Abstract
Conformation-specific antibodies that recognize aggregated proteins associated with several conformational disorders (e.g., Parkinson and prion diseases) are invaluable for diagnostic and therapeutic applications. However, no systematic strategy exists for generating conformation-specific antibodies that target linear sequence epitopes within misfolded proteins. Here we report a strategy for designing conformation- and sequence-specific antibodies against misfolded proteins that is inspired by the molecular interactions governing protein aggregation. We find that grafting small amyloidogenic peptides (6–10 residues) from the Aβ42 peptide associated with Alzheimer’s disease into the complementarity determining regions of a domain (VH) antibody generates antibody variants that recognize Aβ soluble oligomers and amyloid fibrils with nanomolar affinity. We refer to these antibodies as gammabodies for grafted amyloid-motif antibodies. Gammabodies displaying the central amyloidogenic Aβ motif (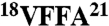 ) are reactive with Aβ fibrils, whereas those displaying the amyloidogenic C terminus (
) are reactive with Aβ fibrils, whereas those displaying the amyloidogenic C terminus (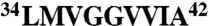 ) are reactive with Aβ fibrils and oligomers (and weakly reactive with Aβ monomers). Importantly, we find that the grafted motifs target the corresponding peptide segments within misfolded Aβ conformers. Aβ gammabodies fail to cross-react with other amyloidogenic proteins and scrambling their grafted sequences eliminates antibody reactivity. Finally, gammabodies that recognize Aβ soluble oligomers and fibrils also neutralize the toxicity of each Aβ conformer. We expect that our antibody design strategy is not limited to Aβ and can be used to readily generate gammabodies against other toxic misfolded proteins.
) are reactive with Aβ fibrils and oligomers (and weakly reactive with Aβ monomers). Importantly, we find that the grafted motifs target the corresponding peptide segments within misfolded Aβ conformers. Aβ gammabodies fail to cross-react with other amyloidogenic proteins and scrambling their grafted sequences eliminates antibody reactivity. Finally, gammabodies that recognize Aβ soluble oligomers and fibrils also neutralize the toxicity of each Aβ conformer. We expect that our antibody design strategy is not limited to Aβ and can be used to readily generate gammabodies against other toxic misfolded proteins.
Keywords: misfolding, beta-amyloid, protein engineering
A hallmark of protein misfolding disorders is that polypeptides of unrelated sequence fold into similar oligomeric and fibrillar assemblies that are cytotoxic (1). The structures of these enigmatic conformers have captured the imagination of many investigators who have sought to explain the molecular basis of proteotoxicity in conformational disorders such as Alzheimer’s disease. Because misfolded proteins are typically refractory to structural methods such as X-ray crystallography and solution NMR, few high-resolution structures of full-length misfolded proteins have been reported (ref. 2 and references therein). The structures of oligomeric intermediates have proven especially difficult to characterize because these conformers are labile, transient, and, in many cases, heterogeneous.
Given the complexity of high-resolution structural analysis of misfolded proteins, alternative biochemical approaches are critical for understanding structure–function relationships of aggregated proteins. A breakthrough in this area has been the development of conformation-specific antibodies that selectively recognize uniquely folded conformers of amyloidogenic proteins (3–13). Indeed, multiple conformation-specific antibodies have been reported that recognize structural features within amyloidogenic oligomers (5) and fibrils (4, 6, 8) in a sequence-independent manner. These and related antibodies have proven invaluable for identifying oligomeric and fibrillar conformers of several disease-linked proteins both in vitro and in vivo (3–13).
The next important step in using antibodies to characterize misfolded proteins is to develop systematic approaches for generating conformation-specific antibodies that recognize sequence-specific epitopes within amyloidogenic proteins. The utility of such antibodies would be even greater if they recognized linear sequence epitopes (instead of discontinuous epitopes) within misfolded proteins because continuous epitopes are easier to identify and provide more direct structural information. To develop conformation-specific antibodies that target linear sequence epitopes, we sought to mimic the natural process of amyloid assembly that is commonly mediated via homotypic interactions between small amyloidogenic peptide segments within misfolded proteins (14–17). We posited that grafting amyloidogenic motifs from the Aβ42 peptide associated with Alzheimer’s disease into the complementarity determining regions (CDRs) of antibodies would generate antibody variants that selectively recognize aggregated Aβ conformers but not Aβ monomers. Moreover, we hypothesized that these antibodies would employ homotypic interactions between the grafted Aβ motifs and the corresponding peptide segments within aggregated Aβ conformers to mediate conformation-specific antibody recognition.
Our hypotheses are motivated by the structure of Aβ fibrils in which amyloidogenic peptide motifs stack in-register with identical motifs from other Aβ molecules (18–21), as well as the ability of Aβ amyloidogenic motifs (by themselves or conjugated to other molecules) to inhibit Aβ aggregation via homotypic interactions (22–25). Our hypotheses are also motivated by the conformation-specific antibodies developed by Williamson and coworkers against the mammalian prion protein (PrP) (26). Although these full-length monoclonal antibodies displaying PrP peptide fragments recognize aggregated PrP conformers, it is unknown whether the antibodies use homotypic interactions to mediate PrP recognition because their binding sites were not determined. Moreover, the PrP-specific recognition of these antibodies is mediated primarily by electrostatic interactions (26, 27). In contrast, we seek to exploit amyloidogenic (nonelectrostatic), homotypic interactions between grafted motifs within antibodies and the corresponding motifs within misfolded polypeptides to mediate conformation- and sequence-specific recognition. Herein, we report the development of a class of grafted amyloid-motif antibodies (which we refer to as gammabodies) that use self-complementary amyloidogenic interactions to recognize conformational epitopes within Aβ oligomers and fibrils in a sequence-specific manner.
Results
Antibody Domains Displaying Amyloidogenic Aβ Motifs Recognize Aggregated Aβ Isoforms in a Conformation-Specific Manner.
To evaluate our hypothesis that antibodies grafted with Aβ amyloidogenic motifs would selectively recognize aggregated Aβ conformers, we first sought to identify an antibody scaffold that is highly stable and tolerant to grafting diverse peptide segments into its CDR loops. We selected an antibody domain (VH) scaffold that is highly soluble and stable, and whose folding is insensitive to mutations in its third CDR (CDR3) loop (28). We find this antibody is well expressed in bacteria (> 5 mg/L), secreted into the bacterial media without cell lysis, highly pure after a single chromatography step (> 95% purity), and stably folded (Fig. S1). Importantly, we confirmed that the wild-type antibody fails to recognize monomeric and aggregated conformers of Aβ and other amyloidogenic proteins (Fig. S1).
We hypothesized that grafting peptides containing amyloidogenic Aβ42 segments (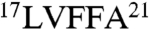 and
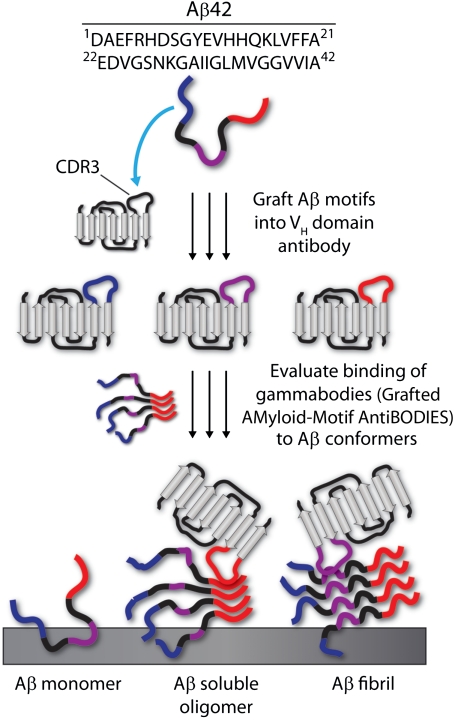
and 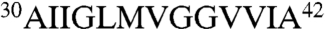 ) (17, 29) would mediate antibody recognition of aggregated Aβ conformers, whereas grafting Aβ segments outside these regions would not. To test this hypothesis, we synthesized a panel of gammabodies in which overlapping 10-mer sequences from Aβ (residues 1–10, 3–12, 6–15, 9–18, 12–21, 15–24, 18–27, 21–30, 24–33, 27–36, 30–39, and 33–42; Table 1) were grafted into CDR3 of the wild-type antibody (Fig. 1). Importantly, all 12 Aβ gammabodies express well in bacteria (> 5 mg/L), and they are soluble and well folded (Fig. S2) in a manner similar to the wild-type antibody (Fig. S1).
) (17, 29) would mediate antibody recognition of aggregated Aβ conformers, whereas grafting Aβ segments outside these regions would not. To test this hypothesis, we synthesized a panel of gammabodies in which overlapping 10-mer sequences from Aβ (residues 1–10, 3–12, 6–15, 9–18, 12–21, 15–24, 18–27, 21–30, 24–33, 27–36, 30–39, and 33–42; Table 1) were grafted into CDR3 of the wild-type antibody (Fig. 1). Importantly, all 12 Aβ gammabodies express well in bacteria (> 5 mg/L), and they are soluble and well folded (Fig. S2) in a manner similar to the wild-type antibody (Fig. S1).
Table 1.
Sequences of the third complementarity determining region (CDR3) of Aβ gammabodies
| Gammabody | CDR3 sequence |
| Aβ1–10 | DAEFRHDSGY |
| Aβ3–12 | EFRHDSGYEV |
| Aβ6–15 | HDSGYEVHHQ |
| Aβ9–18 | GYEVHHQKLV |
| Aβ12–21 | VHHQKLVFFA |
| Aβ15–24 | QKLVFFAEDV |
| Aβ18–27 | VFFAEDVGSN |
| Aβ21–30 | AEDVGSNKGA |
| Aβ24–33 | VGSNKGAIIG |
| Aβ27–36 | NKGAIIGLMV |
| Aβ30–39 | AIIGLMVGGV |
| Aβ33–42 | GLMVGGVVIA |
Fig. 1.
Motif-grafting strategy for designing conformation- and sequence-specific antibody domains against aggregated Aβ conformers. Overlapping Aβ42 peptide segments (4–10 residue peptides) were grafted into the third complementarity determining region (CDR3) of a VH domain antibody (PDB: 3B9V). The binding specificity and affinity of Aβ gammabodies were evaluated against Aβ monomers, soluble oligomers, and fibrils.
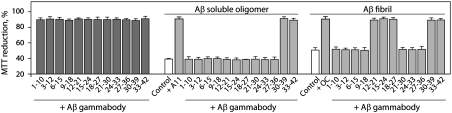
We next sought to evaluate whether each gammabody variant selectively recognized Aβ fibrils and soluble oligomers relative to Aβ monomers. Therefore, we first assembled the Aβ conformers as we described previously (30–32) and deposited each of them on nitrocellulose membranes. We detected each Aβ conformer using sequence-specific monoclonal antibodies that recognize the N terminus (6E10; Aβ residues 1–16), middle region (4G8; Aβ residues 18–22), and C terminus (9F1; Aβ residues 34–39) of Aβ (Fig. 2). We also confirmed that Aβ oligomers and fibrils were specifically recognized by conformation-specific antibodies immunoreactive with oligomeric [A11; ref. (5)] and fibrillar [OC, ref. (8) and WO1, ref. (4)] conformers, respectively.
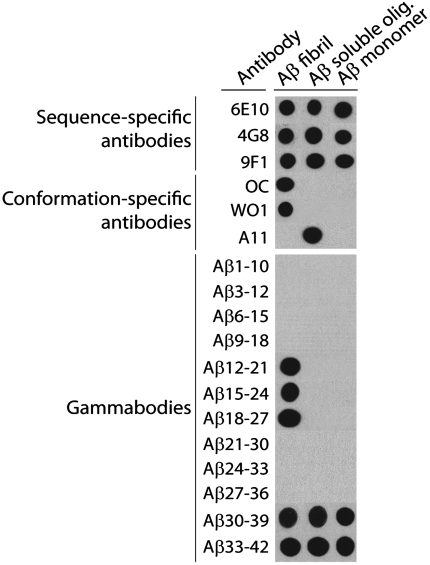
Fig. 2.
Conformation-specific binding activity of Aβ gammabodies. Aβ42 conformers were deposited on nitrocellulose membranes (220 ng), and probed with Aβ gammabodies (6 μM). As loading controls, the same blots were probed with sequence-specific monoclonal antibodies (6E10 specific for Aβ1–17, 4G8 specific for Aβ18–22, and 9F1 specific for Aβ34–39), fibril-specific antibodies (WO1 and OC), and a prefibrillar oligomer-specific antibody (A11).
Having confirmed proper loading of each Aβ conformer, we tested our hypothesis that the grafted antibodies would recognize aggregated Aβ conformers relative to Aβ monomers (Fig. 2). Strikingly, we find that gammabodies displaying Aβ12–21, Aβ15–24, and Aβ18–27 are immunoreactive with Aβ fibrils but not oligomers or monomers. Moreover, we find that antibodies displaying C-terminal Aβ motifs (Aβ30–39 and Aβ33–42) recognize all three Aβ conformers (Fig. 2). In contrast, antibodies displaying hydrophilic Aβ peptides from the N terminus (Aβ1–10, Aβ3–12, Aβ6–15, and Aβ9–18), and between the two amyloidogenic motifs (Aβ21–30, Aβ24–33 and Aβ27–36), do not recognize Aβ.
We sought to further isolate the minimal Aβ peptide motifs that mediate binding to Aβ conformers (Fig. S3). Because the Aβ motif 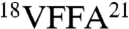 is common to gammabodies that selectively recognize Aβ fibrils (Aβ12–21, Aβ15–24, and Aβ18–27), we synthesized an antibody variant displaying the Aβ16–21 motif. We find that this gammabody selectively recognizes Aβ fibrils in a manner indistinguishable from its parent antibodies (Fig. S3). For the gammabodies displaying C-terminal Aβ segments (Aβ30–39 and Aβ33–42), we find that antibodies displaying six-residue Aβ motifs (Aβ34–39 for the Aβ30–39 gammabody and Aβ37–42 for the Aβ33–42 gammabody) also possess similar binding as their parent antibodies. In contrast, gammabodies displaying shorter Aβ motifs (Aβ36–39 and Aβ39–42) are inactive (Fig. S3).
is common to gammabodies that selectively recognize Aβ fibrils (Aβ12–21, Aβ15–24, and Aβ18–27), we synthesized an antibody variant displaying the Aβ16–21 motif. We find that this gammabody selectively recognizes Aβ fibrils in a manner indistinguishable from its parent antibodies (Fig. S3). For the gammabodies displaying C-terminal Aβ segments (Aβ30–39 and Aβ33–42), we find that antibodies displaying six-residue Aβ motifs (Aβ34–39 for the Aβ30–39 gammabody and Aβ37–42 for the Aβ33–42 gammabody) also possess similar binding as their parent antibodies. In contrast, gammabodies displaying shorter Aβ motifs (Aβ36–39 and Aβ39–42) are inactive (Fig. S3).
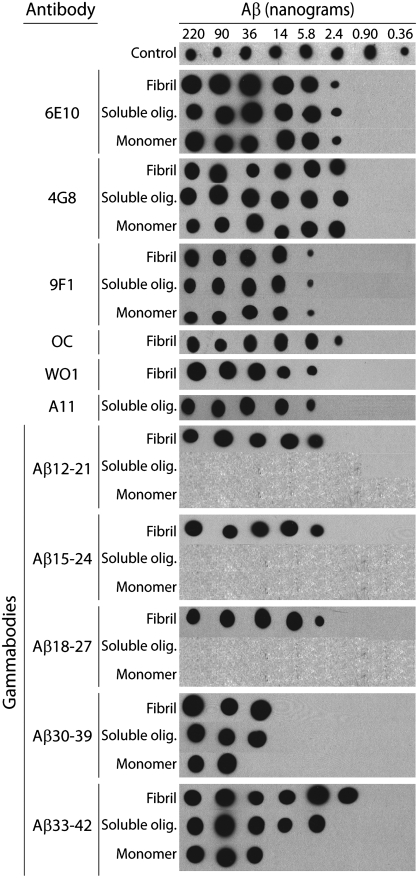
We next investigated the detection sensitivity of the Aβ gammabodies. As a first step toward this aim, we evaluated the binding of each gammabody to immobilized Aβ conformers for a range of Aβ loadings (0.4–220 ng of Aβ; Fig. 3 and Fig. S4) via immunoblot analysis. We find that sequence-specific monoclonal antibodies (6E10, 4G8, 9F1), as well as conformation-specific monoclonal (WO1) and polyclonal (A11 and OC) antibodies detect Aβ at similar loadings (≥2–6 ng Aβ). We confirmed that these results are independent of the concentration of antibody used for detection above 10 nM (Fig. S4). Importantly, we find that the Aβ12–21, Aβ15–24, and Aβ18–27 gammabodies detect fibrils at loadings (≥6 ng Aβ) similar to the monoclonal and polyclonal antibodies (≥2–6 ng Aβ; Fig. 3). Moreover, although the Aβ30–39 and Aβ33–42 gammabodies are reactive with the three Aβ conformers, they display unique detection sensitivities for each Aβ conformer (Fig. 3). The Aβ33–42 gammabody is most sensitive for recognizing fibrils (≥2 ng Aβ) relative to oligomers (≥6 ng Aβ) and monomers (≥36 ng Aβ). Interestingly, the Aβ30–39 gammabody recognizes fibrils and oligomers with equal sensitivity, and it is less sensitive for recognizing each Aβ conformer than the Aβ33–42 gammabody (Fig. 3). We confirmed that these results are independent of gammabody concentration above 300 nM (Fig. S4). We conclude that the detection sensitivity of our designed Aβ gammabodies is similar to monoclonal and polyclonal antibodies generated via immunization, and gammabodies displaying C-terminal Aβ motifs possess conformation-specific Aβ detection sensitivity.
Fig. 3.
Detection sensitivity of Aβ gammabodies for recognizing Aβ conformers. Aβ was deposited on nitrocellulose membranes (0.36–220 ng), and probed with Aβ gammabodies (6 μM). The same blots were also probed with sequence- and conformation-specific monoclonal and polyclonal antibodies, as described in Fig. 2. The loading control was biotinylated Aβ42 monomers detected with peroxidase-conjugated streptavidin.
We next measured the affinity of gammabodies for Aβ fibrils and oligomers using competitive ELISA analysis (33, 34) (Fig. S5). We find those gammabodies that bind to Aβ soluble oligomers and fibrils have dissociation constants between 300–600 nM. Moreover, the relative affinity of each gammabody is consistent with the immunoblot analysis (Fig. 3), because the Aβ33–42 antibody has the highest affinity against fibrils (335 ± 20 nM) and soluble oligomers (420 ± 60 nM), whereas the Aβ30–39 antibody has the lowest binding affinities against both conformers (490 ± 65 nM for fibrils and 595 ± 30 nM for oligomers). Finally, we find that the IC50 values for antibody binding to Aβ oligomers and fibrils are in excellent agreement with the competitive ELISA measurements (Fig. S5). We conclude that Aβ gammabodies display nanomolar binding affinity to Aβ oligomers and fibrils.
Because the grafted Aβ motifs appear to mediate binding to Aβ conformers without assistance from the other antibody CDR loops, we wondered whether these amyloidogenic motifs (without the antibody scaffold) would also bind to Aβ conformers. Therefore, we performed immunoblot analysis using biotinylated Aβ peptide fragments (Aβ10–20, Aβ12–28, Aβ17–28, and Aβ33–42) that overlap with or are identical to the Aβ motifs found to confer binding (Fig. S6). We failed to detect binding of the Aβ peptide fragments to any of the Aβ conformers, even at the highest Aβ loadings (220 ng Aβ). However, we detected binding of biotinylated full-length Aβ42 to fibrils (≥90 ng Aβ; Fig. S7), although it was much less sensitive than the Aβ gammabodies (≥2–6 ng Aβ; Fig. 3). Moreover, we also detected weak binding of biotinylated Aβ42 to soluble oligomers (≥90 ng Aβ) and monomers (≥220 ng Aβ; Fig. S6). We conclude that amyloidogenic Aβ motifs presented within an antibody loop are significantly more immunoreactive with fibrils and oligomers than the motifs presented within Aβ42 monomers or as discrete peptides.
Gammabodies Recognize Aβ Oligomers and Fibrils via Homotypic Interactions Between Amyloidogenic Peptide Motifs.
Given the importance of homotypic interactions between amyloidogenic peptide segments in protein aggregation (15, 19, 35, 36), we hypothesized that the binding of Aβ gammabodies is mediated via homotypic interactions between the Aβ motifs on the antibody surface and the same motifs within aggregated Aβ conformers. This hypothesis would predict that gammabodies displaying the amyloidogenic middle (Aβ15–24) and C-terminal (Aβ33–42) Aβ segments bind to distinct epitopes within Aβ fibrils in a noncompetitive manner. Therefore, we bound each gammabody (Aβ15–24 or Aβ33–42) separately to Aβ fibrils at a saturating antibody concentration and then evaluated the binding of the second gammabody over a range of antibody concentrations (Fig. S7). Importantly, we find that the binding of either gammabody to fibrils does not impact binding of the other gammabody, revealing that the grafted antibodies target unique sites within Aβ fibrils.
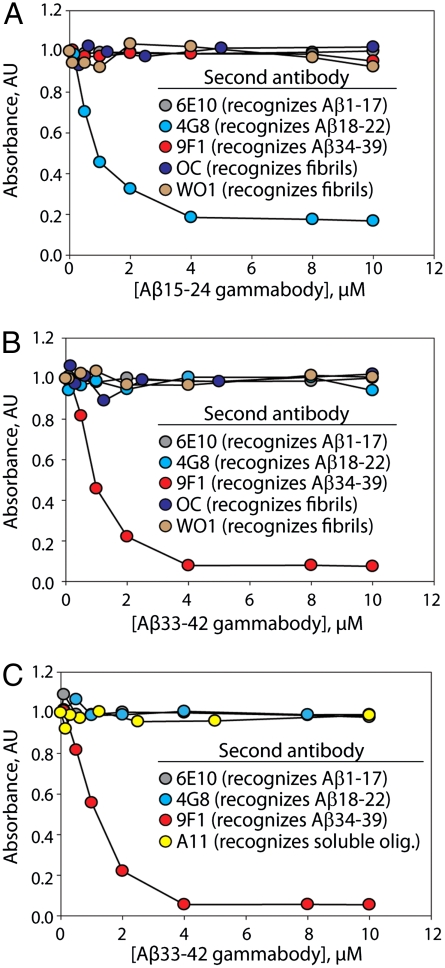
Nevertheless, we sought to further evaluate whether Aβ gammabodies employ homotypic interactions to recognize Aβ fibrils and oligomers. Therefore, we performed additional competitive binding analysis between gammabodies and sequence-specific monoclonal antibodies against Aβ. We hypothesized that gammabodies bound to Aβ conformers would prevent binding of sequence-specific monoclonal antibodies if their Aβ sequence epitopes overlapped. To test this hypothesis, we first bound the Aβ15–24 and Aβ33–42 gammabodies individually to Aβ fibrils over a range of antibody concentrations, and then evaluated binding of each monoclonal antibody (6E10, 4G8, and 9F1; Fig. 4). We find that the binding of the Aβ15–24 gammabody inhibits binding of the monoclonal antibody (4G8) specific for an overlapping sequence (Aβ residues 18–22; Fig. 4A). In contrast, we find that the same gammabody (Aβ15–24) does not inhibit binding of monoclonal antibodies 6E10 and 9F1 specific for nonoverlapping Aβ sequences (Aβ1–17 for 6E10 and Aβ34–39 for 9F1; Fig. 4A). Conversely, binding of the Aβ33–42 gammabody to fibrils interferes with subsequent binding of the monoclonal antibody (9F1) specific for an overlapping sequence (Aβ residues 34–39) but does not interfere with binding of other monoclonal antibodies specific for nonoverlapping Aβ sequences (Fig. 4B). We also find that the Aβ33–42 gammabody binds to the Aβ C terminus within soluble oligomers in a competitive manner with the 9F1 monoclonal antibody (Fig. 4C). Importantly, both Aβ gammabodies are noncompetitive with the fibril-specific (OC and WO1; Fig. 4 A and B) and oligomer-specific (A11; Fig. 4C) antibodies, revealing that they bind to unique conformational epitopes relative to previously identified conformation-specific antibodies (4, 5, 8). We conclude that Aβ gammabodies employ self-interactions between grafted amyloidogenic motifs and the same motifs within Aβ conformers to mediate conformation- and sequence-specific antibody recognition.
Fig. 4.
Aβ gammabodies and monoclonal antibodies bind competitively to Aβ oligomers and fibrils. (A and B) Aβ42 fibrils and (C) soluble oligomers (2.5 μM) were immobilized in microtiter plates, and then (A) Aβ15–24 and (B and C) Aβ33–42 gammabodies were added (0–10 μM). Afterward, monoclonal and polyclonal antibodies (2–5 μM) were bound and detected.
Aβ Gammabodies Are Sequence-Specific.
The grafted Aβ motifs that confer binding activity are hydrophobic and may mediate binding simply based on their amino acid composition rather than their sequence. To further evaluate the specificity of Aβ gammabodies, we scrambled the Aβ motifs within two gammabodies (Aβ12–21 and Aβ33–42) and evaluated binding of the antibody variants to each Aβ conformer (Fig. S8). We find that the scrambled motifs fail to mediate antibody binding to each Aβ conformer, confirming that the amino acid sequence (instead of the amino acid composition) of the grafted Aβ motifs mediates antibody binding.
We performed two additional tests of the specificity of the grafted antibodies. First, we asked whether Aβ gammabodies recognize other amyloidogenic polypeptides (Fig. S9). We assumed that Aβ gammabodies would fail to recognize aggregated polypeptides that lack the cognate amyloidogenic motifs. Indeed, we find that the Aβ12–21 and Aβ33–42 gammabodies fail to recognize fibrils or monomers of several amyloidogenic peptides and proteins (islet amyloid polypeptide, Tau, CsgA, and β2-microglobulin; Fig. S9). We also asked whether Aβ gammabodies would recognize Aβ conformers with the same sensitivity and conformational specificity in the presence of serum and mammalian cell lysate (Fig. S10). Indeed, we find that the detection sensitivity and selectivity of the Aβ12–21 and Aβ33–42 gammabodies are unchanged when Aβ conformers are diluted in serum and cell lysate. We conclude that Aβ amyloidogenic motifs mediate conformation-specific antibody recognition of Aβ conformers in a sequence-specific manner.
Gammabodies Neutralize the Toxicity of Aβ Oligomers and Fibrils.
We next investigated whether our grafted antibodies that recognize Aβ oligomers and fibrils would also inhibit the cellular toxicity of each Aβ conformer. We used a PC12 cell culture assay that we have reported elsewhere (Fig. 5) (30–32). We find that Aβ gammabodies are nontoxic, confirming that the Aβ peptide motifs are benign in the context of the VH domain. Moreover, in the absence of gammabodies, we find that Aβ soluble oligomers are more toxic than Aβ fibrils, as expected (5, 37, 38). Importantly, we find that the Aβ12–21, Aβ15–24, Aβ18–27, Aβ30–39, and Aβ33–42 gammabodies inhibit the toxicity of fibrils (Fig. 5). In contrast, we find that only the Aβ30–39 and Aβ33–42 gammabodies inhibit the toxicity of soluble oligomers. These findings are in excellent agreement with the corresponding immunoblot analysis (Fig. 2) because each grafted antibody that binds to Aβ oligomers and fibrils also neutralizes their toxicity. We conclude that Aβ gammabodies neutralize the toxicity of Aβ oligomers and fibrils in a manner that is strictly dependent on the antibody binding specificity.
Fig. 5.
Aβ gammabodies inhibit the toxicity of Aβ soluble oligomers and fibrils. Aβ42 fibrils and oligomers (12.5 μM) were incubated with Aβ gammabodies (10 μM) and reference conformation-specific antibodies (A11 and OC, 2 nM), diluted 10 times into PC12 cells, and the cell viability [% 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide, or MTT, reduction] was assayed after 2 d (n = 3).
Discussion
Antibodies typically recognize antigens via complementary interactions between multiple antibody loops and continuous or discontinuous sequence epitopes on the target antigen. The complexity of antibody recognition has prevented the design of antibodies that bind to antigens in either a sequence- or conformation-specific manner. We have demonstrated a surprisingly simple design strategy for generating sequence- and conformation-specific antibodies against misfolded Aβ conformers. Our strategy is guided by the structure of Aβ fibrils in which amyloidogenic motifs from one Aβ monomer stack on identical motifs from an adjacent Aβ monomer to form in-register, parallel β-sheets (18–20). We have exploited the same self-complementary interactions between amyloidogenic peptide motifs that govern Aβ aggregation to mediate specific antibody recognition of Aβ oligomers and fibrils.
The fact that Aβ gammabodies use homotypic interactions to recognize Aβ conformers enables us to generate structural hypotheses regarding the conformational differences between Aβ soluble oligomers and fibrils. Because Aβ soluble oligomers mature into fibrils and the central hydrophobic Aβ segment 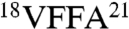 forms β-sheets within fibrils (19, 20), we posit that fibril-specific gammabodies (Aβ12–21, Aβ15–24, and Aβ18–27) recognize the Aβ18–21 motif in a β-sheet conformation. Moreover, because the same gammabodies fail to recognize Aβ oligomers, we posit the conversion of the Aβ18–21 motif into a β-sheet conformation is a key structural change required for Aβ oligomers to convert into fibrils (39, 40). In contrast, we find that gammabodies displaying the hydrophobic C-terminal motif of Aβ display similar (albeit subtly different) immunoreactivity with Aβ fibrils and oligomers, suggesting that these Aβ conformers possess similarly structured C-terminal segments (39–41). Nevertheless, the modest difference in affinity of the Aβ33–42 gammabody for fibrils relative to oligomers suggests that the C terminus of Aβ42 matures structurally when soluble oligomers convert into fibrils (39, 41).
forms β-sheets within fibrils (19, 20), we posit that fibril-specific gammabodies (Aβ12–21, Aβ15–24, and Aβ18–27) recognize the Aβ18–21 motif in a β-sheet conformation. Moreover, because the same gammabodies fail to recognize Aβ oligomers, we posit the conversion of the Aβ18–21 motif into a β-sheet conformation is a key structural change required for Aβ oligomers to convert into fibrils (39, 40). In contrast, we find that gammabodies displaying the hydrophobic C-terminal motif of Aβ display similar (albeit subtly different) immunoreactivity with Aβ fibrils and oligomers, suggesting that these Aβ conformers possess similarly structured C-terminal segments (39–41). Nevertheless, the modest difference in affinity of the Aβ33–42 gammabody for fibrils relative to oligomers suggests that the C terminus of Aβ42 matures structurally when soluble oligomers convert into fibrils (39, 41).
That our grafted antibodies possess well-defined sequence-specific epitopes within Aβ oligomers and fibrils deserves further consideration. Notably, our work represents the most direct identification of conformation-specific antibody binding sites within Aβ oligomers and fibrils to date. Previous efforts to identify the binding sites of conformation-specific antibodies have employed unstructured (or uncharacterized) Aβ peptide fragments as competitor molecules (10, 12). This approach is problematic because unstructured Aβ peptides lack conformation-specific epitopes and aggregated conformers of these peptides may not possess the same conformational epitopes found within aggregated conformers of full-length Aβ42. In contrast, our competitive binding approach using sequence-specific monoclonal antibodies enables facile identification of conformation- and sequence-specific binding sites targeted by Aβ gammabodies. Interestingly, we also found that Aβ gammabodies recognize unique conformational epitopes within Aβ fibrils and soluble oligomers relative to antibodies specific for fibrillar (OC, WO1) and oligomeric (A11) conformers reported previously (4, 5, 8). Our results suggest that Aβ gammabodies recognize linear sequence epitopes in a conformation-specific manner, similar to how Aβ monomers recognize fibrils. In contrast, we speculate that monoclonal (WO1) and polyclonal (A11 and OC) conformation-specific antibodies recognize topological features of fibrils and soluble oligomers involving discontinuous sequences (such as stacks of identical residues along the fibril axis) that do not overlap with those recognized by our grafted antibodies.
We envision many variations of our motif-grafting strategy that should lead to biomolecules with unique conformational specificities and affinities against Aβ oligomers and fibrils relative to the antibodies reported in this work. The autonomy of the Aβ amyloidogenic motifs should allow them to be grafted into proteins other than antibodies possessing appropriate solvent-exposed loops. For example, we expect that fluorescent proteins bearing amyloidogenic motifs in their solvent-exposed loops may be particularly valuable for imaging intracellular and extracellular misfolded proteins. Moreover, we expect that grafting multiple copies of the same amyloidogenic motif or combinations of different motifs into larger antibody fragments (single-chain Fv and Fabs) and full-length antibodies will lead to gammabodies with even higher affinities and unique conformation-specific binding activities relative to those reported here. Finally, we expect that grafting amyloidogenic motifs from other misfolded proteins into diverse antibody formats will lead to similar conformation- and sequence-specific binding affinity as we observed in this work for Aβ. Should our motif-grafting strategy be found to be a general approach for synthesizing conformation-specific antibodies against amyloidogenic proteins, we expect it would lead to a unique class of antibodies for analyzing and targeting misfolded conformers in diverse protein aggregation disorders.
Methods
Preparation of Aβ Conformers.
Aβ soluble oligomers were prepared by dissolving the peptide Aβ42 (American Peptide) in 100% hexafluoroisopropanol (HFIP, Fluka). The HFIP was evaporated and Aβ was dissolved in 50 mM NaOH (1 mg/mL Aβ), sonicated (30 s), and diluted in PBS (25 μM Aβ). The peptide was then centrifuged (22,000 × g for 30 min) and the pelleted fraction (5% of starting volume) was discarded. The supernatant was incubated at 25 °C for 0–6 d without agitation. Aβ fibrils were prepared via the same procedure except that monomers were mixed with preexisting fibrils (10–20 wt% seed) without mixing for 24 h at 25 °C.
Cloning, Expression, and Purification of Gammabodies.
A DNA fragment encoding the parent VH antibody [Protein Data Bank (PDB): 3B9V] with a PelB leader sequence for periplasmic expression and C-terminal tags (3 FLAG tags followed a 7×histidine tag) was created using PCR-based gene synthesis. The parent antibody was ligated into a pET17b plasmid (Novagen) between the NdeI and XhoI restriction sites, and oligonucleotide primers encoding each grafted loop were ligated between the BamHI and NotI restriction sites flanking CDR3. The antibody variants were expressed in bacteria [BL21(DE3)pLysS; Stratagene] for 48 h at 30 °C using autoinduction media (42) supplemented with ampicillin (100 μg/mL) and chloramphenicol (35 μg/mL). Afterward, the cells (without lysis) were pelleted via centrifugation at 3,500 × g, discarded, and the supernatant was incubated overnight with 2.5 mL of Ni-nitrilotriacetate (Ni-NTA) beads (Pierce) at 15 °C with mild agitation. The Ni-NTA beads were collected, and then the antibody was eluted (pH 3, PBS) and neutralized (pH 7). The protein purity was confirmed to be > 95% by SDS/PAGE analysis (10% acrylamide 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)-1,3-propanediol gel; Invitrogen).
Immunoblot Analysis.
Aβ conformers (25 μM) were spotted (2 μL) on nitrocellulose membranes (Hybond ECL; GE Healthcare). The blots were blocked overnight (10% nonfat dry milk in PBS) and probed with each antibody (at the reported concentration). Blots with bound Aβ gammabodies were then probed with anti-FLAG antibody (1∶5,000 dilution; Sigma-Aldrich), and all blots were probed with appropriate horseradish peroxidase-conjugated secondary antibodies.
Affinity Measurements.
The affinities of gammabodies specific for Aβ soluble oligomers and fibrils were measured using competitive ELISA analysis (33). Aβ samples (1–10 μM) were coincubated overnight with a fixed concentration of grafted antibody (0.5–2 μM). The next day the amount of unbound gammabody was quantified by transferring the gammabody-Aβ solutions into 96-well microtiter plates (Nunc Maxisorb; Thermo Fisher) in which the same Aβ conformer of interest had been immobilized (2 μM Aβ). After 30 min, the wells were washed and additional antibodies (anti-FLAG and peroxidase-conjugated secondary antibody) were added and developed. The dissociation constants were calculated based on binding measurements for at least five antigen concentrations in excess of the concentration of Aβ gammabody (33).
Competitive Binding Analysis.
Aβ fibrils and soluble oligomers (2.5 μM) were immobilized in 96-well microtiter plates (Nunc Maxisorb; Thermo Fisher) and blocked overnight (10% milk in PBS). Gammabodies (0–10 μM) were then added to the well plates containing immobilized Aβ and allowed to bind overnight. After removal of unbound antibody, each well was probed with monoclonal (6E10 from Sigma-Aldrich; 4G8 from Covance; 9F1 from Santa Cruz; and WO1 from Ronald Wetzel, University of Pittsburgh, Pittsburgh, PA) and polyclonal (A11; Invitrogen and OC; Millipore) antibodies (1 h). Finally, the bound monoclonal and polyclonal antibodies were detected using the appropriate horseradish peroxidase-conjugated secondary antibody.
Cell Toxicity Assay.
Rat adrenal medulla cells (PC12; ATCC) were cultured in Dulbecco’s Modified Eagle Media (5% fetal bovine serum, 10% horse serum, and 1% penicillin-streptomycin). The cell suspension (90 μL) was incubated in 96-well microtiter plates (CellBIND; Corning) for 24 h. Afterward, Aβ42 and gammabodies (12.5 μM Aβ and 10 μM antibody) were added to microtiter plates (10 μL), and the cells were further incubated for 48 h at 37 °C. The media were then removed, and fresh media (200 μL) and thiazolyl blue tetrazolium bromide (Sigma; 50 μL of 2.5 mg/mL) were added to each well for 3 h at 37 °C. Finally, these solutions were discarded, 250 μL of DMSO was added, and the absorbance was measured at 562 nm. The toxicity values were normalized relative to BSA (12.5 μM).
Supplementary Material
ACKNOWLEDGMENTS.
We greatly appreciate the gift of the WO1 antibody from Dr. Ronald Wetzel (University of Pittsburgh), IAPP fibrils from Dr. Daniel Raleigh (Stony Brook University), Tau fibrils from Dr. Martin Margittai (University of Denver), CsgA fibrils from Dr. Matthew Chapman (University of Michigan), and β2-microglobulin fibrils from Dr. Sheena Radford (University of Leeds). This work was supported by the Alzheimer’s Association (NIRG-08-90967), National Science Foundation (CAREER Award 954450), and Pew Charitable Trust (Pew Scholar Award in Biomedical Sciences).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111232108/-/DCSupplemental.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu Rev Phys Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert MP, et al. Vaccination with soluble Abeta oligomers generates toxicity-neutralizing antibodies. J Neurochem. 2001;79:595–605. doi: 10.1046/j.1471-4159.2001.00592.x. [DOI] [PubMed] [Google Scholar]
- 4.O’Nuallain B, Wetzel R. Conformational Abs recognizing a generic amyloid fibril epitope. Proc Natl Acad Sci USA. 2002;99:1485–1490. doi: 10.1073/pnas.022662599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 6.Habicht G, et al. Directed selection of a conformational antibody domain that prevents mature amyloid fibril formation by stabilizing Abeta protofibrils. Proc Natl Acad Sci USA. 2007;104:19232–19237. doi: 10.1073/pnas.0703793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kayed R, et al. Conformation dependent monoclonal antibodies distinguish different replicating strains or conformers of prefibrillar Abeta oligomers. Mol Neurodegener. 2010;5:57. doi: 10.1186/1750-1326-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kayed R, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kvam E, et al. Conformational targeting of fibrillar polyglutamine proteins in live cells escalates aggregation and cytotoxicity. PLoS One. 2009;4:e5727. doi: 10.1371/journal.pone.0005727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert MP, et al. Monoclonal antibodies that target pathological assemblies of Abeta. J Neurochem. 2007;100:23–35. doi: 10.1111/j.1471-4159.2006.04157.x. [DOI] [PubMed] [Google Scholar]
- 11.Lee EB, et al. Targeting amyloid-beta peptide (Abeta) oligomers by passive immunization with a conformation-selective monoclonal antibody improves learning and memory in Abeta precursor protein (APP) transgenic mice. J Biol Chem. 2006;281:4292–4299. doi: 10.1074/jbc.M511018200. [DOI] [PubMed] [Google Scholar]
- 12.Meli G, Visintin M, Cannistraci I, Cattaneo A. Direct in vivo intracellular selection of conformation-sensitive antibody domains targeting Alzheimer’s amyloid-beta oligomers. J Mol Biol. 2009;387:584–606. doi: 10.1016/j.jmb.2009.01.061. [DOI] [PubMed] [Google Scholar]
- 13.Zameer A, Kasturirangan S, Emadi S, Nimmagadda SV, Sierks MR. Anti-oligomeric Abeta single-chain variable domain antibody blocks Abeta-induced toxicity against human neuroblastoma cells. J Mol Biol. 2008;384:917–928. doi: 10.1016/j.jmb.2008.09.068. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 15.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 16.Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature. 2007;447:556–561. doi: 10.1038/nature05848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci USA. 2010;107:3487–3492. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benzinger TL, et al. Propagating structure of Alzheimer’s beta-amyloid(10–35) is parallel beta-sheet with residues in exact register. Proc Natl Acad Sci USA. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petkova AT, et al. A structural model for Alzheimer’s beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luhrs T, et al. 3D structure of Alzheimer’s amyloid-beta(1–42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torok M, et al. Structural and dynamic features of Alzheimer’s Abeta peptide in amyloid fibrils studied by site-directed spin labeling. J Biol Chem. 2002;277:40810–40815. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- 22.Austen BM, et al. Designing peptide inhibitors for oligomerization and toxicity of Alzheimer’s beta-amyloid peptide. Biochemistry. 2008;47:1984–1992. doi: 10.1021/bi701415b. [DOI] [PubMed] [Google Scholar]
- 23.Fradinger EA, et al. C-terminal peptides coassemble into Abeta42 oligomers and protect neurons against Abeta42-induced neurotoxicity. Proc Natl Acad Sci USA. 2008;105:14175–14180. doi: 10.1073/pnas.0807163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon DJ, Tappe R, Meredith SC. Design and characterization of a membrane permeable N-methyl amino acid-containing peptide that inhibits Abeta1-40 fibrillogenesis. J Pept Res. 2002;60:37–55. doi: 10.1034/j.1399-3011.2002.11002.x. [DOI] [PubMed] [Google Scholar]
- 25.Lowe TL, Strzelec A, Kiessling LL, Murphy RM. Structure-function relationships for inhibitors of beta-amyloid toxicity containing the recognition sequence KLVFF. Biochemistry. 2001;40:7882–7889. doi: 10.1021/bi002734u. [DOI] [PubMed] [Google Scholar]
- 26.Moroncini G, et al. Motif-grafted antibodies containing the replicative interface of cellular PrP are specific for PrPSc. Proc Natl Acad Sci USA. 2004;101:10404–10409. doi: 10.1073/pnas.0403522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solforosi L, et al. Toward molecular dissection of PrPC-PrPSc interactions. J Biol Chem. 2007;282:7465–7471. doi: 10.1074/jbc.M610051200. [DOI] [PubMed] [Google Scholar]
- 28.Barthelemy PA, et al. Comprehensive analysis of the factors contributing to the stability and solubility of autonomous human VH domains. J Biol Chem. 2008;283:3639–3654. doi: 10.1074/jbc.M708536200. [DOI] [PubMed] [Google Scholar]
- 29.Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 30.Ladiwala AR, et al. Resveratrol selectively remodels soluble oligomers and fibrils of amyloid Abeta into off-pathway conformers. J Biol Chem. 2010;285:24228–24237. doi: 10.1074/jbc.M110.133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladiwala AR, Dordick JS, Tessier PM. Aromatic small molecules remodel toxic soluble oligomers of amyloid beta through three independent pathways. J Biol Chem. 2011;286:3209–3218. doi: 10.1074/jbc.M110.173856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ladiwala AR, et al. Polyphenolic glycosides and aglycones utilize opposing pathways to selectively remodel and inactivate toxic oligomers of amyloid beta. Chembiochem. 2011;12:1749–1758. doi: 10.1002/cbic.201100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friguet B, Chaffotte AF, Djavadi-Ohaniance L, Goldberg ME. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 34.Bobrovnik SA. Determination of antibody affinity by ELISA. Theory. J Biochem Biophys Methods. 2003;57:213–236. doi: 10.1016/s0165-022x(03)00145-3. [DOI] [PubMed] [Google Scholar]
- 35.Margittai M, Langen R. Template-assisted filament growth by parallel stacking of tau. Proc Natl Acad Sci USA. 2004;101:10278–10283. doi: 10.1073/pnas.0401911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci U S A. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahlgren KN, et al. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J Biol Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 38.Chromy BA, et al. Self-assembly of Abeta(1–42) into globular neurotoxins. Biochemistry. 2003;42:12749–12760. doi: 10.1021/bi030029q. [DOI] [PubMed] [Google Scholar]
- 39.Yu L, et al. Structural characterization of a soluble amyloid beta-peptide oligomer. Biochemistry. 2009;48:1870–1877. doi: 10.1021/bi802046n. [DOI] [PubMed] [Google Scholar]
- 40.Zhang A, Qi W, Good TA, Fernandez EJ. Structural differences between Abeta(1–40) intermediate oligomers and fibrils elucidated by proteolytic fragmentation and hydrogen/deuterium exchange. Biophys J. 2009;96:1091–1104. doi: 10.1016/j.bpj.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmed M, et al. Structural conversion of neurotoxic amyloid-beta(1–42) oligomers to fibrils. Nat Struct Mol Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.