Abstract
Granzymes (Grs) were discovered just over a quarter century ago. They are produced by cytotoxic T cells and natural killer cells and are released upon interaction with target cells. Intensive biochemical, genetic, and biological studies have been performed in order to study their roles in immunity and inflammation. This review summarizes research on the family of Grs.
Keywords: granzymes, apoptosis, cytotoxic T cells, NK cells
Facts
Granzymes (Grs) are a family of closely related serine proteases expressed in cytotoxic T lymphocytes and natural killer cells.
Grs are packaged in cytoplasmic granules that are exocytosed toward a bound target cell.
Perforin facilitates the transfer of Grs into the target cell cytoplasm.
GrB cleaves substrates at aspartic acid allowing it to activate caspase-3 and initiate apoptosis.
GrA is a tryptase and acts by disruption of the ER-associated SET complex.
Grs cleave a variety of intracellular and extracellular substrates.
Open Questions
What are the implications of species-specific differences in Gr substrate specificities?
Why do mice have so many Grs relative to humans?
Why does Bcl-2 block GrB in vitro killing but not CTL-induced death?
How do tumors inhibit CTL killing?
What are the implications of cleavage of extracellular substrates to disease pathogenesis?
Is GrA inflammatory or lethal or both?
Twenty-five years ago Jürg Tschopp's group1 published a blockbuster paper in cell on the purification and characterization of granzymes (Grs). These are a family of serine proteases found in the cytolytic granules of cytotoxic T lymphocytes and natural killer cells. Jürg's group was able to purify eight members of the family, investigate their substrate specificities, and determine partial amino acid sequences (see Box 1). Two of them corresponded to proteins encoded by cytotoxic cell protease (CCP) genes cloned by the Bleackley laboratory.2 Over the next few years, there was lively debate concerning the naming of these molecules. When Bleackley lab members agreed to call them Grs, it was clear who was the winner. At the time, the conventional wisdom was that another granule protein perforin (a.k.a. cytolysin) was the sole mediator of target cell destruction. The discovery of Grs set the stage for an exciting time in understanding cell-mediated cytotoxicity. Since then, over 3000 papers have been published on Grs. This review is dedicated to the memory of Jürg, not only a trail-blazer, but also a friend.
Table 1. Mouse models of granzymes.
| Mouse | Phenotype and apoptotic response | References |
|---|---|---|
| grB−/−(cluster) | Rapid DNA fragmentation and caspase activation affected | 42,43,44 |
| Moderate susceptibility to MCMV and ectromelia virus | 45,46 | |
| Reduced MHC class I-dependent GVHD | 47 | |
| grB−/− (targeted) | Reduced clearance of allogeneic tumors | 48 |
| Control of gammaherpervirus latent infection | 104 | |
| grA−/− | Apoptotic response not affected | 71 |
| Moderate susceptibility to ectromelia virus | 72 | |
| Control of herpes virus spread to peripheral nervous system | 73 | |
| grA−/− x grB−/− | Highly susceptible to ectromelia virus | 46 |
| Defective clearance of Trypanosoma cruzi | 105 | |
| Control of tumor growth | 106 | |
| grM−/− | Apoptotic response not affected | 85 |
| Mild susceptibility to MCMV |
The Cytotoxic Granule Components
Cytotoxic lymphocytes contain specialized cellular compartments, the secretory lysosomes, which are membrane-bound organelles equipped with cytolytic proteins.3 The primary cytolytic molecules, delivered to target cells upon contact with cytotoxic cells, are perforin and Grs. Currently, the mechanism of perforin-facilitated entry of Grs into target cells remains unresolved. Models proposed include the liberation of Grs from endosomal compartments of the target cells to the cytosol, or a Ca2+-driven plasma membrane repair process initiated by perforin damage.4
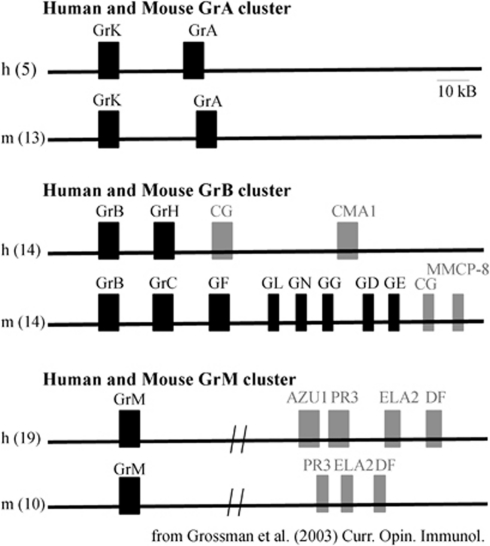
Grs are serine proteases, characterized by a His-Asp-Ser catalytic triad.5 In humans, there are 5 Grs (A, B, H, K, and M) while in mice there are 10 functional Grs (A, B, C, D, E, F, G, K, M and N). The genes are found in clusters among three different chromosomes, the GrA cluster, GrB cluster, and GrM cluster (Figure 1). The GrA cluster, at human chromosome 5 and murine chromosome 5, includes GrA and GrK. Grs A and K are tryptases, cleaving after basic amino acid residues (designated the P1 residue).6, 7
Figure 1.
Genomic organization of serine proteases. Gr genes, found primarily in lymphoid-derived cytotoxic cells are indicated in black. (Mouse GrL is a pseudogene.) Myeloid-derived serine proteases are indicated in grey
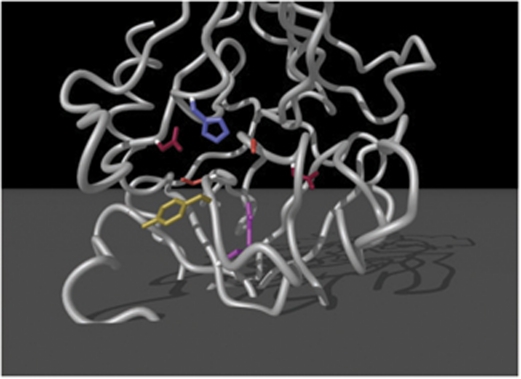
The GrB cluster is found at chromosome 14 of both human and mice but is composed of different enzymes. In humans, the cluster consists of GrB, GrH, cathepsin G (CG), and mast cell chymase-1 (CMA1). In mice, the cluster contains GrB, C, F, N, G, D, E, CG, and CMA1. GrB is the most extensively studied Gr, characterized by the unusual capacity to cleave substrates at aspartic acid residues that is dependent on the presence of an arginine residue within the binding pocket (Figure 2).8 Grs H and GrC–G, are believed to be chymases, and cleave synthetic substrates at hydrophobic residues.5 These proteins are also highly similar at the amino acid level, from 57–61% identity using BLAST alignment analysis.
Figure 2.
The binding pocket structure of GrB. Residues that line the binding pocket are shown in red (Asp), blue (His), orange (Ser), yellow (Tyr), and pink (Arg). The arginine residue at 226 is critical for cleavage of substrates at aspartic acid residues
The GrM cluster is found on chromosome 19 in human and chromosome 10 in mice, and consists of GrM and neutrophil proteases.9 GrM is a metase, preferentially cleaving substrates at methionine, or leucine residues.10 However, human and mouse GrM display divergent substrate specificities.11
Mechanisms of Cytotoxicity and Physiological Roles of Grs
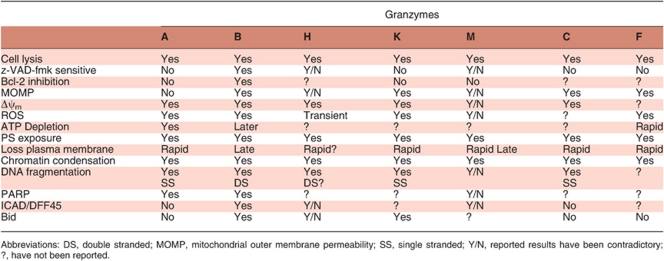
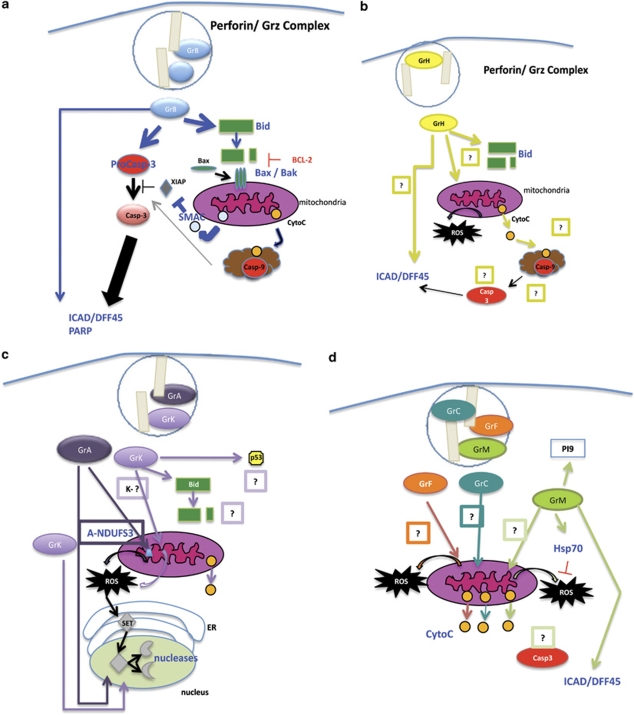
Our understanding of the function of the majority of Grs is very limited. The exception is GrB, which has been established as a potent pro-apoptotic molecule as a result of extensive studies using biochemical techniques and purified or recombinant proteins. The cytotoxic mechanisms of remaining Grs are still active areas of research. Table 1 describes the phenotype of Gr-deficient mice in their ability to induce target cell death and respond to immunological challenges. Table 2 summarizes the features displayed by target cells treated in vitro with purified/ recombinant Grs, and the current models of cytotoxic-mediated cell death are shown in Figures 3(a–d).
Table 2. Summary of granzyme death pathways.
Figure 3.
(a) GrB initiates caspase and mitochondrial cell death pathways, as well as directly processing a number of other pro-apoptotic molecules. (b) GrH cell death induces cell death by targeting the mitochondria and causes DNA through mechanisms that have not been clearly resolved. (c) GrA Cluster – GrA and GrK induce ROS production and the translocation of the SET complex from the ER to the nucleus. DNases from the complex initiate DNA damage. GrK may also target Bid and p53. (d) Grs C, F, and M – GrC, GrF, and GrM induce MOMP through unknown mechanisms. GrM also targets Hsp70 to promote ROS generation and may directly process ICAD/DFF45. GrM has been shown to proteolytically inactivate the GrB inhibitor PI9
Granzyme B
Several groups identified GrB in cytotoxic lymphocytes during the mid-1980s.1, 2, 12, 13, 14 It was also known variously as CCP1 and cytotoxic T lymphocyte associated-1 (CTLA1) before GrB was widely adopted. Initial studies indicated that GrB-induced in vitro cell lysis (51Cr-release), rapid DNA fragmentation, and chromatin condensation.15, 16, 17 Indeed, identification of the DNA fragmenting activity (fragmentin) of GrB was an important piece of the puzzle in understanding the mechanism of CTL-induced death. Later experiments revealed that purified GrB and perforin or adenovirus treatment induced many classical features of apoptosis, such as membrane blebbing, phosphatidylserine exposure, release of cytochrome C, dissipation of Δψm, generation of ROS, and plasma membrane permeability to vital dyes at early timepoints.18, 19
A pivotal piece of information that led to the discovery of the cytotoxic mechanism of GrB was its ability to cleave aspartic acid residues.8 This cleavage specificity is unique among eukaryotic serine proteases. Up to that point, only caspases were known to have this unusual specificity. Subsequently, GrB was found to proteolytically activate a number of caspases in vitro (caspases-2, -3, -6, -7, -8, and -9) and in vivo (caspases-3, -7, -8, and -10).20, 21, 22, 23, 24, 25 These observations led to a model wherein the activation of caspase-3 by GrB was largely responsible for many of the features induced by CTL killing.20, 22, 26, 27 Further studies revealed that GrB could also promote cell death via the mitochondrial pathway.18, 28 These experiments demonstrated that human GrB proteolytically activated Bid at aspartate75, resulting in its translocation to the mitochondrial outer membrane, and the facilitation of oligomerization of Bax/Bak molecules.29, 30, 31 It was initially thought that this resulted in the release of cytochrome C and apoptosome formation. This proved not to be the case, but rather involved inhibitor of apoptosis proteins (IAPs) that are potent antagonists of caspase activity. The release of a mitochondrial intermembrane molecule, SMAC/DIABLO, was later shown to relieve XIAP inhibition of autocatalytic caspase-3 maturation, allowing full activation of caspase-3.32, 33 Thus, in humans rapid GrB-mediated apoptosis is dependent on the integration of GrB proteolytic processing of caspase-3 and mitochondrial outer membrane permeabilization (MOMP). In mice, however, GrB may not efficiently process Bid, and it is speculated that GrB-mediated cell death is due primarily through direct caspase activation.20, 34 Thus, in humans GrB can activate caspases and disrupt mitochondria through Bid, whereas in mice both cell death pathways act via caspases. This would result in different sensitivities to caspase inhibitors between species.
However, in the absence of caspase activation human GrB can still induce cell death through the mitochondria, although largely in the absence of DNA fragmentation.18, 19 Treatment with GrB results in damage to mitochondria detected by the dissipation of Δψm, even in the absence of Bid, Bax, or Bak.35 This implies that unidentified GrB substrates may also be present at the mitochondria. Additional cellular GrB substrates are: ICAD/DFF45, PARP, DNA-PK, α-tubulin, ROCK-1, lamin B, and NUMA.21, 36, 37, 38, 39, 40, 41 The physiological significance of these other GrB substrates in normal killing is unclear. If caspases are blocked, the major short-term consequences of GrB-mediated apoptosis are blocked. This suggests that GrB cleavage of caspases is the pivotal event. However, in certain cases when the caspase pathway is blocked, in for example, virus infection of overexpression of IAPs, then the cleavage of these other proteins could become very important in mediating cell death.
GrB deficiency in mouse models confirmed that the in vitro role of GrB is rapid target cell killing, DNA fragmentation, and caspase activation.42, 43, 44 In vivo, grB−/− knockout mice have shown enhanced susceptibility to mouse cytomegalovirus (MCMV), ectromelia virus, reduced graft versus host disease, and a reduced ability to clear allogeneic tumors.45, 46, 47, 48 However, in various other models, GrB-deficient mice remain competent at viral and tumor clearance, which is likely a reflection of Gr redundancy in these animals.49
Granzyme H
GrH was identified through screening cDNA libraries derived from cytotoxic cells.50, 51 To date, two groups have assessed the cytotoxic potential and mechanism of action of purified recombinant GrH.52, 53 Interestingly, some of their findings were highly disparate. Both groups observed that GrH cytotoxicity resulted in phosphatidylserine exposure, some loss of plasma membrane integrity, chromatin condensation, DNA damage (although they observed different patterns), and mitochondrial damage. However, Fellows et al. proposed a novel pathway that was independent of Bid or ICAD/DFF45 cleavage, caspase activity, did not induce cytochrome C release or DNA laddering, and transiently induced ROS production. In contrast, Zuzen Fan's group observed direct GrH proteolysis of ICAD/DFF45, some sensitivity to zVAD-fmk and the activation of caspase-3, DNA laddering, cytochrome C release, and mitochondrial swelling. They proposed that GrH cytotoxicity was likely mediated by GrH-dependent proteolysis of Bid.
Studies aimed at finding the physiological relevance of GrH have been challenging. As there is no clear GrH homologue expressed in mice, knockout studies cannot be performed. However, cytotoxic studies using a chymase inhibitor with rat NK granules (which express a similar chymase to GrH) diminished YAC-1 cell death compared with untreated samples.54
Alternatively, GrH may have other anti-viral functions, either targeting viral proteins or as an accessory role to other cytotoxic molecules such as GrB. Andrade et al.,55 have reported GrH-proteolytic inactivation of a potent adenovirus-derived GrB inhibitor, LK-100K. In this context, GrH relieved the inhibition of GrB by LK-100K in adenovirus-infected cells, allowing GrB to regain the ability to induce target cell apoptosis. Andrade's group has also reported anti-viral functions of GrH toward an adenovirus DNA replication protein, and a hepatitis-C translation factor La.55, 56
Granzymes C–G
These enzymes are part of the mouse GrB gene cluster, and very little is known about their function. Most of these Grs were identified through N-terminal sequencing of proteins acquired from purified granules, or screening cDNA libraries derived from lymphokine-activated killer cells and CTLs.2, 13, 57, 58
GrC, located just downstream of GrB, is similar to human GrH and has been reported to induce cell death upon its delivery with perforin to target cells.59 YAC-1 exposure to GrC resulted in phosphatidylserine exposure, plasma cell membrane permeability, single-stranded DNA breaks, chromatin condensation, mitochondrial swelling, loss of Δψm, and the release of cytochrome C. GrC did not activate caspases, nor cause direct proteolytic cleavage of ICAD or Bid. Mitochondrial swelling induced by GrC was not sensitive to the classical PTP inhibitor cyclosporine A. Therefore, mechanisms responsible for biochemical changes leading to GrC-induced cell death remain unknown.
Recently, GrF has been reported to induce a novel mechanism of cell death.60 Treatment of YAC-1 target cells with GrF and adenovirus, perforin, or streptolysin O resulted in a form of cell death that was characterized by phosphatidylserine exposure, plasma membrane permeability, mitochondrial swelling and damage, and partial chromatin condensation. Cell death was unimpeded in the presence of caspase inhibitors, and the cells did not show signs of caspase activation, despite the release of cytochrome C from the mitochondria. The investigators reported strong ROS production, and loss of ATP production, implying that GrF-induced mitochondrial dysfunction.
Granzyme A
GrA was first identified as a CTL-specific cDNA clone that was predicted to encode a protease named Hanukah factor.61 Early studies using purified native GrA indicated a role for GrA in cell-mediated cytotoxicity, measured by 51Cr-release, and DNA fragmentation assays.16 Additionally, GrA-stably transfected rat mast cells (RBL-2H3) provided evidence that GrA could induce DNA fragmentation in target cells, although the greatest effects were observed when the cells were co-transfected with GrB.15
Later studies, with the use of recombinant GrA and purified perforin, led to observations that GrA-mediated cell death was characterized by phosphatidylserine exposure, chromatin condensation, single-stranded DNA nicking, the dissipation of Δψm, and ROS production.62 Cell death occurred in the absence of caspase activity, was insensitive to Bcl-2 overexpression, and damage to the mitochondria did not induce the loss of cytochrome C.62 These findings supported the hypothesis that GrA induced a cell death program distinct from other cell-mediated pathways, particularly that of GrB. Substrates of GrA have been identified; lamins A–C, core histones, Ku70 (a double-strand DNA repair enzyme), PARP-1 (DNA repair enzyme), a component of the mitochondrial respiratory complex I, and components the ER-associated oxidative stress response SET complex (composed of SET, pp32, Ape1, HMG2, NM23-H1, and TREX-1).41, 63, 64, 65, 66, 67, 68, 69, 70 The current molecular model of GrA-mediated cell death, proposed by Judy Lieberman's group,67 suggests that GrA enters the mitochondrial matrix and proteolytically inactivates complex I protein NDUFS3. This leads to the generation of mitochondrial superoxide, and loss of Δψm. Reactive oxygen species then induces the translocation of the redox-sensitive ER-associated SET complex to the nucleus.62 There, GrA proteolytically degrades lamins to gain entry to the nucleus, and inactivates SET (an inhibitor of the exonuclease NM23-H1), Ape1 (a base excision repair endonuclease) and HMGB2 (a DNA-binding protein that recognizes distorted DNA), and liberates two nucleases NM23-H1 and TREX-1 to induce DNA damage.63, 64, 65, 66
Studies using grB−/− CTLs have observed target cell death with a few characteristics of purified or recombinant GrA, which were attributed to GrA.47, 62 However, there is strong debate as to whether GrA is truly a cytotoxic protease. Studies of granule-mediated responses in grA−/− knockouts have not shown any defects in response to numerous pathogens.71 The exceptions are the response to poxvirus ectromelia, where mice displayed a delayed clearance of the virus, and during herpes simplex infection, where virus spreads more readily throughout the peripheral nervous system.72, 73 Work recently published by Chris Froelich's group has suggested that GrA can only induce cell death at ‘non-physiological levels', and that the primary function of GrA is actually the elicitation of inflammation.74
Granzyme K
GrK was discovered and purified from LAK cells.75 Initial studies using purified human or rat GrK revealed that GrK-induced cell lysis, DNA damage, and affected the mitochondria of target cells.16, 19 Treatment of target cells with recombinant human GrK and either a cationic liposomal delivery vehicle or adenovirus induced phosphatidylserine exposure, ssDNA nicks, loss of membrane integrity, and chromatin condensation in a manner insensitive to z-VAD-fmk and Bcl-xL overexpression.76 Similarly to GrA, GrK directly cleaved SET, Ape1, HMG2, and induced the translocation of the nuclease NM23-H1 from the ER to the nucleus. Mitochondrial damage was also observed, demonstrated by loss of Δψm, and the generation of ROS.77 However, unlike GrA, GrK was reported to cause MOMP, measured by the release of cytochrome C (although caspase-3 was not activated). The mechanism of MOMP was proposed to be via direct GrK proteolytic activation of Bid. However, the investigators did not ascertain the cleavage site in Bid, nor was GrK-mediated killing evaluated in the absence of Bid.
Recently, Zuzen Fan's group78 has also reported that GrK proteolytically processes p53 to pro-apoptotic forms. This would be a unique pro-apoptotic mechanism among Grs, although caspase-3 has been reported to cleave and activate p53 at alternative residues.79 The mechanism underlying GrK cleavage of p53 and its connection to cell death remains unknown.
Granzyme M
Cytotoxic studies, using recombinant GrM have led to conflicting results. Mark Smyth's group reported that GrM and perforin-induced cell death characterized by rapid lysis (51Cr-release), phosphatidylserine exposure, permeability to PI, chromatin condensation, and cytoplasmic vacuolization. Killing lacked sensitivity to both zVAD-fmk and Bcl-2 overexpression, and did not induce DNA fragmentation, cytochrome C release, ROS production, or loss of Δψm.80 In support of this finding, another group also reported GrM-killing induced cell swelling and permeability to PI, was insensitive to zVAD-fmk, and did not activate caspase-3.81 In contrast, Zuzen Fan's group reported recombinant GrM delivered by a cationic liposome reagent-induced rapid cell lysis (51Cr-release), phosphatidylserine exposure (but remained largely impermeable to PI), and DNA fragmentation. Cell death readouts were also sensitive to zVAD-fmk, as caspase-3 was activated by an undetermined mechanism.82 They reported direct proteolysis of ICAD at serine107 (not a previously reported P1 cleavage residue), and PARP at an unknown cleavage site. Of interest, these results have recently been refuted.81 Fan's group has also reported the release of cytochrome C, ROS production, Δψm dissipation (through a cyclosporine-A-sensitive mechanism), and GrM-proteolysis of heat shock protein 70, a protein that protects cells from ROS damage.83 GrM also disrupts the microtubule network of cells in the presence of zVAD-fmk, and specifically targets ezrin and α-tubulin at numerous cleavage sites.84
Mice recently generated with targeted GrM ablation display normal homeostasis, normal NK cytotoxicity, and an effective response against ectromelia virus. The only alteration in cell-mediated immunity was a partially impaired response to MCMV.85
GrM functions outside of direct killing have also been proposed, as GrM proteolytically inactives a GrB endogenous inhibitor, PI9.86 The investigators proposed a model wherein GrM could assist GrB-induced killing by relieving PI9 inhibition of GrB.
Additional Roles of Grs
Grs share homology with serine proteases derived from myeloid cells, such as neutrophils and mast cells. Serine proteases derived from these cells, such as CMA1s, CG, and neutrophil elastase, have long been implicated in extracellular functions, such as inflammation and tissue remodeling.87, 88 Consequently, in addition to well-established roles in cytotoxicity, it is possible that lymphocyte-derived serine proteases modulate some of these extracellular processes as well.
Elevated levels of soluble GrB and GrB-expressing cells in plasma, synovial fluid, cerebrospinal fluid, and bronchoalveolar lavage are observed in patients suffering from a variety of inflammatory conditions.89 GrB may also be released in vivo from activated cytotoxic cells in the absence of targets, both in active and zymogen forms.90 Furthermore, it has become appreciated that GrB is expressed in the absence of perforin by a variety of non-lymphoid cells.91, 92, 93 Thus, there is great speculation that GrB may have extracellular functions that promote extracellular matrix remodeling, cell death, and inflammation. In support of this theory, reports have demonstrated GrB-mediated proteolysis of extracellular matrix proteins.94, 95, 96 A consequence of this may be perforin-independent cell death of adherent cells, via anoikis, where adherent cells die following detachment from the extracellular matrix. Alternatively, cleavage of these proteins may also influence cellular adhesion and migration processes. Other substrates have included proteins involved in blood clotting, and angiogenesis.94, 97 Additionally, GrB cleaves a number of cell receptors, which may influence their ability to respond to growth and survival signals, as well as promote the production of autoantigens.98, 99
Several studies have suggested an inflammatory role for GrA. Treatment with native human GrA induced the production of inflammatory cytokines from epithelial and fibroblast cells, as well as human monocytes.100 Recently, both purified native and recombinant forms of human and mouse GrA have been observed to induce the release of pro-inflammatory cytokines such IL-1β, TNF- α, and IL-6 from freshly isolated human adherent PBMCs.74 The process did not require perforin, but did require target cell internalization. However, Judy Lieberman's et al.69 has contested these findings, questioning the level of endotoxin contamination in the GrA preparations, which could provoke pro-inflammatory responses in monocyte-like cells. Of note, mouse GrK has also recently been shown to have pro-inflammatory potential.101
Other groups have reported GrA degradation of extracellular proteins, such as collagen type IV, fibronectin, and proteoglycans102 The thrombin receptor is also susceptible to GrA proteolysis, affecting clotting reactions. Physiological evidence of GrA extracellular proteolytic activities originates from the observation that GrA levels are elevated in synovial fluid of rheumatoid arthritis patients.103 Additionally, a number of plasma inhibitors of GrA have been reported, further supporting a potential GrA extracellular function. But, a direct causative effect of GrA and immunopathology has not been clearly established.
Clearly Gr research is still very active a quarter of a century on, and will continue to grow in the years to come. This is just one of the outstanding legacies that Jurg has left to the scientific world.
Jürg Tschopp's contributions to our understanding of granzyme-mediated death.
Identified and sequenced many of the granzyme cDNAs.
Coined the name ‘granzymes' after he had isolated and characterized this novel family of serine proteases from the lytic granules of cytotoxic T lymphocytes.
Purified and characterized perforin, the molecule that facilitates the entry of granzymes into target cells.
Demonstrated that granzymes and perforin exist as a complex with proteoglycans in granules.
Knocked out the perforin gene. This lead to the discovery of a perforin/granzyme-independent pathway that acted via Fas/Fas ligand.
Acknowledgments
Many investigators have contributed to granzyme research. We apologize to those whose work was not cited because of a limitation in the number of references allowed. Work in the authors' laboratories is supported by the Canadian Institutes of Health Research and the Canadian Cancer Society. RCB is a Medical Scientist of Alberta Innovates Health Solutions.
Glossary
- Gr
granzyme
- CG
cathepsin G
- CMA1
mast cell chymase
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- Masson D, Tschopp J. A family of serine esterases in lytic granules of cytolytic T lymphocytes. Cell. 1987;49:679–685. doi: 10.1016/0092-8674(87)90544-7. [DOI] [PubMed] [Google Scholar]
- Lobe CG, Finlay BB, Paranchych W, Paetkau VH, Bleackley RC. Novel serine proteases encoded by two cytotoxic T lymphocyte-specific genes. Science. 1986;232:858–861. doi: 10.1126/science.3518058. [DOI] [PubMed] [Google Scholar]
- Bossi G, Griffiths GM. CTL secretory lysosomes: biogenesis and secretion of a harmful organelle. Semin Immunol. 2005;17:87–94. doi: 10.1016/j.smim.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Cullen SP, Martin SJ. Mechanisms of granule-dependent killing. Cell Death Differ. 2008;15:251–262. doi: 10.1038/sj.cdd.4402244. [DOI] [PubMed] [Google Scholar]
- Kam CM, Hudig D, Powers JC. Granzymes (lymphocyte serine proteases): characterization with natural and synthetic substrates and inhibitors. Biochim Biophys Acta. 2000;1477:307–323. doi: 10.1016/s0167-4838(99)00282-4. [DOI] [PubMed] [Google Scholar]
- Masson D, Nabholz M, Estrade C, Tschopp J. Granules of cytolytic T-lymphocytes contain two serine esterases. EMBO J. 1986;5:1595–1600. doi: 10.1002/j.1460-2075.1986.tb04401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilharm E, Parry MA, Friebel R, Tschesche H, Matschiner G, Sommerhoff CP, et al. Generation of catalytically active granzyme K from Escherichia coli inclusion bodies and identification of efficient granzyme K inhibitors in human plasma. J Biol Chem. 1999;274:27331–27337. doi: 10.1074/jbc.274.38.27331. [DOI] [PubMed] [Google Scholar]
- Odake S, Kam CM, Narasimhan L, Poe M, Blake JT, Krahenbuhl O, et al. Human and murine cytotoxic T lymphocyte serine proteases: subsite mapping with peptide thioester substrates and inhibition of enzyme activity and cytolysis by isocoumarins. Biochemistry. 1991;30:2217–2227. doi: 10.1021/bi00222a027. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Sayers TJ, Wiltrout T, Powers JC, Trapani JA. Met-ase: cloning and distinct chromosomal location of a serine protease preferentially expressed in human natural killer cells. J Immunol. 1993;151:6195–6205. [PubMed] [Google Scholar]
- Rukamp BJ, Kam CM, Natarajan S, Bolton BW, Smyth MJ, Kelly JM, et al. Subsite specificities of granzyme M: a study of inhibitors and newly synthesized thiobenzyl ester substrates. Arch Biochem Biophy. 2004;422:9–22. doi: 10.1016/j.abb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- de Poot SA, Westgeest M, Hostetter DR, Van Damme P, Plasman K, Demeyer K, et al. Human and mouse granzyme M display divergent and species-specific substrate specificities Biochem J2011; 437431–442. [DOI] [PubMed] [Google Scholar]
- Brunet JF, Dosseto M, Denizot F, Mattei MG, Clark WR, Haqqi TM, et al. The inducible cytotoxic T-lymphocyte-associated gene transcript CTLA-1 sequence and gene localization to mouse chromosome 14. Nature. 1986;322:268–271. doi: 10.1038/322268a0. [DOI] [PubMed] [Google Scholar]
- Lobe CG, Havele C, Bleackley RC. Cloning of two genes that are specifically expressed in activated cytotoxic T lymphocytes. Proc Natl Acad Sci USA. 1986;83:1448–1452. doi: 10.1073/pnas.83.5.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Leong LG, Liu CC, Damiano A, Wall DA, Cohn ZA. Isolation and characterization of a serine esterase from cytolytic T cell granules. Cell. 1986;47:183–194. doi: 10.1016/0092-8674(86)90441-1. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Park HL, Henkart PA. Synergistic roles of granzymes A and B in mediating target cell death by rat basophilic leukemia mast cell tumors also expressing cytolysin/perforin. J Exp Med. 1995;181:1037–1046. doi: 10.1084/jem.181.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Kam CM, Powers JC, Aebersold R, Greenberg AH. Purification of three cytotoxic lymphocyte granule serine proteases that induce apoptosis through distinct substrate and target cell interactions. J Exp Med. 1992;176:1521–1529. doi: 10.1084/jem.176.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Kraut RP, Aebersold R, Greenberg AH. A natural killer cell granule protein that induces DNA fragmentation and apoptosis. J Exp Med. 1992;175:553–566. doi: 10.1084/jem.175.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heibein JA, Barry M, Motyka B, Bleackley RC. Granzyme B-induced loss of mitochondrial inner membrane potential (Delta Psi m) and cytochrome c release are caspase independent. J Immunol. 1999;163:4683–4693. [PubMed] [Google Scholar]
- MacDonald G, Shi L, Vande Velde C, Lieberman J, Greenberg AH. Mitochondria-dependent and -independent regulation of granzyme B-induced apoptosis. J Exp Med. 1999;189:131–144. doi: 10.1084/jem.189.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrain C, Murphy BM, Martin SJ. Molecular ordering of the caspase activation cascade initiated by the cytotoxic T lymphocyte/natural killer (CTL/NK) protease granzyme B. J Biol Chem. 2005;280:4663–4673. doi: 10.1074/jbc.M410915200. [DOI] [PubMed] [Google Scholar]
- Andrade F, Roy S, Nicholson D, Thornberry N, Rosen A, Casciola-Rosen L. Granzyme B directly and efficiently cleaves several downstream caspase substrates: implications for CTL-induced apoptosis. Immunity. 1998;8:451–460. doi: 10.1016/s1074-7613(00)80550-6. [DOI] [PubMed] [Google Scholar]
- Darmon AJ, Nicholson DW, Bleackley RC. Activation of the apoptotic protease CPP32 by cytotoxic T-cell-derived granzyme B. Nature. 1995;377:446–448. doi: 10.1038/377446a0. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Amarante-Mendes GP, Shi L, Chuang TH, Casiano CA, O'Brien GA, et al. The cytotoxic cell protease granzyme B initiates apoptosis in a cell-free system by proteolytic processing and activation of the ICE/CED-3 family protease, CPP32, via a novel two-step mechanism. EMBO J. 1996;15:2407–2416. [PMC free article] [PubMed] [Google Scholar]
- Medema JP, Toes RE, Scaffidi C, Zheng TS, Flavell RA, Melief CJ, et al. Cleavage of FLICE (caspase-8) by granzyme B during cytotoxic T lymphocyte-induced apoptosis. Eur J Immunol. 1997;27:3492–3498. doi: 10.1002/eji.1830271250. [DOI] [PubMed] [Google Scholar]
- Van de Craen M, Van den Brande I, Declercq W, Irmler M, Beyaert R, Tschopp J, et al. Cleavage of caspase family members by granzyme B: a comparative study in vitro. Eur J Immunol. 1997;27:1296–1299. doi: 10.1002/eji.1830270535. [DOI] [PubMed] [Google Scholar]
- Atkinson EA, Barry M, Darmon AJ, Shostak I, Turner PC, Moyer RW, et al. Cytotoxic T lymphocyte-assisted suicide. Caspase 3 activation is primarily the result of the direct action of granzyme B. J Biol Chem. 1998;273:21261–21266. doi: 10.1074/jbc.273.33.21261. [DOI] [PubMed] [Google Scholar]
- Darmon AJ, Ley TJ, Nicholson DW, Bleackley RC. Cleavage of CPP32 by granzyme B represents a critical role for granzyme B in the induction of target cell DNA fragmentation. J Biol Chem. 1996;271:21709–21712. doi: 10.1074/jbc.271.36.21709. [DOI] [PubMed] [Google Scholar]
- Pinkoski MJ, Waterhouse NJ, Heibein JA, Wolf BB, Kuwana T, Goldstein JC, et al. Granzyme B-mediated apoptosis proceeds predominantly through a Bcl-2-inhibitable mitochondrial pathway. J Biol Chem. 2001;276:12060–12067. doi: 10.1074/jbc.M009038200. [DOI] [PubMed] [Google Scholar]
- Barry M, Heibein JA, Pinkoski MJ, Lee SF, Moyer RW, Green DR, et al. Granzyme B short-circuits the need for caspase 8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly cleaving Bid. Mol Cell Biol. 2000;20:3781–3794. doi: 10.1128/mcb.20.11.3781-3794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heibein JA, Goping IS, Barry M, Pinkoski MJ, Shore GC, Green DR, et al. Granzyme B-mediated cytochrome c release is regulated by the Bcl-2 family members bid and Bax. J Exp Med. 2000;192:1391–1402. doi: 10.1084/jem.192.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton VR, Davis JE, Cancilla M, Johnstone RW, Ruefli AA, Sedelies K, et al. Initiation of apoptosis by granzyme B requires direct cleavage of bid, but not direct granzyme B-mediated caspase activation. J Exp Med. 2000;192:1403–1414. doi: 10.1084/jem.192.10.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goping IS, Barry M, Liston P, Sawchuk T, Constantinescu G, Michalak KM, et al. Granzyme B-induced apoptosis requires both direct caspase activation and relief of caspase inhibition. Immunity. 2003;18:355–365. doi: 10.1016/s1074-7613(03)00032-3. [DOI] [PubMed] [Google Scholar]
- Sutton VR, Wowk ME, Cancilla M, Trapani JA. Caspase activation by granzyme B is indirect, and caspase autoprocessing requires the release of proapoptotic mitochondrial factors. Immunity. 2003;18:319–329. doi: 10.1016/s1074-7613(03)00050-5. [DOI] [PubMed] [Google Scholar]
- Cullen SP, Adrain C, Luthi AU, Duriez PJ, Martin SJ. Human and murine granzyme B exhibit divergent substrate preferences. J Cell Biol. 2007;176:435–444. doi: 10.1083/jcb.200612025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DA, Scorrano L, Putcha GV, Korsmeyer SJ, Ley TJ. Granzyme B can cause mitochondrial depolarization and cell death in the absence of BID, BAX, and BAK. Proc Natl Acad Sci USA. 2001;98:14985–14990. doi: 10.1073/pnas.261581498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froelich CJ, Hanna WL, Poirier GG, Duriez PJ, D'Amours D, Salvesen GS, et al. Granzyme B/perforin-mediated apoptosis of Jurkat cells results in cleavage of poly(ADP-ribose) polymerase to the 89-kDa apoptotic fragment and less abundant 64-kDa fragment. Biochem Biophy Res Comm. 1996;227:658–665. doi: 10.1006/bbrc.1996.1565. [DOI] [PubMed] [Google Scholar]
- Goping IS, Sawchuk T, Underhill DA, Bleackley RC. Identification of {alpha}-tubulin as a granzyme B substrate during CTL-mediated apoptosis. J Cell Sci. 2006;119 (Pt 5:858–865. doi: 10.1242/jcs.02791. [DOI] [PubMed] [Google Scholar]
- Sebbagh M, Hamelin J, Bertoglio J, Solary E, Breard J. Direct cleavage of II ROCK by granzyme B induces target cell membrane blebbing in a caspase-independent manner. J Exp Med. 2005;201:465–471. doi: 10.1084/jem.20031877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif-Askari E, Alam A, Rheaume E, Beresford PJ, Scotto C, Sharma K, et al. Direct cleavage of the human DNA fragmentation factor-45 by granzyme B induces caspase-activated DNase release and DNA fragmentation. EMBO J. 2001;20:3101–3113. doi: 10.1093/emboj/20.12.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DA, Du C, Xu M, Wang X, Ley TJ. DFF45/ICAD can be directly processed by granzyme B during the induction of apoptosis. Immunity. 2000;12:621–632. doi: 10.1016/s1074-7613(00)80213-7. [DOI] [PubMed] [Google Scholar]
- Zhang D, Beresford PJ, Greenberg AH, Lieberman J. Granzymes A and B directly cleave lamins and disrupt the nuclear lamina during granule-mediated cytolysis. Proc Natl Acad Sci USA. 2001;98:5746–5751. doi: 10.1073/pnas.101329598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusel JW, Wesselschmidt RL, Shresta S, Russell JH, Ley TJ. Cytotoxic lymphocytes require granzyme B for the rapid induction of DNA fragmentation and apoptosis in allogeneic target cells. Cell. 1994;76:977–987. doi: 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- Shresta S, MacIvor DM, Heusel JW, Russell JH, Ley TJ. Natural killer and lymphokine-activated killer cells require granzyme B for the rapid induction of apoptosis in susceptible target cells. Proc Natl Acad Sci USA. 1995;92:5679–5683. doi: 10.1073/pnas.92.12.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo J, Bosque A, Brehm R, Wallich R, Naval J, Mullbacher A, et al. Apoptotic pathways are selectively activated by granzyme A and/or granzyme B in CTL-mediated target cell lysis. J Cell Biol. 2004;167:457–468. doi: 10.1083/jcb.200406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Mullbacher A, Waring P, Tha Hla R, Tran T, Chin S, Stehle T, et al. Granzymes are the essential downstream effector molecules for the control of primary virus infections by cytolytic leukocytes. Proc Natl Acad Sci USA. 1999;96:13950–13955. doi: 10.1073/pnas.96.24.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shresta S, Graubert TA, Thomas DA, Raptis SZ, Ley TJ. Granzyme A initiates an alternative pathway for granule-mediated apoptosis. Immunity. 1999;10:595–605. doi: 10.1016/s1074-7613(00)80059-x. [DOI] [PubMed] [Google Scholar]
- Revell PA, Grossman WJ, Thomas DA, Cao X, Behl R, Ratner JA, et al. Granzyme B and the downstream granzymes C and/or F are important for cytotoxic lymphocyte functions. J Immunol. 2005;174:2124–2131. doi: 10.4049/jimmunol.174.4.2124. [DOI] [PubMed] [Google Scholar]
- Smyth MJ, Street SE, Trapani JA. Cutting edge: granzymes A and B are not essential for perforin-mediated tumor rejection. J Immunol. 2003;171:515–518. doi: 10.4049/jimmunol.171.2.515. [DOI] [PubMed] [Google Scholar]
- Klein JL, Selvakumar A, Trapani JA, Dupont B. Characterization of a novel, human cytotoxic lymphocyte-specific serine protease cDNA clone (CSP-C) Tissue Antigens. 1990;35:220–228. doi: 10.1111/j.1399-0039.1990.tb01787.x. [DOI] [PubMed] [Google Scholar]
- Meier M, Kwong PC, Fregeau CJ, Atkinson EA, Burrington M, Ehrman N, et al. Cloning of a gene that encodes a new member of the human cytotoxic cell protease family. Biochemistry. 1990;29:4042–4049. doi: 10.1021/bi00469a003. [DOI] [PubMed] [Google Scholar]
- Fellows E, Gil-Parrado S, Jenne DE, Kurschus FC. Natural killer cell-derived human granzyme H induces an alternative, caspase-independent cell-death program. Blood. 2007;110:544–552. doi: 10.1182/blood-2006-10-051649. [DOI] [PubMed] [Google Scholar]
- Hou Q, Zhao T, Zhang H, Lu H, Zhang Q, Sun L, et al. Granzyme H induces apoptosis of target tumor cells characterized by DNA fragmentation and Bid-dependent mitochondrial damage. Mol Immunol. 2008;45:1044–1055. doi: 10.1016/j.molimm.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Woodard SL, Jackson DS, Abuelyaman AS, Powers JC, Winkler U, Hudig D. Chymase-directed serine protease inhibitor that reacts with a single 30-kDa granzyme and blocks NK-mediated cytotoxicity. J Immunol. 1994;153:5016–5025. [PubMed] [Google Scholar]
- Andrade F, Fellows E, Jenne DE, Rosen A, Young CS. Granzyme H destroys the function of critical adenoviral proteins required for viral DNA replication and granzyme B inhibition. EMBO J. 2007;26:2148–2157. doi: 10.1038/sj.emboj.7601650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero V, Fellows E, Jenne DE, Andrade F. Cleavage of La protein by granzyme H induces cytoplasmic translocation and interferes with La-mediated HCV-IRES translational activity. Cell Death Differ. 2009;16:340–348. doi: 10.1038/cdd.2008.165. [DOI] [PubMed] [Google Scholar]
- Jenne D, Rey C, Haefliger JA, Qiao BY, Groscurth P, Tschopp J. Identification and sequencing of cDNA clones encoding the granule-associated serine proteases granzymes D, E, and F of cytolytic T lymphocytes. Proc Natl Acad Sci USA. 1988;85:4814–4818. doi: 10.1073/pnas.85.13.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast JA, Pinkoski M, Wolfenden A, Bleackley RC. Structure and evolution of the cytotoxic cell proteinase genes CCP3, CCP4 and CCP5. J Mol Biol. 1991;220:867–875. doi: 10.1016/0022-2836(91)90359-e. [DOI] [PubMed] [Google Scholar]
- Johnson H, Scorrano L, Korsmeyer SJ, Ley TJ. Cell death induced by granzyme C. Blood. 2003;101:3093–3101. doi: 10.1182/blood-2002-08-2485. [DOI] [PubMed] [Google Scholar]
- Shi L, Wu L, Wang S, Fan Z. Granzyme F induces a novel death pathway characterized by Bid-independent cytochrome c release without caspase activation. Cell Death Differ. 2009;16:1694–1706. doi: 10.1038/cdd.2009.101. [DOI] [PubMed] [Google Scholar]
- Gershenfeld HK, Weissman IL. Cloning of a cDNA for a T cell-specific serine protease from a cytotoxic T lymphocyte. Science. 1986;232:854–858. doi: 10.1126/science.2422755. [DOI] [PubMed] [Google Scholar]
- Martinvalet D, Zhu P, Lieberman J. Granzyme A induces caspase-independent mitochondrial damage, a required first step for apoptosis. Immunity. 2005;22:355–370. doi: 10.1016/j.immuni.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, et al. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Fan Z, Beresford PJ, Oh DY, Zhang D, Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112:659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- Fan Z, Beresford PJ, Zhang D, Lieberman J. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol Cell Biol. 2002;22:2810–2820. doi: 10.1128/MCB.22.8.2810-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, Yoshida A, et al. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat Immunol. 2003;4:145–153. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- Martinvalet D, Dykxhoorn DM, Ferrini R, Lieberman J. Granzyme A cleaves a mitochondrial complex I protein to initiate caspase-independent cell death. Cell. 2008;133:681–692. doi: 10.1016/j.cell.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Pasternack MS, Beresford PJ, Wagner L, Greenberg AH, Lieberman J. Induction of rapid histone degradation by the cytotoxic T lymphocyte protease Granzyme A. J Biol Chem. 2001;276:3683–3690. doi: 10.1074/jbc.M005390200. [DOI] [PubMed] [Google Scholar]
- Zhu P, Martinvalet D, Chowdhury D, Zhang D, Schlesinger A, Lieberman J. The cytotoxic T lymphocyte protease granzyme A cleaves and inactivates poly(adenosine 5′-diphosphate-ribose) polymerase-1. Blood. 2009;114:1205–1216. doi: 10.1182/blood-2008-12-195768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Zhang D, Chowdhury D, Martinvalet D, Keefe D, Shi L, et al. Granzyme A, which causes single-stranded DNA damage, targets the double-strand break repair protein Ku70. EMBO Rep. 2006;7:431–437. doi: 10.1038/sj.embor.7400622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebnet K, Hausmann M, Lehmann-Grube F, Mullbacher A, Kopf M, Lamers M, et al. Granzyme A-deficient mice retain potent cell-mediated cytotoxicity. EMBO J. 1995;14:4230–4239. doi: 10.1002/j.1460-2075.1995.tb00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullbacher A, Ebnet K, Blanden RV, Hla RT, Stehle T, Museteanu C, et al. Granzyme A is critical for recovery of mice from infection with the natural cytopathic viral pathogen, ectromelia. Proc Natl Acad Sci USA. 1996;93:5783–5787. doi: 10.1073/pnas.93.12.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira RA, Simon MM, Simmons A. Granzyme A, a noncytolytic component of CD8(+) cell granules, restricts the spread of herpes simplex virus in the peripheral nervous systems of experimentally infected mice. J Virol. 2000;74:1029–1032. doi: 10.1128/jvi.74.2.1029-1032.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metkar SS, Menaa C, Pardo J, Wang B, Wallich R, Freudenberg M, et al. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity. 2008;29:720–733. doi: 10.1016/j.immuni.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Hameed A, Lowrey DM, Lichtenheld M, Podack ER. Characterization of three serine esterases isolated from human IL-2 activated killer cells. J Immunol. 1988;141:3142–3147. [PubMed] [Google Scholar]
- Zhao T, Zhang H, Guo Y, Fan Z. Granzyme K directly processes bid to release cytochrome c and endonuclease G leading to mitochondria-dependent cell death. J Biol Chem. 2007;282:12104–12111. doi: 10.1074/jbc.M611006200. [DOI] [PubMed] [Google Scholar]
- Zhao T, Zhang H, Guo Y, Zhang Q, Hua G, Lu H, et al. Granzyme K cleaves the nucleosome assembly protein SET to induce single-stranded DNA nicks of target cells. Cell Death Differ. 2007;14:489–499. doi: 10.1038/sj.cdd.4402040. [DOI] [PubMed] [Google Scholar]
- Hua G, Wang S, Zhong C, Xue P, Fan Z. Ignition of p53 bomb sensitizes tumor cells to granzyme K-mediated cytolysis. J Immunol. 2009;182:2152–2159. doi: 10.4049/jimmunol.0802307. [DOI] [PubMed] [Google Scholar]
- Sayan BS, Sayan AE, Knight RA, Melino G, Cohen GM. p53 is cleaved by caspases generating fragments localizing to mitochondria. J Biol Chem. 2006;281:13566–13573. doi: 10.1074/jbc.M512467200. [DOI] [PubMed] [Google Scholar]
- Kelly JM, Waterhouse NJ, Cretney E, Browne KA, Ellis S, Trapani JA, et al. Granzyme M mediates a novel form of perforin-dependent cell death. J Biol Chem. 2004;279:22236–22242. doi: 10.1074/jbc.M401670200. [DOI] [PubMed] [Google Scholar]
- Cullen SP, Afonina IS, Donadini R, Luthi AU, Medema JP, Bird PI, et al. Nucleophosmin is cleaved and inactivated by the cytotoxic granule protease granzyme M during natural killer cell-mediated killing. J Biol Chem. 2009;284:5137–5147. doi: 10.1074/jbc.M807913200. [DOI] [PubMed] [Google Scholar]
- Lu H, Hou Q, Zhao T, Zhang H, Zhang Q, Wu L, et al. Granzyme M directly cleaves inhibitor of caspase-activated DNase (CAD) to unleash CAD leading to DNA fragmentation. J Immunol. 2006;177:1171–1178. doi: 10.4049/jimmunol.177.2.1171. [DOI] [PubMed] [Google Scholar]
- Hua G, Zhang Q, Fan Z. Heat shock protein 75 (TRAP1) antagonizes reactive oxygen species generation and protects cells from granzyme M-mediated apoptosis. J Biol Chem. 2007;282:20553–20560. doi: 10.1074/jbc.M703196200. [DOI] [PubMed] [Google Scholar]
- Bovenschen N, de Koning PJ, Quadir R, Broekhuizen R, Damen JM, Froelich CJ, et al. NK cell protease granzyme M targets alpha-tubulin and disorganizes the microtubule network. J Immunol. 2008;180:8184–8191. doi: 10.4049/jimmunol.180.12.8184. [DOI] [PubMed] [Google Scholar]
- Pao LI, Sumaria N, Kelly JM, van Dommelen S, Cretney E, Wallace ME, et al. Functional analysis of granzyme M and its role in immunity to infection. J Immunol. 2005;175:3235–3243. doi: 10.4049/jimmunol.175.5.3235. [DOI] [PubMed] [Google Scholar]
- Mahrus S, Kisiel W, Craik CS. Granzyme M is a regulatory protease that inactivates proteinase inhibitor 9, an endogenous inhibitor of granzyme B. J Biol Chem. 2004;279:54275–54282. doi: 10.1074/jbc.M411482200. [DOI] [PubMed] [Google Scholar]
- Korkmaz B, Moreau T, Gauthier F. Neutrophil elastase, proteinase 3 and cathepsin G: physicochemical properties, activity and physiopathological functions. Biochimie. 2008;90:227–242. doi: 10.1016/j.biochi.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Pejler G, Abrink M, Ringvall M, Wernersson S. Mast cell proteases. Adv Immunol. 2007;95:167–255. doi: 10.1016/S0065-2776(07)95006-3. [DOI] [PubMed] [Google Scholar]
- Boivin WA, Cooper DM, Hiebert PR, Granville DJ. Intracellular versus extracellular granzyme B in immunity and disease: challenging the dogma. Lab Invest. 2009;89:1195–1220. doi: 10.1038/labinvest.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash MD, Bird CH, Bird PI. Active and zymogen forms of granzyme B are constitutively released from cytotoxic lymphocytes in the absence of target cell engagement. Immunol Cell Biol. 2009;87:249–254. doi: 10.1038/icb.2008.98. [DOI] [PubMed] [Google Scholar]
- Hirst CE, Buzza MS, Sutton VR, Trapani JA, Loveland KL, Bird PI. Perforin-independent expression of granzyme B and proteinase inhibitor 9 in human testis and placenta suggests a role for granzyme B-mediated proteolysis in reproduction. Mol Hum Reprod. 2001;7:1133–1142. doi: 10.1093/molehr/7.12.1133. [DOI] [PubMed] [Google Scholar]
- Pardo J, Wallich R, Ebnet K, Iden S, Zentgraf H, Martin P, et al. Granzyme B is expressed in mouse mast cells in vivo and in vitro and causes delayed cell death independent of perforin. Cell Death Differ. 2007;14:1768–1779. doi: 10.1038/sj.cdd.4402183. [DOI] [PubMed] [Google Scholar]
- Tschopp CM, Spiegl N, Didichenko S, Lutmann W, Julius P, Virchow JC, et al. Granzyme B, a novel mediator of allergic inflammation: its induction and release in blood basophils and human asthma. Blood. 2006;108:2290–2299. doi: 10.1182/blood-2006-03-010348. [DOI] [PubMed] [Google Scholar]
- Buzza MS, Dyson JM, Choi H, Gardiner EE, Andrews RK, Kaiserman D, et al. Antihemostatic activity of human granzyme B mediated by cleavage of von Willebrand factor. J Biol Chem. 2008;283:22498–22504. doi: 10.1074/jbc.M709080200. [DOI] [PubMed] [Google Scholar]
- Buzza MS, Zamurs L, Sun J, Bird CH, Smith AI, Trapani JA, et al. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J Biol Chem. 2005;280:23549–23558. doi: 10.1074/jbc.M412001200. [DOI] [PubMed] [Google Scholar]
- Froelich CJ, Zhang X, Turbov J, Hudig D, Winkler U, Hanna WL. Human granzyme B degrades aggrecan proteoglycan in matrix synthesized by chondrocytes. J Immunol. 1993;151:7161–7171. [PubMed] [Google Scholar]
- Mulligan-Kehoe MJ, Drinane MC, Mollmark J, Casciola-Rosen L, Hummers LK, Hall A, et al. Antiangiogenic plasma activity in patients with systemic sclerosis. Arthritis Rheum. 2007;56:3448–3458. doi: 10.1002/art.22861. [DOI] [PubMed] [Google Scholar]
- Gahring L, Carlson NG, Meyer EL, Rogers SW. Granzyme B proteolysis of a neuronal glutamate receptor generates an autoantigen and is modulated by glycosylation. J Immunol. 2001;166:1433–1438. doi: 10.4049/jimmunol.166.3.1433. [DOI] [PubMed] [Google Scholar]
- Loeb CR, Harris JL, Craik CS. Granzyme B proteolyzes receptors important to proliferation and survival, tipping the balance toward apoptosis. J Biol Chem. 2006;281:28326–28335. doi: 10.1074/jbc.M604544200. [DOI] [PubMed] [Google Scholar]
- Sower LE, Klimpel GR, Hanna W, Froelich CJ. Extracellular activities of human granzymes. I. Granzyme A induces IL6 and IL8 production in fibroblast and epithelial cell lines. Cell Immunol. 1996;171:159–163. doi: 10.1006/cimm.1996.0187. [DOI] [PubMed] [Google Scholar]
- Joeckel LT, Wallich R, Martin P, Sanchez-Martinez D, Weber FC, Martin SF, et al. Mouse granzyme K has pro-inflammatory potential Cell Death Differ2011; 181112–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak PP, Spaeny-Dekking L, Kraan MC, Breedveld FC, Froelich CJ, Hack CE. The levels of soluble granzyme A and B are elevated in plasma and synovial fluid of patients with rheumatoid arthritis (RA) Clin Exp Immunol. 1999;116:366–370. doi: 10.1046/j.1365-2249.1999.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J, Thomas DA, Revell PA, Ley TJ, Virgin HWt. Granzymes and caspase 3 play important roles in control of gammaherpesvirus latency. J Virol. 2004;78:12519–12528. doi: 10.1128/JVI.78.22.12519-12528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U, Sobek V, Balkow S, Holscher C, Mullbacher A, Museteanu C, et al. Concerted action of perforin and granzymes is critical for the elimination of Trypanosoma cruzi from mouse tissues, but prevention of early host death is in addition dependent on the FasL/Fas pathway. Eur J Immunol. 2003;33:70–78. doi: 10.1002/immu.200390009. [DOI] [PubMed] [Google Scholar]
- Pardo J, Balkow S, Anel A, Simon MM. Granzymes are essential for natural killer cell-mediated and perf-facilitated tumor control. Eur J Immunol. 2002;32:2881–2887. doi: 10.1002/1521-4141(2002010)32:10<2881::AID-IMMU2881>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]