Abstract
The activity of the dual-specificity receptor kinase, brassinosteroid insensitive 1 (BRI1), reflects the balance between phosphorylation-dependent activation and several potential mechanisms for deactivation of the receptor. In the present report, we elucidate a unique mechanism for deactivation that involves autophosphorylation of serine-891 in the ATP-binding domain. Serine-891 was identified previously as a potential site of autophosphorylation by mass spectrometry, and sequence-specific antibodies and mutagenesis studies now unambiguously establish phosphorylation of this residue. In vivo, phosphorylation of serine-891 increased slowly with time following application of brassinolide (BL) to Arabidopsis seedlings, whereas phosphorylation of threonine residues increased rapidly and then remained constant. Transgenic plants expressing the BRI1(S891A)–Flag-directed mutant have increased hypocotyl and petiole lengths, relative to wild-type BRI1–Flag (both in the bri1-5 background), and accumulate higher levels of the unphosphorylated form of the BES1 transcription factor in response to exogenous BL. In contrast, plants expressing the phosphomimetic S891D-directed mutant are severely dwarfed and do not accumulate unphosphorylated BES1 in response to BL. Collectively, these results suggest that autophosphorylation of serine-891 is one of the deactivation mechanisms that inhibit BRI1 activity and BR signaling in vivo. Many arginine-aspartate (RD)-type leucine-rich repeat receptor-like kinases have a phosphorylatable residue within the ATP-binding domain, suggesting that this mechanism may play a broad role in receptor kinase deactivation.
Keywords: phosphotyrosine, signal transduction, phosphoserine, modification-specific antibodies
The receptor-like kinase (RLK) family in Arabidopsis contains more than 600 members, of which more than 400 are structurally and functionally similar to animal receptor kinases but are evolutionarily distinct (1). Animal receptor kinases are predominantly tyrosine kinases, whereas plant receptor kinases are generally classified as Ser/Thr kinases, although recent work suggests that some plant receptor kinases are dual-specificity kinases that can also autophosphorylate on tyrosine residues (2–5). One of the best-studied plant receptor kinases is BRASSINOSTEROID INSENSITIVE 1 (BRI1), which functions with its coreceptor, BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), in brassinosteroid (BR) signaling (6–11). Current thinking is that BRI1 and BAK1 are in their unphosphorylated forms and inactive in the absence of BR, whereas in the presence of the BR ligand, BRI1 and BAK1 heterodimerize and become activated via auto- and transphosphorylation (12). The BRI1 KINASE INHIBITOR 1 (BKI1) and the BR-signaling kinase 1 (BSK1) may be two of the immediate downstream components that are first phosphorylated by BRI1 (13, 14). BSK then activates the BRI1 SUPPRESSOR 1 (BSU1) phosphatase (15), which in turn inhibits the glycogen synthase 3-like protein kinase, BRASSINOSTEROID INSENSITIVE 2 (BIN2), by dephosphorylation of an essential phosphotyrosine residue (16). The net result is that the transcription factors BRASSINAZOLE-RESISTANT 1 (BZR1) (17, 18) and BRI1-ETHYL METHANESULFONATE SUPPRESSOR 1 (BES1) (19, 20), also known as BZR2 (17) are dephosphorylated and able to move into the nucleus to up- or down-regulate the many genes that are BR regulated (7, 21).
The magnitude and duration of BR signaling will reflect the balance between receptor kinase activation and deactivation mechanisms, but much remains to be learned about both mechanisms. In terms of activation, many arginine-aspartate (RD)-type protein kinases require autophosphorylation of residues within the activation loop (22), and this appears to be the case for both BRI1 (23) and BAK1 (12). Moreover, reciprocal transphosphorylation between BRI1 and BAK1 is essential for enhanced BR signaling in vivo (9, 10, 12). Many phosphorylation sites have been identified on both receptor kinases and the physiological and biochemical functions of some of these modifications have already been elucidated. For example, residues located in the activation loops that must be phosphorylated for kinase activity, such as threonine-1049 of BRI1 (23–25) and threonine-455 in BAK1 (12), are essential for BR signaling in vivo. Whereas autophosphorylation of activation loop residues is required for activity, both BRI1 and BAK1 each have at least one phosphorylation site that appears to inhibit kinase activity. With BRI1, threonine-872 has been identified as an in vivo phosphorylation site (23) and preventing phosphorylation by substitution of alanine at this site dramatically increases autophosphorylation and peptide kinase activity of recombinant Flag–BRI1 cytoplasmic domain (23). Accordingly, expression of the T872A mutant of BRI1–Flag in the bri1-5 weak allele background appeared to rescue the dwarf phenotype to an even greater extent than the wild-type BRI1–Flag. However, analysis of the impact on plant growth was restricted to the T1 generation and needs further study with more advanced generations of these lines (23), and in addition, the behavior of the phosphomimetic T872D/E needs to be determined before the role of phosphorylation of threonine-872 can be discerned. With BAK1, serine-286 in the juxtamembrane (JM) domain has been identified as an in vitro autophosphorylation site and, whereas substitution with alanine had no effect on autophosphorylation of the recombinant protein, substitution with aspartate to mimic the phosphorylated state resulted in complete loss of autophosphorylation activity and ability to transphosphorylate/activate BRI1 in vitro (12). Furthermore, overexpression of the S286D phosphomimetic mutant in bri1-5 caused a severe dominant-negative phenotype, consistent with the lack of kinase activity in vitro (12). However, phosphorylation at the serine-286 site has not been detected in vivo, and the role in BR signaling, if any, is not clear. Nonetheless, phosphorylation of sites on receptor kinases that inhibit activity could play a role in receptor deactivation along with other mechanisms such as dephosphorylation by protein phosphatase 2A (PP2A) (26) and/or endocytosis (27, 28). We produced several modification-specific antibodies to monitor the phosphorylation of selected autophosphorylation sites that may function in receptor deactivation. Threonine-872 in the JM domain was of particular interest because as mentioned above, phosphorylation of this site may inhibit kinase activity (23). We also selected Ser-891 in the kinase domain because this residue was previously identified as a possible autophosphorylation site (24) that we speculated might inhibit kinase activity because of its location within the ATP-binding domain. The results of the present study support this notion and provide unique insights into the deactivation of the BRI1 receptor kinase, which may have broad application to other plant receptor kinases.
Results
Autophosphorylation of BRI1 at the Serine-891 Site Inhibits Kinase Activity.
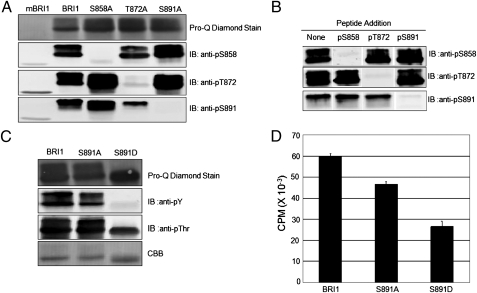
In previous studies, serine-891 was identified as a potential autophosphorylation site but could not be distinguished from serine-887, which was present on the same tryptic phosphopeptide, and moreover, it was not clear whether either site was phosphorylated in vivo (23, 24). We developed custom antibodies to determine whether phosphorylation occurs at the serine-891 site because of the location of this residue in the glycine-rich loop (G-loop) of subdomain I (GS891 GGFG), which is involved in binding and positioning of the substrate ATP. Hence, the potential for autophosphorylation of serine-891 to inhibit kinase activity seemed reasonable and warranted investigation. As shown in Fig. 1A, serine-891 was indeed phosphorylated in the recombinant Flag–BRI1 cytoplasmic domain (CD) as the custom anti-pS891 antibodies cross-reacted strongly with the native sequence protein but not the S891A-directed mutant. For comparison, we also developed modification-specific antibodies to monitor phosphorylation at the serine-858 and threonine-872 sites, which are known to occur in vitro and in vivo (23). The specificity of these antibodies was similarly demonstrated by strong cross-reaction with recombinant Flag–BRI1 CD but not the site-directed mutant where the corresponding serine or threonine was substituted with alanine. Moreover, peptide competition experiments also confirmed the specificity of all three modification-specific antibodies as cross-reactivity was only blocked by the appropriate phosphopeptide (Fig. 1B). None of the custom antibodies cross-reacted with the kinase-inactive mBRI1(K911E) (Fig. 1A), and as illustrated for the anti-pS891 antibodies even with higher concentrations of the BRI1 protein in Fig. S1, indicating that all were strictly phosphorylation specific. Thus, the sequence and modification specificity of these antibodies was established and autophosphorylation on serine-858 and threonine-872 was confirmed and serine-891 was established in vitro.
Fig. 1.
Recombinant Flag–BRI1 protein autophosphorylates on serine-891 and the S891D phosphomimetic site-directed mutant has reduced auto- and transphosphorylation activity. (A) Modification-specific antibodies confirm autophosphorylation of recombinant Flag–BRI1 protein (1.5 μg protein) at the serine-858, threonine-872, and serine-891 sites. Note the lack of cross-reactivity of the antibodies with the corresponding site-directed alanine-substitution mutant lacking the phosphorylation site. mBRI1 is the K911E-directed mutant of BRI1, which is devoid of kinase activity (23). (B) Demonstration of the specificity of the modification-specific antibodies by phosphopeptide competition. Recombinant protein (1.5 μg protein) was probed with the indicated antibodies that had been preincubated at 4 °C for 1 h with no additions (control) or 30 μg of the indicated phosphopeptide. Elimination of the immunoblot signal by only the antigen peptide confirms the sequence specificity of the antibodies. (C) Autophosphorylation of the S891A- and S891D-directed mutants. (D) Peptide kinase assays were performed using the SP11 peptide (sequence: GRJRRIASVEJJK, where J is norleucine). Reaction mixtures contained 80 μg mL−1 SP11 peptide and 0.1 mM [32P] ATP (150 DPM nmol−1) and reactions were incubated for 10 min at room temperature. Values are means of three determinations ± SEM.
To determine whether autophosphorylation of serine-891 inhibits kinase activity, we compared the autophosphorylation and peptide kinase activity of recombinant Flag–BRI1 with the S891A- and S891D-directed mutants. As shown in Fig. 1C, both wild-type Flag–BRI1- and the S891A-directed mutant displayed the characteristic doublet band on SDS/PAGE and both bands were strongly autophosphorylated on threonine and tyrosine (and presumably serine) residues. In marked contrast, the Flag–BRI1(S891D)-directed mutant completely lacked the hyperphosphorylated upper band and was totally devoid of autophosphorylation on tyrosine residues. Likewise, transphosphorylation activity of the recombinant proteins using the SP11 synthetic peptide substrate was substantially reduced in the S891D phosphomimetic mutant compared with the wild type or S891A mutant (Fig. 1D), suggesting that phosphorylation at the serine-891 site may reduce but not completely inhibit activity.
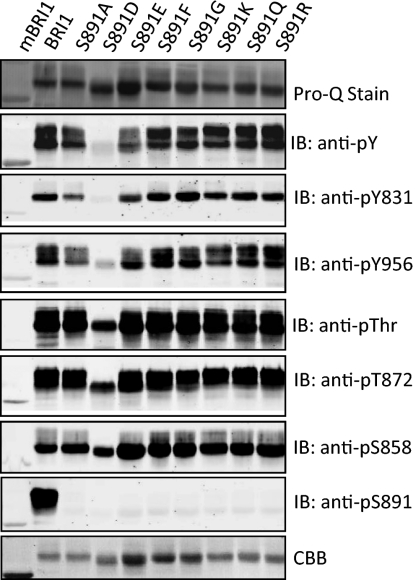
We produced recombinant Flag–BRI1 with additional substitutions for serine-891 to further characterize the structural constraints of the ATP-binding domain. As shown in Fig. 2, the most dramatic effects on BRI1 autophosphorylation were observed when serine-891 was substituted with aspartate, which eliminated autophosphorylation on tyrosine residues, and strongly reduced autophosphorylation on threonine and serine residues occurred but at a reduced level compared with wild-type Flag–BRI1 (Fig. 2). Substitution of a glutamate for serine-891 reduced tyrosine autophosphorylation but had little apparent effect on autophosphorylation on threonine residues. Clearly, negative charge is a factor, but not the only one involved in affecting autophosphorylation specificity. However, in terms of impact on tyrosine autophosphorylation, substitution of the two acidic residues had similar effects. As illustrated in Fig. S2, when phosphotyrosine content was normalized for BRI1 protein, the S891D and S891E mutants both had strongly reduced phosphorylation on tyrosine residues and were distinct from the other directed mutants where a small, neutral (glycine), hydrophobic (phenylalanine), polar (glutamine), or basic (lysine, arginine) residue was substituted for serine-891. It is also important to note that staining of the recombinant proteins with ProQ Diamond phosphoprotein stain was nearly identical among the site-directed mutants (Fig. 2), suggesting that serine autophosphorylation was the prominent modification and was relatively unaffected by the different substitutions, including the acidic substitutions (S891D and S891E) that inhibited tyrosine autophosphorylation. Collectively, these results indicate that the ATP-binding domain is least tolerant of negative charge at the 891 position and therefore suggest that the reduced activity of the S891D mutant, and to a lesser extent the S891E mutant, reflects the action of acidic residues as phosphomimetics. The corollary is that phosphorylation of serine-891 in the wild-type BRI1 protein would be expected to inhibit kinase activity, in particular on tyrosine residues.
Fig. 2.
Autophosphorylation of recombinant Flag-BRI1 proteins with various substitutions at the 891 position. Immunopurified proteins (1.5 μg protein) were analyzed by staining or immunoblotting as indicated.
Phosphorylation of Serine-891 Occurs in Vivo and Plays a Role in BR Signaling.
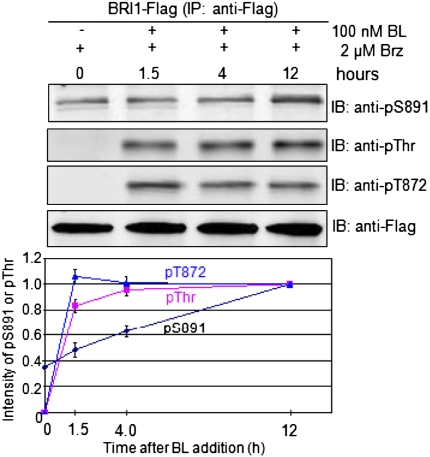
Importantly, phosphorylation of serine-891 could be demonstrated to occur in vivo (Fig. 3). Following addition of the BL biosynthesis inhibitor, brassinazole (Brz) (29), to Arabidopsis seedlings expressing BRI1–Flag, threonine phosphorylation was completely removed after 4 d of treatment but some phosphorylation of serine-891 was still apparent, indicating that specific phosphosites on BRI1 were dephosphorylated to different extents in vivo. Whether this reflects differences in rates of dephosphorylation or continued phosphorylation in the presence of Brz, presumably catalyzed by other protein kinases, is not clear and will be examined in future studies. Subsequent addition of brassinolide (BL) to Brz-treated seedlings resulted in a gradual time-dependent increase in serine-891 phosphorylation that was generally linear over the 12-h time course of the experiment. In marked contrast, phosphorylation of BRI1 on threonine residues was very rapidly restored following addition of BL. The phosphothreonine content reached a maximum level within 4 h and thereafter remained constant, and similar results were obtained for phosphorylation at the threonine-872 site (Fig. 3).
Fig. 3.
Rapid phosphorylation of BRI1–Flag on threonine residues but slow phosphorylation at the serine-891 site following addition of BL to Brz-treated seedlings. Transgenic Arabidopsis plants expressing BRI1–Flag in the bri1-5 background were grown in shaking liquid culture in continuous light. On day 6, the cultures were treated with 2 μM Brz and on day 11, 100 nM BL was added and samples were collected at 0, 1.5, 4, and 12 h after BL addition. Microsomal membranes were isolated and BRI1–Flag was immunopurified before SDS/PAGE and immunoblotting. Values in the plot are means from three independent experiments ± SEM.
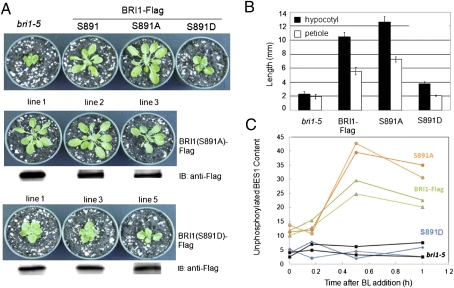
To determine the physiological significance of phosphorylation at the serine-891 site, we transformed the weak bri1-5 mutant with the native sequence, full-length BRI1–Flag or the site-directed mutant S981A, to eliminate phosphorylation at this site, or S981D, as a constitutive phosphomimetic (Fig. 4). As expected, the dwarf phenotype of the bri1-5 mutant was fully rescued by expression of the native sequence protein. Likewise, expression of the BRI1(S891A)–Flag-directed mutant rescued growth, and plants were slightly larger than those expressing the wild-type BRI1–Flag, whereas expression of the phosphomimetic BRI1(S891D)–Flag increased growth of the bri1-5 mutant only slightly. Three independent transgenic lines expressing the S891A- or S891D-directed mutant are shown in Fig. 4A, where nearly equal expression of the transgenes is documented by immunoblotting with anti-Flag antibodies. The visual differences in growth were also evident in measurements of hypocotyl and petiole lengths that decreased in the order BRI1(S891A) –Flag > BRI1–Flag > BRI1(S891D) –Flag ∼ bri1-5 (Fig. 4B). As another physiological readout of BRI1-mediated signaling, we monitored accumulation of the unphosphorylated form of BES1 in response to exogenous BL. A representative immunoblot is presented as Fig. S3, and the results of two independent experiments are quantified in Fig. 4C. Consistent with previous results (20, 30, 31), unphosphorylated BES1 accumulated in plants expressing wild-type BRI1–Flag to reach a maximum level within 30 min of treatment, whereas no accumulation was observed in bri1-5 plants. Importantly, the S891D-directed mutant plants gave a similar response to the bri1-5 plants, whereas the plants expressing the S891A-directed mutant responded to an even greater extent than the native sequence BRI1–Flag plants. Collectively, these results are consistent with the notion that phosphorylation of serine-891 inhibits BRI1 kinase activity in vivo and therefore functions as a deactivation mechanism.
Fig. 4.
Phosphorylation of BRI1 at the serine-891 site affects plants growth. (A, Top) Wild-type BRI1–Flag and the S891A-directed mutant rescue the dwarf phenotype of bri1-5 plants, whereas the S891D-directed mutant does not. (Middle and Bottom) Three independent transgeninc lines of each directed mutant, and the immunoblots show equal expression of the BRI1–Flag transgene. (B) Hypocotyl and petiole lengths measured 6 d after seed germination for plants grown in soil with a short day photoperiod (8 h light/16 h dark). Values are means of measurements from 10 seedlings ± SEM. (C) The S891A-directed mutant accumulates more unphosphorylated BES1 protein in response to exogenous BL treatment, whereas the S891D mutant is unresponsive. Values plotted are from two independent experiments. A representative immunoblot from which these results were obtained is presented in Fig. S2.
Autophosphorylation of BRI1–Flag in Vivo.
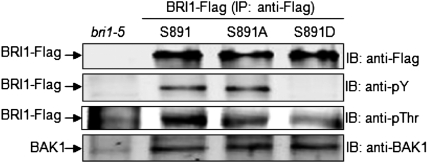
We examined the phosphorylation status of wild-type BRI1–Flag and the S891A/D-directed mutants in plants grown in soil. The BRI1–Flag proteins were immunoprecipitated with anti-Flag antibodies and the phosphorylation status of the BRI1 protein was analyzed by immunoblotting. As shown in Fig. 5, the native sequence BRI1–Flag protein contained phosphotyrosine and phosphothreonine and endogenous BAK1 protein was coimmunoprecipitated with the protein. Very similar results were obtained for the BRI1(S891A)–Flag protein. However, in marked contrast, the S891D-directed mutant protein lacked detectable phosphotyrosine and had reduced phosphorylation on threonine residues relative to the native sequence (or S891) BRI1–Flag or S891A-directed mutant. These results are reminiscent of the autophosphorylation profiles observed for the corresponding recombinant proteins (Fig. 1C) and the physiological readouts of plants expressing the S891D-directed mutant. Interestingly, the amount of endogenous BAK1 protein that coimmunoprecipitated with the BRI1–Flag proteins was similar for the native sequence protein (S891) and the S891A and S891D mutants, suggesting that the BRI1:BAK1 interaction per se was not altered by any of the substitutions at the 891 position.
Fig. 5.
Transgenic Arabidopsis plants expressing BRI1(S891D)–Flag have reduced autophosphorylation on threonine residues and are devoid of tyrosine phosphorylation, but interact with endogenous BAK1 similar to the native sequence and S891A-directed mutant. A second independent biological replicate yielded similar results. Plants were grown for 25 d in soil under short photoperiod conditions (8 h light/16 h dark).
Discussion
The most important result of the present study is the demonstration that phosphorylation of BRI1 at serine-891 in the ATP-binding domain occurs in vivo, which attenuates signaling and thus appears to function as a deactivation mechanism. Phosphorylation at the serine-891 site was established with sequence- and phosphorylation-specific antibodies, and the functional significance of this modification was assessed by site-directed mutagenesis to alanine, to eliminate phosphorylation, or aspartate, to mimic constitutive phosphorylation. Results with the corresponding mutants (i.e., S891A and S891D) expressed as recombinant proteins indicated that the serine-to-aspartate substitution strongly inhibits both autophosphorylation of BRI1 (Fig. 1C) and transphosphorylation of the SP11 synthetic peptide (Fig. 1D). Interestingly, the glycine-rich G-loop of BRI1 could accommodate a wide range of amino acids at the 891 position without impact on autophosphorylation (Fig. 2), indicating that the region was not a highly constrained structure that could not tolerate substitutions. Thus, the inhibition of kinase activity by acidic substitutions for serine-891 likely reflects their action as phosphomimetics rather than simply causing nonspecific alterations. However, aspartate had a greater effect than glutamate, indicating that factors in addition to negative charge are important. Nonetheless, given that an acid residue at the 891 position inhibits autophosphorylation, in particular on tyrosine residues (Fig. S2), it is reasonable to expect that phosphorylation of the naturally occurring serine residue at this position in wild-type BRI1 would also have an inhibitory effect on autophosphorylation.
The notion that phosphorylation of serine-891 is one of the deactivation mechanisms that reduces BR signaling in vivo is suggested by several lines of evidence. First, expression of BRI1(S891A)–Flag in the bri1-5 background resulted in plants with elongated petioles and hypocotyls relative to plants expressing native-sequence BRI1–Flag, whereas expression of the S891D-directed mutant had the opposite effect and resulted in severely dwarfed plants with short petioles and hypocotyls (Fig. 4). Second, exogenous BL induced greater accumulation of the unphosphorylated form of BES1 in plants expressing the S891A-directed mutant compared with native sequence BRI1–Flag (Fig. 4C), which is consistent with the phenotypic analysis of these plants. Third, the slow time-dependent phosphorylation of BRI1 at the serine-891 site was distinct from the more rapid phosphorylation on threonine residues, which is associated with activation of kinase activity (Fig. 3). Interestingly, phosphorylation at the threonine-872 site followed similar kinetics to overall phosphothreonine with rapid phosphorylation following BL addition (Fig. 3). Thus, if phosphorylation of threonine-872 inhibits BRI1 kinase activity as speculated (23), the effect is distinct from the phosphorylation of serine-891, which inhibits BRI1 kinase activity and attenuates BR signaling in vivo, likely in a time-dependent manner. It is also interesting that phosphorylation at the serine-891 site is not completely removed after 4 d of treatment with Brz, whereas threonine phosphosites are completely dephosphorylated (Fig. 3). Whether these responses reflect different rates of dephosphorylation by endogenous protein phosphatases remains an important question for future studies. However, the fact that the phosphoserine-891 site is not dephosphorylated in vivo as completely as phosphothreonine sites has implications for subsequent cycles of activation/deactivation of this relatively long-lived receptor kinase and likely explains the enhanced signaling phenotype of the S891A-directed mutant. Thus, the deactivation associated with serine-891 phosphorylation seems to play a role in attenuation of signaling rather than maintaining the inactive state of BRI1 in the absence of its hormone ligand.
One might suspect that phosphorylation of a residue within the ATP-binding domain would cause steric or electrostatic interference with ATP binding or positioning and thereby inhibit kinase activity. Indeed, cyclin-dependent protein kinases (CDKs) are regulated in part by inhibitory phosphorylation of conserved residues within the active site. Detailed studies with cyclin-dependent kinase 2 (CDK2) demonstrated that phosphorylation of threonine-14 and tyrosine-15 within the glycine-rich loop (GEGTYGVVY) inhibits kinase activity by interfering with peptide substrate binding and alters, but does not prevent, ATP binding (32). With the CDKs, the inhibitory phosphorylation within the ATP-binding domain is catalyzed by other kinases, whereas with BRI1 the inhibitory modification is an autophosphorylation reaction. However, the possibility that other kinases also contribute in vivo to phosphorylation of BRI1 at the serine-891 site cannot be ruled out. At the position corresponding to serine-891 in BRI1 (GSGXXGXV), 16 other RD-type leucine-rich receptor–receptor-like kinases (LRR–RLKs) in Arabidopsis have a phosphorylatable residue at this position (Table S1), suggesting that this mechanism may occur in other receptor kinases. Fifty-eight LRR-RLKs have a basic residue at the position corresponding to serine-891 in BRI1. If a positive side chain is tolerated at this position, then it is reasonable that a negatively charged phosphoserine would not be tolerated. One other interesting point to note is that 24 other receptor kinases have an acidic residue at this position, which suggests that the intrinsic kinase activity of these receptors may be at least partially down-regulated. However, 23 of the 24 receptor kinases have a glutamate residue, which at least within the BRI1 sequence, caused less inhibition than did an aspartate residue. Thus, to the known mechanisms of deactivation of receptor kinases such as endocytosis and dephosphorylation of essential residues, we can also now include phosphorylation of inhibitory sites such as serine-891 in the ATP-binding domain of BRI1, although future studies will be required to determine the relative timing and importance of each of these mechanisms.
Materials and Methods
Plant Growth and Transformation.
Arabidopsis thaliana ecotype Ws-2 was used as the wild type. Seeds were surface sterilized, kept at 4 °C for 2 d, and then sown on germination medium [one-quarter strength Murashige and Skoog salt and vitamins medium (PhytoTechnology Laboratories), supplemented with 0.8% (wt/vol) agar, 30 mg L−1 hygromycin B (Sigma-Aldrich) and 2% (wt/vol) sucrose, pH 5.7]. For phenotypic analysis between wild type, bri1-5 allele, and tyrosine site-directed mutants, the concentration of agar was 1.2% and plates were kept at 22 °C with an 8-h photoperiod for 9 d. Seedlings were then transferred to soil and grown in the same temperature and light conditions to analyze phenotype. BRI1–Flag and mBRI1–Flag transgenic lines in the bri1-5 mutant background were as previously described (2). The same BRI1–Flag construct was used to generate the site-directed mutants, S891A and S891D, and each construct was transformed individually into the bri1-5 mutant using the floral dip method (33). All plants were grown in soil for 35 d after seed germination under short day conditions (8 h light/16 h dark) as previously described.
Site-Directed Mutagenesis of Flag–BRI1 Cytoplasmic Domain and Recombinant Protein Studies.
After sequencing to confirm mutated regions, BRI1 CD and its mutations were expressed in BL21(DE3)pLysS cells (Novagen). Cultures were induced with 1 mM IPTG at 25 °C for 16 h and the soluble recombinant protein produced was purified using anti-Flag M2 affinity gel (Sigma-Aldrich). After elution from the beads, the protein solution was dialyzed against a 1,000× volume of buffer containing 20 mM Mops, pH 7.5, and 1 mM DTT. Peptide substrate phosphorylation assays were performed as described (24) using the SP11 peptide (sequence: GRJRRIASVEJJK, where J is norleucine; Bethyl Laboratories) and the filter binding assay.
Preparation of Microsomal Membranes and Immunopurification of BRI1–Flag.
Microsomal membranes were isolated, solubilized with Triton X-100, and BRI1-Flag was immunopurified using anti-Flag M2 affinity gel (Sigma) as previously described (23).
Immunoblot Analysis.
Immunoprecipitated full-length BRI1–Flag or recombinant Flag–BRI1 cytoplasmic domain proteins were subjected to SDS/PAGE followed by transfer to PVDF membranes and immunoblot analysis using anti-Flag antibodies (1:1,000 dilution), antiphosphothreonine antibodies (1:500 dilution), antiphosphotyrosine antibodies (1:500 dilution), or custom antibodies [anti-pS891 (1:2,000 dilution), anti-pS858 (1:3,000 dilution), or anti-pT872 (1:3,000 dilution) antibodies]. The custom antibodies were produced against the following sequences: pS891, DSLIGpSGGFGD; pS858, KEALpSINLAA; and pT872, PLRKLpTFADL). The antibodies were produced by GenScript and sequentially affinity purified using the nonphosphorylated and then the phospho-containing antigen peptides. Immunoblots were scanned using an Odyssey infrared imaging system (LI-COR Bioscience) for visualization. Blots stained with ProQ Diamond phosphoprotein stain (Invitrogen) were scanned using a Typhoon Molecular Dynamics phosphor/fluorescence imager.
Supplementary Material
Acknowledgments
This work was supported in part by National Science Foundation Grants IOS-1022177, MCB-0740211, and MCB-1021363 and the Department of Agriculture–Agricultural Research Service.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108321109/-/DCSupplemental.
References
- 1.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh M-H, et al. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:658–663. doi: 10.1073/pnas.0810249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlova R, et al. Identification of in vitro phosphorylation sites in the Arabidopsis thaliana somatic embryogenesis receptor-like kinases. Proteomics. 2009;9:368–379. doi: 10.1002/pmic.200701059. [DOI] [PubMed] [Google Scholar]
- 4.Oh MH, Clouse SD, Huber SC. Tyrosine phosphorylation in brassinosteroid signaling. Plant Signal Behav. 2009;4:1–4. doi: 10.4161/psb.4.12.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh MH, Wu X, Clouse SD, Huber SC. Functional importance of BAK1 tyrosine phosphorylation in vivo. Plant Signal Behav. 2011;6:400–405. doi: 10.4161/psb.6.3.14337. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Gendron JM, Wang Z-Y. Multiple mechanisms modulate brassinosteroid signaling. Curr Opin Plant Biol. 2007;10:436–441. doi: 10.1016/j.pbi.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 8.Clouse SD. Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23:1219–1230. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 10.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Jaillais Y, et al. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25:232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mora-García S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim T-W, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 18.He J-X, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 20.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 21.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 22.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: Structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, et al. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh MH, et al. Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol. 2000;124:751–766. doi: 10.1104/pp.124.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, et al. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev Cell. 2005;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Wu G, et al. Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal. 2011;4:ra29. doi: 10.1126/scisignal.2001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geldner N, Robatzek S. Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 2008;147:1565–1574. doi: 10.1104/pp.108.120287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russinova E, et al. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asami T, et al. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 2000;123:93–100. doi: 10.1104/pp.123.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 31.Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007;21:1598–1602. doi: 10.1101/gad.1561307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welburn JP, et al. How tyrosine 15 phosphorylation inhibits the activity of cyclin-dependent kinase 2-cyclin A. J Biol Chem. 2007;282:3173–3181. doi: 10.1074/jbc.M609151200. [DOI] [PubMed] [Google Scholar]
- 33.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.