Abstract
The centromere is the genetic locus that organizes the proteinaceous kinetochore and is responsible for attachment of the chromosome to the spindle at mitosis and meiosis. In most eukaryotes, the centromere consists of highly repetitive DNA sequences that are occupied by nucleosomes containing the CenH3 histone variant, whereas in budding yeast, a ∼120-bp centromere DNA element (CDE) that is sufficient for centromere function is occupied by a single right-handed histone variant CenH3 (Cse4) nucleosome. However, these in vivo observations are inconsistent with in vitro evidence for left-handed octameric CenH3 nucleosomes. To help resolve these inconsistencies, we characterized yeast centromeric chromatin at single base-pair resolution. Intact particles containing both Cse4 and H2A are precisely protected from micrococcal nuclease over the entire CDE of all 16 yeast centromeres in both solubilized chromatin and the insoluble kinetochore. Small DNA-binding proteins protect CDEI and CDEIII and delimit the centromeric nucleosome to the ∼80-bp CDEII, only enough for a single DNA wrap. As expected for a tripartite organization of centromeric chromatin, loss of Cbf1 protein, which binds to CDEI, both reduces the size of the centromere-protected region and shifts its location toward CDEIII. Surprisingly, Cse4 overproduction caused genome-wide misincorporation of nonfunctional CenH3-containing nucleosomes that protect ∼135 base pairs and are preferentially enriched at sites of high nucleosome turnover. Our detection of two forms of CenH3 nucleosomes in the yeast genome, a singly wrapped particle at the functional centromere and octamer-sized particles on chromosome arms, reconcile seemingly conflicting in vivo and in vitro observations.
Keywords: Saccharomyces cerevisiae, chromatin immunoprecipitation, chromosome segregation
The centromere is the genetic locus that specifies the location of the kinetochore, the complex proteinaceous structure that attaches to spindle microtubules for regular segregation to the poles at mitosis and meiosis (1). Every chromosome must have one and only one centromere, because acentric and dicentric chromosomes are lost, leading to aneuploidy and cell death. This stringent requirement for a single centromere has led to the expectation that centromeres would be defined by DNA sequence, and indeed this is the case in the budding yeast, Saccharomyces cerevisiae, where each of the 16 centromeres consists of a ∼120-bp sequence that is entirely responsible for centromere specification (2). However, a common feature of centromeres in multicellular eukaryotes is that they are embedded in highly repetitive satellite DNA, which has made their molecular study difficult (3). Furthermore, the existence of neocentromeres that entirely lack centromeric satellites indicates that specific DNA sequences are not necessary for centromere function (4).
Despite the fundamental differences between budding yeast and multicellular eukaryotes with respect to sequence determinants of centromere identity, there are common protein determinants. Most important is the histone variant, CenH3 (CENP-A in mammals and Cse4 in yeast), which replaces histone H3 in centromeric nucleosomes and is essential for recruitment of the other structural components of the kinetochore (5). It has been previously shown that the Cse4 nucleosome wraps DNA in a right-handed orientation (6, 7), consistent with in vivo observations of heterotypic tetrameric nucleosomes in Drosophila (8) and humans (9). However, several studies have shown that CenH3 nucleosomes are left-handed octamers in vitro (10–14).
To help reconcile these findings, we have characterized yeast centromeric particles using a single base-pair resolution mapping method (15). This mapping revealed that the centromere DNA element (CDE) is well protected in intact particles that also contain histone H2A, but is preferentially cleaved internally at sites of binding for sequence-specific factors, termed CDEI and CDEIII (16), where we mapped distinct particles that, respectively, correspond to the known binding sites for the Cbf1 protein and the Centromere Binding Factor 3 (CBF3) complex. Using a yeast strain containing multiple copies of Cse4, we found that Cse4-containing particles incorporate at canonical nucleosome positions throughout the genome, and are enriched at sites of rapidly turning over nucleosomes. The existence of two Cse4 nucleosomal species, a stable particle with a single DNA wrap at centromeres and an unstable octamer in chromosome arms, supports a general model in which unstable CenH3 nucleosomes are rapidly turned over on chromosome arms to maintain one and only one centromere per chromosome.
Results
Cse4-Containing Chromatin Particles Map Precisely to Functional Centromeres.
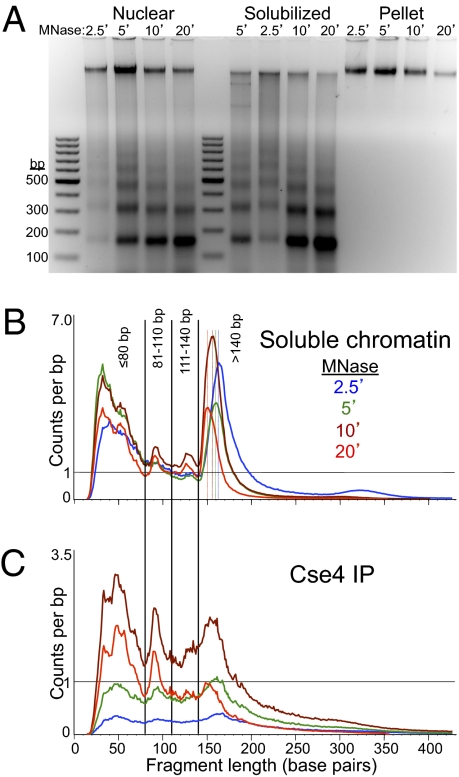
We performed MNase digestion of crude yeast nuclei from log-phase cells grown in rich medium according to a standard protocol (17), except that we included a needle extraction step to gently but thoroughly solubilize MNase-digested chromatin (15). This procedure resulted in essentially complete recovery of MNase nucleosome ladder DNA (Fig. 1A) and solubilization of most of Cen3 by quantitative PCR (qPCR) (Fig. S1). In the experiment shown in Fig. 1, we used MNase digestion times of 2.5, 5, 10, and 20 min, and followed this by ChIP of FLAG-Cse4. We applied a modified protocol for Solexa library preparation that results in recovery and sequencing of particles down to ∼25 bp (15). After paired-end library preparation and sequencing on an Illumina HiSeq 2000 instrument, we obtained on average ∼45 million mapped paired-end reads for solubilized chromatin (the input for ChIP). These mapped fragments showed two prominent size features: a broad distribution of fragments ranging in size from ∼20–80 bp, with a peak at ∼30 bp, and a narrow distribution corresponding to nucleosome-sized DNA fragments (Fig. 1B). The nucleosomal peaks showed stepwise reductions in average size as expected for an MNase series, ranging from 163 bp (2.5 min) to 151 bp (20 min). For the ChIP material, we obtained on average ∼23 million mapped paired-end reads. These reads showed a broader size distribution, with indistinct nucleosome-sized peaks, and a broad peak at ∼50 bp for 10- and 20-min digestions (Fig. 1C). A sharp peak at ∼90-bp seen in the 10- and 20-min samples for both the soluble chromatin input and the ChIP is attributable to internal cleavage of canonical nucleosomes (15). In the analyses described below, we used all mapped fragments regardless of size.
Fig. 1.
Paired-end sequencing of soluble chromatin and Cse4 ChIP yields distinct size classes of MNase-protected particles. (A) Agarose gel analysis of MNase time-point samples showing DNA from whole nuclei extracted from strain SBY5146 after MNase digestion (Nuclear), DNA from soluble chromatin after needle extraction and pooling of extracts (Solubilized), and DNA from the insoluble residue after solubilization (Pellet). The gradual reduction in size of protected DNA fragments can be seen as jogs in the solubilized chromatin samples loaded out-of-order (5, 2.5, 10, and 20 min) in the middle set of lanes. Size distributions of mapped paired-end reads for Solubilized chromatin (B) and FLAG-Cse4 ChIP (C), showing the size classes chosen for further analysis.
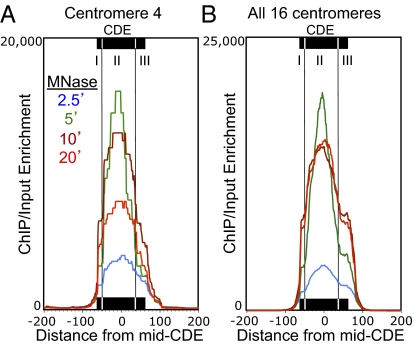
We first mapped the ratio of Cse4/Input using as metric the paired-end read count density (15). For all 16 chromosomes, the maximum Cse4/Input occupancy was over the centromere, consistent with a previous genome-wide mapping study (18). However, that study used cross-linking and sonication before immunoprecipitation (X-ChIP), resulting in a low resolution map, with enrichment extending >300 bp to either side of the center of functional centromeres (Fig. S2). In striking contrast, our native chromatin mapping of Cse4/Input at a centromere (Cen4) shows it to be confined primarily to the Cen4 CDE (Fig. 2A). We attribute the broad distribution of Cse4 obtained by X-ChIP in part to the large size of sonicated fragments and in part to the cross-linking of centromeric nucleosomes to flanking nucleosomes and other proteins. The nearly precise mapping of Cse4 ChIP material to Cen4 that we observed using native chromatin delimits the span of the Cse4 nucleosome to the functional centromere sequence. Interestingly, the ChIP/Input signal is nonuniform, showing about twice the protection from MNase over CDEII than over CDEI and CDEIII for all samples.
Fig. 2.
ChIP maps the Cse4 nucleosome to CDEII. Mapped paired-end reads for Cse4 ChIP displayed as ChIP/Input ratios at 1-bp resolution. (A) MNase series of FLAG-Cse4 ChIP for Cen4. (B) Same as A, but for all 16 centromeres aligned around the mid-CDE. See also Fig. S2 for a comparison with Cse4 X-ChIP data.
To ascertain the generality of this result, we aligned ChIP/Input profiles for all 16 yeast centromeres around the midpoint of each CDE and averaged the signal over each base pair. This analysis confirmed that the clear pattern seen for Cen4 for all four samples in the MNase series is general, with nearly precise protection of the functional centromere. Our analysis also confirms a previous report of precise positioning of MNase-protected particles over all 16 CDEs (Fig. S3) (19). The greatest ChIP signal was centered over CDEII, with distinct shoulders on either side corresponding to partial MNase protection of CDEI and CDEIII (Fig. 2B). CDEI corresponds to the 8-bp consensus sequence for Cbf1, a conserved general transcription factor that is found at a large number of sites throughout the yeast genome, including centromeres (20, 21). CDEIII corresponds to a 26-bp consensus sequence that is the binding site for CBF3, a multisubunit complex that is specific for budding yeast centromeres (22). CDEII is conserved for AT-richness and length (from 78 to 86 bp), but otherwise has no distinguishing features (23). Because of the documented presence of Cbf1 and the CBF3 complex at the CDE, we interpret the higher ChIP signal for Cse4 over CDEII relative to CDEI and CDEIII as strong evidence for a well-positioned Cse4-containing particle precisely over CDEII.
Distinct Particles over CDEI and CDEIII Flank the Cse4 Nucleosome.
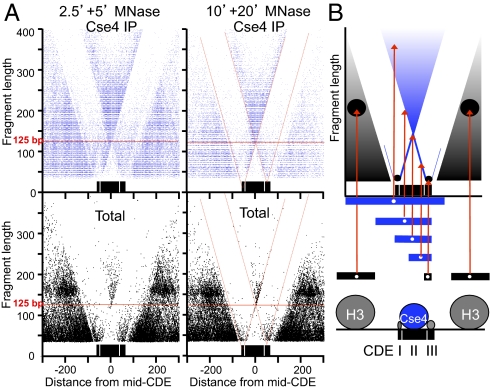
To obtain independent confirmation our ChIP/Input occupancy mapping of CDEs, we analyzed the size distribution of MNase-protected DNA around centromeres by plotting the length of each mapped fragment on the y axis versus the distance of its midpoint location from the midpoint of the CDE on the x axis (a “V-plot”) (15). Strikingly, we observed a clear V-shaped pattern centered precisely over the midpoint of the aligned CDEs from all centromeres (Fig. 3A). The sharp edges of the V map midpoints of fragments that are cleaved precisely at one edge of the CDE and extend beyond the opposite edge, and the vertex maps the midpoint of fragments that are cleaved precisely on both sides of the CDE (diagramed in Fig. 3B). The vertex corresponds to a fragment size of ∼120 bp, which is the average size of annotated CDEs (http://www.yeastgenome.org), indicating that the CDE is almost perfectly protected from MNase digestion.
Fig. 3.
V-plot mapping reveals subnucleosome particles at CDEI and CDEIII. (A) The distance from the midpoint of each fragment to the midpoint of the CDE that maps ±300 bp of a mid-CDE is represented by a dot, where its position on the x axis is the distance between its midpoint and the midpoint of the CDE, and its position on the y axis represents its fragment length. Cse4 immunoprecipitation (blue) and total soluble chromatin (black) maps are shown for data from 2.5- and 5-min MNase digestion time points (Left) and from 10- and 20-min MNase digestion time points (Right). The diagonals are marked with red lines (Right) to show that they intersect at the same positions within CDEI and CDEIII for both Cse4 ChIP and Total soluble chromatin. (B) Diagram showing the position of dots placed on the V-plot (red arrows) for fragments of various sizes and map positions. Below is a model of particles that protect DNA from MNase digestion deduced from the midpoint maps.
Each diagonal of a V-plot represents a single sharply defined cleavage on one side of a particle and random cleavage on the other side (15). In the case of CDEs, we observed an additional pair of V-shaped patterns, one over CDEI and the other over CDEIII (Fig. 3A). These patterns were generated by cleavages between CDEI and CDEII and between CDEII and CDEIII, respectively. These internal cleavages were too rare to generate detectable numbers of CDEII-only fragments, which implies that each element of the CDE is protected by one of three closely packed particles that block MNase from cleaving between them. Interestingly, these Vs in the Cse4 ChIPs fade below ∼50 bp, suggesting that MNase digestion released these two protected particles from association with Cse4. Consistent with this interpretation, protected fragments of the expected size and in the expected position can be observed in total chromatin. Extrapolation of diagonals to the vertex of each V results in a minimum protected size of ∼10 bp directly over CDEI and of ∼20 bp directly over CDEIII for both the Cse4 ChIP and the total chromatin (red dotted diagonal lines in Fig. 3A). The identification of distinct protected particles over both CDEI and CDEIII in soluble chromatin indicates that the distinct shoulders observed in the density plots (Fig. 2) represent partial protection by Cbf1 and the CBF3 complex that is independent of association with Cse4.
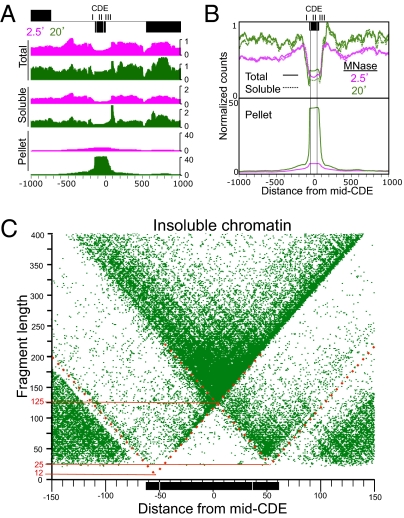
Centromeric sequences were recovered in the solubilized chromatin fraction used for ChIP at approximately two-thirds the level of that from total nuclei (Fig. 4 A and B), as if kinetochore attachment rendered a subset of these sequences relatively insoluble. To investigate this possibility, we sequenced libraries made from pellet-extracted DNA (15), which we found to be enriched ∼100-fold for the CDE in the 20-min digestion sample relative to the total nuclear DNA (Fig. 4 A and B). To determine the minimally protected region of the insoluble kinetochore, we displayed the data from the pellet fraction as V-plots (Fig. 4C and Fig. S4A). The patterns were nearly identical to those for Cse4 ChIP (Fig. S4 B and C), which demonstrates that the tripartite structure of centromeric chromatin can be observed without ChIP, based exclusively on reduced kinetochore solubility.
Fig. 4.
Kinetochore enrichment in insoluble chromatin. (A) Occupancy maps of total nuclei and soluble and insoluble (pellet) fractions for the 2-kb region spanning Centromere 4 showing changes that occur between 2.5- (magenta) to 20-min (green) MNase digestion. (B) An occupancy map for the same region showing the average of all 16 centromeres aligned at the midpoints of their CDEs. Based on the partitioning of total 20-min MNase-treated nuclear DNA into soluble and insoluble chromatin fractions, we estimate that approximately one-third of centromeric chromatin is insoluble and that insoluble centromeric chromatin is enriched ∼100-fold relative to other insoluble chromatin in the pellet. (C) The central 300 bp of the V-plot for insoluble chromatin for all 16 aligned centromeres shows a V-plot that is nearly identical to those seen for ChIP from soluble chromatin (Fig. 3 and Fig. S4 B and C). Dotted red lines indicate approximate extensions of diagonals, where each intersection maps the midpoint location of the protected region on the x axis and the minimal protected region on the y axis (solid red lines).
Loss of Cbf1 Reduces the Size and Shifts the Location of the Centromere-Protected Region.
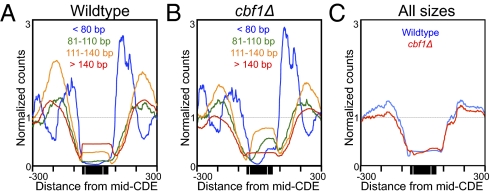
It remained formally possible that a single Cse4-containing particle spans the entire CDE, but the Cbf1 protein and the CBF3 complex bind DNA that is exposed on the nucleosomal surface. Surface binding is unlikely considering that both Cbf1 and CBF3 sharply bend DNA (24, 25), and also that Cbf1 binding excludes canonical nucleosomes (15, 21). To directly test the possibility that the Cse4 nucleosome occupies the entire CDE, we asked whether loss of one of the flanking particles causes the expected loss of protection of the centromeric element that the particle occupies. The CBF3 complex is essential for viability and for Cse4 nucleosome localization; however, Cbf1-null mutations are viable. Previously, Kent et al. (21) had performed paired-end DNA sequencing on MNase-protected fragments in both wild-type and cbf1Δ strains. We remapped their raw data to the yeast genome such that all mappable fragment sizes were included, and constructed average centromere density plots. For analysis, we separated the paired-end reads into four size classes: ≤80 bp, 81–110 bp, 111–140 bp, and >140 bp. We observed that loss of Cbf1 caused a striking reduction of 22 bp in the median size distribution of fragments that map to the CDE, with increases in ≤80-bp, 81- to 110-bp, and 111- to 140-bp size classes relative to the >140-bp size class (Fig. 5 A and B), with no noticeable change in overall occupancy (Fig. 5C). We also observed a conspicuous shift of the median fragment center, 10-bp closer to the CDEIII side of CDEII, with encroachment of ≤80-bp particles into CDEI. It is possible that the continued occupancy of CDEI is caused by the presence of other DNA-binding proteins in yeast that bind to the same CACGTG consensus sequence as Cbf1 (26). The reduction and shift in protection seen in most cells are as expected if Cbf1 protects CDEI in wild-type, but that loss of Cbf1 allows MNase better accessibility despite occupation of CDEI by other small particles. Alternatively, loss of Cbf1, which helps to exclude H3 nucleosomes in its vicinity (15, 21), might have reduced protection of the CDE, allowing transient occupancy by an H3 nucleosome in a small subset of cells.
Fig. 5.
Loss of Cbf1 reduces the size and shifts the location of the centromere-protected region. (A and B) Occupancy maps of DNA size classes from MNase-protected particles using data obtained from SRA SRR058444 and SRR058445 (21), comparing wild-type to cbf1Δ strains to illustrate the reduction in size distribution with loss of Cbf1. (C) No net change in centromeric occupancy occurs with loss of Cbf1.
Small Particles and Well-Positioned Nucleosomes Flank the CDE.
V-plots revealed moderate enrichment of subnucleosome-sized particles in total chromatin immediately flanking CDEs (Fig. 3A). In the Cse4 ChIP material, we also observed strong enrichment of ≤80-bp fragments centered ∼50 bp on either side of the CDEs, which were rapidly depleted with increasing MNase digestion (Fig. S5). Most subnucleosomal particles mapped elsewhere in the yeast genome are relatively stable to MNase digestion (15), which suggests that whatever is protecting CDE-flanking DNA on both sides might span the CDE. The subnucleosomal particles on either side of the CDE are themselves flanked by well-positioned H3 nucleosomes (Fig. 3A and Fig. S5B). The fact that all centromeres have these positioned nucleosomes at approximately the same distance from the CDE on both sides confirms and extends previous studies showing that centromeres are flanked by phased nucleosomes (17, 27).
H2A Is as Abundant at Centromeres as It Is Genome-Wide.
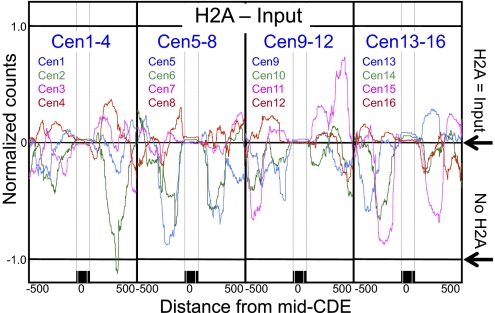
Previous studies have reported depletion of H2A and H2B nucleosomes at yeast centromeres, suggesting that the only histones in the Cse4 nucleosome are Cse4 and H4 (28, 29). To determine the composition of Cse4 nucleosomes in our chromatin preparations, we performed ChIP of FLAG-tagged H2A followed by paired-end DNA sequencing. If Cse4 nucleosomes were deficient in H2A, then we would expect that normalized counts for the H2A ChIP would be fewer than for the corresponding input DNA over centromeres. Rather, we found that at all 16 centromeres, the H2A ChIP signal was equal to that for input DNA (Fig. 6). The precise positioning of centromeric nucleosomes is evident from these profiles, insofar as flanking regions showed wide variations in H2A occupancy, with almost no variation within the CDE region or between centromeres.
Fig. 6.
The Cse4 nucleosome contains H2A. ChIP-seq profile of the difference in normalized counts between H2A and Input over all 16 centromeres after ChIP of H2A-FLAG from strain SBY2688, showing that H2A = Input (i.e., H2A − Input = 0). If H2A were absent from centromeres, H2A − Input would equal −1 (No H2A). Fragments larger than 200 bp were excluded to avoid any contribution from dinucleosomes. Similar centromeric results were obtained by plotting published data of crosslinked chromatin followed by ChIP-chip (Fig. S6C). The expected depletion of H2A at the H2A.Z-enriched Gal4 UAS is confirmed in Fig. S6A. The variability in flanking nucleosomes is probably because of the variable abundance of H2A.Z in canonical nucleosomes.
To confirm that our measurements were sufficiently sensitive to detect differences in H2A occupancy, we examined the single nucleosome occupying the Gal4 UAS in cells grown in glucose, which had been shown to be depleted of H2A because of the high relative abundance of the H2A.Z variant (30). Our H2A ChIP data showed clear depletion of H2A from this nucleosome relative to neighboring nucleosomes, confirming the sensitivity of our profiling assay (Fig. S6A). H2A is similarly depleted from nucleosomes immediately flanking centromeres (Fig. S6B), suggesting that these nucleosomes are also enriched for H2A.Z. For further confirmation of the abundance of H2A at centromeres, we similarly analyzed published X-ChIP-chip H2A data from the six centromeres represented on the microarray used in that study, and obtained a very similar profile to that for our native ChIP-seq data from all 16 centromeres (Fig. S6B).
Misincorporated Cse4 Particles Are Enriched at Sites of Rapid Turnover.
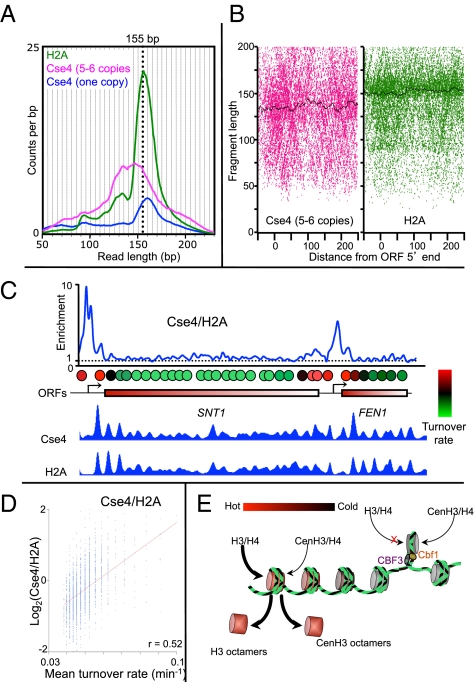
To directly compare the incorporation of Cse4 to that of a canonical histone, we performed ChIP using a strain expressing both FLAG-tagged H2A and five to six copies of Myc-tagged Cse4 (Fig. S7). Although FLAG-H2A yielded a nucleosomal size distribution expected for the degree of MNase digestion used (∼155 bp), DNA from Cse4-Myc nucleosomes immunoprecipitated from the same solubilized chromatin displayed a broader size distribution, with a peak at ∼135 bp that is not evident in control ChIPs from a single-copy Cse4 strain (Fig. 7A). These ∼135-bp protected Cse4 particles are phased at canonical nucleosomal positions in highly expressed genes (Fig. 7B), indicating that they correspond to mislocalized Cse4 nucleosomes. Notably, their size distribution matches that of partially unwrapped left-handed CenH3 octamers produced in vitro from purified components (Fig. S8A) (10, 13). Therefore, it is likely that excessive amounts of Cse4 led to the formation of conventional left-handed octameric particles in vivo that were deposited as nucleosomes genome-wide, in contrast to the much smaller Cse4 particle confined to the ∼80-bp CDEII.
Fig. 7.
Overproduced CenH3 particles occupy canonical nucleosome positions and protect ∼135 bp. Size distributions (A) of mapped fragments for FLAG-H2A (magenta) and Cse4-Myc (green) ChIPs, including a FLAG-Cse4 size distribution from a control (blue). See also Fig. S8A. (B) V-plots show nucleosomal fragments from the 20 most highly expressed genes aligned at their 5′ ORF ends. Black lines indicate median fragment sizes. (C) Overproduced Cse4-Myc and H2A over the SNT1-FEN1 region showing enrichment of Cse4/H2A (Top), an aligned hot nucleosome map reproduced from Dion et al. (31) (Middle), and enrichment of Cse4 and H2A (Bottom). (D) Scatterplot of ChIP signals versus turnover rates for Cse4/H2A. (E) Model in which CenH3 hemisomes are stably held in place by Cbf1 and CBF3, whereas unstable CenH3 octamers are rapidly evicted from chromosome arms and targeted for degradation.
We asked where the larger Cse4 particles in the overproduction strain were assembled by mapping them genome-wide. We found that Cse4-Myc and FLAG-H2A ChIP peaks precisely coincided, which implies that overproduced Cse4-Myc nucleosomes are incorporated in place of H3 nucleosomes genome-wide (Fig. 7C). However, the profiles were quantitatively different. For example, by taking the ratio of Cse4-Myc to FLAG-H2A over the SNT1-FEN1 region, which has been previously characterized with respect to its rate of turnover (31), we found that peaks of Cse4 incorporation corresponded closely to sites of rapidly turning-over (“hot”) nucleosomes (Fig. 7C) and around 5′ ORFs genome-wide (Fig. S8B). We also compared Cse4 and H2A abundances to nucleosome turnover rates globally, and found that Cse4 was in general enriched at sites of high turnover, whereas H2A was depleted from these same sites (Pearson's r = 0.52) (Fig. 7D). Similar findings of Cse4 misincorporation at sites of hot nucleosomes have been reported for strains that overproduce Cse4 as a result of mutations in components of the nucleosome assembly apparatus (32). Despite the fact that Cse4-Myc is the only Cse4 copy present in this overproducing strain, the misincorporated particles have evidently not formed functional centromeres, as the strain grew normally.
Discussion
We have used native ChIP and paired-end sequencing to map Cse4 nucleosomes to centromeres at high resolution. We found that Cse4 nucleosomes are confined to the central ∼80-bp CDEII region of functional centromeres, tightly flanked by distinct small particles over CDEI and CDEIII. This finding confirms that DNA wraps only once around a Cse4-containing core (33), as implied by our previous study showing that Cse4 nucleosomes induce positive supercoiling at functional centromeres in vivo (6). Our findings are consistent with the observation that singly wrapped CenH3 particles also occupy Drosophila and human centromeres (8, 9), suggesting that this organization is a universal feature of centromeric nucleosomes. Singly wrapped tetrameric nucleosomes are universal for archaea (34), as if centromeres have retained the ancestral nucleosomal organization.
Our native ChIP-seq analysis also showed that H2A is present at all 16 yeast centromeres at the same level as over the rest of the genome. We confirmed this result by analyzing X-ChIP data from a published study (35). Because that study was performed on formaldehyde cross-linked chromatin by the same laboratory that previously reported deficiency of H2A over centromeres (28), we might attribute the different outcomes to the use of sonication in the first study to fragment and solubilize DNA, versus the use of MNase in the second study (35). It is possible that sonication caused loss of poorly cross-linked nucleosomes, and differences in the degree of cross-linking of different histones (8) might have resulted in discrepant ChIP efficiencies between them. The excellent concordance between our study using native chromatin and that of Luk et al. (35) using cross-linked chromatin confirms that H2A is as abundant over centromeres as it is over the entire genome, as expected for a centromeric particle containing all four histones. Our findings are consistent with a report that the Mif2 kinetochore-specific protein coimmunopurifies with Cse4, H4, H2A, and H2B, but not detectably to H3, when purified without cross-linking or enzymatic DNA fragmentation (36).
The low recovery of Cse4 nucleosomes isolated using standard MNase digestion protocols that is evident from our work and that of others (19) (Fig. S9) confirms previous findings of centromere hypersensitivity to MNase digestion in Schizosaccharomyces pombe (37) and Drosophila (8). This low recovery also raises questions about reports of what appear to be conventional CenH3 octameric nucleosomes isolated from diverse eukaryotes (38, 39), because these particles were extracted under conditions that led to depletion of yeast centromeric chromatin, which we might attribute to catastrophic loss by cleavage within the singly wrapped CenH3 particle. Our finding that nonfunctional octamer-like Cse4-containing particles are present at noncentromeric sites of high turnover provides a possible alternative explanation for the immunoprecipitation of octameric nucleosomes in some studies (38, 39), but tetramers in others (8, 9). The ∼90% AT-richness of CDEII might be an adaptation to prevent formation of these aberrant particles at functional centromeres, which would explain why octameric Cse4 nucleosomes fail to assemble on CDEs in vitro (12, 14, 40).
Misincorporation of Cse4 at sites of hot nucleosomes have been reported for strains that overproduce Cse4 as a result of mutations in components of the nucleosome assembly apparatus (32). Cse4 nucleosomes are also enriched at promoters of highly transcribed genes in strains that do not overproduce Cse4 (14, 18) (Fig. S8C), suggesting that high turnover is a normal mechanism for evicting misincorporated CenH3 from chromosome arms (11) (Fig. 7E). At centromeres, multiple factors would favor incorporation of a single stable tetramer, including exclusion of H3 octamers by Cbf1 (15, 21), the 90% AT-richness of CDEII, which resists assembly of Cse4 octamers (14, 40), and the recruitment of Cse4 by the adjacent CBF3 complex (41). In multicellular eukaryotes, heterochromatin condensation would help to stabilize CenH3 tetramers by preventing nucleosome turnover (42). Our detection of two distinct forms of Cse4 particles, one at centromeres and the other on chromosome arms, thus reconciles seemingly conflicting reports of left-handed CenH3 octamers produced in vitro (10–14) and of right-handed wrapping (6) and heterotypic CenH3 tetramers (8, 9) observed in vivo.
Materials and Methods
Strains used in this study are listed in Table S1. Preparation of yeast nuclei, MNase digestion, and DNA extraction steps were performed as described (15). ChIP was performed as previously described (17).
Paired-end libraries were prepared using either our modified protocol (15) or the standard Illumina protocol, followed by at least 20 rounds of paired-end sequencing in an Illumina HiSeq 2000 by the Fred Hutchinson Cancer Research Center Genomics Shared Resource. Data were processed and mapped to Version 61 of the yeast genome using Novoalign (www.novocraft.com), as previously described (15). SRA SRR058444 and SRR058445 data were mapped similarly with Bowtie (http://bowtie-bio.sourceforge.net/) using default parameters. Solexa data analysis was performed as previously described (15), except that the fraction of mapped reads spanning each base pair was multiplied by the total number of base pairs in the reference sample to give a normalized count for that base pair. To construct V-plots, a table of fragment midpoint and length pairs was displayed using the scatterplot function of Kaleidograph (version 4.1; Synergy Software).
Supplementary Material
Acknowledgments
We thank Max Press for help with yeast chromatin preparation protocols, Sue Biggins for strains and helpful discussions, Take’ Furuyama and Paul Talbert for comments on the manuscript, Andy Marty and Jeff Delrow for paired-end Illumina sequencing advice and discussions of library preparation protocols, and J. J. Miranda for advice.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE29298).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118898109/-/DCSupplemental.
References
- 1.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 3.Rudd MK, Willard HF. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Warburton PE. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004;12:617–626. doi: 10.1023/B:CHRO.0000036585.44138.4b. [DOI] [PubMed] [Google Scholar]
- 5.De Wulf P, Earnshaw WC. The Kinetochore. Berlin: Springer; 2009. [Google Scholar]
- 6.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang CC, Chang KM, Cui H, Jayaram M. Histone H3-variant Cse4-induced positive DNA supercoiling in the yeast plasmid has implications for a plasmid origin of a chromosome centromere. Proc Natl Acad Sci USA. 2011;108:13671–13676. doi: 10.1073/pnas.1101944108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci USA. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tachiwana H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 11.Conde e Silva N, et al. CENP-A-containing nucleosomes: Easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 12.Dechassa ML, et al. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kingston IJ, Yung JS, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2011;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henikoff JG, Belsky J, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci USA. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Densmore L, Payne WE, Fitzgerald-Hayes M. In vivo genomic footprint of a yeast centromere. Mol Cell Biol. 1991;11:154–165. doi: 10.1128/mcb.11.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefrançois P, et al. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole HA, Howard BH, Clark DJ. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc Natl Acad Sci USA. 2011;108:12687–12692. doi: 10.1073/pnas.1104978108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemmerich P, et al. Interaction of yeast kinetochore proteins with centromere protein/transcription factor Cbf1. Proc Natl Acad Sci USA. 2000;97:12583–12588. doi: 10.1073/pnas.97.23.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent NA, Adams S, Moorhouse A, Paszkiewicz K. Chromatin particle spectrum analysis: A method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res. 2011;39:e26. doi: 10.1093/nar/gkq1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 23.Fitzgerald-Hayes M. Yeast centromeres. Yeast. 1987;3:187–200. doi: 10.1002/yea.320030306. [DOI] [PubMed] [Google Scholar]
- 24.Niedenthal RK, Sen-Gupta M, Wilmen A, Hegemann JH. Cpf1 protein induced bending of yeast centromere DNA element I. Nucleic Acids Res. 1993;21:4726–4733. doi: 10.1093/nar/21.20.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pietrasanta LI, et al. Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc Natl Acad Sci USA. 1999;96:3757–3762. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacIsaac KD, et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloom KS, et al. Chromatin conformation of yeast centromeres. J Cell Biol. 1984;99:1559–1568. doi: 10.1083/jcb.99.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 29.Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Floer M, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 32.Lopes da Rosa J, Holik J, Green EM, Rando OJ, Kaufman PD. Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics. 2011;187:9–19. doi: 10.1534/genetics.110.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 34.Sandman K, Reeve JN. Archaeal histones and the origin of the histone fold. Curr Opin Microbiol. 2006;9:520–525. doi: 10.1016/j.mib.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Luk E, et al. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell. 2010;143:725–736. doi: 10.1016/j.cell.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westermann S, et al. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi K, et al. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erhardt S, et al. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gent JI, et al. Distinct influences of tandem repeats and retrotransposons on CENH3 nucleosome positioning. Epigenetics Chromatin. 2011;4:3. doi: 10.1186/1756-8935-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao H, et al. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalal Y, Bui M. Down the rabbit hole of centromere assembly and dynamics. Curr Opin Cell Biol. 2010;22:392–402. doi: 10.1016/j.ceb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.