SUMMARY
Numerous chromatin regulators are required for embryonic stem (ES) cell self-renewal and pluripotency, but few have been studied in detail. Here, we examine the roles of several chromatin regulators whose loss affects the pluripotent state of ES cells. We find that Mbd3 and Brg1 antagonistically regulate a common set of genes by regulating promoter nucleosome occupancy. Furthermore, both Mbd3 and Brg1 play key roles in the biology of 5-hydroxymethylcytosine (5hmC): Mbd3 co-localizes with Tet1 and 5hmC in vivo, Mbd3 knockdown preferentially affects expression of 5hmC-marked genes, Mbd3 localization is Tet1-dependent, and Mbd3 preferentially binds to 5hmC relative to 5-methylcytosine in vitro. Finally, both Mbd3 and Brg1 are themselves required for normal levels of 5hmC in vivo. Together, our results identify an effector for 5hmC, and reveal that control of gene expression by antagonistic chromatin regulators is a surprisingly common regulatory strategy in ES cells.
Keywords: embryonic stem cells, pluripotency, self-renewal, nucleosome remodeling, histone deacetylase, BAF, Brg1, NURD, Mbd3, 5-hydroxymethylcytosine, Tet1, polycomb
Introduction
In human and mouse embryonic stem (ES) cells, a number of transcriptional regulators are essential to maintain the pluripotent state. Several transcription factors are required for pluripotency, including Oct4, Sox2 and Nanog, which function as “master regulators” of the ES cell transcriptional network (Young, 2011). Along with sequence-specific transcription factors, many chromatin regulators also play essential roles in ES cell gene regulation, self-renewal, and differentiation. Several protein complexes that catalyze covalent modification of histones have important roles in ES cells, including proteins involved in histone methylation, acetylation, and ubiquitylation (Niwa, 2007; Surface et al., 2010). Compared to most somatic cells, ES cells exhibit unusual patterns of histone modifications, notably the “bivalent” juxtaposition of a mark associated with active genes (H3K4me3) with a repressive mark (H3K27me3) near the promoters of developmentally regulated genes (Azuara et al., 2006; Bernstein et al., 2006). The chromatin modifying complexes creating these marks, MLL/SET1 Complex and Polycomb Repressive Complex 2 (PRC2), respectively, are highly conserved and have important roles in development (van Lohuizen, 1998). It has been proposed that H3K4me3 and H3K27me3 have opposing effects on gene expression at some promoters, or “poise” genes for future regulatory changes (Bernstein et al., 2006).
ATP-dependent nucleosome remodeling factors, which modify the spacing or subunit composition of nucleosomes, also play important roles in ES cells (Fazzio and Panning, 2010; Keenen and de la Serna, 2009). BAF (Brahma/Brg1 Associated Factor) complexes are a family of ATP-dependent nucleosome remodeling factors that share homology to yeast SWI/SNF complex and function to both activate and silence transcription by remodeling nucleosomes near promoters and enhancers (Clapier and Cairns, 2009). In ES cells, a single BAF complex, esBAF, predominates (Ho et al., 2009b). Homozygous knockout (KO) or knockdown (KD) of any of several BAF subunits results in defects in ES cell self-renewal and pluripotency, highlighting their critical roles in maintaining the ES cell gene expression pattern (Gao et al., 2008; Ho et al., 2011; Ho et al., 2009b; Kidder et al., 2009; Yan et al., 2008).
Conversely, NURD (Nucleosome Remodeling and Deacetylase) complexes are chromatin remodeling factors that utilize nucleosome remodeling and histone deacetylase activities to create repressive chromatin structure (Denslow and Wade, 2007). KD or KO of the gene encoding the NURD subunit Mbd3 in ES cells results in a defect in differentiation, as well as altered developmental potency (Kaji et al., 2006; Kaji et al., 2007; Zhu et al., 2009). Mbd3 is one of four proteins named “Mbd” for “methyl-CpG binding domain”, based on the homology of these proteins to the methylcytosine binding domain in MeCP2 (Hendrich and Bird, 1998). However, whereas mammalian Mbd1, Mbd2 and Mbd4 bind to cytosine-methylated substrates in vitro, Mbd3 does not (Hendrich and Bird, 1998; Zhang et al., 1999), raising the question of what role the methyl binding domain plays in Mbd3 biology. NURD complexes containing either Mbd2 or Mbd3 (hereafter Mbd2/NURD and Mbd3/NURD), or both, have been purified from mammalian cells (Feng and Zhang, 2001; Le Guezennec et al., 2006), suggesting these complexes may be targeted to regions of the genome with distinct epigenetic marks.
Recently, members of the Tet family of proteins (Tet1, Tet2, and Tet3) have been shown to carry out hydroxylation of 5-methylcytosine (5mC) to generate 5-hydroxymethylcytosine (5hmC) (Ito et al., 2010; Tahiliani et al., 2009). Knockdown of Tet1 and Tet2 in ES cells leads to defects in differentiation (Koh et al., 2011), while Tet1 knockdown also leads to defects in self-renewal (Ito et al., 2010). Despite these defects in KD cells, Tet1 KO mice are viable and fertile (Dawlaty et al., 2011).
While it is commonly believed that hydroxymethylation primarily serves as an intermediate stage in cytosine demethylation (Bhutani et al., 2011), the relatively high steady-state levels of 5hmC observed in several contexts (for example, during global “demethylation” of paternal DNA after fertilization (Inoue and Zhang, 2011; Iqbal et al., 2011)) suggest that hydroxymethylation may also serve a specific regulatory function, since an intermediate in an enzymatic demethylation pathway would not be expected to persist for hours or days. Interestingly, Mbd3 KD in ES cells has defects similar to those of Tet1 KD ES cells, exhibiting increased expression of some trophectoderm markers (Kaji et al., 2006; Zhu et al., 2009). These data suggest the possibility that Mbd3 may bind hydroxymethylated regions of DNA and regulate genes whose regulatory sequences are enriched for this modification.
Here, we investigate functional interactions among six chromatin regulators necessary for ES cell self-renewal using genomic expression analysis, identifying a large set of overlapping targets of Mbd3 and Brg1. Genes activated by Brg1 and repressed by Mbd3 were significantly associated with both proteins. Mapping of Mbd3 showed that it is strongly enriched at Polycomb target genes, and gene expression changes were highly similar in Mbd3 KD and Suz12 KD cells, revealing that Mbd3 plays a role in regulation of bivalent genes in ES cells. Furthermore, we found that Mbd3 binding strongly overlaps with Tet1 binding profiles, that Mbd3-bound genes are enriched for DNA marked with 5hmC, and that KD of Tet1 abrogated Mbd3 binding to its genomic targets. Mbd3 binds to DNA harboring 5hmC, but not 5mC, in vitro, suggesting that Mbd3’s requirement for Tet1 to bind chromatin in vivo is mediated by direct binding to 5hmC. Finally, we find that Mbd3 and Brg1 are required for normal bulk levels of 5hmC in ES cells. Together, these data identify a novel effector function for 5hmC in vivo, indicating that it is not simply a demethylation intermediate, and further identify a positive feedback loop in which Mbd3 is both dependent upon 5hmC for chromatin binding and is necessary for normal levels of 5hmC within the genome.
RESULTS
Brg1 and Mbd3 have opposing effects on expression of shared target genes
To better understand chromatin regulation of the ES cell transcriptional regulatory network, we sought to identify the transcriptional targets and functional relationships among six chromatin regulators with important roles in ES cell self-renewal and pluripotency: Tip60, p400, Suz12 (involved in H3K27 methylation), Ash2l (involved in H3K4 methylation), Mbd3 and Brg1. We measured genome-wide mRNA changes upon knockdown (KD) of each factor alone or in all pair-wise combinations (to identify synergistic or epistatic relationships) in murine ES cells (Table S1). Consistent with previous data (Fazzio et al., 2008b), KD of Tip60 or p400, alone or in combinations with other factors, exhibited similar changes in gene expression (Figures 1A, S1A), while Brg1 and Mbd3 KD profiles were poorly correlated with these gene expression effects. Principal component analysis (PCA) confirmed that KD of Tip60 or p400 elicited the strongest changes in gene expression (Figure 1B).
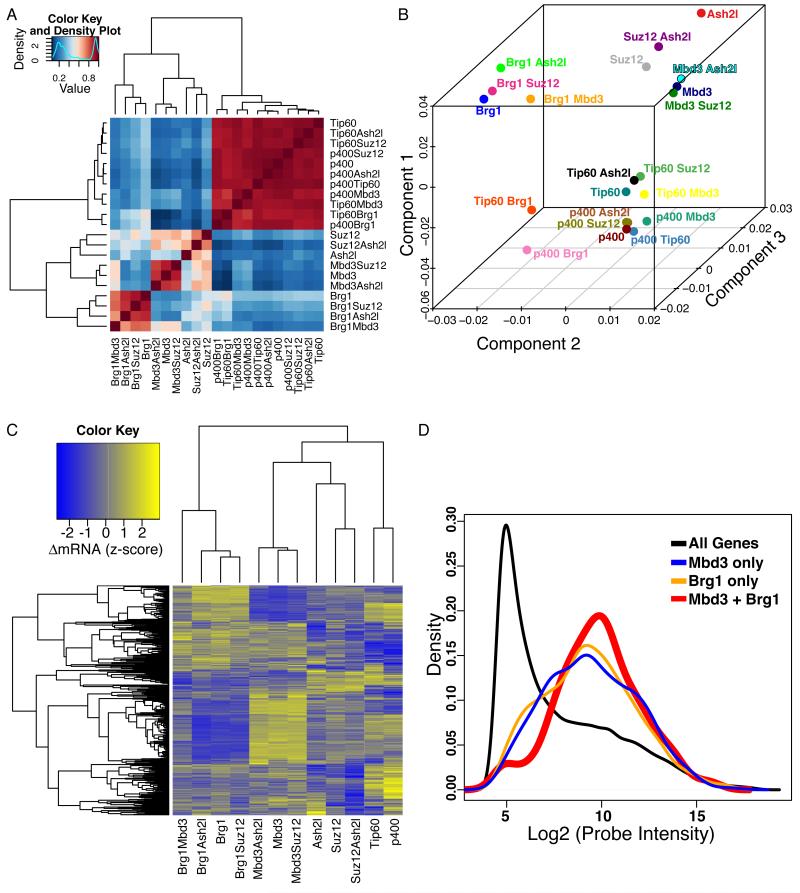
Figure 1. Antagonistic effects of Brg1 and Mbd3 on gene expression in mES cells.
(A) Gene expression data for single and double knockdowns of Brg1, Mbd3, Ash2l, Suz12, p400, and Tip60. Heatmap shows pairwise Pearson correlation coefficients for the 21 datasets. Four major clusters emerge, roughly corresponding to Brg1, Mbd3, Ash2l/Suz12, and Tip60/p400.
(B) Principal Component Analysis. Genes significantly misregulated (adjusted p-value < 0.01) in any data set from (A) were subjected to principal component analysis. Shown are individual data sets plotted along three most prominent principal components, which account for 87% of the total variance in gene expression.
(C) Mbd3 and Brg1 antagonistically regulate a common set of genes. Unsupervised clustering of genes misregulated in both the Mbd3 KD and Brg1 KD datasets (adjusted p-value < 0.05). Clustering was performed on data sets containing either Mbd3 KD or Brg1 KD (or both), as well as the Tip60, p400, Suz12 and Ash2l single KD datasets for contrast.
(D) Genes regulated by both Mbd3 and Brg1 tend to be moderately to highly expressed. Shown are mRNA abundance distributions (expressed as log2 of microarray probe intensity) for all genes, and for genes regulated by either or both Mbd3 and Brg1.
Surprisingly, Brg1 KD and Mbd3 KD affected the same principal component as one another but in opposite directions, suggesting that these two factors oppositely regulate expression of a common set of targets. Indeed, the overlap of genes misregulated upon Brg1 or Mbd3 KD was highly significant (p < 2.2e-16). As genes that were similarly up- or down-regulated in both Mbd3 KD and Brg1 KD cells were also nonspecifically affected by other KDs (Figure S1B), we therefore we focused on the large group of genes upregulated upon Mbd3 KD and downregulated upon Brg1 KD (Figure 1C). We validated several of these common Brg1/Mbd3 targets by quantitative RT-PCR (RTqPCR) (Figure S1C), and found that expression of these genes was also altered in independent Mbd3 or Brg1 KDs using non-overlapping esiRNAs (Figure S1D) and in KDs of additional subunits of either the Mbd3/NURD or BAF complexes (Figure S1E). We next searched for epistatic effects between Mbd3 and Brg1. Double KD of Brg1 and Mbd3 resulted in a more wild type mRNA profile than either single KD, indicating that these proteins play antagonistic roles at their common target genes (Figures 1C and S1C).
What features do Mbd3/Brg1 target genes share in common? In normal ES cells, Mbd3/Brg1 target genes are expressed at moderate to high levels (Figure 1D). Shared targets between these complexes were enriched for a number of functional categories, several of which relate to cellular adhesion and signaling (Table S2). Among the common targets of Brg1 and Mbd3, we noted genes encoding signaling molecules with important functions in ES cell self-renewal or differentiation, including Wnt3a and Tgfb1 (Figure S1C). These findings suggest that Mbd3/NURD and BAF complexes function in opposition to fine-tune the expression of a set of genes required for ES cell viability or self-renewal.
Mbd3 binds just downstream of the TSS
Are the joint Brg1/Mbd3-regulated genes direct targets of these complexes? While Brg1 has been mapped genome-wide in ES cells (Ho et al., 2009a), Mbd3 localization is currently unknown. We therefore carried out ChIP-Seq for Mbd3 in murine ES cells, finding that Mbd3 localized largely to promoters (Figures 2A-C, S2, S3A-B), with only weak localization to enhancers (Figures S4A-B). Genes upregulated upon Mbd3 KD were associated with somewhat higher levels of Mbd3 than were unaffected genes (Figure S3C), although, as is commonly observed with chromatin regulators (Fan et al., 2005; Jiang et al., 2011; Rando and Chang, 2009; Wu et al., 2011b), only a small subset of Mbd3-bound loci are transcriptionally affected by Mbd3 loss (Figure 2A).
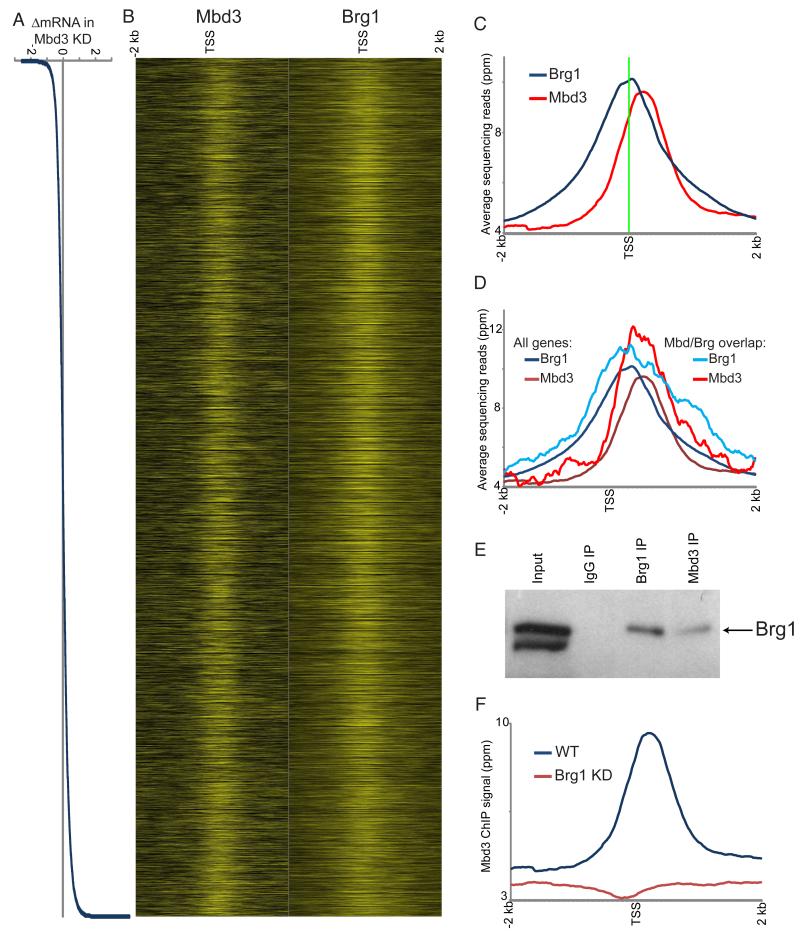
Figure 2. Genome-wide localization of Mbd3.
(A) Mbd3 KD effects on gene expression. Genes are sorted by change in mRNA abundance in Mbd3 KD, shown here in Log2.
(B) Mbd3 was mapped across the genome in ES cells by ChIP-Seq. Left panel: Mbd3 mapping data for 4 kb surrounding the transcriptional start sites (TSS) of 17,992 genes for which Mbd3KD gene expression data were available, with heatmap yellow saturating at 20 ppm normalized abundance. Right panel: published data for Brg1 (Ho et al., 2009a). In both panels, genes are sorted as in (A).
(C) Mbd3 binds downstream of Brg1. Averaged data for all genes in (B) are shown relative to the TSS.
(D) Mbd3 and Brg1 physically associate with antagonistically-regulated genes. Mbd3 and Brg1 data are shown for all genes as in C, or only for genes significantly repressed by Mbd3 and activated by Brg1.
(E) Mbd3 and Brg1 physically associate. Western blots for Brg1 following immunoprecipitation with the indicated antibodies.
(F) Brg1 is required for Mbd3 localization. Mbd3 was mapped genome-wide in Brg1 KD cells, and data for all genes are averaged and plotted as in (C).
Comparing Mbd3 maps to prior maps of Brg1 binding (Ho et al., 2009a), we found significant overlap between the binding sites of the two factors (Figure 2B, S4C). We validated Brg1 and Mbd3 binding at several common target promoters in ES cells by qPCR (Figure S3B). Interestingly, Mbd3 binding typically occurred ~100-200 bp downstream of the average peak of Brg1 (Figure 2C), suggesting that these complexes bind in an oriented fashion with respect to the direction of transcription (see below). Furthermore, both Mbd3 and Brg1 were significantly associated with their antagonistically-regulated target genes (Figure 2D, Figure S4D).
How are Brg1 and Mbd3 recruited to common promoters? We first asked whether Brg1 and Mbd3 physically interact in vivo using IP-Westerns. Immunoprecipitation using anti-Mbd3 antibody, but not control IgG, co-precipitated Brg1 (Figure 2E), indicating that these proteins interact in ES cells, as observed in K562 cells (Mahajan et al., 2005) and suggested by mass spectrometry of esBAF (Ho et al., 2009b). To further probe the functional relationship between Brg1 and Mbd3, we mapped Mbd3 genome-wide in Brg1 KD ES cells (Figure 2F), finding that Mbd3 localization was lost upon Brg1 KD. Together, these results indicate that Brg1 and Mbd3 directly associate with and regulate a common set of target genes in ES cells.
Mbd3 binding is enriched at bivalent genes
We next compared Mbd3 binding to previously published ChIP-Seq datasets in murine ES cells. Figure 3A shows the difference in Mbd3 binding levels between genes bound by a particular factor and those unbound, as defined in (Kim et al., 2010). Mbd3 binding was substantially higher at targets of the Polycomb proteins Ezh1, Suz12, Phc1, and Eed, as well as the histone modifications associated with most Polycomb targets in ES cells: H3K27me3 and H3K4me3 (Figures 3A-C). Furthermore, similar to Polycomb localization, Mbd3 localization was strongest at CpG-rich promoters and weakest at CpG-poor promoters (Figure 3D). Finally, Mbd3 was localized to all four Hox clusters, as previously observed for several PRC1 and PRC2 subunits (Boyer et al., 2006) (Figure S3A).
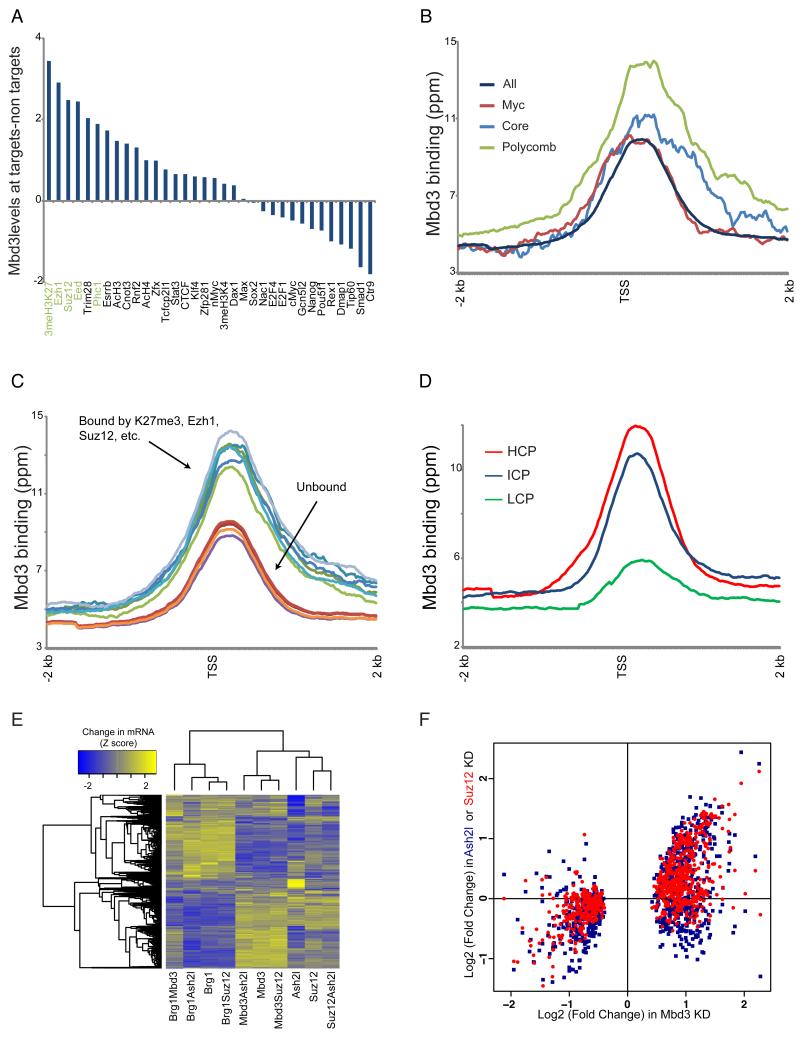
Figure 3. Mbd3 directly regulates Polycomb target genes.
(A) Mbd3 binding at Polycomb target genes. For 34 mapped factors with thresholded binding defined in (Kim et al., 2010), Mbd3 binding levels (mean ChIP signal for 1 kb centered on the +200 position) were calculated for bound and unbound subsets of genes. Genes annotated as unbound by all 34 factors were removed from this analysis. Factors are sorted according to the relative Mbd3 binding at factor targets relative to nontargets.
(B) Mbd3 binding at three ES cell “modules.” Average Mbd3 binding for all genes, and for the three modules defined in (Kim et al., 2010), is plotted relative to the TSS.
(C) Mbd3 binding at Polycomb targets and non-targets. For various Polycomb-related marks, averaged Mbd3 profile is shown for bound and unbound genes.
(D) Mbd3 binds preferentially to high-CpG promoters. Mbd3 binding data are averaged for high, intermediate, and low CpG (HCP, ICP, and LCP) promoters, as defined in (Weber et al., 2007).
(E) Mbd3 KD affects Polycomb targets. Clustered mRNA data for KD of Suz12, Ash2l, Mbd3, or Brg1, and assorted double knockdowns. Genes significantly misregulated in any of the included datasets are shown.
(F) Scatterplot of Mbd3 KD gene expression vs. Ash2l KD and Suz12 KD. Only genes showing significant misregulation in Mbd3 KD are shown.
These data suggest that Mbd3 may function with Polycomb and/or MLL/SET1 complexes to regulate gene expression. Interestingly, we noted that the gene expression profile of Mbd3 KD ES cells strongly correlated with that of Suz12 KD ES cells, and to a lesser extent with that of Ash2l KD ES cells (Figure 1A), indicating that both H3K4me3 and H3K27me3 affect many “bivalent” genes similarly, and that Mbd3 may also regulate these genes. Indeed, most genes significantly up- or down-regulated upon Mbd3 KD were changed in the same direction upon KD of either Suz12 or Ash2l (Figures 3E-F), although the effects of Suz12 or Ash2l on gene expression were generally less severe. Thus, Mbd3 is directly associated with a significant fraction of Polycomb-bound genes, and contributes to their repression.
To attempt to understand why only a subset of Mbd3-bound genes were affected transcriptionally by Mbd3 knockdown, we carried out a multivariate regression analysis (see Experimental Procedures). Linear combinations of genome-wide binding data for 38 factors or modifications were optimized to best explain the effects of Mbd3 KD on expression of Mbd3/Brg1 common targets (Figures S5A-B). The full model exhibited a high correlation (R = 0.40) between predicted and measured effects of Mbd3 KD on gene expression. The strongest predictors of Mbd3 effects on gene expression included Tet1 and 5-hydroxymethylcytosine localization (see below). Beyond these, strong predictors of Mbd3 function included H3K27me3 (individual R=0.29), Mbd3 (R=0.13), and transcription factors such as Esrrb, E2F1, and Klf4, whereas core pluripotency factors such as Oct4, Sox2, and Nanog contributed little (Figure S5B and data not shown). These data are consistent with proteomics studies showing that Esrrb physically associates with the BAF and Mbd3/NURD complexes (van den Berg et al., 2010), and suggest that genes regulated by this transcription factor are particularly sensitive to Mbd3 function.
Opposing chromatin remodeling functions of Mbd3 and Brg1 regulate recruitment of RNA Polymerase II
How do Mbd3 and Brg1 control gene expression at their common target genes? NURD complexes contain both a Swi2/Snf2-related ATPase (Mi-2) and a pair of histone deacetylase subunits (Hdac1 and Hdac2). We therefore analyzed nucleosome occupancy (measured by H3 abundance) and H4 acetylation at Mbd3/Brg1 targets by ChIP-qPCR. Genes repressed by Mbd3 exhibited decreased H3 occupancy and increased H4 acetylation upon Mbd3 KD (Figures 4A-B), suggesting that the Mbd3 complex represses target genes by deacetylating and stabilizing nucleosomes at promoters. Conversely, Brg1 knockdown resulted in variable effects on H4 acetylation, but consistent increases in H3 occupancy (Figures 4A-B). Thus, we find that Brg1 and Mbd3 antagonistically control nucleosome occupancy at target genes, with Brg1-mediated nucleosome loss associated with gene activation, and competing nucleosome stabilization and deacetylation by Mbd3 associated with gene repression.
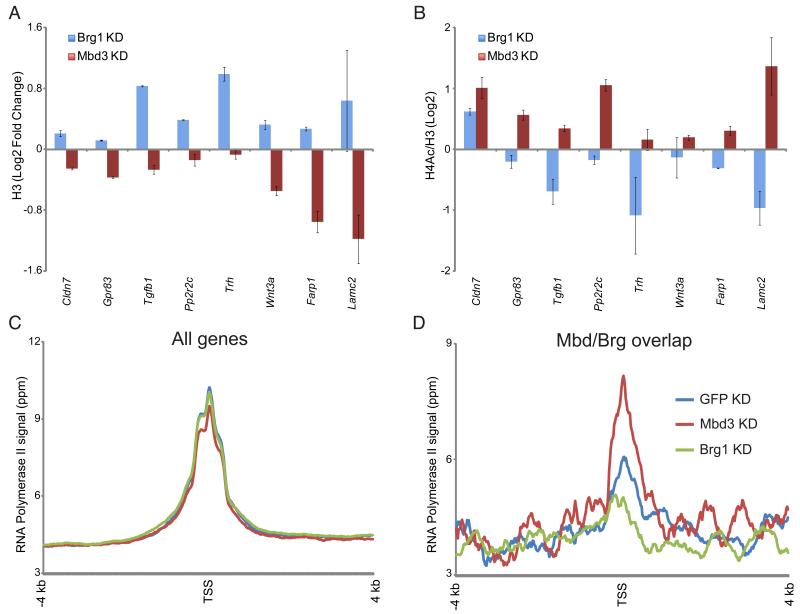
Figure 4. Mbd3 and Brg1 regulate chromatin structure and transcription initiation at target genes.
(A) H3 occupancy at Mbd3/Brg1 targets. H3 occupancy is plotted for control KD, Mbd3 KD, and Brg1 KD.
(B) H4 acetylation at Mbd3/Brg1 targets. As in (A), for H4 acetylation. Data are normalized to nucleosome occupancy (H3 ChIP).
(C) Genome-wide RNA Polymerase II (Pol II) mapping. Average TSS-aligned profiles of Pol II occupancy is shown for all genes for control, Brg1 KD, and Mbd3 KD cells.
(D) Pol II levels at the 5′ end correlate with mRNA abundance changes in Brg1 and Mbd3 KDs. As in (C), but only for genes repressed by Mbd3 and activated by Brg1.
How do these changes in chromatin architecture affect transcription? Many genes in eukaryotes are regulated by transcriptional pausing (Guertin et al., 2010). Given the localization of Mbd3 downstream of the TSS (Figure 2C), we considered the hypothesis that Mbd3 could play a role in enforcing a transcriptional pause. We therefore carried out whole-genome RNA Polymerase II (Pol II) mapping in EGFP KD, Mbd3 KD, and Brg1 KD ES cells. Overall, genome-wide Pol II localization was similar in all three KDs (Figure 4C), exhibiting in each case the expected promoter-proximal peaks corresponding to bidirectional transcription (Core et al., 2008; Seila et al., 2008). We next focused on the group of genes repressed by Mbd3 and activated by Brg1 (Figure 4D). If Mbd3 functioned solely to arrest Pol II, then mRNA increases in Mbd3 KD should be reflected in a loss of 5′ Pol II and an increase in downstream Pol II. Instead, we find that the 5′ peak of Pol II increases at these target genes in the Mbd3 KD, and decreases in the Brg1 KD, mirroring the effects of each KD on mRNA levels. Since the global Pol II profiles are nearly identical for all three KDs (Figure 4C), these data are consistent with a primary role for Mbd3 and Brg1 in regulating recruitment of Pol II specifically to target promoters, although an additional role in regulating Pol II pausing cannot be ruled out.
Knockdown of Tet1 impairs Mbd3 recruitment to target genes
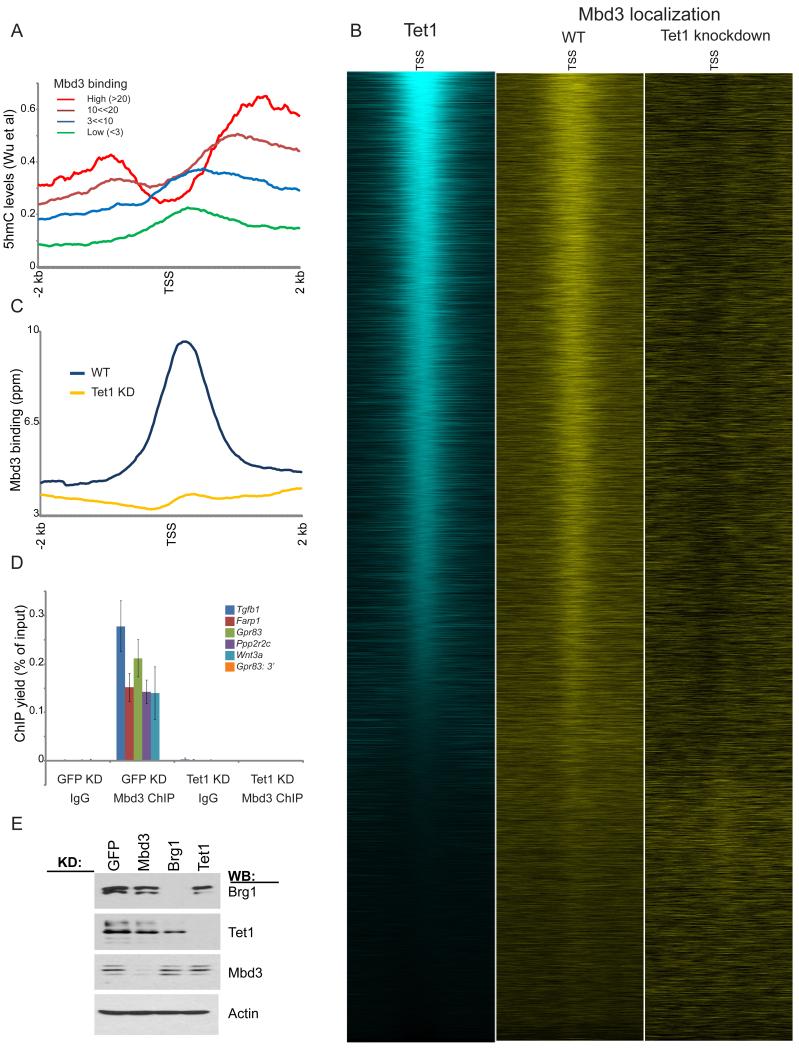
Despite being named Methyl Binding Domain based on its homology to the methyl binding domain from MeCP2, mammalian Mbd3 does not appear to associate with cytosine-methylated DNA (Hendrich and Bird, 1998; Zhang et al., 1999). Interestingly, we noted that one prominent phenotype of Mbd3 null or KD ES cells — upregulation of genes involved in trophectoderm differentiation — shares similarities to that of ES cells depleted of Tet1 (Ito et al., 2010; Koh et al., 2011), which converts 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC). We therefore speculated that Mbd3 might be regulated by 5hmC, rather than 5mC. To test this hypothesis, we compared Mbd3 localization to maps of Tet1 in murine ES cells (Wu et al., 2011b). Similar to Tet1 localization, Mbd3 was found at CpG-rich promoters, and correlated with Polycomb localization patterns (Figures 3C-D). High levels of 5hmC accompany Mbd3-associated genes in ES cells (Wu et al., 2011a) (Figure 5A), and Mbd3 and Tet1 localization patterns exhibited strong overlap (Figure 5B). Furthermore, genes repressed by Mbd3 were associated with significantly higher levels of Tet1 and 5hmC than genes unaffected by Mbd3 KD (Figures S5B-C).
Figure 5. Mbd3 associates with hydroxymethylated regions of the genome.
(A) Mbd3-bound genes are associated with high levels of hydroxymethylation. Average hydroxymethylation data from (Wu et al., 2011a) are averaged for genes with the indicated levels of Mbd3 binding.
(B) Mbd3 colocalizes with Tet1. Left panel: Tet1 (Wu et al., 2011b) mapping data are shown for all named genes, sorted by Tet1 binding levels. Middle and right panels: Mbd3 localization is shown for control and Tet1 KD ES cells.
(C) Mbd3 binding data for control and Tet1 KD are plotted as in Figures 2C-D.
(D) qPCR validation of Mbd3 binding at 6 selected target loci in GFP or Tet1 KD. Shown are mean +/− sem.
(E) Knockdowns of Tet1, Brg1, and Mbd3 do not affect protein levels of the other remaining factors. Knockdowns of the various factors were assayed by Western blot, as indicated.
See also Figure S5.
We therefore asked whether cytosine hydroxymethylation functions in Mbd3 localization. Since, in ES cells, Tet1 KD significantly decreases hydroxymethylation levels, we carried out genome-wide mapping of Mbd3 in Tet1 KD ES cells. Strikingly, Mbd3 was completely delocalized in Tet1 KD cells, despite normal levels of Mbd3 protein in this knockdown (Figures 5B-E). Thus, hydroxymethylation, or some other aspect of Tet1 function, is necessary for Mbd3 association with target genes.
Mbd3 binds 5hmC- but not 5mC-containing DNA in vitro
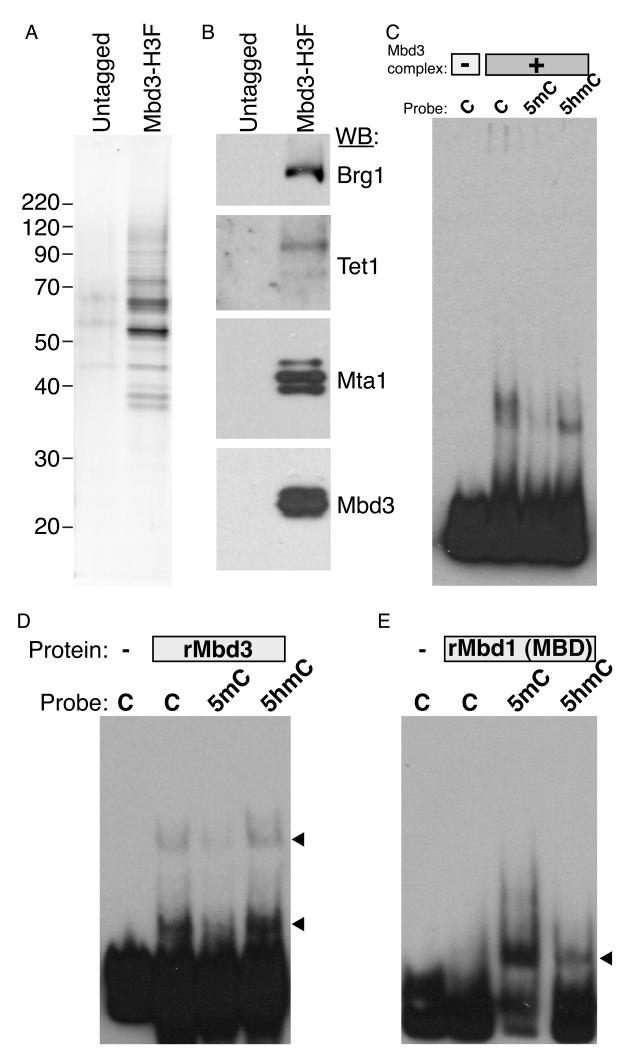
Our results suggest that hydroxymethylation is not solely an intermediate state in a cytosine demethylation pathway, but that it also plays a distinct role in gene regulation via Mbd3 recruitment. As Mbd3 does not specifically bind to 5mC in vitro, we considered the hypothesis that it might instead bind to 5hmC. To test this, we purified Mbd3/NURD complex from ES cells (Figures 6A, S6A-B), finding that this complex contained low levels of Tet1 (Figure 6B), identifying a direct physical link between Tet1 and Mbd3.
Figure 6. Mbd3 directly binds to hydroxymethylated DNA in vitro.
(A) Silver stain showing tandem affinity purification from untagged or Mbd3-6His-3XFLAG ES cells.
(B) Western Blot of purifications described in (A) for indicated proteins. Mta1 is a component of the Mbd3/NURD complex. Preliminary mass spec results also identified most other known NURD subunits in this purification (not shown).
(C) EMSA assay using Mbd3/NURD complex and DNA probes containing unmodified cytosine (C), methylated cytosine (5mC) or hydroxymethylated cytosine (5hmC).
(D-E) EMSA assay using recombinant mouse Mbd3 (D) or recombinant Mbd1 methyl-binding domain (E), and various modified probes as in (C).
See also Figure S6.
We carried out electrophoretic mobility shift assays (EMSAs) using probes with unmodified cytosine (C), 5mC, or 5hmC. As shown in Figure 6C, Mbd3/NURD complex had little effect on the methylated probe, but strongly shifted the 5hmC probe, whereas an untagged purification did not shift any of the probes (Figure S6C). Interestingly, Mbd3/NURD also shifted the unmodified “C” probe, but the shifted band was broader than the uniform shift of the 5hmC probe. These data suggest that Mbd3/NURD complex employs distinct modes of binding to unmodified and hydroxymethylated DNA.
To determine whether Mbd3/NURD’s specificity for 5hmC over 5mC was due to Mbd3, we tested recombinant Mbd3 for binding to the same probes. Consistent with the results obtained using the full complex, recombinant Mbd3 exhibited strongest binding to hydroxymethylated DNA probes, with comparable but slightly reduced binding to unmodified probe, and dramatically lower binding to methylated probe (Figures 6D, S6D). As a control, recombinant methyl binding domain from Mbd1 specifically shifted the methylated probe (Figure 6E), as expected (Hendrich and Bird, 1998; Ohki et al., 2001). Together, these data confirm the hypothesis that Mbd3 and Mbd3 complexes do not bind to DNA harboring 5mC, but can bind 5hmC-marked DNA in a manner qualitatively distinct from that of DNA containing unmodified cytosine.
Knockdown of Mbd3 or Brg1 affects global 5hmC levels
Given the physical interaction between Mbd3 and Tet1, we next asked if Mbd3 affected 5hmC in vivo. Compared to control cells, Tet1 KD ES cells exhibited reduced 5hmC levels by dot blotting and thin layer chromatography (Figures 7A-C), as previously reported (Koh et al., 2011; Wu et al., 2011a) (Ficz et al., 2011). To our surprise, both Mbd3 KD and Brg1 KD ES cells also exhibited strong reductions in global 5hmC levels as assayed by dot blotting (Figures 7A, S7). This effect of Mbd3 KD on bulk 5hmC levels was independently quantified by thin layer chromatography (Figures 7B-C). To extend these results to in vivo targets of Mbd3, we measured the enrichment of 5hmC (Song et al., 2011) at several Mbd3 target genes. We observed similar losses of 5hmC at six loci in Tet1 KD and in Mbd3 KD cells (Figure 7D). Finally, we compared 5hmC staining of individual cells by immunofluorescence in control cells to cells depleted of Tet1, Mbd3, or Brg1. Consistent with the above results, we observed noticeably reduced 5hmC levels in most Tet1, Mbd3 and Brg1 KD cells compared to control (Figure 7E). Therefore, we conclude that Mbd3 and Brg1 function globally to establish or maintain normal levels of 5hmC in ES cells.
Figure 7. Mbd3 is required for global hydroxymethylation in vivo.
(A) Dot blots of 5hmC. Top panel shows positive (5hmC) and negative (5mC) controls for antibody specificity. Bottom panels show dilution series of genomic DNA isolated from the indicated knockdown ES cells. Mbd3 and Brg1 KDs have similar effects to Tet1KD on bulk 5hmC levels.
(B) Thin layer chromatography of radioactively end-labeled bases from MspI-digested genomic DNA (Ficz et al., 2011) from the indicated knockdowns.
(C) Quantitation of 5hmC levels (normalized to levels of T) measured as in (B) for 5 independent replicate experiments. Columns show mean +/− sem.
(D) Tet1 and Mbd3 knockdowns have similar effects on hydroxymethylation of target gene promoters. Hydroxymethylated DNA was isolated by capture of biotin-glucosylated 5hmC-containing DNA fragments (Song et al., 2011), and fold enrichment over input was assessed by qPCR at the indicated loci and expressed as fold change relative to 5hmC levels in control (EGFP) KD cells. Data represent mean +/− sem.
(E) Immunofluorescence imaging of Mbd3 and 5hmC. Immunofluorescence images are pseudocolored blue (DAPI), green (Mbd3), and red (5hmC) for the indicated KDs – top panel shows 5hmC data only, bottom panel shows all 3 colors.
See also Figure S7.
DISCUSSION
The epigenetic control of stem cell pluripotency and self-renewal has been subject to intense scrutiny in recent years. Extensive attention has been given to specific coactivators/corepressors such as Polycomb Repressive Complexes, whereas other important chromatin regulators have received less attention. Here, we focused on Brg1 and Mbd3, components of coactivator and corepressor complexes, the mechanisms by which they antagonistically regulate a group of common target genes, and their role in the biology of 5hmC.
Antagonistic control of gene expression by BAF and Mbd3/NURD complexes
Several hundred genes were antagonistically regulated by Brg1 and Mbd3, showing increased expression in Mbd3KD, decreased expression in Brg1KD, and wild type-like expression in double KDs. These results are of great interest in the context of the “bivalent” chromatin architecture seen in ES cells, in which the activation-associated histone mark H3K4me3 and the repression-associated H3K27me3 mark co-occur at a large number of “master regulator” genes involved in cell fate decisions (Bernstein et al., 2006). However, reduction of H3K4me3 in ES cells by KD of Ash2l (Fazzio et al., 2008b) or Dpy-30 (Jiang et al., 2011) does not cause self-renewal defects, nor does H3K4me3 appear to be important for expression of most genes in ES cells (Jiang et al., 2011) (Table S1). Indeed, our results with Ash2l and Suz12 knockdowns show that reduction of either H3K4me3 or H3K27me3 results, “paradoxically”, in similar effects on gene expression (Figures 3E-F). Conversely, we show here that Mbd3 and Brg1 exhibit opposing functional effects on RNA Polymerase II recruitment and gene expression at several hundred genes. Together, these data suggest that the juxtaposition of opposing histone modifications or chromatin regulators near the regulatory sequences of shared target genes is a common regulatory strategy in ES cells.
Mbd3/NURD plays a central role in 5-hydroxymethylcytosine biology
The inability of Mbd3 to bind specifically to 5mC (Hendrich and Bird, 1998; Zhang et al., 1999) raises the question of what function the “methyl binding domain” of this protein serves. We noted that ES cells depleted of Tet1, which catalyzes the hydroxylation of 5mC to form 5hmC, exhibit phenotypes similar to those of Mbd3 KD ES cells (Kaji et al., 2006; Koh et al., 2011; Zhu et al., 2009). Intriguingly, one of the few sequence differences in the methyl binding domain between human/mouse Mbd3 and remaining Mbd family members is a substitution of a phenylalanine for a tyrosine (numbered Y34/F34 in human Mbd1/Mbd3). This sequence change is largely responsible for Mbd3’s lack of 5mC binding in vitro (Saito and Ishikawa, 2002). In the structure of Mbd1 bound to methylated DNA, Y34 is located immediately adjacent to the 5-methyl group of 5mC (Ohki et al., 2001) (Figure S6E). Thus, loss of the hydroxyl group (Y34F) at this position in Mbd3 could almost perfectly compensate for the additional hydroxyl group in 5hmC relative to 5mC, thus allowing direct binding of Mbd3 to this modification in a manner structurally analogous to the binding of remaining Mbd members to 5mC.
Consistent with this hypothesis, we found that Mbd3 localization patterns are similar to Tet1 localization patterns, and Mbd3 is enriched at genes with high levels of hydroxymethylation (Wu et al., 2011b). Like Tet1, Mbd3 is associated largely with CpG-rich promoters bound by Polycomb, and is required for normal expression of many of these targets. Mbd3 localization requires Tet1, suggesting that hydroxymethylation plays a role in Mbd3 recruitment in vivo. Finally, we found that Mbd3 preferentially binds to 5hmC-containing probes relative to 5mC-containing probes. Thus, our data are most consistent with a model in which Tet1-catalyzed hydroxymethylation serves to recruit Mbd3/NURD complex, and thus Mbd3/NURD may be an effector that mediates some of the effects of hydroxymethylation on gene expression.
Interestingly, we found that purified Mbd3/NURD also bound to unmodified probe in vitro, but that this binding was qualitatively distinct from binding to the 5hmC probe: the shifted band was more discrete and of higher mobility when the probe was hydroxymethylated. One potential explanation for this could be the binding location of the complex on the probe, as probe bound at either end should have higher mobility than internally bound probe. At present it is unclear to us why Tet1KD, which results in incomplete loss of 5hmC in vivo, almost completely abolishes Mbd3 localization. Given the increase in 5mC observed in Tet1KD cells at loci that lose 5hmC (Ficz et al., 2011; Wu et al., 2011a), and our observation that 5mC inhibits Mbd3 binding, we speculate that genomic regions with 5mC intermixed with 5hmC might be unfavorable for Mbd3 binding. Future studies will dissect the details of how Mbd3/NURD differentially interacts with unmodified and hydroxymethylated DNA, and with hemi-modified or symmetrically modified DNA.
Finally, we found that KD of either Mbd3 or Brg1 results in reduction of bulk levels of 5hmC in vivo. This could occur via several possible mechanisms: as two examples, Mbd3 could bind to a region of hydroxymethylated DNA and recruit Tet enzymes to hydroxylate adjacent methylcytosines, or Mbd3 could bind to hydroxymethylated loci and protect them from further steps in a demethylation pathway. Either way, the extensive interdependency between these factors – Tet1 is necessary for Mbd3 localization, Mbd3 is necessary for cytosine hydroxymethylation – is reminiscent of other co-dependencies in chromatin pathways. For instance, Polycomb is required for H2A.Z incorporation at promoters of developmental genes in ES cells, and H2A.Z is in turn required for Polycomb binding (Creyghton et al., 2008). Dissecting the detailed mechanistic basis for such interdependencies is a challenging goal for studies on chromatin regulation.
Together, these data describe the first downstream effector that regulates expression of 5hmC-marked genes, suggesting that 5hmC plays a role in gene regulation beyond serving simply as an intermediate in a demethylation pathway. It will be of great interest in future studies to determine whether Mbd3 also plays a role in 5hmC biology in other contexts such as early embryos (Inoue and Zhang, 2011; Iqbal et al., 2011), imprinting (Reese et al., 2007), or neurons (Kriaucionis and Heintz, 2009). Finally, our discovery of an interdependent regulatory network consisting of 5hmC and two antagonistic chromatin regulators suggests that control of gene expression by opposing chromatin regulators is a common regulatory strategy in pluripotent ES cells.
EXPERIMENTAL PROCEDURES
RNAi-Mediated Knockdown
RNA interference using endoribonuclease III digested siRNAs (esiRNAs) was performed as described previously (Fazzio et al., 2008b), using E14 mouse ES cells. All KDs were performed for 48 hours to achieve effective KD, but avoid some indirect effects of prolonged loss of chromatin regulators. Stable Mbd3 KD lines were made by infection of ES cells with lentiviral shRNA vectors from the TRC library (Open Biosystems).
Expression Profiling
Array hybridizations were performed at the Sandler Asthma Basic Research (SABRE) Center Functional Genomics Core Facility as described previously (Fazzio et al., 2008a). For each single and double KD, a linear model was fit to the comparison to estimate the mean log2 (fold change) in 2 biological replicates and to calculate a moderated t-statistic, B statistic, false discovery rate and p-value for each probe. Adjusted p-values were produced as decribed previously (Holm, 1979). All procedures were carried out using functions in the R package limma (Gentleman et al., 2004; Smyth, 2004) or made4 (Culhane et al., 2005) in R/Bioconductor. Enrichment of Gene Ontology terms was performed with DAVID 6.7 (Dennis et al., 2003). mRNA data are available at GEO, accession GSE31008.
ChIP-Seq
Chromatin immunoprecipitation and deep sequencing library construction were performed using minor modifications of established protocols (Barski et al., 2009; Lee et al., 2006) (Supplementary Experimental Procedures). Data are available at GEO, accession GSE31690.
Multivariate linear regression model
Multivariate linear regression was used to predict factors that regulate Mbd3/Brg1-target genes. 38 ChIP-seq datasets were divided into 40 bp bins surrounding the TSS of each gene that changed oppositely upon Mbd3 KD and Brg1 KD, with another bin representing the gene body. Binding of each of the 38 factors was compared to gene expression changes (see Supplementary Experimental Procedures for details).
Mbd3/NURD Purification
An ES cell line with a C-terminal 6-Histidine-3X FLAG tag placed just upstream of the Mbd3 stop codon was constructed as described in the Supplementary Experimental Procedures. Mbd3/NURD complex was purified from ~ 4 × 108 of these cells by sequential affinity purification steps using FLAG-M2 Agarose followed by TALON Agarose beads.
EMSA Assays
EMSA assays were performed on 5% polyacrylamide 0.5X TBE gels. Biotinylated probes corresponding to +529 - +628 of the Tgfb1 gene were made by PCR using Phusion polymerase and NTPs containing either unmodified, methylated, or hydroxymethylated dCTP. The PCR primer sequences for probe construction were: biotin-TGCCTCTTGAGTCCCTCGCATC and AGTGGGTGTTCTTAAATAGGGGAGCT. Binding reactions were performed in 1X Binding Buffer (LightShift Chemiluminescent EMSA Kit; Thermo Scientific) with 100 mM KCl, 8% glycerol, 0.02% NP-40, 5 mM MgCl2, 0.85 μg BSA, 30 ng yeast genomic DNA, 1 mM ATP, and with or without 1 μl purified Mbd3/NURD or recombinant Mbd3, as indicated. After electrophoresis, samples were transferred to charged Nylon membrane and probed with streptavidin-HRP according to the instructions in the EMSA kit.
Dot Blotting
2-fold serial dilutions of genomic DNA were denatured in 0.4 M NaOH/10mM EDTA at 95C for 10 min, then added to an equal volume of cold 2M ammonium acetate (pH 7.0). Denatured DNA samples were spotted onto nitrocellulose. The membrane was washed with 2XSSC buffer and then UV cross-linked. Membrane was blocked with 5% non-fat milk for 1 hr and incubated with rabbit anti-5hmC, detected by HRP-conjugated secondary antibody and enhanced chemiluminescence.
Immunofluorescence
Cells were resuspended in PBS, cytospun onto glass slides, fixed in 4% paraformaldehyde for 10 min at room temperature, washed twice with PBS, permeabilized in 2% TritonX-100-PBS for 10 min, and blocked with 5% FBS and 0.1% TritonX-100 in PBS for 15 min at room temperature. Cells were incubated with primary antibody diluted 1:500 with 5% FBS and 0.1% Triton X-100 in PBS for 30 minutes at room temperature, washed 3 × 5 min with PBS, and incubated for 30 min at room temperature with secondary antibody diluted 1:200 with 5% FBS and 0.1% Triton X-100 in PBS. DAPI was added in mounting medium (Vectashield, H1500).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Rando and Fazzio labs for critical reading of the manuscript, J. Benanti and P. Kaufman for technical advice, C. Li for assistance with TLC, and J.M. Bishop, in whose lab some of these experiments were performed. We thank C. He and C. Song for the generous gift of reagents for isolation of 5hmC. TGF is supported in part by grant CA140854 from NCI and is a Pew Scholar in the Biomedical Sciences. OJR is supported in part by grant GM088618 from NIGMS, and by the Harold and Leila Mathers Charitable Foundation. ZW is supported by NSF grant DBI-0850008. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. OY, TGF, and OJR designed all experiments. OY and TGF carried out mapping experiments. OY performed 5hmC quantitation, TGF carried out gene expression and gel shift experiments, and LE and PBC performed experiments in Figure 4A-B. TGF analyzed gene expression data. OJR, XD, JHH, RL, and ZW analyzed localization data. OJR, OY, and TGF wrote the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, Zhao K. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–1751. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culhane AC, Thioulouse J, Perriere G, Higgins DG. MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics. 2005;21:2789–2790. doi: 10.1093/bioinformatics/bti394. [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. Chromatin regulation Tip(60)s the balance in embryonic stem cell self-renewal. Cell Cycle. 2008a;7:3302–3306. doi: 10.4161/cc.7.21.6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008b;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Panning B. Control of embryonic stem cell identity by nucleosome remodeling enzymes. Curr Opin Genet Dev. 2010;20:500–504. doi: 10.1016/j.gde.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011 doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin MJ, Petesch SJ, Zobeck KL, Min IM, Lis JT. Drosophila heat shock system as a general model to investigate transcriptional regulation. Cold Spring Harb Symp Quant Biol. 2010;75:1–9. doi: 10.1101/sqb.2010.75.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009a;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009b;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Inoue A, Zhang Y. Replication-Dependent Loss of 5-Hydroxymethylcytosine in Mouse Preimplantation Embryos. Science. 2011 doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES Cell-Fate Specification by Regulation of H3K4 Methylation within Bivalent Domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- Keenen B, de la Serna IL. Chromatin remodeling in embryonic stem cells: regulating the balance between pluripotency and differentiation. J Cell Physiol. 2009;219:1–7. doi: 10.1002/jcp.21654. [DOI] [PubMed] [Google Scholar]
- Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan MC, Narlikar GJ, Boyapaty G, Kingston RE, Weissman SM. Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodeling complex at the beta-globin locus control region. Proc Natl Acad Sci U S A. 2005;102:15012–15017. doi: 10.1073/pnas.0507596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. Open conformation chromatin and pluripotency. Genes Dev. 2007;21:2671–2676. doi: 10.1101/gad.1615707. [DOI] [PubMed] [Google Scholar]
- Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, Shirakawa M. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese KJ, Lin S, Verona RI, Schultz RM, Bartolomei MS. Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3. PLoS Genet. 2007;3:e137. doi: 10.1371/journal.pgen.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Ishikawa F. The mCpG-binding domain of human MBD3 does not bind to mCpG but interacts with NuRD/Mi2 components HDAC1 and MTA2. J Biol Chem. 2002;277:35434–35439. doi: 10.1074/jbc.M203455200. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen M. Functional analysis of mouse Polycomb group genes. Cell Mol Life Sci. 1998;54:71–79. doi: 10.1007/s000180050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011a;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun Y. Eve, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011b doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, Piao Y, Aiba K, Matoba R, Wang W, Ko MS. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Fang J, Li Y, Zhang J. Mbd3, a component of NuRD/Mi-2 complex, helps maintain pluripotency of mouse embryonic stem cells by repressing trophectoderm differentiation. PLoS One. 2009;4:e7684. doi: 10.1371/journal.pone.0007684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.