Abstract
Skeletal myogenesis is precisely regulated by multiple transcription factors. Previously, we demonstrated that enhancer of polycomb 1 (Epc1) induces skeletal muscle differentiation by potentiating serum response factor (SRF)-dependent muscle gene activation. Here, we report that an interacting partner of Epc1, ret finger protein (RFP), blocks skeletal muscle differentiation. Our findings show that RFP was highly expressed in skeletal muscles and was downregulated during myoblast differentiation. Forced expression of RFP delayed myoblast differentiation, whereas knockdown enhanced it. Epc1-induced enhancements of SRF-dependent multinucleation, transactivation of the skeletal α-actin promoter, binding of SRF to the serum response element, and muscle-specific gene induction were blocked by RFP. RFP interfered with the physical interaction between Epc1 and SRF. Muscles from rfp knockout mice (Rfp−/−) mice were bigger than those from wild-type mice, and the expression of SRF-dependent muscle-specific genes was upregulated. Myotube formation and myoblast differentiation were enhanced in Rfp−/− mice. Taken together, our findings highlight RFP as a novel regulator of muscle differentiation that acts by modulating the expression of SRF-dependent skeletal muscle-specific genes.
Keywords: ret finger protein, muscle differentiation, serum response factor, enhancer of polycomb1
Skeletal muscle is critical for the generation of active force, the maintenance of posture, and body shape. Muscle regeneration from myoblasts is important for the repair and maintenance of skeletal muscles after muscle injury or dystrophy. Myogenesis from the precursor cells to the skeletal muscle lineage consists of multiple steps, and each of these steps is tightly organized by extrinsic and intrinsic signaling pathways. Thus, not only for understanding the regeneration of injured skeletal muscles, but also for furthering therapeutic approaches to muscle atrophy, a hot issue in current research is how the proliferation and differentiation of myoblasts are regulated. To ultimately provide important clues for therapeutics, a comprehensive understanding of how various signaling mechanisms collaborate to regulate the gene expression and epigenetic programs for myoblast differentiation is needed.
The TRIM (tripartite motif) protein family (also known as the RBCC protein family) has the common structures of three zinc-binding domains, a RING, a B-box type 1, and a B-box type 2, followed by a coiled-coil region.1, 2 Although little is known about the molecular and biological mechanisms mediated by the TRIM genes, the proteins in this family are known to be involved in diverse cellular processes, such as differentiation, cell growth, and even some human diseases including cancers.3, 4 Ret finger protein (RFP), also known as TRIM27, was first discovered in transformed mouse fibroblasts that were transfected with genomic DNA from a human lymphoma.5 RFP was found as a fusion protein of RFP/ret by DNA rearrangement.6, 7 Like other TRIM family proteins such as TIF-18 and the promyelocytic leukemia (PML) gene,9, 10 RFP becomes oncogenic when its TRIM27 is recombined with the tyrosine kinase domain from the c-ret proto-oncogene.7 Although its biological and functional roles are yet to be defined, RFP can physically interact with several proteins, such as PML,11 int-6,12 and enhancer of polycomb 1 (Epc1), a chromatin protein.13
Recently, we investigated the mechanism of skeletal muscle differentiation and reported that Epc1 binds to serum response factor (SRF) and activates SRF/SRE (serum response element)-dependent genes by recruiting p300, which then induces skeletal muscle differentiation.14 Epc1 is required for the full activation of those SRF-dependent muscle-specific genes. We carried out this study to further clarify the functional role of Epc1 in skeletal muscle differentiation by characterizing its binding partners. We performed yeast two-hybrid screening with the human adult skeletal cDNA library as prey and found that Epc1 recruited RFP. We also elucidated that RFP reduces the SRF-dependent expression of skeletal muscle genes by interfering with the interaction of Epc1 and SRF. In addition, we clearly demonstrated that muscle regeneration and myoblast differentiation are enhanced in rfp knockout mice (Rfp−/−) mice. Although many research groups have established clear positive relations between RFP and the development of certain types of cancers, the functional role of the protein in skeletal muscle has not been described. We herewith report RFP as a novel negative regulator of Epc1/SRF-dependent skeletal muscle differentiation.
Results
RFP, a binding partner of Epc1, is highly expressed in neonatal skeletal muscles
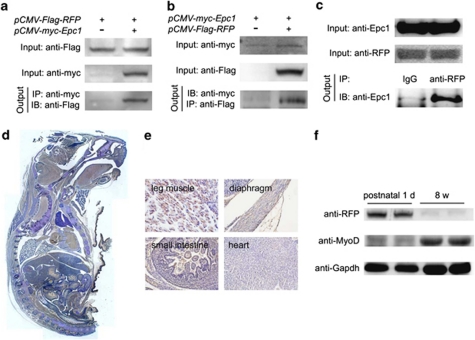
To investigate further the functional role of Epc1 in skeletal muscle differentiation, we tried to find binding partners of Epc1 in skeletal muscle. We used a yeast two-hybrid technique to screen a mouse skeletal muscle cDNA library with full-length Epc1 as bait and identified ∼200 clones that were positive to His and β-galactosidase. The most frequent hit was RFP, and the interaction was confirmed in the mammalian cells (Figures 1a–c). Although the physical interaction between Epc1 and RFP has been shown in other cell types,13 we confirmed whether this interaction takes place in muscle cells. The physical interaction of endogenous RFP and Epc1 was reproduced in C2C12 myoblast cells (Figure 1c).15 Both proteins were colocalized in the nucleus of C2C12 cells (Supplementary Figure 1).
Figure 1.
RFP, a binding partner of Epc1, is highly expressed in neonatal skeletal muscles. (a) Epc1 recruits RFP in 293T cells. pCMV-Flag-RFP constructs with either pcDNA3.1 or pCMV-myc-Epc1 were transiently transfected into 293 T cells, and anti-myc antibodies were used for immunoprecipitation. (b) Reverse immunoprecipitation in H9c2, myoblast cells. (c) Physical interaction of endogenous Epc1 and RFP was demonstrated in C2C12 cells. (d) Immunohistochemistry analysis showing the distribution of RFP in a sagittal section from a postnatal day 1 mouse. RFP expression was visualized as brown color. (e) RFP is expressed in skeletal muscles such as leg muscle and diaphragm but is barely detected in cardiac and intestinal smooth muscles. (f) Western blot analysis showing RFP was highly expressed in neonatal skeletal muscles but was dramatically decreased in adult
Next we checked whether RFP is expressed in skeletal muscles. First, we used sagittal sections of postnatal day 1 mice for immunohistochemistry (Figure 1d). RFP was prominently expressed in leg muscle and diaphragm, all of which contain striated skeletal muscle. However, interestingly, RFP was not expressed in cardiomyocytes or intestinal smooth muscles (Figure 1e). We also compared the expression of RFP in neonatal and adult mouse skeletal muscles by western blot analysis with anti-RFP antibody. We observed abundant immunoreactivity of the 58 kDa protein in the neonatal skeletal muscles (Figure 1f), but not in adult muscles.
RFP inhibits myoblast differentiation
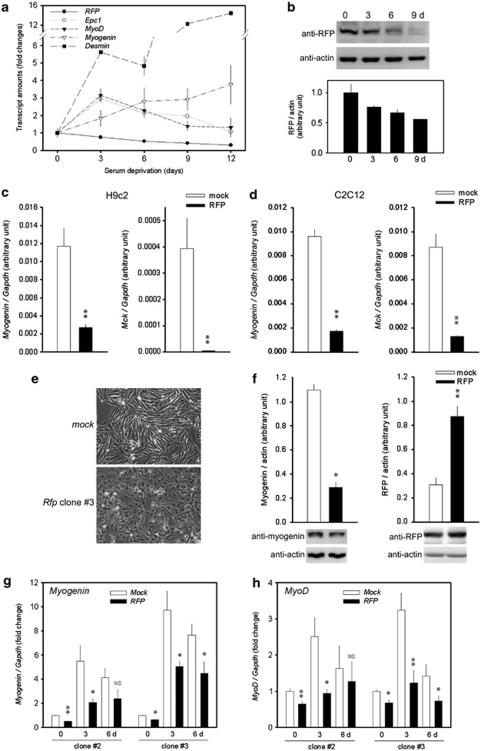
We further investigated the changes in RFP expression during differentiation of C2C12 or H9c2 cells. In this study, serum starvation by substituting growth medium with differentiation medium was enough to lead C2C12 cells to have properties of differentiated skeletal myocytes, as demonstrated by the increase in skeletal muscle-specific gene transcripts such as myogenin, MyoD, and desmin (Figure 2a). The expression of Epc1 was also increased by serum deprivation as previously described.16 Interestingly, the expression of Rfp was downregulated during the differentiation of C2C12 cells (Figure 2a) and in primary cultured myoblasts (Figure 2b).
Figure 2.
RFP inhibits skeletal muscle differentiation. (a) Transcripts for Rfp, Epc1, MyoD, myogenin, and desmin were detected by qRT-PCR. C2C12 cells were forced to undergo differentiation into skeletal myocytes by replacing growth medium with differentiation medium. The expression of Rfp was gradually reduced during myoblast differentiation, whereas that of Epc1 was increased. Muscle-specific genes such as myogenin, MyoD, and desmin were upregulated by serum deprivation. The results from 4∼6 samples were shown. (b) The changes in Rfp protein amounts were also shown in primary cultured myoblasts cells isolated from the hamstring muscles of mice. Lower panel: quantification results from three independent sets of western blots. (c) Decreased myogenin and Mck transcript levels in RFP-overexpressing H9c2 cell lines. Transcript levels of myogenin and Mck were measured by qRT-PCR. (d) Transcripts for myogenin and Mck were measured by qRT-PCR in RFP-overexpressing C2C12 cells. (e) Impaired myotube formation of RFP-expressing cells. Mock- or RFP- expressing H9c2 cells were grown in differentiation media for 6 days. Mock-cells were elongated by serum deprivation, whereas RFP-cells were not. (f) Serum deprivation failed to induce myogenin in RFP-expressing cell lines. Band intensities from three independent sets of western blot analysis were averaged. (g and h) Transcript levels of myogenin (g) and MyoD (h) in two clones of RFP-overexpressing H9c2 cells were measured by qRT-PCR. NS, not significant. *P<0.05, **P<0.01, compared with mock-transfected cells
The downregulation of Rfp in differentiating skeletal muscle led us to speculate that RFP may participate in maintenance of the myoblasts by inhibiting muscle differentiation. Therefore, to further investigate the role of RFP, we generated stable cell lines overexpressing RFP by utilizing H9c2 and C2C12 cells. Although H9c2 cells are originated from cardiomyoblasts, it is known that serum deprivation without addition of retinoic acid leads the cells to differentiate into skeletal myocytes but not cardiomyocytes.17 In the cells maintained in growth medium, the expression of myogenin and Mck (muscle creatine kinase) was significantly reduced in RFP-overexpressing H9c2 cells (Figure 2c), and this finding was reproduced in C2C12 cells (Figure 2d).
We next challenged the RFP-overexpressing cell lines with serum deprivation. In mock-transfected cell lines, serum deprivation for 6 days induced elongation of H9c2 cells to form myotubes. However, it failed to induce elongation in RFP-overexpressing cells (Figure 2e, lower panel). We also examined the myogenin expression levels in clone #2 cells by western blot analysis; the myogenin protein amount was significantly reduced in RFP cell lines (Figure 2f, left panel). RFP expression in the established stable cell lines was 2.5∼3-fold greater than in the mock-cell lines (Figure 2f, right panel).
Failure of differentiation was further confirmed by quantitative RT-PCR to detect myogenin (Figure 2g) and MyoD (Figure 2h). In mock-transfected cell lines, serum deprivation caused an increase in the expression of these muscle-specific genes. However, the induction of those genes was blunted in both clones #2 and #3, which overexpress RFP.
RFP abolishes Epc1-mediated potentiation of the SRF effect by interfering with their interaction
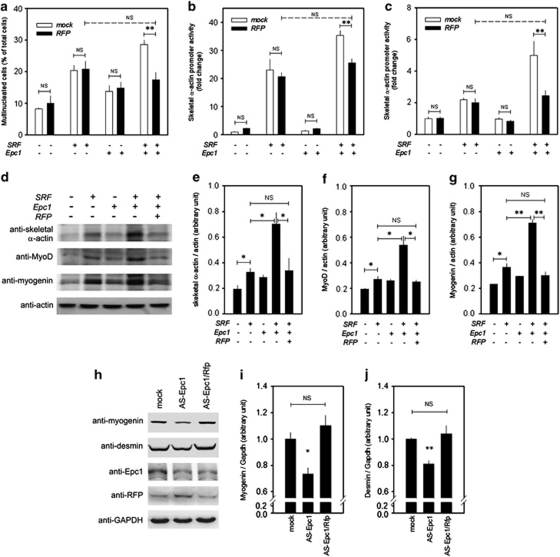
Previously, we showed that Epc1 synergistically potentiates SRF-induced muscle differentiation. The present results that RFP, which interacts with Epc1 (Figures 1a–c), retards muscle differentiation (Figure 2) prompted us to study the mechanisms of RFP's association with both Epc1 and SRF. We first checked the effects of RFP on multinucleation induced by SRF and Epc1. RFP itself did not affect the basal level of multinucleation of H9c2 cells (Figure 3a, first versus second column). As reported previously,14 SRF induced an increase in the multinucleation of the myoblasts (third column), which was further potentiated by co-transfection of Epc1 (seventh column). Co-transfection of RFP did not affect the SRF-induced multinucleation (fourth column, compared with third column). However, co-transfection of RFP reduced the Epc1/SRF-induced multinucleation (seventh versus eighth column).
Figure 3.
RFP blocks Epc1-induced potentiation of SRF-dependent differentiation of skeletal myoblasts. (a) Epc1 potentiated SRF-induced multinucleation of H9c2 cells in differentiation medium. However, the potentiation was completely blocked by co-transfection of RFP. Note that multinucleation after RFP transfection returned to that of the SRF-level. (b) Epc1-induced potentiation of SRF-transactivation was completely absent after co-transfection of RFP in 293T cells. (c) The same results of promoter analysis were obtained from C2C12 myoblast cells. (d) Representative western blot analysis showing that Epc1-induced potentiation of SRF-mediated expression of muscle-specific genes was blocked by RFP. (e and g) Quantification results from three independent western blots of skeletal α-actin (e), MyoD (f), and myogenin (g). NS, not significant. (h) Western blot analysis of Epc1- or Epc1/Rfp- knockdown C2C12 cells. The knockdown cells were prepared with antisense technology. Expression of myogenin and desmin was decreased in antisense Epc1 cells and it was recovered in antisense Epc1/Rfp cells. (i) Quantification results from three western blots of myogenin. (j) Quantification results from western blots of desmin. *P<0.05, **P<0.01
In our previous report,14 we confirmed the Epc1-induced potentiation of SRF-dependent myoblast differentiation by promoter analysis with the skeletal α-actin minimal promoter. Here, we investigated the effects of RFP on the potentiation effect with the promoter in 293T cells, a human embryonic kidney cell line. The potentiation effect of Epc1 over the SRF (seventh column) was blocked by co-transfection of RFP (eighth column, Figure 3b). The level of transactivation after RFP co-transfection (eighth column) was the same as with the transactivation of SRF alone (third column). We repeated the experiments in C2C12 cells, and obtained the same results (Figure 3c). Transfection of RFP did not repress the basal activity of the skeletal α-actin or myogenin promoter in either C2C12 or 293T cells (Supplementary Figures 2a–d).
The changes in the protein amounts of skeletal α-actin after co-transfection of SRF, Epc1, and RFP were further investigated by western blot analysis (Figure 3d, upper panel). The SRF-induced increase in the skeletal α-actin protein amount (second lane) was further increased by co-transfection of Epc1 (fourth lane). However, co-transfection of RFP reduced the amount of protein expression (fifth lane). The changes in the skeletal α-actin protein amount were quantified after three independent sets of immunoblot analysis (Figure 3e).
We next studied the muscle-specific transcription factors that are induced during myoblast differentiation. The increase in MyoD and myogenin induced by SRF (Figure 3d, second lanes in the second and third immunoblots) was further potentiated by co-transfection of Epc1 (fourth lanes), which was blocked by co-transfection of RFP (fifth lanes). These changes in protein amounts were quantified (Figure 3f for MyoD and Figure 3g for myogenin).
We previously reported that muscle differentiation markers are reduced in Epc+/− mice. Thus, we sought to determine whether muscle differentiation impairment induced by knockdown of Epc1could be restored by double knockdown of Epc1 and Rfp in C2C12 cells. The reduction of myogenin and desmin in Epc1 knockdown C2C12 cells was recovered in the double knockdown cells (Figures 3h–j).
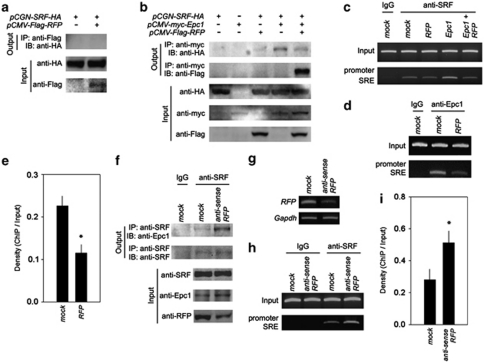
Our previous study showed that Epc1 interacts with SRF and enhances the binding of SRF to the SRE in the skeletal α-actin promoter.14 First, we assumed that Epc1/SRF/RFP may form a complex and that SRF may interact with RFP. We therefore checked whether RFP can directly interact with SRF. However, we were not able to demonstrate the physical interaction of the proteins in our experimental condition (Figure 4a). This unexpected result suggests that RFP may compete with SRF for binding to Epc1. To test our hypothesis, we checked whether RFP can affect the interaction between Epc1 and SRF. As reported previously, Epc1 successfully pulled down SRF (Figure 4b, fourth lane in the uppermost panel). Interestingly, however, co-transfection of RFP reduced the interaction of both proteins (Figure 4b, fourth versus fifth lane in the uppermost panel). The physical interaction between Epc1 and RFP was shown again (fifth lane in the second immunoblot).
Figure 4.
RFP squelched Epc1 from SRF, which resulted in the reduction of SRF-dependent transactivation. (a) SRF did not associate with RFP. (b) RFP blocked binding of Epc1 to SRF. Uppermost panel: Epc1 successfully pulled down SRF (fourth lane). However, co-transfection of RFP reduced the interaction of both proteins (fifth lane). Second panel: the physical interaction between Epc1 and RFP was shown again. Third to fifth panels: the expression of individual proteins was confirmed. (c) RFP blocked binding of SRF to the SRE in the presence of Epc1. ChIP assay revealed that Epc1 potentiated binding of SRF to the SRE of the proximal skeletal α-actin promoter. However, co-transfection of RFP reduced its binding. (d) Binding of Epc1 to the SRE in skeletal α-actin promoter was also significantly reduced by co-transfection of RFP. (e) Quantification results of ChIP results of (d). (f) Reduction in RFP amounts caused increased association between SRF and Epc1 (uppermost panel). Anti-sense RFP C2C12 cell lines were established and used for the immunoprecipitation. Mock-transfected cell lines were used for control. (g) Decreased transcript level of RFP in anti-sense RFP cell lines. (h) ChIP assay showed that reduction of RFP caused the increase in the binding of SRF to the SRE. (i) Quantification of the ChIP assay. *P<0.05
On the basis of our previous report,14 we also postulated that RFP may affect the potentiation effects of Epc1. We performed chromatin immunoprecipitation (ChIP) analysis with anti-SRF antibody and amplified the promoter region flanking the SRE in the skeletal α-actin promoter with H9c2 cells. RFP itself did not significantly alter the binding of SRF to the SRE in the absence of exogenous Epc1 (Figure 4c, second versus third lane in the lower gel). As reported previously,14 Epc1 potentiated the binding of SRF to the SRE (second versus fourth lane). However, co-transfection of RFP reduced the binding of SRF to the SRE (fourth versus fifth lane).
Epc1 binds indirectly to the SRE by interacting through SRF.14 Therefore, we next checked whether RFP affects the indirect association of Epc1 with the SRE by ChIP analysis with anti-Epc1 antibody. Binding of Epc1 to the SRE was significantly reduced when RFP was co-transfected (second versus third lane in lower panel in Figures 4d and e).
We tried to check whether binding of SRF to either Epc1 or SRE is increased in the absence of RFP. Anti-sense RFP C2C12 cells showed a significant reduction of RFP protein (Figure 4f, lowermost panel) and transcript (Figure 4g). As expected, the reduction in RFP resulted in an increased association between SRF and Epc1 in C2C12 cells (Figure 4f, uppermost panel). The decrease in RFP also resulted in a significant increase in binding of SRF to the SRE (lower panel in Figures 4h and i). Taken together, these results suggest that RFP negatively regulates SRF by interfering with the potentiation effects of Epc1 on SRF.
Muscle mass and muscle-specific gene expression are enhanced in Rfp−/− mice
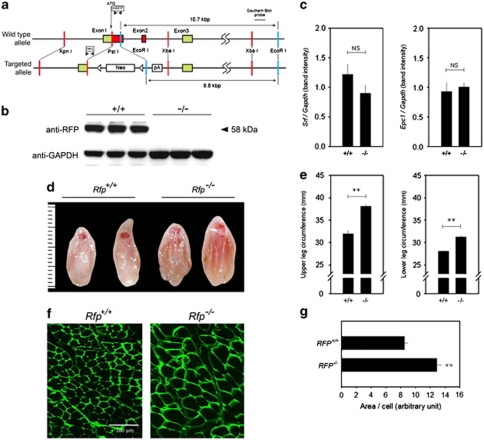
To investigate further the functional roles of RFP in muscle differentiation, we generated rfp−/− (Figure 5a; Supplementary Figures 3a–c). Rfp−/− mice were fertile and did not show significant phenotypic alterations in growth, movement, or glucose tolerance, as reported previously.18 No RFP protein was detected in the skeletal muscles of Rfp−/− mice (Figure 5b), and ablation of Rfp did not result in changes in the transcript levels of SRF and Epc1 (Figure 5c). Interestingly, however, the hamstring muscles of adult Rfp−/− mice were bigger than those of wild-type mice (Figures 5d–e).
Figure 5.
Muscular phenotypes in rfp−/−. (a) Schematic diagram showing the genetic locus of Rfp (wild-type allele; top) and the knockout strategy (bottom). The Rfp gene consists of 8 coding exons, and the first 3 are shown in the figure. The structure of the targeted Rfp allele is shown (bottom), as is the location of the Southern blot probes. mRFP: primer sets to detect wild-type allele. neo: primer sets for targeted allele. (b) Rfp protein was not detected in hamstring muscles of three Rfp−/− mice (right three lanes). (c) Srf and Epc1 transcript levels in the skeletal muscles of Rfp−/− and wild-type mice. (d) Entire thighs were excised and shown. A ruler is shown on the left (unit=mm). (e) The circumferences of the upper and lower legs (n=6–8). (f) Immunohistochemical analysis of hamstring muscle from wild-type and Rfp−/− mice. Wheat germ agglutinin staining showed the increase in myofiber size in Rfp−/− mice compared with Rfp wild-type mice. (g) Myofiber size was determined by measuring myofiber area and number per field. NS, not significant. **P<0.01
To further determine whether the larger muscles of the Rfp−/− mice were due to myofiber hypertrophy or to myofiber hyperplasia, we examined myofiber size and myofiber number in the hamstring muscle. Compared with the wild-type mice, Rfp−/− mice showed a significant increase in myofiber area, as demonstrated either by wheat germ agglutinin staining (Figures 5f and g) or by hematoxylin–eosin staining (Supplementary Figure 4). However, no apparent proliferating cell nuclear antigen-positive cells were observed in the null mice, which suggests that myofiber hyperplasia was not evident in the skeletal muscles from adult Rfp−/− mice (data not shown).
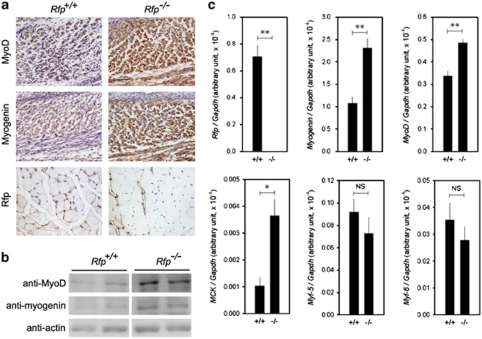
Immunohistochemical analysis of 18.5 day Rfp−/− embryos revealed that the expression of MyoD and myogenin was enhanced (Figure 6a). RFP protein was not detected in skeletal muscles from adult Rfp−/− mice (Figure 6a, lowermost panel). The expression of myogenin, MyoD, and Mck was significantly increased in the muscle from adult knockout mice (Figures 6b and c). Interestingly, we did not see alterations in the gene expression of Myf-5 and Myf-6, both of which are SRF-independent muscle-regulatory factors (Figure 6c).
Figure 6.
Muscle-specific gene expression is enhanced in Rfp−/− mice skeletal muscles. (a) The expression of MyoD and myogenin in 18.5 day embryo back muscles. Rfp protein was present in the muscles of wild-type but was absent in adult Rfp−/− skeletal muscles. (b) Protein amounts of MyoD and myogenin were increased in adult Rfp−/− skeletal muscles. In each panel, pictures were taken from the same gel. (c) Quantitative real-time RT-PCR results showing that transcript levels of SRF-dependent genes were increased in Rfp−/− mice, whereas those of SRF-independent ones (Myf-5 and Myf-6) were not. NS, not significant. *P<0.05, **P<0.01
Skeletal myoblast differentiation is enhanced in Rfp−/− mice
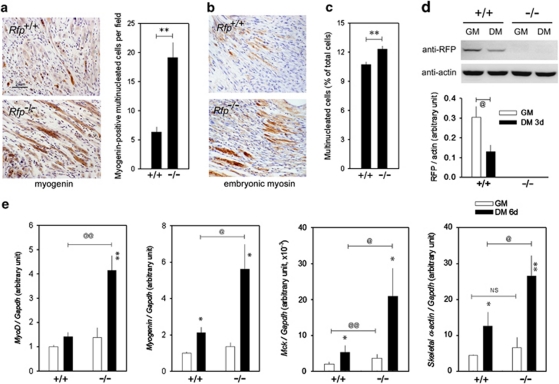
We used an in vivo wound-healing assay to examine the capacity of Rfp−/− mice to regenerate skeletal muscle. Cross-sectional slides were obtained from the hamstring muscles at the suture sites 1 week after surgery. Compared with that in the wild-type mice, myotube formation and muscle fiber differentiation were enhanced in Rfp−/− mice (Figure 7a, left panel). Counting of myogenin-positive myotubules and differentiating muscle fibers further confirmed the potentiation of muscle regeneration in the Rfp−/− mice (Figure 7a, right panel). The expression of embryonic myosin, a hallmark of muscular regeneration,19 was significantly increased in skeletal muscles from Rfp−/− mice (Figure 7b).
Figure 7.
Muscle regeneration and myoblast differentiation in Rfp−/− mice. (a) In vivo wound-healing assay showing enhanced myotube formation in Rfp−/− mice. The sections were collected 1 week after cutting and suture of hamstring muscles and were analyzed by immunohistochemistry analysis with myogenin. Note that myogenin-positive multinucleated myotubes were well developed in the wound of Rfp−/− mice (left lower panel). Myogenin-positive myotubes and differentiating muscle fibers were increased in Rfp−/− mice (right panel). (b) Increased embryonic myosin-immunoreactivity in Rfp−/− mice skeletal muscles. (c) Serum deprivation for 6 days induced greater multinucleation in the isolated myoblasts obtained from rfp−/−. (d) Serum deprivation-induced muscle-specific gene expression in myoblasts in knockout mice myoblasts. Western blot analysis showing the changes in Rfp protein amounts after serum deprivation for 3 days (upper panel). Quantification of the changes in Rfp protein amounts from four independent sets of experiments (lower panel). (e) Quantification of qRT-PCR for the transcripts of MyoD myogenin, Mck and skeletal α- actin. Muscle differentiation was induced by substituting growth medium (GM) with differentiation medium (DM) for 6 days. The induction of these genes was enhanced in the myoblasts from rfp−/−. NS, not significant. @,*P<0.05, @@,**P<0.01
We next checked the capacity of the myoblast cells to differentiate. Multinucleation induced by differentiation medium for 6 days was more prominent in myoblasts from Rfp−/− mice than in those from wild-type (Figure 7c). Serum deprivation for 3 days reduced RFP protein amounts in wild-type cells (Figure 7d). Increases in the amounts of MyoD, myogenin, Mck, and skeletal α-actin transcripts by 6 day serum deprivation were enhanced in myoblasts isolated from Rfp−/− mice (Figure 7e).
Discussion
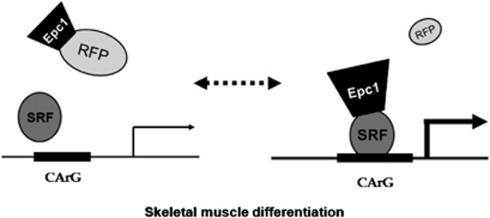
In this study, we uncovered functional and biological roles of RFP (TRIM27), a relatively uncharacterized member of the TRIM protein family, in skeletal myogenesis. We clearly showed that RFP inhibits muscle differentiation by altering SRF-dependent transcriptional activity. The mechanism we propose by which RFP acts in muscle differentiation is shown in Figure 8. During myoblast differentiation, a reduction in RFP expression results in its loss of binding to Epc1, which increases the binding of Epc1 to SRF and then enhances SRF-dependent muscle–gene transcription. Our findings, which show that RFP works as a novel modulator of myogenesis by modulating the interaction of Epc1 and SRF, provide new insights into the molecular mechanisms of gene expression during myogenesis.
Figure 8.
Suggested mechanism by which RFP regulates SRF-induced myogenesis. In the undifferentiated state, RFP inhibits Epc1-induced potentiation of the SRF-dependent transcriptional activation of muscle-specific genes. When a differentiation signal comes to the myoblasts, however, RFP is downregulated, which results in the relief of Epc1 from the inhibition and then in the potentiation of SRF-dependent skeletal muscle differentiation
RFP was first described to be associated with the nuclear matrix with high DNA-binding activity. RFP is a component of PML nuclear bodies and interacts with other protein family members of PML, other B-box family members,11, 20, 21 and other diverse proteins.22, 23, 24, 25, 26, 27 RFP even interacts with itself for homodimerization.28 By use of yeast two-hybrid screening, we found that RFP interacts with Epc1. An interaction between Epc1 and RFP in testis was established in a previous report.13 We found in this study that RFP is abundant in skeletal muscle, where Epc1 is also highly expressed,16 and that the interaction between RFP and Epc1 takes place in skeletal myoblast cells. These findings led us to postulate that the interaction may have a biological function in skeletal muscles.
Indeed, we next showed that RFP expression was gradually downregulated during myoblast differentiation induced by serum deprivation, which was in contrast with the reciprocal increase in the expression of Epc1. We also observed that the reduced expression in muscle-specific genes in Epc1 knockdown cells was recovered by double knockdown of RFP and Epc1. This result further suggests that RFP reversely regulates Epc1-mediated muscle differentiation.
Although it should be examined during the embryonic period when skeletal myogenesis is vigorous, the bigger muscles in the Rfp−/− mice than in the wild-type mice might have been caused by skeletal myofiber hypertrophy rather than myofiber hyperplasia. Considering that MyoD and myogenin are also involved in muscular hypertrophy,29 the apparent hypertrophy in Rfp−/− mice may be caused by an increase in those transcription factors.
RFP is known to have intrinsic transcription repressor function, which is dependent on its coiled-coil and RFP domains,13 and several different mechanisms of RFP-induced transcriptional repression have been proposed, largely dependent on its binding partners. For example, Shimono et al.22 reported that RFP forms a transcriptional repressor complex by its association with Mi-2β and BRG1. RFP can also enhance the transcriptional repression by MBD2 and MBD4 by its direct association.25 Thus, we first postulated that the delay in muscle differentiation might be caused by direct transcriptional modulation of muscle-specific genes, and we studied whether RFP directly inhibited the transcriptional activity of muscle-specific genes, such as skeletal α-actin or myogenin. However, it is not likely that those mechanisms are involved in the RFP-induced transcriptional modulation of Epc1/SRF, because RFP did not repress the basal transcriptional activity. Interestingly, however, RFP did inhibit Epc1/SRF-induced skeletal α-actin promoter activation. Of note, RFP abolished the Epc1-induced potentiation effect of SRF (Figure 3). RFP disrupted the binding of Epc1 to SRF by direct association with Epc1 (Figure 4). These results suggest that the competition between RFP and SRF for binding to Epc1 may be the mechanism by which RFP modulates SRF-dependent transcription. Although RFP failed to directly repress transcription in the present study, it is still noteworthy that RFP interacts with MyoD to inhibit its transcriptional activation of downstream genes in a trichostatin A-sensitive manner.30 This interaction suggests that inhibition of MyoD-dependent transcriptional regulation of myogenesis would be at least involved in the RFP-mediated inhibition of muscle–gene regulation.
Induction of skeletal muscle growth and differentiation is of major interest in the treatment of muscular dystrophy. One example of a therapeutic target is myostatin, a TGFβ superfamily protein, which strongly inhibits skeletal muscle growth31 and even induces cachexia.32 Thus, inhibition of myostatin or its receptors either by neutralizing antibody33 or by soluble decoy receptors34 would be useful for muscle regeneration and thereby muscular dystrophies. As RFP is highly expressed in certain types of cancers and it is closely associated with tumor progression and metastasis,35, 36 it has been suggested that ablation of RFP and its binding partners may be an excellent target for tumor therapeutics.37 In addition to targets for anticancer therapeutics, our findings emphasize that RFP may be an excellent target for muscle diseases.
Materials and Methods
All procedures were performed according to Chonnam National Medical School Research Institutional Animal Care and Use Committee approved procedures.
Plasmid constructs
The pCMV-myc-mouse Epc1 and skeletal α-actin promoter–reporter constructs were described previously.14 pFlag-CMV-RFP was also described previously.13
Antibodies, cell cultures, and transfection study
Epc1 antibody was described previously.16 Antibodies were used to recognize MyoD (C-20, Sc-304), myogenin (M-225, Sc-576), skeletal α-actin (Alpha sr-1, Sc-58671), MHC3 (embryonic myosin, Sc-53091), Gapdh (Sc-32233) (all of the above were from Santa Cruz Biotechnology (Santa Cruz, CA, USA)), desmin (ab6322; Abcam, Cambridge, UK), wheat germ agglutinin (W11261, Invitrogen, Eugene, OR, USA), and RFP (#01-8791; American Research Products, Inc., Belmont, MA, USA). H9c2, Cos7, 293T, and C2C12 cells were obtained from the Korean Cell Line Bank (KCLB, Seoul, Korea) and were maintained with DMEM containing 10 or 15% fetal bovine serum for C2C12 cells (growth medium). To induce differentiation, growth medium was replaced with differentiation medium of 1% fetal bovine serum for H9c2 or 2% horse serum for C2C12 cells. To establish stable cell lines, pCMV-Flag-RFP and vector were transfected into H9c2 or C2C12 cells. The cells were treated with 5 μg/ml blasticidine (Invitrogen, Carlsbad, CA, USA), and positive colonies were selected 2 weeks later.
To establish Epc1/Rfp double antisense cell lines, pcDNA3.1-V5-His- antisense Epc1 and pcDNA6/myc-HisA-antisense Rfp were transfected to C2C12 cells. The cells were treated with 5 μg/ml blasticidine and 150 μg/ml G418, and positive colonies were selected 2 weeks later.
For transient transfection of RFP and Epc1, pFlag-CMV-RFP and/or CMV-myc-Epc1 was introduced to 293T cells by use of Fugene 6 Transfection Reagent (Roche Diagnostics Corp., Indianapolis, IN, USA) or to C2C12 or H9c2 cells by use of Lipofectamine Plus reagent (Invitrogen).
Promoter analysis, immunoprecipitation, western blot, fluorescent immunocytochemistry, quantitative RT-PCR, and histology
These procedures were described previously.16 The antibodies for western blot analysis were anti-Flag (1 : 500), anti-myc (1 : 500), anti-RFP (1 : 500), anti-Epc1 (1 : 200), anti-MyoD (1 : 1000), anti-myogenin (1 : 1000), anti-skeletal α-actin (1 : 1000), and anti-actin (1 : 2000). The primer sequences for the RT-PCR reaction will be provided upon request. The RT-PCR amplification products were confirmed by sequencing. For immunohistochemistry, primary antibody dilutions of Rfp (1 : 500), embryonic myosin (1 : 100), myogenin (1 : 50), MyoD (1 : 100), and wheat germ agglutinin (1 : 200) were used. The muscle specimens were fixed in 4% paraformaldehyde, washed, dehydrated, paraffin-embedded, cut into 6-μm sections, and mounted. Immunohistochemistry was performed by using LSAB2 system-HRP kit (Dako, Carpinteria, CA, USA), according to the manufacturer's instructions. The sections were counterstained with Harris hematoxylin.
Morphometric analysis
The myofiber area and number in hamstring muscle of RFP null and wild mice were measured respectively from the fluorescence images of anti-wheat germ agglutinin-stained sections using Image Pro Plus (MediaCybernetics, Bethesda, MD, USA). The relative value of myofiber area/number in each group is expressed as mean±S.E.M.
ChIP assays
ChIP assays were described previously.14 Briefly, cells were treated with 1% formaldehyde for 10 min. The sonicated chromatin was immunoprecipitated with anti-SRF or anti-Epc1 antibody and was then recovered with protein A agarose/salmon sperm DNA beads. After reversing the cross-links, chromatin was subjected to proteinase K digestion and the DNA was purified. Primers used for PCR reactions were rat skeletal α-actin promoter (−192∼+5) and mouse skeletal α-actin promoter (−184∼+22).
Real-time RT-PCR
After reverse transcriptase reaction, 1 μl of cDNA was subjected to real-time PCR amplification by using the Rotor-Gene 3000 (Corbett Research, Mortlake, NSW, Australia) with a SyberGreen MasterMix (Applied Biosystems Inc., Foster City, CA, USA) according to the manufacturer's recommendations. For one experimental condition, the reaction was performed in triplicate with three or more independent cDNA samples.
Animal experiments
The investigation conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996). The experimental protocol was approved by the Chonnam National Medical School Research Institutional Animal Care and Use Committee. Mice were housed individually in plastic cages in a temperature-controlled room.
Rfp−/− mice and in vivo wound-healing assay
Rfp−/− were generated by conventional gene-targeting methods. Homologous recombination of the gene-targeting vector at the Rfp locus (Figure 5a, wild-type allele; top) was designed to insert a Neo cassette into the position of 10 amino acids after the start codon, which results in disruption of the remaining first coding exon and the entire second exon. The structure of the targeted Rfp allele (bottom) is shown (Figure 5a). The circumferences of the upper and lower legs were measured 9 mm proximal and 11 mm distal to the lower margin of the patella, respectively. The in vivo wound-healing assay was performed as described previously,16 and the sections were obtained 1 week after surgery.
Myoblast isolation
Primary skeletal myoblasts from either wild-type or rfp−/− were prepared as described previously with slight modifications. After mincing, muscle tissues from hind limbs were treated with collagenase (2 mg/ml; Worthington, Lakewood, NJ, USA) and the cell suspension was filtered through a 100-μm and then a 70-μm sieve (Cell Strainer Nylon, BD Falcon, San Jose, CA, USA). After preplating, the myoblasts were grown in 15% fetal bovine serum in DMEM supplemented with 5 nM basic fibroblast growth factor (Biosource, Camarillo, CA, USA). The isolated myoblasts underwent differentiation by replacing the growth medium (15% fetal bovine serum) with differentiation medium (2% horse serum).
Statistical analysis
Data are presented as mean±S.E.M. Student's t-tests were used to determine statistical significance.
Acknowledgments
This study was supported by a grant of the Korea Healthcare technology R&D Project, Ministry for Health, Welfare and Family Affairs (A084869), by the Korea Research Foundation (KRF-2007-313-E00126), and by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MEST, #2010-0015012). J-R Kim was supported by a Korea Research Foundation Grant (KRF-2008-532-E00003). H Joung was supported in part by the Brain Korea 21 Project through Chonnam National University Medical School.
Glossary
- ChIP
chromatin immunoprecipitation
- Epc1
enhancer of polycomb1
- Mck
muscle creatine kinase
- RFP
ret finger protein
- rfp−/−
rfp knockout mice
- SRE
serum response element
- SRF
serum response factor
- Trim27
tripartite motif
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Piacentini
Supplementary Material
References
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem Cell Biol. 1998;76:351–358. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- Grignani F, Fagioli M, Alcalay M, Longo L, Pandolfi PP, Donti E, et al. Acute promyelocytic leukemia: from genetics to treatment. Blood. 1994;83:10–25. [PubMed] [Google Scholar]
- Le Douarin B, Zechel C, Garnier JM, Lutz Y, Tora L, Pierrat P, et al. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Cooper GM. ret transforming gene encodes a fusion protein homologous to tyrosine kinases. Mol Cell Biol. 1987;7:1378–1385. doi: 10.1128/mcb.7.4.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Ritz J, Cooper GM. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell. 1985;42:581–588. doi: 10.1016/0092-8674(85)90115-1. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Inaguma Y, Hiai H, Hirose F. Developmentally regulated expression of a human “finger”-containing gene encoded by the 5′ half of the ret transforming gene. Mol Cell Biol. 1988;8:1853–1856. doi: 10.1128/mcb.8.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klugbauer S, Rabes HM. The transcription coactivator HTIF1 and a related protein are fused to the RET receptor tyrosine kinase in childhood papillary thyroid carcinomas. Oncogene. 1999;18:4388–4393. doi: 10.1038/sj.onc.1202824. [DOI] [PubMed] [Google Scholar]
- Chang KS, Stass SA, Chu DT, Deaven LL, Trujillo JM, Freireich EJ. Characterization of a fusion cDNA (RARA/myl) transcribed from the t(15;17) translocation breakpoint in acute promyelocytic leukemia. Mol Cell Biol. 1992;12:800–810. doi: 10.1128/mcb.12.2.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow J, Goddard AD, Sheer D, Solomon E. Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science. 1990;249:1577–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- Cao T, Duprez E, Borden KL, Freemont PS, Etkin LD. Ret finger protein is a normal component of PML nuclear bodies and interacts directly with PML. J Cell Sci. 1998;111 (Part 10:1319–1329. doi: 10.1242/jcs.111.10.1319. [DOI] [PubMed] [Google Scholar]
- Morris-Desbois C, Bochard V, Reynaud C, Jalinot P. Interaction between the Ret finger protein and the Int-6 gene product and co-localisation into nuclear bodies. J Cell Sci. 1999;112 (Part 19:3331–3342. doi: 10.1242/jcs.112.19.3331. [DOI] [PubMed] [Google Scholar]
- Shimono Y, Murakami H, Hasegawa Y, Takahashi M. RET finger protein is a transcriptional repressor and interacts with enhancer of polycomb that has dual transcriptional functions. J Biol Chem. 2000;275:39411–39419. doi: 10.1074/jbc.M006585200. [DOI] [PubMed] [Google Scholar]
- Kim JR, Kee HJ, Kim JY, Joung H, Nam KI, Eom GH, et al. Enhancer of polycomb1 acts on serum response factor to regulate skeletal muscle differentiation. J Biol Chem. 2009;284:16308–16316. doi: 10.1074/jbc.M807725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo C, Jang BG, Jo SA. MEK1 plays contrary stage-specific roles in skeletal myogenic differentiation. Cell Signal. 2009;21:1910–1917. doi: 10.1016/j.cellsig.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Kee HJ, Kim JR, Nam KI, Park HY, Shin S, Kim JC, et al. Enhancer of polycomb1, a novel homeodomain only protein-binding partner, induces skeletal muscle differentiation. J Biol Chem. 2007;282:7700–7709. doi: 10.1074/jbc.M611198200. [DOI] [PubMed] [Google Scholar]
- Menard C, Pupier S, Mornet D, Kitzmann M, Nargeot J, Lory P. Modulation of L-type calcium channel expression during retinoic acid-induced differentiation of H9C2 cardiac cells. J Biol Chem. 1999;274:29063–29070. doi: 10.1074/jbc.274.41.29063. [DOI] [PubMed] [Google Scholar]
- Kato T, Shimono Y, Hasegawa M, Jijiwa M, Enomoto A, Asai N, et al. Characterization of the HDAC1 complex that regulates the sensitivity of cancer cells to oxidative stress. Cancer Res. 2009;69:3597–3604. doi: 10.1158/0008-5472.CAN-08-4368. [DOI] [PubMed] [Google Scholar]
- Karsch-Mizrachi I, Travis M, Blau H, Leinwand LA. Expression and DNA sequence analysis of a human embryonic skeletal muscle myosin heavy chain gene. Nucleic Acids Res. 1989;17:6167–6179. doi: 10.1093/nar/17.15.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Duprez E, Borden KL, Freemont PS. Surface residue mutations of the PML RING finger domain alter the formation of nuclear matrix-associated PML bodies. J Cell Sci. 1997;110 (Part 18:2197–2205. doi: 10.1242/jcs.110.18.2197. [DOI] [PubMed] [Google Scholar]
- Tezel G, Nagasaka T, Iwahashi N, Asai N, Iwashita T, Sakata K, et al. Different nuclear/cytoplasmic distributions of RET finger protein in different cell types. Pathol Int. 1999;49:881–886. doi: 10.1046/j.1440-1827.1999.00957.x. [DOI] [PubMed] [Google Scholar]
- Shimono Y, Murakami H, Kawai K, Wade PA, Shimokata K, Takahashi M. Mi-2 beta associates with BRG1 and RET finger protein at the distinct regions with transcriptional activating and repressing abilities. J Biol Chem. 2003;278:51638–51645. doi: 10.1074/jbc.M309198200. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Shimono Y, Kawai K, Murakami H, Urano T, Niwa Y, et al. PIAS proteins are involved in the SUMO-1 modification, intracellular translocation and transcriptional repressive activity of RET finger protein. Exp Cell Res. 2005;308:65–77. doi: 10.1016/j.yexcr.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt M, Ellis M, Weekes DB, Bull JJ, Eilers M, Vivanco MD, et al. Selective ablation of retinoblastoma protein function by the RET finger protein. Mol Cell. 2005;18:213–224. doi: 10.1016/j.molcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Fukushige S, Kondo E, Gu Z, Suzuki H, Horii A. RET finger protein enhances MBD2- and MBD4-dependent transcriptional repression. Biochem Biophys Res Commun. 2006;351:85–92. doi: 10.1016/j.bbrc.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Townson SM, Kang K, Lee AV, Oesterreich S. Novel role of the RET finger protein in estrogen receptor-mediated transcription in MCF-7 cells. Biochem Biophys Res Commun. 2006;349:540–548. doi: 10.1016/j.bbrc.2006.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha J, Han KJ, Xu LG, He W, Zhou Q, Chen D, et al. The Ret finger protein inhibits signaling mediated by the noncanonical and canonical IkappaB kinase family members. J Immunol. 2006;176:1072–1080. doi: 10.4049/jimmunol.176.2.1072. [DOI] [PubMed] [Google Scholar]
- Cao T, Borden KL, Freemont PS, Etkin LD. Involvement of the rfp tripartite motif in protein-protein interactions and subcellular distribution. J Cell Sci. 1997;110 (Part 14:1563–1571. doi: 10.1242/jcs.110.14.1563. [DOI] [PubMed] [Google Scholar]
- Ishido M, Kami K, Masuhara M. Localization of MyoD, myogenin and cell cycle regulatory factors in hypertrophying rat skeletal muscles. Acta Physiol Scand. 2004;180:281–289. doi: 10.1046/j.0001-6772.2003.01238.x. [DOI] [PubMed] [Google Scholar]
- Bloor AJ, Kotsopoulou E, Hayward P, Champion BR, Green AR. RFP represses transcriptional activation by bHLH transcription factors. Oncogene. 2005;24:6729–6736. doi: 10.1038/sj.onc.1208828. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- Zimmers TA, Davies MV, Koniaris LG, Haynes P, Esquela AF, Tomkinson KN, et al. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296:1486–1488. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]
- Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LA, Ahima RS, et al. Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002;420:418–421. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- Ohsawa Y, Hagiwara H, Nakatani M, Yasue A, Moriyama K, Murakami T, et al. Muscular atrophy of caveolin-3-deficient mice is rescued by myostatin inhibition. J Clin Invest. 2006;116:2924–2934. doi: 10.1172/JCI28520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto H, Kato T, Enomoto A, Nakamura N, Shimono Y, Jijiwa M, et al. Expression of Ret finger protein correlates with outcomes in endometrial cancer. Cancer Sci. 2009;100:1895–1901. doi: 10.1111/j.1349-7006.2009.01278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezel GG, Uner A, Yildiz I, Guler G, Takahashi M. RET finger protein expression in invasive breast carcinoma: relationship between RFP and ErbB2 expression. Pathol Res Pract. 2009;205:403–408. doi: 10.1016/j.prp.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Tezel G, Shimono Y, Murakumo Y, Kawai K, Fukuda T, Iwahashi N, et al. Role for O-glycosylation of RFP in the interaction with enhancer of polycomb. Biochem Biophys Res Commun. 2002;290:409–414. doi: 10.1006/bbrc.2001.6161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.