Abstract
Animals often use sex pheromones for mate choice and reproduction. As for other signals, the genetic control of the emission and perception of sex pheromones must be tightly coadapted, and yet we still have no worked-out example of how these two aspects interact. Most models suggest that emission and perception rely on separate genetic control. We have identified a Drosophila melanogaster gene, desat1, that is involved in both the emission and the perception of sex pheromones. To explore the mechanism whereby these two aspects of communication interact, we investigated the relationship between the molecular structure, tissue-specific expression, and pheromonal phenotypes of desat1. We characterized the five desat1 transcripts—all of which yielded the same desaturase protein—and constructed transgenes with the different desat1 putative regulatory regions. Each region was used to target reporter transgenes with either (i) the fluorescent GFP marker to reveal desat1 tissue expression, or (ii) the desat1 RNAi sequence to determine the effects of genetic down-regulation on pheromonal phenotypes. We found that desat1 is expressed in a variety of neural and nonneural tissues, most of which are involved in reproductive functions. Our results suggest that distinct desat1 putative regulatory regions independently drive the expression in nonneural and in neural cells, such that the emission and perception of sex pheromones are precisely coordinated in this species.
Keywords: sensory communication, pleiotropy, hydrocarbon, oenocyte
The evolution and maintenance of sensory communication in animals is a fundamental biological problem. The genetic control of the signal and its reception must be tightly coadapted, especially in interindividual sexual communication to ensure sexual isolation (1–3). However, the basic genetic architecture of prezygotic sexual isolation remains largely unexplored in most natural populations (4, 5), and there is very little empirical evidence for tight genetic linkage between the emission and reception of a sensory signal (6–9). Theoritical prediction and experimental studies assume that the “emission/reception coupling” depends on the inheritance of separate genes found on the same or on different chromosomes and linked with a high probability (linkage desequilibrium (10–14), whereas the “single-gene” hypothesis seems very unlikely (4), as the tissues involved in the emission and perception of sensory signals are usually different (15).
Like in many animals, Drosophila melanogaster flies use sex pheromones to detect potential mates and reproduce (16, 17). Most of these pheromones include fatty acid-derived hydrocarbons present on the fly cuticle (cuticular hydrocarbons, CHs), which are thought to be mostly perceived by contact with the taste hairs covering the tarsi and proboscis (18, 19). In D. melanogaster, CHs differ between the sexes, both for their occurrence and for their behavioral effect on male courtship. The predominant CHs of wild-type females tend to increase male intraspecific courtship and mating [7,11 dienes (20–22)], whereas the male principal CH reduces male-male courtship [7-tricosene (7-T) (23, 24)]. Female and male CHs share a double-bond on carbon 7, which depends on desat1, a gene coding multiple transcripts, all giving rise to a single desaturase enzyme (25, 26).
We found a transposable PGal4 element inserted in the regulatory region of desat1 that drastically altered the production and the perception of sex pheromones in D. melanogaster flies (26, 27). The defects induced in the two phenotypes could be dissociated following unprecise deletion of the P-Gal4 sequence (27) and RNA deregulation induced by EP-elements inserted in various putative regulatory regions (9, 28). Because the alteration in the two phenotypes was not always coincidental, this finding suggests that the desat1 gene has pleiotropic—separate—effects on pheromonal communication. As very few single genes have been reported to control both the emission and reception of sensory signals [(7); see also the discussion in refs. 5 and 13], we aim at understanding how a single gene can be involved in such different aspects of pheromonal communication. We characterized the molecular structure of desat1 and its five transcripts and used the five putative regulatory regions—each one corresponding to a different transcript—to build driver transgenes. These drivers were used to target: (i) GFP, to visualize the pattern of tissue expression in male and female flies, and (ii) desat1-RNAi, to measure the consequence of desat1 down-regulation on pheromone production and perception.
Results
We aimed to find a relationship among the molecular structure, the tissue expression, and the two principal pheromonal phenotypes (production/discrimination) of desat1.
Molecular Characterization and Dissection of desat1.
We characterized the structure of desat1 gene in the Canton-S (Cs) wild-type strain and isolated five transcripts, which only diverged for their first noncoding exon (successively: RA, RC, RE, RB, RD) (Fig. 1A and Tables S1 and S2). The transcription initiation and splicing sites of the first alternative exon for the RE transcript were very different from the sites initially predicted (http://flybase.org/), as they are located at 294-bp and 305-bp downstream, respectively. The other transcripts showed a gap of several nucleotides for their transcription initiation starting site (13-bp and 9-bp upstream for RC and RD transcripts) and for their splicing site (25-bp upstream for RB and 11-bp downstream for RD) compared with those predicted by the genome project. Only the RA transcript perfectly matched the prediction. Except for the first exon, all transcripts included the identical downstream exons (2–5) and the same splicing sites, resulting in an identical coding sequence translated into a Δ9 desaturase enzyme. Transcripts showed no sex difference.
Fig. 1.
Structure of the desat1 gene and transgenes. (A) The desat1 gene includes five alternative first exons (shown as shaded boxes and indicated RA, RC, RE, RB and RD), all of which combine with exons 2–5 (corresponding to the coding region) to produce five transcripts all translated into the Desat1 desaturase. (B) Transgenes with the desat1 regulatory regions were built with: (i) the sequence of each region combined with its exon [PRR(RA) to PRR(RD)], and (ii) with the putative regulatory region either complete PRR(-6908) or deleted of the most distal regions down to the minimal regulatory region PRR(-660). These PRR sequences were fused to a reporter sequence corresponding either to the GFP sequence to visualize desat1 expression or to the Gal4 sequence to target UAS transgenes.
Because all transcripts encode the same unique product, we postulated that the “pheromone production” and “pheromone discrimination” phenotypes associated with desat1 depend on the distinct tissular expression of transcripts. To study the tissue expression corresponding to each transcript, we built transgenic lines with the different desat1 putative regulatory regions (PRR) (Fig. 1B and Table S3). These lines contained the sequence of each putative regulatory region plus the first alternative exon fused either to the GFP or to the Gal4 sequence [single-exon transgenes: PRR(RA) to PRR(RD)-GFP or -Gal4]. Other transgenes were made with the complete region [PRR(-6908)] or with shorter regions—resulting from the progressive deletion of the most distal regions—down to the minimal one: PRR(-660).
Expression of desat1 in the Abdomen.
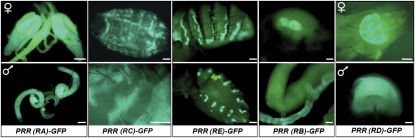
First, we used the single-exon PRR(desat1)-GFP transgenes to visualize the tissue expression driven by each putative regulatory region. These trangenic drivers induced nonoverlapping expression in the fly abdomen (Fig. 2; see also the matrix of expression, Table S4). More specifically, PRR(RA)-GFP and PRR(RD)-GFP drove expression in sexually dimorphic tissues: PRR(RA)-GFP targeted testis and ovaries (with an strong signal in the germinarium), whereas PRR(RD)-GFP targeted a vaginal moon-shaped structure and the male ejaculatory bulb (EjB). PRR(RE)-GFP induced a strong expression in the dorsal and ventral oenocytes and a weaker expression in the accessory glands. PRR (RC)-GFP targeted the fat body (FB), whereas PRR(RB)-GFP was expressed in the rectal papilla, the Malpighi tubules, and the midgut. Overall, we found the GFP expression targeted by single-exon PRR to be coherent with that driven by longer PRR-GFP transgenes (Table S4).
Fig. 2.
Expression of desat1 in the adult abdomen. The pictures show GFP expression driven by the five single-exon PRR(desat1)-GFP transgenes. PRR(RA)-GFP and PRR(RD)-GFP females and males showed sexually dimorphic expression in ovaries and testis [PRR(RA)] and in a vaginal moon-shaped structure and in the EjB [PRR(RD)]. PRR(RE)-GFP drove expression in dorsal (Upper) and ventral oenocytes (Lower). PRR(RC)-GFP targeted the FB; PRR(RB)-GFP was expressed in the rectal papilla (Upper) and in the Malpighi tubules (Lower). [Scale bars: 100 μm for PRR(RA)-GFP, 200 μm for PRR(RC)- and PRR(RE)-GFP, and 10 μm for PRR(RB)- and PRR(RD)-GFP.]
Expression of desat1 in the Head.
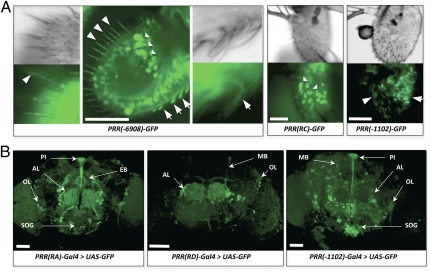
Using the complete set of transgenes, we detected a prominent GFP expression in the third antennal segment and in the brain. The complete desat1 regulatory region, PRR(-6908)-GFP, induced a strong antennal expression in large epidermal cells, trichoid sensilla, and large basiconic sensilla (Fig. 3A and Table S4). The expression in large epidermal cells was exclusively targeted by the PRR(RC)-GFP transgene, whereas the signal in the trichoid and large basiconic sensilla was targeted by PRR(-1102)-GFP [but by neither PRR(RD)-GFP nor PRR(-660)-GFP drivers].
Fig. 3.
Expression of desat1 in the adult antenna and brain. GFP expression in the third antennal segment (A) and in the brain (B) driven by different PRR(desat1) transgenes. (A) Antennal expression driven both by the complete PRR [PRR(-6908)-GFP] and by the fat body PRR [PRR(RC)-GFP] was detected in the giant epidermal cells (indicated by triangles). Expression driven both by the PRR(-6908)-GFP and by the PRR(-1102)-GFP transgenes was detected in the trichoid (arrowhead) and large basiconic sensilla (arrows). (Scale bars, 50 μm.) (B) In the brain, GFP expression was targeted by the PRR(RA)-Gal4, PRR(RD)-Gal4, and PRR(-1102)-Gal4 transgenes. More specifically, expression was detected in the PI [PRR(RA), PRR(-1102)], in the EB [PRR(RA)], AL (three transgenes), SOG [PRR(RA); PRR(-1102)], and OL (three transgenes). Moreover, PRR(-1102)-Gal4 induced a strong expression in the PI, SOG, and scattered cells of the central brain and OLs. The two latter PRR(desat1)-Gal4 drivers also induced a faint expression in the MBs. (Scale bars, 100 μm.)
Three PRR-GFP transgenes induced a partly overlapping expression in the brain (Fig. 3B and Table S4). PRR(RA)-GFP targeted the pars intercerebralis (PI; rich in neurosecretory neurons), the ellipsoid body (EB; a part of the central complex), clustered cells in the lateral region of the antennal lobes (ALs), and scattered cells in the suboesophageal ganglia (SOG) and in the optic lobes (OLs). The brain expression pattern driven by PRR(RD)-Gal4 and PRR(-1102)-Gal4 showed some similarities and differences. Both drivers targeted clustered AL cells, with a somewhat similar or overlapping pattern to that targeted by PRR(RA)-Gal4. Moreover, PRR(-1102)-Gal4 induced a strong expression in the PI, SOG, and scattered cells of the central brain and OLs. The two latter PRR(desat1)-Gal4 drivers also induced a faint expression in the mushroom bodies (MBs).
Targeting desat1 RNAi to Induce Specific Pheromonal Defects.
To establish a structure–function relationship between the tissues targeted by each putative regulatory region and its effect on pheromonal phenotypes, each PRR(desat1)-Gal4 driver was combined with a UAS-desat1 RNAi transgene. The effect of desat1 down-regulation was measured on sex pheromone production and on sex pheromone discrimination in PRR(desat1)-Gal4 > UAS-desat1 RNAi transgenic males. To check the efficiency of desat1 RNAi, we measured the Desat1 protein level in PRR(RC)-Gal4> UAS-desat1 RNAi using both histology and immunoblotting methods (Fig. S1).
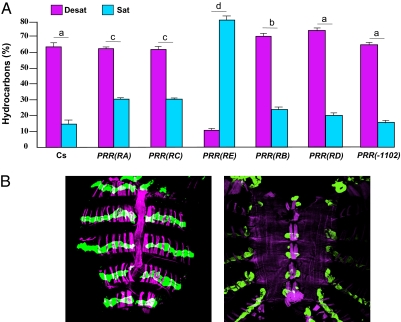
We first assessed the desat1 RNAi effect on the production of CHs and compared the ratio for the total percentage of desaturated and saturated linear CHs in transgenic males (Desat and Sat, respectively) (Fig. 4A). Compared with wild-type Cs males where the Desat/Sat ratio was strongly biased in favor of Desat CHs (4.12), PRR(RE)-Gal4 > UAS-desat1 RNAi males showed a dramatically altered ratio biased in favor of Sat CHs (0.13). Moreover, and compared with control flies, the ratio was decreased in PRR(RA)-, and PRR(RC)-Gal4 > UAS-desat1 RNAi males and, to a lesser extent, in PRR(RB)-Gal4 > UAS-desat1 RNAi males [one-way ANOVA with Tukey–Kramer honestly significant difference (HSD): F(6,142) = 134.4; P < 0.0001]. A similar effect on CHs was found in transgenic female flies (Fig. S2).
Fig. 4.
Down-regulation of desat1 expression and cuticular hydrocarbons. (A) The expression of the UAS-desat1 RNAi transgene was targeted in different tissues with six PRR(desat1)-Gal4 transgenes. Bars indicate the mean (± SEM) total proportion of desatured (Desat) and linear saturated (Sat) CHs produced by individual 5-d-old males of the six transgenic genotypes and control Cs males. We compared the Desat/Sat ratio between genotypes. The different letters above each pair of bars indicate the significant differences (one-way ANOVA with a Tukey–Kramer HSD post hoc test). The ratio was drastically affected in PRR(RE)-Gal4 > UAS-desat1 RNAi flies. n = 15–32, except for RD = 9. (B) The GFP expression in the abdomen of PRR(RE)-Gal4 > UAS-GFP flies (Left: dorsal view; Right: ventral view) was detected in the oenocytes (labeled in green by anti-GFP antibody). The magenta color indicates the presence of muscles labeled by phalloidin. (Scale bars, 100 μm.)
To precisely assess the cells targeted by PRR(RE)-Gal4, the flies carrying this transgene were mated with flies carrying UAS-GFP: it clearly labeled dorsal and ventral oenocytes that have been implicated in pheromonal production (Fig. 4B), but its expression was not detected in the central nervous system (Fig. S3).
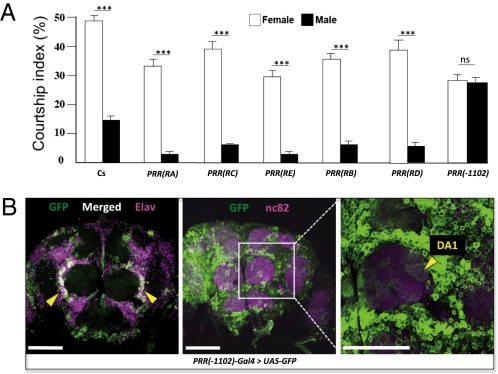
Next, we measured the desat1 RNAi effect on male ability to discriminate sex pheromones. Single transgenic tester males were simultaneously presented to control female and male target flies and their courtship index to each target measured (CIf and CIm, respectively) (Fig. 5A). The comparison between CIf and CIm indicates the ability of males to discriminate sex pheromones (in wild-type Cs males: CIf >>> CIm ; t = 4.78; df = 76 ; P < 0.0001). Most transgenic lines showed such a high ability (t = 5.12–8.44; df = 86–96; P < 0.0001), with the exception of PRR(-1102)-Gal4 > UAS-desat1 RNAi males, which completely lost their ability to discriminate sex pheromones (CIf = CIm; t = 0.33; df = 102 ; P = not significant).
Fig. 5.
Down-regulation of desat1 expression and male discrimination of sex pheromones. (A) The expression of the UAS-desat1 RNAi transgene was targeted in different tissues with six PRR(desat1)-Gal4 transgenes (Fig. 1). Bars represent the mean (± SEM) courtship index shown by individual 5-d-old control (Cs) and transgenic males to a target control female (empty bars) and to a target control male (filled bars). The discrimination ability of each male genotype was statistically assessed by the courtship index difference to both targets (Student t test; ***P < 0.001; ns = not significant; n = 32–54). (B) GFP expression in a frontal view of the brain of PRR(-1102)-Gal4 > UAS-GFP male flies. (Left) The expression of the GFP (Desat1; green color) show a colabeling with the neuronal marker Elav (magenta) in cell bodies lateral to the antennal lobes (white; arrow heads). A magnified view of the central brain (Center: expression of GFP shown in green and nc82, a neuropil marker, shown in magenta) reveals that neural expression in the left antennal lobe (Right) is located in the DA1 glomerulus (arrowhead). (Scale bars, 50 μm.)
Because male pheromonal discrimination was strongly affected when desat1 down-regulation was targeted by PRR(-1102)-Gal4, we mapped the cells targeted by this transgene. In the brain of PRR(-1102)-Gal4 > UAS-GFP males, a group of neurons was targeted in the lateral side of ALs (Fig. 5B). A careful observation revealed that some of these neurons arborize in the DA1 (dorsal) glomerulus, which is implicated in pheromonal perception (29).
Discussion
We previously found that desat1 separately affects the two principal aspects of pheromonal communication: this pleiotropy phenomenon was related to the quantitative variation of distinct transcripts (9, 27). Here, we showed that the separate control of sex-pheromone emission and reception depends on the effect of distinct putative regulatory regions targeting either nonneuronal cells necessary for pheromone biosynthesis or neurons involved in pheromone perception. If, in several moths species, the emergence of new olfactory signals depends on desaturase enzymes, their involvement in the reception of these signals has not been shown (30). For exemple, in the European Corn Borer Ostrinia nubilalis, the system of pheromonal communication depends on factors segregating with different chromosomes and with distinct loci on the same autosome (12). The pheromonal differences between O. nubilalis and Ostrinia furnacalis result from the activation of an ancestral desaturase gene in O. furnacalis—inactivated in all other Ostrinia species—combined with the presence of rare males able to respond to the new pheromone (31). However, the effect of the ancestral desaturase gene in the ability of O. furnacalis males to respond to this new compound remains unknown, and this contrasts with the multiple pheromonal aspects controled by the desat1 gene in D. melanogaster.
In agreement with other studies (32–34), our data clearly pinpoint the role of oenocytes in the production of sex pheromones, and the additive effect—to a lesser extent—of other tissues on this process. The moderate effect induced by PRR(RA)-Gal4 > UAS-desat1 RNAi on pheromone production indicates that the PI exerts its action through a neurohormonal control, as suggested by other studies (9, 35–37). The moderate changes in pheromone production induced by PRR(RC)-Gal4 > UAS-desat1 RNAi suggest that FB also contribute to CH synthesis, as previously hypothezised (33). One of the primary functions of desat1 may be to regulate lipid metabolism in the fly (38), either directly [with the FB and oenocytes (38, 39)] or through the hormonal control [of the PI (40, 41)]. This gene also targets tissues involved in the regulation of water content (Malpighi tubule and rectal papilla) and this may be linked to the fact that CHs also serve to regulate water content in the fly body (42).
Targeted down-regulation of desat1 also induced clear-cut effects on pheromonal discrimination: among transgenic lines, only PRR(-1102)-Gal4 > UAS-desat1 RNAi males showed a strikingly altered pheromonal discrimination. Interestingly, this driver targeted the neural cells potentially involved in pheromonal perception and, more particularly, in the neurons housed by trichoid sensilla (43), which project to a sexually dimorphic glomerulus (DA1) (44)]. This glomerulus is implicated in the perception of cis-vaccenyl acetate (45, 46), a male-specific volatile pheromone produced in the EjB (47), modulating sexual behavior (18, 29). Both trichoid and basiconic sensilla could also respond to other sex pheromones (43, 48), such as 7-T (a male-predominant CH) thought to be perceived by the antenna (49).
Apart from pheromonal communication, the desat1 gene is also expressed in a variety of tissues modulating other aspects of reproduction such as the maturation of gametes (by testis, accessory glands, and ovaries), and their release by male (EjB) and female flies [PI (50) and accessory glands (51)]. Because some of desat1-positive tissues also produce sex signals [cis-vaccenyl acetate in the EjB (47); CHs in the oenocytes and FB (32–34); sex peptides in accessory glands (52)], a possible coevolutionary process may have shaped some desat1 regulatory regions to allow expression in chemosensory and brain neurons responsive to sex pheromones. Our next challenge will consist of precisely unraveling the function of desat1 in these neurons in relation with their role in pheromonal perception.
Materials and Methods
Strains.
All D. melanogaster strains were raised on yeast/cornmeal/agar medium and kept at 24 ± 0.5 °C with 65 ± 5% humidity on a 12-h light-dark cycle. To generate transgenic flies, all constructs were injected in w1118 mutant strain embryos according to a standard procedure (53). We used the Canton-S (Cs) strain as a control, the UAS-mCD8::GFP transgenic line (UAS-GFP) to visualize PRR(desat1)-Gal4 expression patterns (54) (Bloomington Stock Center), and three UAS-desat1-RNAi transgenic lines to knock down desat1 expression (VDRC; #33338, 47141, 47142; Data shown here were obtained with the #33338 line). Fly lines carrying these UAS-fusion transgenes were combined with those carrying PRR(desat1)-Gal4 transgenes established in our laboratory using standard techniques and genetic tools (55, 56). To enhance the effects of the PRR(desat1)-Gal4 > UAS-desat1 RNAi system, the pheromonal and behavioral phenotypes were always measured in flies homozygous for both the PRR(desat1)-Gal4 drivers and the UAS-desat1 RNAi reporter transgene. No pheromonal alteration was detected in parental flies homozygous for either of these transgenes.
Molecular Characterization of desat1 Transcripts.
Total RNA was extracted from 30 5-d-old sexed flies using TRIzol (Invitrogen) following the manufacturer's protocol. RNA was subsequently treated with DNase I for 15 min at 37 °C. The first strand cDNA was synthesized by SuperScript II RT enzyme (Invitrogen) for 50 min at 42 °C from 1.5 μg RNA using primer oligo(dT) to characterize the different desat1 transcripts or a primer localized in the last exon for the 5′ RACE-PCR. Amplification products were ligated in pGEM-T (Promega) and maintained in Escherichia coli DH-5α cells. The positive clones analyzed by restriction were sequenced. Each analysis was repeated with three independent RNA biological extracts. The putative transcription start sites were identified as the first nucleotide immediately adjacent to the polynucleotide tail (for more details, see Tables S1 and S2).
Construction of GFP and Gal4 Fusion Vectors.
The 6,908-bp DNA fragment containing the whole 5′ cis-regulatory region of the desat1 gene was amplified by Hot Start PCR reactions (20 cycles) from Cs genomic DNA (100 ng) using Platinium Taq DNA polymerase High Fidelity (Invitrogen). The amplification products were cloned in pGEM-T (Promega) and maintained in E. coli DH-5α cells. We selected a single clone without mutation, to serve as a template for the different constructs (pGEM-6908/+4).
To obtain the regulatory sequences used to build the plasmids, we used two alternative approaches (see Table S3 for details). Plasmids were named according to their exons and regulatory sequences: PRR(RA), PRR(RC), PRR(RE), PRR(RB), and PRR(RD). In some plasmids, the coding sequence of cytoplasmic eGFP contained in the KpnI-SpeI DNA fragment of the pGreen Pelican was replaced by the Gal4 coding sequence amplified by PCR from plasmid pPTGAL (57).
Hydrocarbons.
CHs were extracted from 5-d-old intact individual flies by gas chromatography following a brief wash in hexane according to standard procedures (58). Analyses were performed with a Varian CP3380 chromatograph, equipped with a Cp-sil 25-m capillary column with hydrogen as the carrier gas. All of the D. melanogaster predominant CHs have already been identified and characterized (20, 59). We analyzed 24 CHs in female flies and 14 in male flies, all with a chain length ranging between 23 and 29 carbons (26). Each CH was characterized by its percentage relatively to the sum of all CHs.
For the sake of clarity, we only show the sum of unsaturated CHs (Desat: 9-T + 7-T + 5-T + 9-P + 7-P + 5-P for males and with the 7,11-PD + 9,13-HD + 7,11-HD + 9,13-ND + 7,11-ND for female flies) and the sum of saturated linear CHs (Sat: n-C23 + n-C25 + n-C27 + n-C29). For Desat CH nomenclature, numbers indicate the position of double bonds and letters the compound name: T, tricosene; P, pentacosene; HD, heptacosadiene; ND, nonocosadiene. Saturated linear alkanes are named according to their carbon chain (e.g.: n-C23 is n-tricosane). To compare the ratio between the total levels of Desat and Sat CHs between genotypes, we used a one-way ANOVA normalized with a Box-Cox transformation (optimized λ-value: 0.843 and 0.613 in males and females, respectively) with a Tukey–Kramer HSD post hoc test.
Behavior.
The simultanous discrimination of single tester males was measured during a 5-min period 1 to 4 h after lights on. Tests were carried out under a dim red light (25 W with a Kodak Safe-light filter n°1) to remove all visual stimuli (60) and with two decapitated target flies (male vs. female) to remove acoustic and behavioral signals (61). All flies were isolated 0–4 h after eclosion under CO2 anesthesia. Tester male flies (i.e., the sexual responses of which to target flies were measured) were held individually in fresh glass food vials for 5 d before testing. Wild-type target flies were similary isolated and held in groups of five for the same period. All tests were performed at 24 ± 0.5 °C with 65 ± 5% humidity. For each genotype, tests were performed over several days. Tester males were individually aspirated (without anesthesia) under a watch glass used as a courtship observation chamber (1.6 cm3). After the 5-min period necessary for the male to habituate to the chamber, the two target flies were introduced and the observation period started. During the test, we precisely measured the total duration of male courtship expressed as the courtship index toward the target female (CIf) and the target male (CIm). Courtship index is the proportion of time that the male spends in active courtship (tapping, wing vibration, licking, and attempting copulation). To assess the male ability to discriminate the sex of target flies, individual CIms and CIfs were compared with a Student t test.
Histology.
The expression patterns revealed by PRR(desat1)-GFP and PRR(desat1)-Gal4 > UAS-GFP flies were examined. No qualitative difference in the expression pattern was detected between PRR(desat1)-GFP and PRR(desat1)-Gal4 > UAS-GFP flies, provided that they contained the same desat1 fragment. Note that GFP expressed from the direct fusion PRR(desat1)-GFP transgene is cytoplasmic in contrast to that produced by the UAS-mCD8::GFP transgene driven by PRR(desat1)-Gal4. Moreover, because the pUAS vector (used to build the UAS-fusion transgenes) contained the hsp70 minimal promoter, PRR(desat1)-Gal4 > UAS-GFP did not show expression in the germ line (62), contrary to direct fusion PRR(desat1)-GFP transgenes, which allowed us to visualize GFP expression in the gonads.
Brains were dissected out from 5-d-old flies in PBS and fixed with 4% PFA/PBS for 1 h at room temperature. After several washes with 0.3% Triton-X100/PBS (PBT), brains were blocked with normal goat serum (10% in PBT) overnight at room temperature and incubated for 72–96 h at 4 °C with the primary antibodies. Mouse monoclonal anti-nc82 (1/10; Developmental Studies Hybridoma Bank) and rabbit polyclonal anti-GFP (1/1,000; Molecular Probes) were used as the primary antibodies. After washing in PBT, samples were incubated 48–72 h at 4 °C in a mixture of secondary antibodies containing Alexa Fluor 633 goat anti-mouse IgG and Alexa Fluor 488 goat anti-rabbit IgG (1/400 each; Molecular Probes).
Abdominal tissues were dissected from the 5-d-old flies in PBS, and fixed with 4% PFA/PBS for 20 min at room temperature. After washing and blocking with normal goat serum (10% in PBT) for 12 h, the abdominal tissues were incubated for 24–48 h at 4 °C with rabbit polyclonal anti-GFP (1/1,000; Molecular Probes). Following washes in PBT, tissus were stained with a solution containing Alexa Fluor 488 goat anti-rabbit IgG (1/500; Molecular Probes) and Texas Red-conjugated phalloidin (1/200; Molecular Probes).
Tissues were mounted in 80% (vol/vol) glycerol after four 30 min washes in PBT. Immunofluorescence was observed using a Leica TCS SP2 AOBS confocal microscope. Images were scanned at 1-μm section intervals and processed with Adobe Photoshop software.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for their comments on the manuscript, and Claude Everaerts for help with the statistics. This work was supported in part by the Centre National de la Recherche Scientifique, the Burgundy Regional Council, and by the Agence Nationale de la Recherche (Insect Aversive Learning) (F.B., B.H., I.C., S.C., S.D., and J.-F.F.); an Institutional Program for Young Reseacher Overseas Visits from the Japan Society for the Promotion of Science (T.N.); Ministry of Education, Culture, Sports, Science, and Technology (MEXT) Grant 1802012 (to D.Y.); the Tohoku Neuroscience Global Centers of Excellence program (MEXT) (D.Y.); and the Strategic Japanese-French Cooperative Program (Centre National de la Recherche Scientifique-Japan Science and Technolgy; Structure and Function of Biomolecules) (D.Y. and J.-F.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 5.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109166108/-/DCSupplemental.
References
- 1.Endler JA. Some general comments on the evolution and design of animal communication systems. Philos Trans R Soc Lond B Biol Sci. 1993;340:215–225. doi: 10.1098/rstb.1993.0060. [DOI] [PubMed] [Google Scholar]
- 2.Hauser MD. The Evolution of Communication. Cambridge: MIT Press; 1996. [Google Scholar]
- 3.Andersson M, Simmons LW. Sexual selection and mate choice. Trends Ecol Evol. 2006;21:296–302. doi: 10.1016/j.tree.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Moehring AJ, et al. Quantitative trait loci for sexual isolation between Drosophila simulans and D. mauritiana. Genetics. 2004;167:1265–1274. doi: 10.1534/genetics.103.024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kronforst MR, et al. Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proc Natl Acad Sci USA. 2006;103:6575–6580. doi: 10.1073/pnas.0509685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blows MW. Evolution of the genetic covariance between male and female components of mate recognition: An experimental test. Proc Biol Sci. 1999;266:2169–2174. doi: 10.1098/rspb.1999.0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukamachi S, et al. Dual control by a single gene of secondary sexual characters and mating preferences in medaka. BMC Biol. 2009;7:64. doi: 10.1186/1741-7007-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw KL, Lesnick SC. Genomic linkage of male song and female acoustic preference QTL underlying a rapid species radiation. Proc Natl Acad Sci USA. 2009;106:9737–9742. doi: 10.1073/pnas.0900229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houot B, Bousquet F, Ferveur JF. Regulation of desat1 affects both pheromone emission and detection in Drosophila melanogaster. Genetics. 2010;185:1297–1309. doi: 10.1534/genetics.110.117226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lande R. Models of speciation by sexual selection on polygenic traits. Proc Natl Acad Sci USA. 1981;78:3721–3725. doi: 10.1073/pnas.78.6.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noor MAF, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci USA. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Löfstedt C, Hansson BS, Roelofs W, Bengtsson BO. No linkage between genes controlling female pheromone production and male pheromone response in the European corn borer, Ostrinia nubilalis Hübner (Lepidoptera; Pyralidae) Genetics. 1989;123:553–556. doi: 10.1093/genetics/123.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smadja C, Butlin RK. On the scent of speciation: The chemosensory system and its role in premating isolation. Heredity. 2009;102:77–97. doi: 10.1038/hdy.2008.55. [DOI] [PubMed] [Google Scholar]
- 14.Gould F, et al. Sexual isolation of male moths explained by a single pheromone response QTL containing four receptor genes. Proc Natl Acad Sci USA. 2010;107:8660–8665. doi: 10.1073/pnas.0910945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boake CR. Coevolution of senders and receivers of sexual signals: Genetic coupling and genetic correlations. Trends Ecol Evol. 1991;6:225–227. doi: 10.1016/0169-5347(91)90027-U. [DOI] [PubMed] [Google Scholar]
- 16.Wyatt TD. Pheromones and Animal Behaviour: Communication by Smell and Taste. Cambridge, UK: Cambridge Unive Press; 2003. [Google Scholar]
- 17.Ferveur JF. Cuticular hydrocarbons: Evolution and role in the pheromonal communication of Drosophila. Behav Genet. 2005;35:279–295. doi: 10.1007/s10519-005-3220-5. [DOI] [PubMed] [Google Scholar]
- 18.Jallon JM. A few chemical words exchanged by Drosophila during courtship and mating. Behav Genet. 1984;14:441–478. doi: 10.1007/BF01065444. [DOI] [PubMed] [Google Scholar]
- 19.Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: A review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- 20.Antony C, Jallon JM. The chemical basis for sex recognition in Drosophila melanogaster. J Insect Physiol. 1982;28:873–880. [Google Scholar]
- 21.Savarit F, Sureau G, Cobb M, Ferveur JF. Genetic elimination of known pheromones reveals the fundamental chemical bases of mating and isolation in Drosophila. Proc Natl Acad Sci USA. 1999;96:9015–9020. doi: 10.1073/pnas.96.16.9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcillac F, Ferveur JF. A set of female pheromones affects reproduction before, during and after mating in Drosophila. J Exp Biol. 2004;207:3927–3933. doi: 10.1242/jeb.01236. [DOI] [PubMed] [Google Scholar]
- 23.Ferveur JF, Sureau G. Simultaneous influence on male courtship of stimulatory and inhibitory pheromones produced by live sex-mosaic Drosophila melanogaster. Proc Biol Sci. 1996;263:967–973. doi: 10.1098/rspb.1996.0143. [DOI] [PubMed] [Google Scholar]
- 24.Lacaille F, et al. An inhibitory sex pheromone tastes bitter for Drosophila males. PLoS ONE. 2007;2:e661. doi: 10.1371/journal.pone.0000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wicker-Thomas C, Henriet C, Dallerac R. Partial characterization of a fatty acid desaturase gene in Drosophila melanogaster. Insect Biochem Mol Biol. 1997;27:963–972. doi: 10.1016/s0965-1748(97)00077-5. [DOI] [PubMed] [Google Scholar]
- 26.Marcillac F, Bousquet F, Alabouvette J, Savarit F, Ferveur JF. A mutation with major effects on Drosophila melanogaster sex pheromones. Genetics. 2005a;171:1617–1628. doi: 10.1534/genetics.104.033159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcillac F, Grosjean Y, Ferveur JF. A single mutation alters production and discrimination of Drosophila sex pheromones. Proc Biol Sci. 2005b;272:303–309. doi: 10.1098/rspb.2004.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rørth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- 30.Baker TC. Mechanism for saltational shifts in pheromone communication systems. Proc Natl Acad Sci USA. 2002;99:13368–13370. doi: 10.1073/pnas.222539799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roelofs WL, et al. Evolution of moth sex pheromones via ancestral genes. Proc Natl Acad Sci USA. 2002;99:13621–13626. doi: 10.1073/pnas.152445399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferveur JF, et al. Genetic feminization of pheromones and its behavioral consequences in Drosophila males. Science. 1997;276:1555–1558. doi: 10.1126/science.276.5318.1555. [DOI] [PubMed] [Google Scholar]
- 33.Savarit F, Ferveur JF. Genetic study of the production of sexually dimorphic cuticular hydrocarbons in relation with the sex-determination gene transformer in Drosophila melanogaster. Genet Res. 2002;79:23–40. doi: 10.1017/s0016672301005481. [DOI] [PubMed] [Google Scholar]
- 34.Billeter JC, Atallah J, Krupp JJ, Millar JG, Levine JD. Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature. 2009;461:987–991. doi: 10.1038/nature08495. [DOI] [PubMed] [Google Scholar]
- 35.Pennanech M, Ferveur JF, Pho DB, Jallon JM. Insect fatty acid related pheromones: A review of their biosynthesis, hormonal regulation and genetic control. Ann Soc Entomol Fr. 1991;27:245–263. [Google Scholar]
- 36.Wicker C, Jallon JM. Hormonal control of sex pheromone biosynthesis in Drosophila melanogaster. J Insect Physiol. 1995;41:65–70. [Google Scholar]
- 37.Arienti M, Antony C, Wicker-Thomas C, Delbecque JP, Jallon JM. Ontogeny of Drosophila melanogaster female sex-appeal and cuticular hydrocarbons. Integr Zool. 2010;5:272–282. doi: 10.1111/j.1749-4877.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 38.Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- 40.Belgacem YH, Martin JR. Neuroendocrine control of a sexually dimorphic behavior by a few neurons of the pars intercerebralis in Drosophila. Proc Natl Acad Sci USA. 2002;99:15154–15158. doi: 10.1073/pnas.232244199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Géminard C, Rulifson EJ, Léopold P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 2009;10:199–207. doi: 10.1016/j.cmet.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 42.Gibbs AG. Water-proofing properties of cuticular lipids. Am Zool. 1998;38:471–482. [Google Scholar]
- 43.van der Goes van Naters W, Carlson JR. Receptors and neurons for fly odors in Drosophila. Curr Biol. 2007;17:606–612. doi: 10.1016/j.cub.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondoh Y, Kaneshiro KY, Kimura K, Yamamoto D. Evolution of sexual dimorphism in the olfactory brain of Hawaiian Drosophila. Proc Biol Sci. 2003;270:1005–1013. doi: 10.1098/rspb.2003.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 46.Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 47.Butterworth FM. Lipids of Drosophila: A newly detected lipid in the male. Science. 1969;163:1356–1357. doi: 10.1126/science.163.3873.1356. [DOI] [PubMed] [Google Scholar]
- 48.Stocker RF, Gendre N. Courtship behavior of Drosophila genetically or surgically deprived of basiconic sensilla. Behav Genet. 1989;19:371–385. doi: 10.1007/BF01066165. [DOI] [PubMed] [Google Scholar]
- 49.Grillet M, Dartevelle L, Ferveur JF. A Drosophila male pheromone affects female sexual receptivity. Proc Biol Sci. 2006;273:315–323. doi: 10.1098/rspb.2005.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boulétreau-Merle J. Destruction of the pars intercerebralis in drosophila melanogaster: effect on the fecundity and the stimulation through copulation (Translated from French) J Insect Physiol. 1976;22:933–940. doi: 10.1016/0022-1910(76)90074-3. [DOI] [PubMed] [Google Scholar]
- 51.Heifetz Y, Lung O, Frongillo EA, Jr, Wolfner MF. The Drosophila seminal fluid protein Acp26Aa stimulates release of oocytes by the ovary. Curr Biol. 2000;10:99–102. doi: 10.1016/s0960-9822(00)00288-8. [DOI] [PubMed] [Google Scholar]
- 52.Ram KR, Wolfner MF. Sustained post-mating response in Drosophila melanogaster requires multiple seminal fluid proteins. PLoS Genet. 2007;3:e238. doi: 10.1371/journal.pgen.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 54.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 55.Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. San Diego, CA: Academic Press; 1992. [Google Scholar]
- 56.Greenspan RJ. Fly Pushing: The theory and Practice of Drosophila Genetics. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2004. [Google Scholar]
- 57.Sharma Y, Cheung U, Larsen EW, Eberl DF. PPTGAL, a convenient Gal4 P-element vector for testing expression of enhancer fragments in Drosophila. Genesis. 2002;34:115–118. doi: 10.1002/gene.10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferveur JF. Genetic control of pheromones in Drosophila simulans. I. Ngbo, a locus on the second chromosome. Genetics. 1991;128:293–301. doi: 10.1093/genetics/128.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Everaerts C, Farine JP, Cobb M, Ferveur JF. Drosophila cuticular hydrocarbons revisited: Mating status alters cuticular profiles. PLoS ONE. 2010;5:e9607. doi: 10.1371/journal.pone.0009607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boll W, Noll M. The Drosophila Pox neuro gene: Control of male courtship behavior and fertility as revealed by a complete dissection of all enhancers. Development. 2002;129:5667–5681. doi: 10.1242/dev.00157. [DOI] [PubMed] [Google Scholar]
- 61.Ferveur JF, Störtkuhl KF, Stocker RF, Greenspan RJ. Genetic feminization of brain structures and changed sexual orientation in male Drosophila. Science. 1995;267:902–905. doi: 10.1126/science.7846534. [DOI] [PubMed] [Google Scholar]
- 62.Rørth P. Gal4 in the Drosophila female germline. Mech Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.