Abstract
HTLV-1 is endemic in Brazil and HIV/HTLV-1 coinfection has been detected, mostly in the northeast region. Cosmopolitan HTLV-1a is the main subtype that circulates in Brazil. This study characterized 17 HTLV-1 isolates from HIV coinfected patients of southern (n=7) and southeastern (n=10) Brazil. HTLV-1 provirus DNA was amplified by nested PCR (env and LTR) and sequenced. Env sequences (705 bp) from 15 isolates and LTR sequences (731 bp) from 17 isolates showed 99.5% and 98.8% similarity among sequences, respectively. Comparing these sequences with ATK (HTLV-1a) and Mel5 (HTLV-1c) prototypes, similarities of 99% and 97.4%, respectively, for env and LTR with ATK, and 91.6% and 90.3% with Mel5, were detected. Phylogenetic analysis showed that all sequences belonged to the transcontinental subgroup A of the Cosmopolitan subtype, clustering in two Latin American clusters.
The human T-lymphotropic virus type 1 (HTLV-1) was first described in 1980, and infects about 10–20 million people worldwide. Since then several epidemiological, clinical, and laboratory studies have been conducted around the world.1 The most frequent subtype circulating in the world is the HTLV-1a (Cosmopolitan).2 In South America, the prevalence of HTLV infection ranges from 0.1% to 5% in the general population.2 Brazil emerges as first in the number of HTLV-infected individuals in the world, estimated at 2.5 million. HTLV-1 infection is detected mostly in northeastern Brazil, probably as a consequence of the slave trade during the seventeenth, eighteenth, and nineteenth centuries,3 and HIV/HTLV-1 coinfection in this region is more frequent than HIV/HTLV-2 coinfection.
The HTLV-1 retrovirus has a conserved genome, with only 0.1% to 6.9% genetic diversity among strains. The env and long terminal repeat (LTR) regions have the highest variability,4 and are useful for viral subtype characterization. Mostly individuals remain asymptomatic, but this infection may cause a wide spectrum of diseases, including leukemia (adult T cell leukemia/lymphoma, ATL) and neurodegenerative disorders (HTLV-1-associated myelopathy/tropical spastic paraparesis, HAM/TSP).1 In Brazil, the majority of HTLV-1 molecular characterization studies were conducted in patients presenting ATL and HAM/TSP and in asymptomatic blood donors. Studies of HTLV-1 in HIV-coinfected patients are limited and are attributed to HTLV-1a, the main strain also circulating in HIV/HTLV-1-coinfected individuals.3,5–8
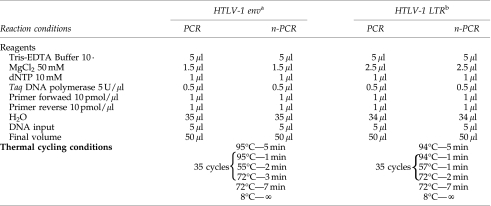
The present study was conducted to add information concerning HTLV-1 strains that circulate in HIV-coinfected patients from the south and southeast regions of Brazil, thought to have more cases of HIV/HTLV-2 coinfection.9,10 Seventeen HIV/HTLV-1-coinfected patients attending Aids Reference Centers, seven (one woman and six men) from Londrina-LO (south), and 10 (seven women and three men) from São Paulo-SP (southeast), were enrolled in this analysis. HTLV genomic DNA was extracted from peripheral blood leukocytes (PBLs). Nested polymerase chain reaction (n-PCR) was performed to amplify a 763-bp env and 766-bp LTR region of HTLV-1, using protocols previously described (Tables 1 and 2)6,11 in which some primers have undergone minor nucleotide modifications and thermal cycling conditions were adjusted. Env and LTR amplified products were sequenced in an ABI 3100 Genetic Analyser (Applied Biosystems, USA), with the same primers used in the n-PCR.
Table 1.
Primers Employed in Polymerase Chain Reaction Assays (env and LTR)
| Molecular assay | Primer | Sequence 5′–3′ and product size | Positiona |
|---|---|---|---|
| PCR envb | D498 forward | ATG GGT AAG TTT CTB GCC (18 bp) | 5202–5219 |
| D500 reverse | TTA CAG GGA TGA CTB AGG (18 bp) | 6668–6651 | |
| n-PCR envb | LUI 7 forward | CCG TCT CCA GYC CMT MCT GGA (21 bp) | 5547–5567 |
| LUI 8 reverse | CCT CGT CTR TTY TGG GCW GCA (21 bp) | 6309–6289 | |
| PCR LTRc | LTR-I.03 forward | GGC TTA GAG CCT CCC AGT GA (20 bp) | 57–76 |
| LTR-I.02 reverse | CGC GGA ATA GGG CTA GCG CT (20 bp) | 864–845 | |
| n-PCR LTRc | LTR-I.03 forward | GGC TTA GAG CCT CCC AGT GA (20 bp) | 57–76 |
| LTR-I.04 reverse | GCC TAG GGA ATA AAG GGG CG (20 bp) | 822–803 |
Primer nucleotide position is provided as aligned with ATK (HTLV-1-infected cell line; GenBank Accession number J02029). bp, base pairs; n-PCR, nested polymerase chain reaction.
Adapted from Caterino-de-Araujo et al.11
Adapted from Laurentino et al.6
Primers were employed to detect HTLV-1 provirus DNA in peripheral blood leucocytes of HIV coinfected patients from the south (Londrina-LO) and the southeast (São Paulo-SP) regions of Brazil.
Table 2.
PCR Assays (env and LTR) Protocol
 |
All sequencing chromatograms obtained were assembled and manually edited with Sequencher 4.7 software. Multiple alignments were performed using the Clustal W multiple-sequence alignment tool from BioEdit Sequence Alignment Editor version 7.0.5.3 software, with a reference set available in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank). HTLV-1 subtyping was primarily screened by NCBI-Genotyping (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi) and REGA-Subtyping (http://www.bioafrica.net/rega-genotype/html/subtypinghtlv.html) tools websites. Neighbor-joining (NJ) and maximum-likelihood (ML) phylogenetic trees were constructed based in appropriate nucleotide substitution models determined by Modeltest v3.7 (TrN + G model for env and GTR + G model for LTR), using PAUP v4b10 software. Bootstrapping was performed using the stepwise addition algorithm for 1.000 replicates. The Mel5 (HTLV-1c) sequence was used as the outgroup. MEGA4 software was used to estimate nucleotide distances.
Env sequences (705 bp) from 15 isolates and LTR sequences (731 bp) from 17 isolates were obtained. Molecular analysis of env and LTR sequences disclosed, respectively, nucleotide similarities of 99.5% and 98.8% among sequences, 99% and 97.4% with ATK, and 91.6% and 90.3% with Mel5.
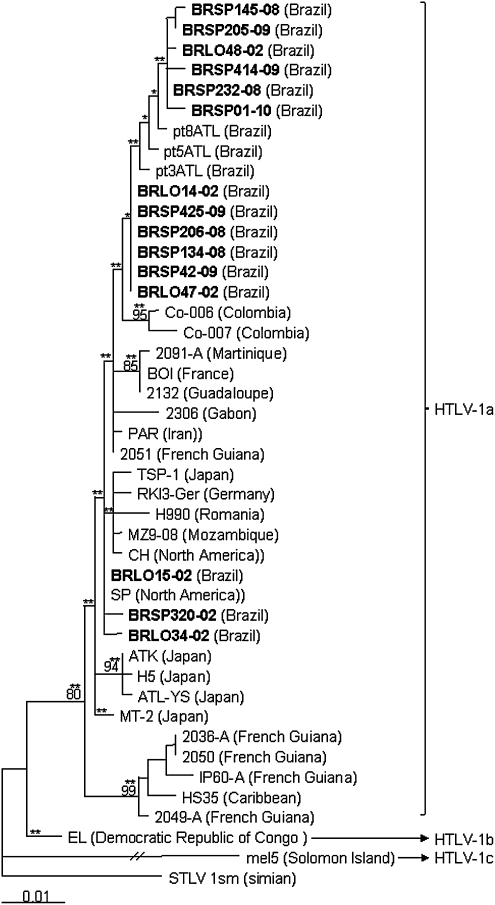
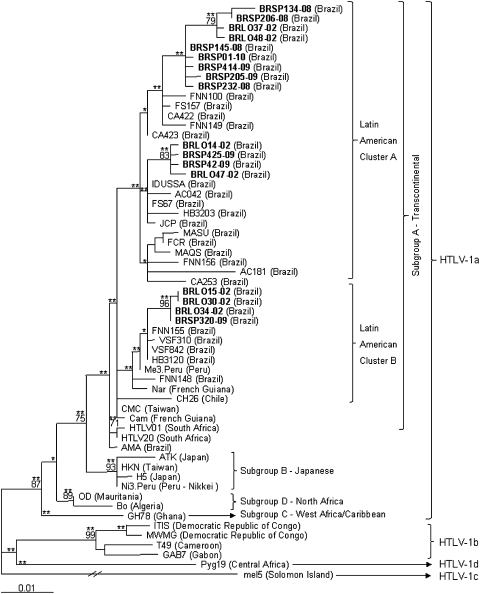
Phylogenetic analysis of a segment of 705 bp of 44 HTLV-1 env sequences available in the GenBank database including the 15 new sequences from the south and southeast regions of Brazil were used to construct the env phylogenetic tree (Fig. 1). A segment of 692 bp of 59 HTLV-1 LTR sequences available in GenBank including the 17 new sequences of this study was used to construct the LTR phylogenetic tree (Fig. 2).
FIG. 1.
Phylogenetic tree constructed by the maximum-likelihood method using the PAUP v4b10 software for the partial env region of 705 bp (position 5614–6318 relative to the ATK prototype) of HTLV-1 of 44 isolates, including sequences from the south and southeast regions of Brazil (GenBank AN HM770426-HM770440) in bold, generated with the TrN + G model. Bootstrap values above 70% and zero length using the likelihood ratio test with p<0.001 (**) and p≤0.05 (*) in key branches are depicted. The HTLV-1 Mel5 isolate was used as the outgroup.
FIG. 2.
Phylogenetic tree constructed by the maximum-likelihood method using the PAUP v4b10 software for the partial LTR region of 692 bp (position 75–766 relative to the ATK prototype) of HTLV-1 of 59 isolates, including sequences from the south and southeast regions of Brazil (GenBank AN JF271836-JF271852) in bold, generated with the GTR + G model. Bootstrap values above 70% and zero length using the likelihood ratio test with p<0.001 (**) and p≤0.05 (*) in key branches are depicted. The HTLV-1 Mel5 isolate was used as the outgroup.
Four sequences that showed the highest similarities with ATK clustered separately in the trees (p≤0.001 for env and LTR, bootstrap=96% for LTR); they had C386T, T594C and C595T nucleotide substitutions in the LTR region (BRLO14-02, BRLO30-02, BRLO34-02, and SP320-09). All except these four sequences had mutations T5637G, T5730C, and A6120G in env and T500A, A549G, G676A, T766C, and T767G in the LTR regions. Six sequences presented the amino acid change V1981I and clustered together in the env tree (BRLO48-02, BRSP145-08, BRSP205-09, BRSP232-08, BRSP414-09, and BRSP01-10).
According to the NCBI-Genotyping and the REGA-Subtyping websites all isolates from the present study belonged to the Cosmopolitan HTLV-1a subtype, the same subtype detected by phylogenetic analysis (bootstrap value of 80% for env and 87% for LTR, and a p<0.001 for both). In addition, all isolates belonged to the Transcontinental subgroup A (supported by a p<0.001) and clustered in two Latin American clusters (A and B) in the LTR tree (supported by a p<0.001 in ML analyses).
These phylogenetic results are in accordance with the results obtained in HTLV-1 strains characterized from the north and northeast regions of Brazil. In fact, nucleotide similarities in LTR sequences of 99.2% and 98.9%, respectively, were detected in strains isolated from individuals from Cruz das Almas and Salvador cities (southeastern Brazil), and of 99.3% and 97.3%, respectively, in strains isolated from individuals from Fortaleza and Rio Branco cities (northern Brazil).12,13
Molecular analysis of HTLV-1 isolates from HIV-1/HTLV-1-coinfected individuals from northern Brazil disclosed the Cosmopolitan HTLV-1a subtype circulating in four women from Feira de Santana, state of Bahia,6 and in two men from Belém, state of Para.7 Corroborating these and the present study, phylogenetic analysis of HTLV-1 isolates from several populations in Brazil confirms HTLV-1a, Transcontinental subgroup A as the most prevalent in the country,8,12,13 and suggests a post-Columbian introduction of HTLV-1 into the Brazilian population in the state of Bahia and maybe in other places in Latin America.3,7,12,14
Indeed, the majority of the sequences from this study clustered into Latin American Cluster A, although some sequences clustered in Latin American Cluster B. The well-divided clusters of Latin American could suggest different introductions of HTLV-1 in Brazil, and also in HIV/HTLV-1-coinfected individuals (both from the south and southeast regions). Still, the high level of similarities among sequences leads us to speculate on a recent introduction of HTLV-1 in Brazil.
In conclusion, these data show that the Cosmopolitan HTLV-1a subtype is also frequent in HIV coinfected patients from southern and southeastern Brazil, suggesting spreading of HTLV-1 in the country. The mutations observed should be monitored in the context of molecular epidemiology. Moreover, this is the first study of the molecular characterization of HTLV-1 sequences from Londrina, southern Brazil, providing insight into the HTLV-1 dynamic epidemic in this country.
Sequence Data
The GenBank accession numbers for the HTLV-1 sequences included in the phylogenetic study are as follows: env: ATK (J02029), TSP-1 (M86840), MT-2 (M37747), HS35(D13784), CH (M69044), 2036-A (AY604874), pt3ATL (U81866), pt5ATL (U81867), pt8ATL (U81868), 2306 (AY604896), MZ9-08 (HM770441), EL (M67514), Co-006 (AF405343), Co-007 (AF405344), 2049-A (AY604875), 2050 (AY604876), 2051 (AY604877), 2091-A (AY604885), 2132 (AY604887), BOI (L36905), ATL-YS (U19949), H5 (M37301), H990 (U81862), IP60-A (AY604934), PAR (AY604935), RK13-Ger (AF042071), SP (M69044), Mel5 (L02534), STLV-1-sm (U94516); LTR: ATK (J02029), pyg19 (L76310), IT IS (Z32527), MWMG (Z31662), T49 (L76305), GAB7 (L76311), GH78 (D23693), OD (U12805), BO (U12804), Ni3.Peru (Y16485), H5 (M37299), HTLV01 (DQ005556), HTLV20 (DQ005564), CH26 (D23690), FNN148 (DQ005548), Nar (AF063820), Me3.Peru (Y16480), FNN155 (DQ005551), Cam (AF063819), IDUSSA (DQ005555), FNN156 (DQ005552), MAQS (X88876), MASU (X88877), JCP (X88875), HKN (X88874), FCR (X88873), CMC (X88872), AMA (X88871), FNN100 (DQ005547), FNN149 (DQ005549), CA253 (EU108722), CA423 (EU108724), CA422 (EU108721), HB3120 (DQ471206), HB3203 (DQ471205), FS157 (GU225732), FS67 (GU225731), VSF842 (EF672337), VSF310 (EF672336), AC042 (EU392160), AC181 (EU392159), Mel5 (L02534).
The GenBank accession numbers of the HTLV-1 fragments sequenced in our laboratory and included in the phylogenetic analysis are as follows: env: BRLO14-02 (HM770426), BRLO15-02 (HM770427), BRLO34-02 (HM770428), BRLO47-02 (HM770429), BRLO48-02 (HM770430), BRSP01-10 (HM770431), BRSP134-08 (HM770432), BRSP145-08 (HM770433), BRSP205-09 (HM770434), BRSP206-08 (HM770435), BRSP232-08 (HM770436), BRSP320-09 (HM770437), BRSP42-09 (HM770438), BRSP414-09 (HM770439), BRSP425-09 (HM770440); LTR: BRLO14-02 (JF271836), BRLO15-02 (JF271837), BRLO30-02 (JF271838), BRLO34-02 (JF271839), BRLO37-02 (JF271840), BRLO47-02 (JF271841), BRLO48-02 (JF271842), BRSP134-08 (JF271843), BRSP145-08 (JF271844), BRSP206-08 (JF271845), BRSP232-08 (JF271846), BRSP42-09 (JF271847), BRSP205-09 (JF271848), BRSP320-09 (JF271849), BRSP414-09 (JF271850), BRSP425-09 (JF271851), BRSP01-10 (JF271852).
Acknowledgments
This study was supported by Ministério da Ciência e Tecnologia/Conselho Nacional de Desenvolvimento Científico e Tecnológico (MCT/CNPq), Brazil (Universal Grant 481040/2007-2), a fellowship to A.C.A. (Grant 303328/2009-6), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil (Ph.D. fellowship to M.C.M.), and Instituto Adolfo Lutz (Grant 39/07).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Proietti FA. Carneiro-Proietti ABF. Catalan-Soares BC. Murphy EL. Global epidemiology of HTLV-1 infection and associated diseases. Oncogene. 2005;24:6058–6068. doi: 10.1038/sj.onc.1208968. [DOI] [PubMed] [Google Scholar]
- 2.Carneiro-Proietti AB. Catalan-Soares BC. Proietti FA. GIPH (Interdisciplinary HTLV-I/IIResearch Group): Human T cell lymphotropic viruses (HTLV-I/II) in South America: Should it be a public health concern? J Biomed Sci. 2002;9:587–595. doi: 10.1159/000067286. [DOI] [PubMed] [Google Scholar]
- 3.Alcantara LC. de Oliveira T. Gordon M, et al. Tracing the origin of Brazilian HTLV-1 as determined by analysis of host and viral genes. AIDS. 2006;20(5):780–782. doi: 10.1097/01.aids.0000216383.14808.13. [DOI] [PubMed] [Google Scholar]
- 4.Ratner L. Philpott T. Trowbridge DB. Nucleotide sequence analysis of isolates of human T-lymphotropic virus type I of diverse geographical origins. AIDS Res Hum Retroviruses. 1991;7(11):923–941. doi: 10.1089/aid.1991.7.923. [DOI] [PubMed] [Google Scholar]
- 5.Yamashita M. Veronesi R. Menna-Barreto M, et al. Molecular epidemiology of human T-cell leukemia virus type I (HTLV-1) Brazil: The predominant HTLV-1s in South America differ from HTLV-l of Japan and Africa, as well as those of Japanese immigrants and their relatives in Brazil. Virology. 1999;261(1):59–69. [PubMed] [Google Scholar]
- 6.Laurentino RV. Lopes IGL. Azevedo VN, et al. Molecular characterization of human T-cell lymphotropic virus coinfecting human immunodeficiency virus 1 infected patients in the Amazon region of Brazil. Mem Inst Oswaldo Cruz. 2005;100(4):371–376. doi: 10.1590/s0074-02762005000400006. [DOI] [PubMed] [Google Scholar]
- 7.Rego FFA. Mota-Miranda A. Santos ES. Galvão-Castro B. Alcantara LC. Seroprevalence and molecular epidemiology of HTLV-1 isolates from HIV coinfected women in Feira de Santana, Bahia, Brazil. AIDS Res Hum Retroviruses. 2010;26(12):1333–1339. doi: 10.1089/aid.2009.0298. [DOI] [PubMed] [Google Scholar]
- 8.Kashima S. Alcântara LC. Takayanagui OM, et al. Distribution of human T cell lymphotropic vírus type 1 (HTLV-1) subtypes in Brazil: Genetic characterization of LTR and Tax region. AIDS Res Hum Retroviruses. 2006;22(10):953–959. doi: 10.1089/aid.2006.22.953. [DOI] [PubMed] [Google Scholar]
- 9.Etzel A. Shibata GY. Rozman M. Jorge MLSG. Damas CD. Segurado AAC. HTLV-1 and HTLV-2 infections in HIV-infected individuals from Santos, Brazil: Seroprevalence and risk factors. J Acquir Immune Defic Syndr. 2001;26:185–190. doi: 10.1097/00042560-200102010-00015. [DOI] [PubMed] [Google Scholar]
- 10.Morimoto HK. Caterino-de-Araujo A. Morimoto AA, et al. Seroprevalence and risk factors for human T-cell lymphotropic virus type 1 and 2 infection in human immunodeficiency virus (HIV)-infected patients attending AIDS Referral Center Health Units in Londrina and other communities in Paraná, Brazil. AIDS Res Hum Retroviruses. 2005;21(4):256–262. doi: 10.1089/aid.2005.21.256. [DOI] [PubMed] [Google Scholar]
- 11.Caterino-de-Araujo A. Favero A. Santos-Fortuna E. Suleiman J. Chieco-Bianchi L. Calabrò ML. HTLV-I/HTLV-II coinfection in AIDS patient from São Paulo, Brazil. AIDS Res Hum Retroviruses. 2000;16:715–719. doi: 10.1089/088922200308710. [DOI] [PubMed] [Google Scholar]
- 12.Magalhães T. Mota-Miranda AC. Alcantara LC. Olavarria V. Galvão-Castro B. Rios-Grassi MF. Phylogenetic, molecular analysis of HTLV-1 isolates from a medium sized town in northern of Brazil: Tracing a common origin of the virus from the most endemic city in the country. J Med Virol. 2008;80(11):2040–2045. doi: 10.1002/jmv.21278. [DOI] [PubMed] [Google Scholar]
- 13.Mota-Miranda AC. Araújo SP. Dias JP. Colin DD. Kashima S. Covas DT. Tavares-Neto J. Galvão-Castro B. Alcantara LC. HTLV-1 infection in blood donors from the Western Brazilian Amazon region: Seroprevalence and molecular study of viral isolates. J Med Virol. 2008;80(11):1966–1971. doi: 10.1002/jmv.21300. [DOI] [PubMed] [Google Scholar]
- 14.Rego FFA. Alcantara LC. Moura Neto JP. Miranda AC. Pereira Ode S. Gonçalves Mde S. Galvão-Castro B. HTLV type 1 molecular study in Brazilian villages with African characteristics giving support to the post-Columbian introduction hypothesis. AIDS Res Hum Retroviruses. 2008;24(5):673–677. doi: 10.1089/aid.2007.0290. [DOI] [PubMed] [Google Scholar]