0 | 0 | 0 | 0 | 3 | 4.3 | – | – | – | – | cytosol | 7 |
| 23 | At2g18960 | gi|15224264 (+1) | AHA1 (Arabidopsis H+ ATPase 1) | 104 | 0 | 0 | 3 | 3 | 0 | 0 | ° + | – | – | – | plasma membrane, vacuole | 13 |
| 24 | At2g42520 | gi|15227951 (+1) | DEAD box RNA helicase, putative | 68 | 2 | 3.3 | 7 | 7.3 | 0 | 0 | – | – | – | – | peroxisome | 3 |
| 25 | At4g13940 | gi|15236376 (+6) | SAHH1 (S-adenosyl-l-homocysteine hydrolase), HOG1 HOMOLOGY-DEPENDENT GENE SILENCING 1, MATERNAL EFFECT EMBRYO ARREST 58, MEE58 | 53 | 0 | 0 | 2 | 2 | 0 | 0 | – | – | – | plasma membrane, vacuole | 20 | |
| 26 | At3g06810 | gi|110739702 (+2) | IBR3 (IBA-response 3) | 92 | 0 | 0 | 0 | 0 | 2 | 3.5 | – | – | – | + | peroxisome | 1 |
| 27 | At5g59870 | gi|15238549 | HTA6 (histone H2A protein) | 16 | 0 | 0 | 0 | 0 | 2 | 6 | – | – | – | – | nucleus * | 2 |
| 28 | At5g60980 | gi|30697452 (+1) | nuclear transport factor 2 (NTF2) family protein/RNA recognition motif (RRM)-containing protein | 49 | 0 | 0 | 2 | 5 | 0 | 0 | – | – | – | – | nucleus | 0 |
| 29 | At5g47210 | gi|145334757 (+2) | nuclear RNA-binding protein, putative | 32 | 0 | 0 | 2 | 9 | 0 | 0 | – | – | – | ° – | nucleus | 3 |
| 30 | At3g63460 | gi|30695804 (+2) | WD-40 repeat family protein | 120 | 0 | 0 | 2 | 1.9 | 0 | 0 | – | – | – | ° − | cell wall | 2 |
| 31 | At3g13920 | gi|21555870 | EIF4A1 (Eukaryotic translation initiation factor 4A1) | 47 | 0 | 0 | 0 | 0 | 2 | 5.8 | – | – | – | ° − | cytosol * | 14 |
| 32 | At2g23350 | gi|15227815 (+1) | PAB4 (Poly(A) binding protein 4) | 72 | 0 | 0 | 2 | 3.6 | 0 | 0 | – | – | – | – | nucleus | 5 |
| 33 | At2g20580 | gi|18399399 | ATRPN1A (26S Proteasome regulatory subunit S2 1A) | 98 | 0 | 0 | 2 | 2.8 | 0 | 0 | – | – | – | – | nucleus | 3 |
| 34 | At3g10640 | gi|12322781 (+1) | VPS60.1 | 25 | 0 | 0 | 2 | 5 | 0 | 0 | – | – | – | – | ESCRT-III | 0 |
| 35 | At2g45960 | gi|145331417 | PIP1B (plasma membrane intrinsic protein 1;2) | 32 | 0 | 0 | 2 | 6.3 | 0 | 0 | + | – | – | °– | plasma membrane | 8 |
| * found in the nucleus of other proteomic analyses | 2 | 0 | 0 | 1 | ||||||||||||
| ° found in membrane fractions | ||||||||||||||||
Proteins are sorted by abundance of identified peptides. Annotation to cellular compartments and the occurrence in other proteomic analyses are indicated. MW, molecular weight; IP, immunoprecipitation; TMD, transmembrane domain; SP, signal peptide; GPI, GPI anchor; N-Myr, myristoylation site.
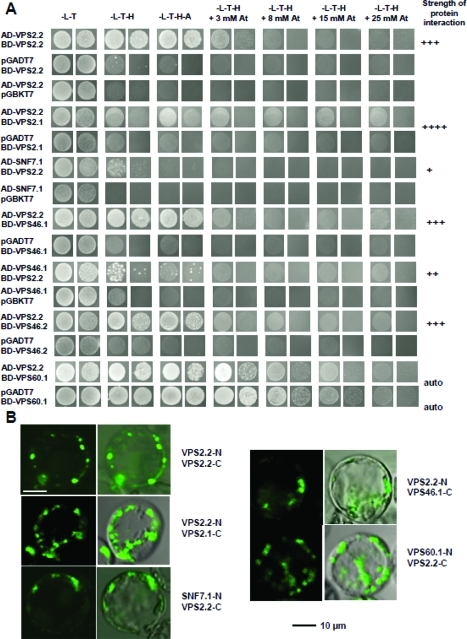
Not surprisingly two ESCRT-III components (AtVPS2.1, AtSNF7.1) have been successfully pulled down indicating that the C-terminal GFP-tag did not affect the interaction. Yet three ESCRT-III associated proteins AtVPS46.1, AtVPS46.2 and AtVPS60.1 were identified as interactors which have previously not been described to interact with AtVPS2.2 homologues in other systems. To support our findings of the biochemically identified interactions yeast to hybrid (Y2H) and bifluorescence complementation analyses (BiFC) in Arabidopsis protoplasts were carried out and supported some of these novel interactions (Figure 3, Supplemental Figure 1A, Supporting Information). The results indicate that the composition of ESCRT-III complexes might vary and/or diverse ESCRT-III complexes exist with different composition in plants. ESCRT-III components are encoded in the Arabidopsis genome by at least two and the VPS2 gene family by three genes.27 Mutant analyses show that AtVPS2.1 is embryo lethal while mutants of AtVPS2.2 and AtVPS2.3 have root growth defects.30 In contrast to AtVPS2.1, AtVPS2.2 and AtVPS2.3 did not show a Microtubule Interacting and Transport (MIT)-Interacting Motif (MIM)-dependent interaction with the deubiquitinating enzyme AMSH3. Furthermore in contrast to AtVPS2.1, AtVPS2.2 and AtVPS2.3 did not accumulate in class E compartments induced by overexpression of a dominant-negative AtVPS4/AtSKD1.30
Figure 3.
In vivo interaction studies of AtVPS2.2 with known and novel interacting partners of the ESCRT-III complex. (A) Yeast two hybrid analyses showing growth assays for homo- and heterodimerization and the autoactivation of the AtVPS60.1 DNA-binding domain fusion. The identity of the cotransformants is indicated on the left, the selective media above and the strength of the interactions on the right. Shown are spotted undiluted and 1:10 diluted cultures. (B) BiFC interactions in Arabidopsis protoplasts demonstrating homo- and heterodimerizations of AtVPS2.2 with AtVPS2.1, AtSNF7.1, AtVPS46.1 and AtVPS60.1. The interactions label vesicles in the cytoplasm and at the plasma membrane.
AtVPS2.2 Interactors Are Enriched in Membrane Fractions
To elucidate if AtVPS2.2 and its interactors are enriched in specific subcellular compartments, the identified proteins were categorized based on gene ontology annotations (Table 1). Also secondary structure predictions and post-translational modification motifs characteristic for subcellular localizations were used for the classification such as transmembrane domains, signal peptides and myristoylation signatures (Table 1). None of the AtVPS2.2-GFP interactors had signals for GPI-anchors, prenylation or palmitoylation. Only the IBA-response 3 protein contains a myristoylation motif and is annotated as peroxisomal protein.
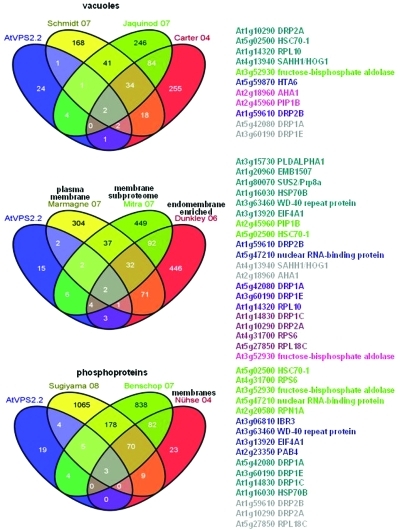
Although the classical function of the ESCRT-III complex in the biogenesis of MVBs suggests that AtVPS2.2-GFP interactors might be targeted to lytic compartments, only four proteins are annotated to vacuoles. Since these proteins are known to be membrane associated they probably locate at the tonoplast. However, 11 AtVPS2.2 interactors have also been identified in five published proteomic studies of isolated vacuoles (Figure 4).55−57 None of these are classical luminal vacuolar proteins.
Figure 4.
Subcellular localization of AtVPS2.2 interacting proteins. Venn diagrams of the overlap between AtVPS2.2-GFP interactors and (phosphor-) proteins that have been isolated from membranes and vacuoles. The colored AGI codes of the proteins correspond to the overlapping sectors. The first authors of the publications used for comparison are also color coded.
According to the gene ontology annotation eight interactors out of the 35 proteins (23%) are assigned to the plasma membrane/cell plate and/or tonoplast (Table 1). They include a member of the water channel protein family, AtPIP1B, and the H+ ATPase 1, AtAHA1. Comparison to published proteomic studies revealed that 20 (57%) interactors have been previously isolated in membrane fractions (Figure 4).58−60 Half of them were identified also in phosphoproteomes.61−63
Apart from the two integral membrane proteins which might be substrates of the ESCRT machinery, six dynamin-related proteins (DRP1A, DRP1B, DRP1C, DRP1E, DRP2A, DRP2B) and PLDα1 were pulled down (Table 1). PLDs interact and stimulate dynamin GTP-activity64 but interactions of ESCRT-III components with both PLDs and dynamins have not been shown yet.
In eukaryotic cells PLDs functions include but are not limited to the endocytic and exocytic pathways where they for example are involved in the generation and fusion of secretory vesicles.65−67 In mammalian cells PLDs could be found at sites of the plasma membrane where exocytosis occurs.68−70 PLDs facilitate the bending of membranes due to the cone-shaped geometry of their hydrolysis product phosphatidic acid (PA) which induces a negative curvature.71,72 Furthermore, the PLD products, phosphatidylserine and PAs, create an acidic environment at the cytoplasmic leaflet of the plasma membrane73,74 where the basic surface of ESCRT-III subunits and associated proteins can electrostatically bind. Whitley et al.75 identified a specific lysine (K49) on human CHMP3/VPS24 that binds phosphatidylinositol l (3,5)-bisphosphate (PtdIns(3,5)P2) and which is highly conserved in all SNF7-domain containing ESCRT-III subunits and associated proteins.27 Thus it is likely that PLD modified membranes are attracting ESCRT-III subunits.
AtPLDα1 is a plant-specific PLD with a polybasic PtdIns(4,5)P2-binding motif, a G-protein interacting motif and a C2 domain which mediates Ca2+-dependent phospholipid-association. AtPLDα1 is present on intercellular membranes and translocates between membranes and the cytosol in response to stress.76 AtPLDα1 binds in the same manner as dynamin to PtdIns(4,5)-bisphosphate and it is likely that both are recruited to the same membrane domains.
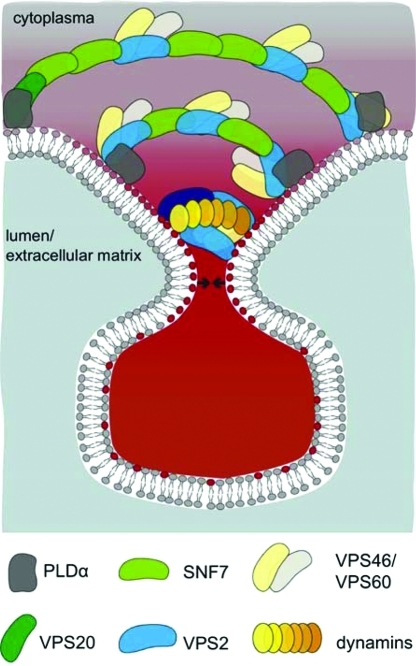
Dynamin oligmers have been visualized with electron microscopy on necks of endocytotic vesicles. The classical view of dynamin mediated membrane fission is that upon narrowing and stretching dynamin oligomeres membranes loose their stability and abscission occurs. Recently it has been shown that long dynamin oligomers are not necessary for membrane fission and it is postulated that limited dynamin assembly rearrange local lipids in such a way that nonleaky fission events can occur.77 Therefore it is possible that dynamins support ESCRT-III mediated membrane narrowing and lipid rearrangements at the neck (Figure 7). We cannot exclude that the interaction of an ESCRT-III subunit with dynamins is only restricted to the plant specific member of the VPS2 protein family.27
Figure 7.
Model how ESCRT-III might mediate vesicle budding and fission in conjunction with dynamins and PLDα. Several dynamins assemble and interact with the ESCRT-III subunit AtVPS2.2 on membrane domains on the inner side of the neck which is acidified by PLDα. This association supports membrane narrowing and lipid rearrangements at the neck of budding vesicles away from the cytoplasm so that membrane fission and fusion occurs. Since AtVPS2.2-GFP interacts also with other ESCRT-III components we propose that at least in plants the large membrane scaffolding complex consists of a mixture of AtSNF7, AtVPS2 and the associated AtVPS46 and AtVPS60 proteins. Dark green, AtVPS20; middle green, AtSNF7; blue, AtVPS2; dark blue, AtVPS24; light yellow, AtVPS46; light gray, AtVPS60; dark gray, PLDα; yellow to orange, dynamins; gray, neutral lipids; red, acidified lipids. Arrows indicate the forces that upon constriction of ESCRT-III and dynamins mediate membrane fission and fusion.
AtVPS2.2-GFP Localizes in the Nucleus and Crosstalk Might Exist with Regulators of Gene Expression and RNA Maturation and Processing
An unexpected high fraction of seven AtVPS2.2 interactors are annotated to the nucleus and five more have been isolated in proteome studies of isolated nuclei (Figure 5).78,79 Although AtVPS2.2-GFP localizes primarily to vesicles of different sizes and at distinct domains of the plasma membrane (Figure 5B–D,F, Supplemental Figure S1B, Supporting Information), weak GFP signals are also detected in nuclei of root meristems (Figure 5C,D,F). We found AtVPS2.2 specific interactions with nuclear proteins such as histone H2A and five RNA binding proteins including ATP-dependent helicases (EMB1507, EIFA1), a poly (A) binding protein (PABP), a spliceosome subunit (SUS2, abnormal suspensor 2), the nuclear transport factor 2 (NTF2) and a putative nuclear RNA-binding protein. These interactions and the localization of AtVPS2.2 raise the possibility that AtVPS2.2 might function in gene regulation as well.
This possibility is further substantiated by earlier findings with the human ESCRT-III associated protein CHMP1/VPS46. CHMP1 has been localized in the nucleus and interacts with the transcriptional repressor polycomb-like protein (Pcl) on condensed chromatin suggesting its role in gene silencing.17 Furthermore the human homologue of VPS2, CHMP2, is also known as breast adenocarcinoma 2 (BC-2) marker due to its strong upregulation. BC-2 has been identified in the nuclear matrix to both diffuse and discrete speckles.80 Overexpression of CHMP2/BC-2 induces condensed chromatin regions where a PcG complex protein, a predominantely plasma membrane localized small Ras family GTPase, Rac1, and the phosphorylated histone 3 are recruited.81 Based on these findings and similar to CHMP1, AtVPS2.2 probably participates also in nuclear events as well.
AtVPS2.2-GFP Interacts with Extracellular Proteins
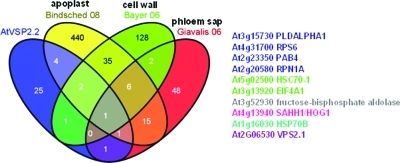
Ten of the AtVPS2.2-GFP interactors have also been isolated in proteome studies aimed to enrich for extracellular proteins of the cell wall, the apoplast and the phloem sap (Figure 6).54,82,83 None of these extracellular AtVPS2.2-GFP interactors belong to classical secretory proteins. However ESCRT proteins have been recently identified in the proteome of secreted vesicles of archaeal Sulfolobus species.16 One explanation is that the mechanism of budding of vesicles in archeae is similar to the release of exosomes and viruses. In both events the ESCRT machinery is involved.84 Exosomes are well studied in mammals and function in cell–cell communication.85 Even mRNA and miRNA (referred to as exosomal shuttle RNA or esRNA) are transported via exosomes between cells.86,87 The exocarta database (http://exocarta.ludwig.edu.au,88) which includes 75 studies on exosome proteomes revealed that apart from the classical exosomal markers related to the ESCRT machinery, TSG101/VPS23 and ALIX also the ESCRT-I components VPS23; VPS28; VPS37A, B, C, D; the ESCRT-II components VPS22, VPS25, VPS36 and the ESCRT-III core and associated proteins CHMP6/VPS20, CHMP2A and B, CHMP3/VPS24, CHMP5/VPS60 and CHMP1A and B and VPS4A and B and VTA1 have been isolated from exosomes. Interestingly all of the other noncanonically secreted proteins of our proteome analyses such as PLDα1, a proteasome subunit (RNP1A/PSMD2), a fructose-biphosphate aldolase, an S-adenosyl-l-homocysteine hydrolase (SAHH1/HOG1), heat shock proteins (HSC70–1, HSP70B) and RNA binding proteins (eIF4A1, RPS6) are also present in the exocarta database.
Figure 6.
Atypical subcellular localization of AtVPS2.2 interacting proteins. Venn diagrams of the overlap between AtVPS2.2-GFP interactors and proteins that have been isolated from and annotated to the apoplast, cell wall and phloem sap. The colored AGI codes of the proteins correspond to the overlapping sectors. The first authors of the publications used for comparison are also color coded.
In plants secreted ESCRT-III components and interactors have been identified in the apoplastic fluid89 and the phloem sap.54 On the microscopic level paramural vesicles share some characteristics with MVBs and exosomes.90 Such paramural vesicles have recently been detected at fungal penetration sites and during plugging of plasmodesmata between hypersensitive cells and intact neighboring cells.91,91 However, further studies are needed for a conclusive demonstration of the involvement of an ESCRT-mediated secretion in plants.
Working Model for the Involvement of AtVPS2.2 Interactors in Membrane Bending and Scission
Vesicle-mediated transport in the secretory and endocytic pathways involves the assembly of coat protein complexes on the cytoplasmic face of a lipid bilayer.92 The ESCRT machinery might function similarly by recruiting and concentrating membrane proteins into microdomains of vesicle buds that are sequestered away from the cytoplasm. While ESCRT-I and II induces initial membrane buds, ESCRT-III subunits form large membrane scaffolding protein complexes of filamentous and spiral appearance which push membranes away from the cytoplasm creating tubes with dome-like caps at the neck.49,93 The order and ratio of ESCRT-III subunits is only loosely defined and starts with the myristoylated and membrane inserted VPS20 subunit. Continuous self-assembly of ESCRT-III subunits SNF7, VPS24 and VPS2 inside the tube and neck stabilizes and narrows the membranes and ultimately leads to membrane scission.94
In this study, we have identified and established novel partners of the ESCRT-III component AtVPS2.2-GFP, in particular dynamins and PLDα1 which might assist in pinching off vesicular packages from membranes to deliver cargoes to other subcellular or even extracellular compartments.
Similar to ESCRT-subunits, dynamins assemble on membrane surfaces and are able to scaffold membranes into cylinders.77,95,96 It is known that dynamins bind the cytoplasmic side of the neck upon endocytosis. Endocytosis is a budding process into the cytoplasm and reverse to the process mediated by the ESCRT-III machinery which buds away from the cytoplasm. Nevertheless, we find that dynamins are interacting with the ESCRT-III subunit VPS2.2. Therefore we propose that ESCRT-III and dynamins assemble together on membrane domains that have been modified by AtPLDα1 thereby supporting membrane narrowing and lipid rearrangements at the neck of budding vesicles away from the cytoplasm. It is possible that dynamins bind to the inner side of the ESCRT machinery induced neck which is acidified by PLDα and therefore serves as landing platform for the basic surface of ESCRT-III subunits. This membrane association might support membrane narrowing and lipid rearrangements at the neck of budding vesicles away from the cytoplasm so that membrane fission and fusion occurs. Since AtVPS2.2-GFP interacts also with other ESCRT-III components we propose that at least in plants the large membrane scaffolding complex consists of a mixture of SNF7, VPS2 and the associated VPS46 and VPS60 proteins.
Conclusions
The present study is the first in vivo proteome approach to identify ESCRT-III interactors. The analysis of the immunoprecipitated proteins from root extracts of Arabidopsis plants expressing a functional AtVPS2.2-GFP fusion and the additional protein–protein interaction studies are consistent with a role for AtVPS2.2 in ESCRT-III complexes. Both microscopic studies and interactors annotated to and identified in the nucleus and the extracellular spaces indicate that AtVPS2.2 might be involved in processes beyond the classical roles of the ESCRT machinery. However, further experiments are necessary to confirm this hypothesis. The overrepresentation of dynamins and the copurification of a PLDα is an intriguing starting point for the characterization of proteins that may associate with the ESCRT-III machinery on membranes. These results provide the basis for future studies on the mechanism and functional significance of the interaction between the ESCRT-III subunit AtVPS2.2 and its identified partners in plants.
Acknowledgments
We are grateful to Klaus Harter for the BiFC vectors and the trans-Golgi network (TGN) marker VHAa1-mRFP line, Wilfried Rozhon and Claudia Jonak for the pGreenII029-35S-GFP-RL vector, Margit Menges, James A.H. Murray and Bayer CropScience for the MM2d suspension culture, Gerhard Adam for the Y2H vectors and yeast strain, and Juan Antonio Torres-Acosta for help with the establishment of the Y2H system in our laboratory. We thank Sonja Frosch for technical help in sample preparation for mass spectrometry measurements. We are thankful to Cornelia Konlechner and Helga Kudler for critically reading the manuscript. This work was supported by grants from the FWF (P16410–B12, P17888–B14) to M.-T.H.
Supporting Information Available
Supplemental Figure S1, Single scans through protoplasts transformed with either empty BiFC vectors as negative controls or with AtVPS2.2-GFP and stained with the membrane selective dye FM4–64.Supplemental Table S1, AtVPS2.2-GFP interacting proteins identified in three independent pull down screens. Proteins are sorted by abundance of identified peptides. Indicated are the AGI and accession numbers, molecular weight, number of unique peptides, number of assigned peptides and sequence coverage.Supplemental Table S2, Overview of the primer pairs used for cloning of the AtVPS2.2-GFP construct and the Y2H and BiFC interaction analyses. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
§ Department of Botany, University of Wisconsin, 430 Lincoln Drive, Madison, Wisconsin 53706, United States
Supplementary Material
References
- Gruenberg J.; Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell. Biol. 2004, 5 (4), 317–23. [DOI] [PubMed] [Google Scholar]
- Russell M. R.; Nickerson D. P.; Odorizzi G. Molecular mechanisms of late endosome morphology, identity and sorting. Curr. Opin. Cell. Biol. 2006, 18 (4), 422–8. [DOI] [PubMed] [Google Scholar]
- Katzmann D. J.; Odorizzi G.; Emr S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 2002, 3 (12), 893–905. [DOI] [PubMed] [Google Scholar]
- Piper R. C.; Katzmann D. J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gassart A.; Geminard C.; Hoekstra D.; Vidal M. Exosome secretion: the art of reutilizing nonrecycled proteins?. Traffic 2004, 5 (11), 896–903. [DOI] [PubMed] [Google Scholar]
- Fevrier B.; Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16 (4), 415–21. [DOI] [PubMed] [Google Scholar]
- Lee Y. S.; Pressman S.; Andress A. P.; Kim K.; White J. L.; Cassidy J. J.; Li X.; Lubell K.; Lim do H.; Cho I. S.; Nakahara K.; Preall J. B.; Bellare P.; Sontheimer E. J.; Carthew R. W. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 2009, 11 (9), 1150–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings D. J.; Ciaudo C.; Erhardt M.; Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11 (9), 1143–9. [DOI] [PubMed] [Google Scholar]
- Hurley J. H.; Hanson P. I. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat. Rev. Mol. Cell. Biol. 2010, 11 (8), 556–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel S.; Macheboeuf P.; Martinelli N.; Weissenhorn W. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem. Sci. 2011, 36 (4), 199–210. [DOI] [PubMed] [Google Scholar]
- Rusten T. E.; Stenmark H. How do ESCRT proteins control autophagy?. J. Cell Sci. 2009, 122 (Pt 13), 2179–83. [DOI] [PubMed] [Google Scholar]
- Wollert T.; Hurley J. H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464 (7290), 864–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahriari M.; Keshavaiah C.; Scheuring D.; Sabovljevic A.; Pimpl P.; Hausler R. E.; Hulskamp M.; Schellmann S. The AAA-type ATPase AtSKD1 contributes to vacuolar maintenance of Arabidopsis thaliana. Plant J. 2011, 64 (1), 71–85. [DOI] [PubMed] [Google Scholar]
- Barajas D.; Jiang Y.; Nagy P. D. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 2009, 5 (12), e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindas A. C.; Karlsson E. A.; Lindgren M. T.; Ettema T. J.; Bernander R. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. U.S.A. 2008, 105 (48), 18942–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellen A. F.; Albers S. V.; Huibers W.; Pitcher A.; Hobel C. F.; Schwarz H.; Folea M.; Schouten S.; Boekema E. J.; Poolman B.; Driessen A. J. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 2009, 13 (1), 67–79. [DOI] [PubMed] [Google Scholar]
- Stauffer D. R.; Howard T. L.; Nyun T.; Hollenberg S. M. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J. Cell Sci. 2001, 114 (Pt 13), 2383–93. [DOI] [PubMed] [Google Scholar]
- Muziol T.; Pineda-Molina E.; Ravelli R. B.; Zamborlini A.; Usami Y.; Gottlinger H.; Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell 2006, 10 (6), 821–30. [DOI] [PubMed] [Google Scholar]
- Bajorek M.; Schubert H. L.; McCullough J.; Langelier C.; Eckert D. M.; Stubblefield W. M.; Uter N. T.; Myszka D. G.; Hill C. P.; Sundquist W. I. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 2009, 16 (7), 754–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J.; Chen X. W.; Davies B. A.; Saltiel A. R.; Katzmann D. J.; Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol. Biol. Cell 2009, 20 (15), 3514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D.; Saksena S.; Emr S. D. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 2008, 15 (4), 578–89. [DOI] [PubMed] [Google Scholar]
- Wollert T.; Wunder C.; Lippincott-Schwartz J.; Hurley J. H. Membrane scission by the ESCRT-III complex. Nature 2009, 458 (7235), 172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S.; Obita T.; Pobbati A. V.; Perisic O.; Samson R. Y.; Bell S. D.; Williams R. L. Evolution and assembly of ESCRTs. Biochem. Soc. Trans. 2009, 37 (Pt 1), 151–5. [DOI] [PubMed] [Google Scholar]
- Lata S.; Schoehn G.; Solomons J.; Pires R.; Gottlinger H. G.; Weissenhorn W. Structure and function of ESCRT-III. Biochem. Soc. Trans. 2009, 37 (Pt 1), 156–60. [DOI] [PubMed] [Google Scholar]
- Saksena S.; Wahlman J.; Teis D.; Johnson A. E.; Emr S. D. Functional reconstitution of ESCRT-III assembly and disassembly. Cell 2009, 136 (1), 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill S. A.; Hanson P. I. Activation of Human VPS4A by ESCRT-III Proteins Reveals Ability of Substrates to Relieve Enzyme Autoinhibition. J. Biol. Chem. 2010, 285 (46), 35428–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter V.; Hauser M. T. Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 2006, 11 (3), 115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas T. J.; Sliwinski M. K.; Martinez D. E.; Preuss M.; Ebine K.; Ueda T.; Nielsen E.; Odorizzi G.; Otegui M. S. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 2007, 19 (4), 1295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C.; Reyes F. C.; Buono R.; Sliwinski M. K.; Haas T. J.; Otegui M. S. The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 2009, 21 (3), 749–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiarimpa A.; Anzenberger F.; Schlager N.; Neubert S.; Hauser M.-T.; Schwechheimer C.; Isono E. The Arabidopsis Deubiquitinating Enzyme AMSH3 Interacts with ESCRT-III Subunits and Regulates Their Localization. Plant Cell 2011, 23 (8), 3025–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T.; Saksena S.; Ghazi-Tabatabai S.; Gill D. J.; Perisic O.; Emr S. D.; Williams R. L. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 2007, 449 (7163), 735–9. [DOI] [PubMed] [Google Scholar]
- Kozlov M. M.; McMahon H. T.; Chernomordik L. V. Protein-driven membrane stresses in fusion and fission. Trends Biochem. Sci. 2010, 35 (12), 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]; Field M. C.; Sali A.; Rout M. P. On a bender—BARs, ESCRTs, COPs, and finally getting your coat. J. Cell Biol. 2011, 193 (6), 963–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton M. D.; Skalicky J. J.; Kieffer C.; Karren M. A.; Ghaffarian S.; Sundquist W. I. ESCRT-III recognition by VPS4 ATPases. Nature 2007, 449 (7163), 740–4. [DOI] [PubMed] [Google Scholar]
- Yorikawa C.; Takaya E.; Osako Y.; Tanaka R.; Terasawa Y.; Hamakubo T.; Mochizuki Y.; Iwanari H.; Kodama T.; Maeda T.; Hitomi K.; Shibata H.; Maki M. Human Calpain 7/PalBH Associates with a Subset of ESCRT-III-related Proteins in its N-terminal Region and Partly Localizes to Endocytic Membrane Compartments. J. Biochem. 2008, 143 (6), 731–45. [DOI] [PubMed] [Google Scholar]
- Yang D.; Rismanchi N.; Renvoise B.; Lippincott-Schwartz J.; Blackstone C.; Hurley J. H. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 2008, 15 (12), 1278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue S. M.; Mattei S.; Saksena S.; Emr S. D. Novel Ist1-Did2 Complex Functions at a Late Step in Multivesicular Body Sorting. Mol. Biol. Cell 2008, 19 (2), 475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke G. J.; McMahon H. T. The dynamin superfamily: universal membrane tubulation and fission molecules?. Nat. Rev. Mol. Cell. Biol. 2004, 5 (2), 133–47. [DOI] [PubMed] [Google Scholar]
- Müller S.; Fuchs E.; Ovecka M.; Wysocka-Diller J.; Benfey P. N.; Hauser M. T. Two new loci, PLEIADE and HYADE, implicate organ-specific regulation of cytokinesis in Arabidopsis. Plant Physiol. 2002, 130 (1), 312–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M. T.; Morikami A.; Benfey P. N. Conditional root expansion mutants of Arabidopsis. Development 1995, 121 (4), 1237–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J.; Bent A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16 (6), 735–43. [DOI] [PubMed] [Google Scholar]
- Menges M.; Hennig L.; Gruissem W.; Murray J. A. Cell cycle-regulated gene expression in Arabidopsis. J. Biol. Chem. 2002, 277 (44), 41987–2002. [DOI] [PubMed] [Google Scholar]
- Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001, 127 (4), 1466–75. [PMC free article] [PubMed] [Google Scholar]
- Hauser M. T.; Adhami F.; Dorner M.; Fuchs E.; Glossl J. Generation of co-dominant PCR-based markers by duplex analysis on high resolution gels. Plant J. 1998, 16 (1), 117–25. [DOI] [PubMed] [Google Scholar]
- Mortz E.; Krogh T. N.; Vorum H.; Gorg A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 2001, 1 (11), 1359–63. [DOI] [PubMed] [Google Scholar]
- Karsai A.; Müller S.; Platz S.; Hauser M. T. Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 2002, 32 (4), 790–2. [DOI] [PMC free article] [PubMed] [Google Scholar]; 794 - 6.
- Ito T.; Kuwahara S.; Yokota T. Automatic and manual latex agglutination tests for measurement of cholera toxin and heat-labile enterotoxin of Escherichia coli. J Clin Microbiol 1983, 17 (1), 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M.; Chaban C.; Schutze K.; Batistic O.; Weckermann K.; Nake C.; Blazevic D.; Grefen C.; Schumacher K.; Oecking C.; Harter K.; Kudla J. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 2004, 40 (3), 428–38. [DOI] [PubMed] [Google Scholar]
- Oliveros J. C.VENNY: An interactive tool for comparing lists with Venn Diagrams; http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Hanson P. I.; Roth R.; Lin Y.; Heuser J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008, 180 (2), 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U. K.; Stuchell M.; Muller B.; Ward D. M.; Chung H. Y.; Morita E.; Wang H. E.; Davis T.; He G. P.; Cimbora D. M.; Scott A.; Krausslich H. G.; Kaplan J.; Morham S. G.; Sundquist W. I. The protein network of HIV budding. Cell 2003, 114 (6), 701–13. [DOI] [PubMed] [Google Scholar]
- Strack B.; Calistri A.; Craig S.; Popova E.; Gottlinger H. G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 2003, 114 (6), 689–99. [DOI] [PubMed] [Google Scholar]
- Shim S.; Kimpler L. A.; Hanson P. I. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 2007, 8 (8), 1068–79. [DOI] [PubMed] [Google Scholar]; Howard T. L.; Stauffer D. R.; Degnin C. R.; Hollenberg S. M. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 2001, 114 (Pt 13), 2395–404. [DOI] [PubMed] [Google Scholar]
- Leung K. F.; Dacks J. B.; Field M. C. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 2008, 9 (10), 1698–716. [DOI] [PubMed] [Google Scholar]
- Giavalisco P.; Kapitza K.; Kolasa A.; Buhtz A.; Kehr J. Towards the proteome of Brassica napus phloem sap. Proteomics 2006, 6 (3), 896–909. [DOI] [PubMed] [Google Scholar]
- Carter C.; Pan S.; Zouhar J.; Avila E. L.; Girke T.; Raikhel N. V. The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 2004, 16 (12), 3285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U. G.; Endler A.; Schelbert S.; Brunner A.; Schnell M.; Neuhaus H. E.; Marty-Mazars D.; Marty F.; Baginsky S.; Martinoia E. Novel tonoplast transporters identified using a proteomic approach with vacuoles isolated from cauliflower buds. Plant Physiol. 2007, 145 (1), 216–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquinod M.; Villiers F.; Kieffer-Jaquinod S.; Hugouvieux V.; Bruley C.; Garin J.; Bourguignon J. A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell. Proteomics 2007, 6 (3), 394–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmagne A.; Ferro M.; Meinnel T.; Bruley C.; Kuhn L.; Garin J.; Barbier-Brygoo H.; Ephritikhine G. A high content in lipid-modified peripheral proteins and integral receptor kinases features in the arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 2007, 6 (11), 1980–96. [DOI] [PubMed] [Google Scholar]
- Dunkley T. P.; Hester S.; Shadforth I. P.; Runions J.; Weimar T.; Hanton S. L.; Griffin J. L.; Bessant C.; Brandizzi F.; Hawes C.; Watson R. B.; Dupree P.; Lilley K. S. Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. U.S.A. 2006, 103 (17), 6518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S. K.; Gantt J. A.; Ruby J. F.; Clouse S. D.; Goshe M. B. Membrane proteomic analysis of Arabidopsis thaliana using alternative solubilization techniques. J Proteome Res 2007, 6 (5), 1933–50. [DOI] [PubMed] [Google Scholar]
- Benschop J. J.; Mohammed S.; O’Flaherty M.; Heck A. J.; Slijper M.; Menke F. L. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 2007, 6 (7), 1198–214. [DOI] [PubMed] [Google Scholar]
- Nuhse T. S.; Stensballe A.; Jensen O. N.; Peck S. C. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 2004, 16 (9), 2394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N.; Nakagami H.; Mochida K.; Daudi A.; Tomita M.; Shirasu K.; Ishihama Y. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 2008, 4, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. S.; Kim I. S.; Park J. B.; Lee M. N.; Lee H. Y.; Suh P. G.; Ryu S. H. The phox homology domain of phospholipase D activates dynamin GTPase activity and accelerates EGFR endocytosis. Nat. Cell Biol. 2006, 8 (5), 477–84. [DOI] [PubMed] [Google Scholar]
- Jenkins G. M.; Frohman M. A. Phospholipase D: a lipid centric review. Cell. Mol. Life Sci. 2005, 62 (19–20), 2305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haucke V.; Di Paolo G. Lipids and lipid modifications in the regulation of membrane traffic. Curr. Opin. Cell Biol. 2007, 19 (4), 426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G. Phospholipase D in endocytosis and endosomal recycling pathways. Biochim. Biophys. Acta 2009, 1791 (9), 845–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaLonde M.; Janssens H.; Yun S.; Crosby J.; Redina O.; Olive V.; Altshuller Y. M.; Choi S. Y.; Du G.; Gergen J. P.; Frohman M. A. A role for Phospholipase D in Drosophila embryonic cellularization. BMC Dev. Biol. 2006, 6, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale N.; Caumont A. S.; Chasserot-Golaz S.; Du G.; Wu S.; Sciorra V. A.; Morris A. J.; Frohman M. A.; Bader M. F. Phospholipase D1: a key factor for the exocytotic machinery in neuroendocrine cells. EMBO J. 2001, 20 (10), 2424–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.; Altshuller Y. M.; Hou J. C.; Pessin J. E.; Frohman M. A. Insulin-stimulated plasma membrane fusion of Glut4 glucose transporter-containing vesicles is regulated by phospholipase D1. Mol. Biol. Cell 2005, 16 (6), 2614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman E. E.; Chupin V.; Fuller N. L.; Kozlov M. M.; de Kruijff B.; Burger K. N.; Rand P. R. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry 2005, 44 (6), 2097–102. [DOI] [PubMed] [Google Scholar]
- Verkleij A. J.; De Maagd R.; Leunissen-Bijvelt J.; De Kruijff B. Divalent cations and chlorpromazine can induce non-bilayer structures in phosphatidic acid-containing model membranes. Biochim. Biophys. Acta 1982, 684 (2), 255–62. [DOI] [PubMed] [Google Scholar]
- McLaughlin S. The electrostatic properties of membranes. Annu. Rev. Biophys. Biophys. Chem. 1989, 18, 113–36. [DOI] [PubMed] [Google Scholar]
- Murray D.; Arbuzova A.; Hangyas-Mihalyne G.; Gambhir A.; Ben-Tal N.; Honig B.; McLaughlin S. Electrostatic properties of membranes containing acidic lipids and adsorbed basic peptides: theory and experiment. Biophys. J. 1999, 77 (6), 3176–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley P.; Reaves B. J.; Hashimoto M.; Riley A. M.; Potter B. V.; Holman G. D. Identification of mammalian Vps24p as an effector of phosphatidylinositol 3,5-bisphosphate-dependent endosome compartmentalization. J. Biol. Chem. 2003, 278 (40), 38786–95. [DOI] [PubMed] [Google Scholar]
- Fan L.; Zheng S.; Cui D.; Wang X. Subcellular distribution and tissue expression of phospholipase Dalpha, Dbeta, and Dgamma in Arabidopsis. Plant Physiol. 1999, 119 (4), 1371–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov P. V.; Akimov S. A.; Evseev A. I.; Schmid S. L.; Zimmerberg J.; Frolov V. A. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell 2008, 135 (7), 1276–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle A. F.; Clark G. P.; Boon R.; Lewandowska D.; Lam Y. W.; Andersen J.; Mann M.; Lamond A. I.; Brown J. W.; Shaw P. J. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell 2005, 16 (1), 260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae M. S.; Cho E. J.; Choi E. Y.; Park O. K. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 2003, 36 (5), 652–63. [DOI] [PubMed] [Google Scholar]
- Keesee S. K.; Obar R.; Wu Y.-J.. Materials and methods for detection of breast cancer. US-Patent 1999, US005914238A.
- Hodges E.; Redelius J. S.; Wu W.; Hoog C. Accelerated discovery of novel protein function in cultured human cells. Mol. Cell. Proteomics 2005, 4 (9), 1319–27. [DOI] [PubMed] [Google Scholar]
- Bayer E. M.; Bottrill A. R.; Walshaw J.; Vigouroux M.; Naldrett M. J.; Thomas C. L.; Maule A. J. Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 2006, 6 (1), 301–11. [DOI] [PubMed] [Google Scholar]
- Bindschedler L. V.; Palmblad M.; Cramer R. Hydroponic isotope labelling of entire plants (HILEP) for quantitative plant proteomics; an oxidative stress case study. Phytochemistry 2008, 69 (10), 1962–72. [DOI] [PubMed] [Google Scholar]
- Keller S.; Sanderson M. P.; Stoeck A.; Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 2006, 107 (2), 102–8. [DOI] [PubMed] [Google Scholar]
- Simons M.; Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21 (4), 575–81. [DOI] [PubMed] [Google Scholar]
- Valadi H.; Ekstrom K.; Bossios A.; Sjostrand M.; Lee J. J.; Lotvall J. O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9 (6), 654–9. [DOI] [PubMed] [Google Scholar]
- Mathivanan S.; Ji H.; Simpson R. J. Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 2010, 73 (10), 1907–20. [DOI] [PubMed] [Google Scholar]
- Mathivanan S.; Simpson R. J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9 (21), 4997–5000. [DOI] [PubMed] [Google Scholar]
- Regente M.; Corti-Monzon G.; Maldonado A. M.; Pinedo M.; Jorrin J.; de la Canal L. Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett. 2009, 583 (20), 3363–6. [DOI] [PubMed] [Google Scholar]
- An Q.; van Bel A. J.; Huckelhoven R. Do plant cells secrete exosomes derived from multivesicular bodies?. Plant Signal. Behav. 2007, 2 (1), 4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Q.; Huckelhoven R.; Kogel K. H.; van Bel A. J. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol. 2006, 8 (6), 1009–19. [DOI] [PubMed] [Google Scholar]
- Barr F. A.; Shorter J. Membrane traffic: do cones mark sites of fission?. Curr. Biol. 2000, 10 (4), R141–4. [DOI] [PubMed] [Google Scholar]
- Lata S.; Schoehn G.; Jain A.; Pires R.; Piehler J.; Gottlinger H. G.; Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science 2008, 321 (5894), 1354–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrikant G.; Lata S.; Riches J. D.; Briggs J. A.; Weissenhorn W.; Kozlov M. M. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput. Biol. 2009, 5 (11), e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil T. J.; Schmid S. L. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell 2008, 135 (7), 1263–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S.; Damke H.; Schmid S. L. Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J. Cell Biol. 2000, 150 (5), 1137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.