Abstract
Captive breeding programs are widely used for the conservation and restoration of threatened and endangered species. Nevertheless, captive-born individuals frequently have reduced fitness when reintroduced into the wild. The mechanism for these fitness declines has remained elusive, but hypotheses include environmental effects of captive rearing, inbreeding among close relatives, relaxed natural selection, and unintentional domestication selection (adaptation to captivity). We used a multigenerational pedigree analysis to demonstrate that domestication selection can explain the precipitous decline in fitness observed in hatchery steelhead released into the Hood River in Oregon. After returning from the ocean, wild-born and first-generation hatchery fish were used as broodstock in the hatchery, and their offspring were released into the wild as smolts. First-generation hatchery fish had nearly double the lifetime reproductive success (measured as the number of returning adult offspring) when spawned in captivity compared with wild fish spawned under identical conditions, which is a clear demonstration of adaptation to captivity. We also documented a tradeoff among the wild-born broodstock: Those with the greatest fitness in a captive environment produced offspring that performed the worst in the wild. Specifically, captive-born individuals with five (the median) or more returning siblings (i.e., offspring of successful broodstock) averaged 0.62 returning offspring in the wild, whereas captive-born individuals with less than five siblings averaged 2.05 returning offspring in the wild. These results demonstrate that a single generation in captivity can result in a substantial response to selection on traits that are beneficial in captivity but severely maladaptive in the wild.
Keywords: fisheries, genetics, parentage, rapid evolution, salmon
Captive breeding programs are commonly used for the conservation of endangered species and, more recently, for the restoration of declining populations (1–4). Mounting evidence suggests that captive-born individuals released into the wild can have substantially lower fitness than their wild-born counterparts and that these fitness declines can occur after only a few generations in captivity (5–8). Identifying the mechanisms that cause reduced fitness in the wild is vital for deciding if, when, and how captive breeding programs should be applied for conservation and management purposes (5, 7). Explanations for the rapid fitness declines (8–12) include environmental effects of captive rearing (including heritable epigenetic effects), inbreeding among close relatives, relaxed natural selection, and unintentional domestication selection (adaptation to the novel environment). Each of these mechanisms creates subtle but testable differences in patterns of reproductive success.
Environmental effects of captive rearing, for example, could produce differences in fitness between captive-born and wild-born individuals but would not create differences in fitness among individuals that experienced identical captive environments (12, 13). Relaxed natural selection in captivity is a compelling hypothesis because it can manifest in a myriad of forms. Lack of mate choice, for example, could result in combinations of immune-related genes that do not maximize fitness (14, 15). Nevertheless, theoretical analyses suggest that for relaxed natural selection to cause a rapid fitness decline, the population must have a high standing mutational load or spend many generations in captivity (9). Unintentional domestication selection, on the other hand, can rapidly reduce fitness in the wild, especially if multiple traits are under selection (10, 16).
If unintentional domestication selection is occurring, we expect to observe two unique patterns. First, captive-born individuals should perform better in captivity than wild-born individuals. Second, there should be a tradeoff among the wild-born broodstock: Those with the greatest fitness in a captive environment will produce offspring that perform the worst in the wild. These predictions are not expected under relaxed natural selection because individuals with fit and unfit genotypes (when expressed in the wild) would perform identically in a captive environment where that genetic variation is selectively neutral. We test these competing explanations with a detailed pedigree analysis of a wild steelhead (Oncorhynchus mykiss) population that was supplemented with captive-reared individuals.
Billions of captive-reared salmon are intentionally released into the wild each year in efforts to increase fishery yields, mitigate environmental disturbances, and bolster severely declining populations (17–19). Steelhead from the Hood River in Oregon are listed as threatened under the US Endangered Species Act (20), and part of their recovery plan includes supplementation with juvenile fish produced in a captive breeding program (i.e., fish hatchery). For winter-run steelhead from this population, we constructed three-generation pedigrees from 15 run-years by genotyping 12,700 fish at eight highly polymorphic microsatellite loci. Steelhead en route to their spawning grounds in the Hood River were first passed over the Powerdale Dam, which was a complete barrier to migrating fish (SI Materials and Methods). Because of this barrier, we were able to obtain samples of every returning fish that spawned in the wild. Previous work from this system documented that captive-born fish with two wild parents averaged 85% of the reproductive success of their wild-born counterparts (6). However, the mechanism responsible for the documented fitness decline remained unknown.
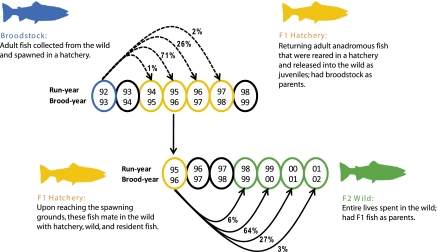
In this captive breeding program, ocean-returning wild-born and first-generation hatchery adults were collected from the wild and spawned in a hatchery (hereafter, “broodstock”; Fig. 1). Their offspring (hereafter, “F1” fish) were reared in a hatchery environment and released near wild-spawning habitat as juveniles. After release, the F1 fish went out to sea, returned as adults, and spawned in the wild. The progeny of F1 fish (hereafter, “F2” fish) spent their entire lives in the wild. Using parentage analyses, we assigned F1 hatchery fish back to their broodstock parents. We again used parentage analysis to assign the returning, wild-born F2 fish back to their F1 parents. We calculated the individual reproductive success of each wild-born broodstock fish in the hatchery as the total number of its F1 progeny that returned as reproductively mature adults. We also calculated the per capita F1 reproductive success in the wild as the average number of returning F2 progeny for all F1 fish assigned to a given broodstock. We performed separate analyses for each F1 run-year and brood-year in addition to individual analyses for broodstock females, broodstock males, and broodstock pairs.
Fig. 1.
Illustration of steelhead run-timing and study design. Broodstock fish were first collected from the wild and spawned in captivity. The F1 offspring were reared in a hatchery until becoming juveniles; at that time, they were released into the wild near spawning grounds. F1 juveniles went out to sea and subsequently returned to spawn as adults in the wild. All returning adult fish were sampled at the Powerdale Dam en route to their spawning grounds. Broodstock reproductive success was measured as the number of returning adult F1 offspring assigned to broodstock with parentage analysis. The reproductive success of F1 fish was measured by assigning F2 fish back to their F1 parents. F2 fish spent their entire lives in the wild. Each F1 hatchery run-year consists of fish from multiple brood-years. In run-years 1995–1999, a small number of returning F1 hatchery fish were used as broodstock. Notice that hatchery fish return, on average, 1 y earlier than wild-born fish owing to accelerated growth in the freshwater phase of the life cycle (1 y in the hatchery vs. an average of 2 y in the wild).
Results
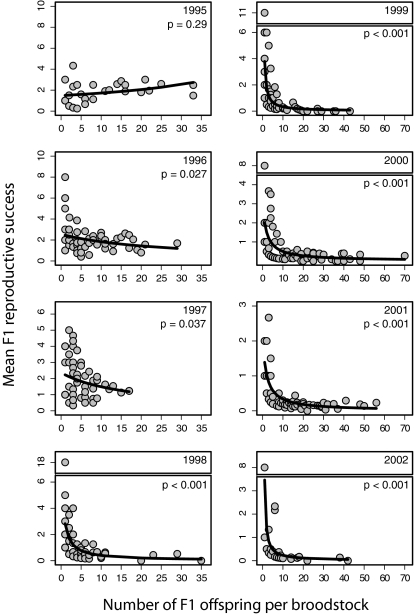
In 7 of the 8 F1 run-years we examined, we found a substantial tradeoff between performance in the hatchery and performance in the wild (Fig. 2 and Table S1). For broodstock families that produced five or more returning offspring (the median), their F1 offspring averaged 71% lower per capita reproductive success in the wild than did F1 progeny from broodstock families having fewer than five offspring. Thus, fish with trait values associated with success in the captive environment produced large numbers of hatchery-reared F1 offspring that survived to become reproductively mature adults. However, the F1 fish from those large families had low per capita reproductive success in the wild. We observed the same tradeoff in F1 reproductive success regardless of whether we considered male and female broodstock separately or considered broodstock pairs (Fig. S1 and Table S2). The first run-year of returning F1 hatchery fish, 1995, did not show the tradeoff observed in subsequent years. However, in the year that these F1 fish were released into the wild, only a handful of broodstock fish were used (62% fewer than in other years) and substantially fewer smolts were released (4,600 vs. a mean of 52,700 in other years), which may have reduced the selection pressure and created less opportunity for selection (Tables S1 and S3).
Fig. 2.
Reproductive success of captive-spawned broodstock plotted against the per capita reproductive success of their F1 offspring (gray circles). A tradeoff between the reproductive success of fish in captivity (broodstock) and the subsequent reproductive success of their offspring in the wild is shown in 7 of 8 F1 run-years. P values are shown for the slope of a GLM (fitted regression line drawn in black). Notice that the axes are scaled to each plot and that large y-axis values are scaled independently (boxes) for run-years 1998, 1999, 2000, and 2002.
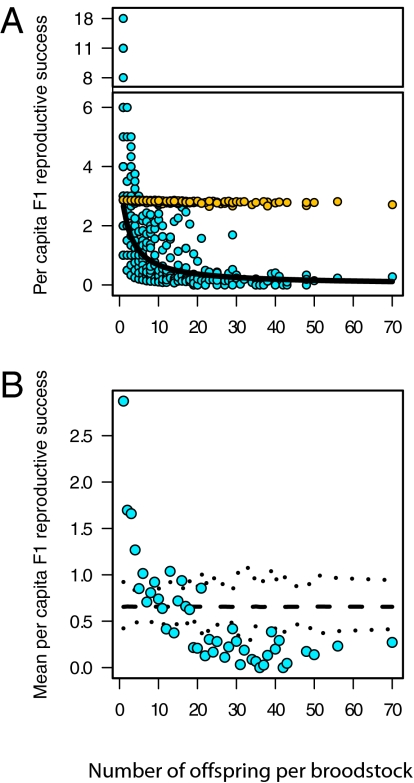
The documented tradeoff is consistent with a rapid response to domestication selection. One alternate explanation is that F1 fish from large families were more likely to mate with relatives, and subsequently exhibit lower per capita fitness, owing to inbreeding depression. Using our known pedigree and simulated random pairings of returning hatchery fish with a high species-specific genetic load (11 lethal equivalents) (21), we calculated the relative fitness reduction for F1 fish as a function of family size (i.e., number of siblings). Our calculations demonstrate that inbreeding among related hatchery fish cannot explain the rapid decline in per capita reproductive success of F1 fish associated with increased family size (Fig. 3A). We also fit a generalized linear model (GLM) to establish that the slope of our explanatory variable was significantly different from zero (P < 0.001; Fig. 3A and Table S1) and used a randomization test to demonstrate that the documented mean per capita F1 reproductive success cannot be explained by the increased variance associated with smaller sample sizes for broodstock with fewer offspring (P < 0.001; Fig. 3B and Table S1). Both the GLM and the randomization procedure illustrate that relaxed natural selection cannot be the predominate cause of fitness differences between hatchery and wild fish because we would expect a flat slope (compare with Fig. 3A) and most points to lie within the confidence intervals (compare with Fig. 3B). These tests remained highly significant (P < 0.001) when the aberrant F1 run-year was included (i.e., 1995; Table S2).
Fig. 3.
Reproductive success of captive-spawned broodstock plotted against the per capita reproductive success of their F1 offspring. Results are pooled for the 7 F1 run-years for which a substantial tradeoff was documented (compare with Fig. 2). (A) Blue circles show the per capita reproductive success for each F1 family. The solid black line represents the fitted GLM. Orange points represent the expected reduction in per capita F1 reproductive success attributable to inbreeding as a function of F1 family size and cannot explain the documented pattern. (B) Blue circles illustrate the mean per capita F1 reproductive success (i.e., for broodstock families of a given size). The dashed lines represent the mean and 95% confidence intervals for expected values calculated with random sampling of observed F1 reproductive success. Notice that the observed pattern is consistent with unintentional domestication selection but is not consistent with relaxed natural selection, where we would expect a flat slope and points to lay largely within the confidence intervals.
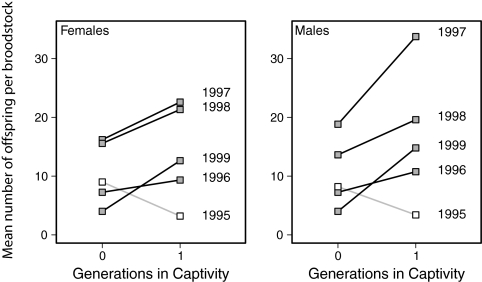
Because we also expect unintentional domestication selection to cause rapid adaptation to captivity, we next examined whether first-generation hatchery fish (i.e., F1 fish) had higher reproductive success than wild fish when used as broodstock in captivity. For 5 run-years, a portion of broodstock matings consisted of pairings between wild fish and F1 hatchery fish (6). The remaining broodstock matings were restricted to crosses between two wild fish. We again measured reproductive success as the total number of returning adult offspring. In 4 of 5 run-years, the F1 hatchery broodstock had nearly twice the reproductive success of wild broodstock (Fig. 4), which is a pattern consistent with adaptation to captivity. As before, the first-generation hatchery broodstock collected from the first F1 return year (1995), for which few hatchery smolts were released (Table S3), showed no evidence for adaptation to captivity (compare with Fig. 2).
Fig. 4.
Comparison of reproductive success between broodstock of wild ancestry and broodstock that were first-generation hatchery fish. Reproductive success was measured as the number of returning offspring. All F1 hatchery broodstock were mated with a wild fish. Wild broodstock reproductive success was only considered for matings between two wild fish. For 4 of 5 y examined (gray boxes), the number of broodstock offspring was greater for F1 hatchery broodstock than for wild fish (females: P < 0.012; males: P < 0.020). This pattern demonstrates rapid adaptation to the captive environment. No difference was detected in 1995 (open boxes), which corresponded with the return year for which far fewer hatchery smolts had been released (compare with Fig. 2).
Discussion
Under a model of domestication selection, certain traits are unintentionally selected on in the hatchery and this selection causes a high variance in reproductive success among the broodstock (Fig. S2). However, the trait values associated with success in the captive environment are detrimental in the wild, resulting in low reproductive success in the wild by fish from families that were successful in captivity. Because substantial selection coefficients on a single trait are required to explain this pattern, we speculate that multiple, and possibly correlated, traits are unintentionally selected on (10). Previous work has suggested that growth rate may be under strong selection in hatcheries because steelhead are released as yearling smolts, whereas fish in the wild generally take 2 y or longer to smolt (22). Other possible contenders for selected traits include egg size, fecundity, physiological processes associated with smoltification, and individual behaviors (e.g., predator avoidance) (10, 22–24). For the F1 run-year in which selection was not detected (1995), the smolts were reared in low-density conditions and the release size of the smolts was considerably larger than in other years (Table S3), which points to crowding as a possible selection pressure (25).
Although we have demonstrated rapid adaptation to captivity in a steelhead population, other taxa may not respond in an equivalent fashion. Factors that exacerbate adaptation to captivity include strong selection pressures, large effective population sizes, high genetic diversity, and multiple generations in captivity (7, 9). Thus, the expected amount of genetic adaptation to captivity depends on both the intrinsic genetic composition of the designated broodstock and the design and implementation of the captive breeding program. Steelhead are genetically diverse, highly fecund (thousands of eggs per female), and often spawned using dozens of families (26). Other highly fecund organisms, such as plants, invertebrates, amphibians, and other fishes, may also respond quickly to domestication selection. Captive populations of less fecund animals, such as some mammals and birds, may not adapt as quickly to captivity provided that they are not kept in captivity for many generations. One unusual feature of captive breeding via supplementation hatcheries is that only the early life history stages are kept in captivity. After release into the wild, there is very high mortality (often >95%), and thus ample opportunity for directional and purifying natural selection. Whether the oceanic phase somehow enhances the domestication effect is not clear. It is certainly possible that phenotypic variation generated during the captive, juvenile phase (e.g., body size at release) could be under intense viability selection at sea (22).
This study illustrates that domestication selection (i.e., adaptation to a novel environment) can cause a rapid fitness decline of hatchery fish in the wild. Although more complex mechanisms may also be involved (e.g., relaxed purifying selection, heritable epigenetic effects), the documented patterns of reproductive success indicate that domestication selection is a primary explanation. We conclude that (i) the wild population contained the requisite genetic variation for rapid adaptation to captivity and (ii) less than one generation in captivity (i.e., fertilization through smolting) generated selection intensities necessary to produce a rapid, genetically based fitness reduction in the wild. Understanding that unintentional selection in captivity can cause rapid fitness declines has important conservation and management implications: Determining which traits are under selection and whether captive breeding programs can be modified to mitigate those selection pressures will be the next big challenge for improving the science of captive breeding.
Materials and Methods
Parentage Analysis.
Extensive details on this study system, management practices, laboratory methods, steelhead life history, and reproductive success can be found elsewhere (27–30) (SI Materials and Methods). To calculate the reproductive success of broodstock parents, we first used parentage analysis to assign F1 hatchery fish back to their broodstock parents. Detailed records on broodstock pairings in the hatchery allowed for assignment to both parents. We used genotypes of the known broodstock pairs sorted by the year in which they were spawned as the putative parents. Genotypes of the F1 hatchery fish, sorted by brood-year, were used as the putative offspring. Because there can be some error associated with the aging of scales, we also used hatchery fish ± 1 brood-year as putative offspring. Our analyses revealed that less than 3% (139 of 4,653) of hatchery fish had been assigned via scale aging to the incorrect brood-year, which is not always the case with wild-born fish (31). We used Mendelian exclusion to assign hatchery fish to their broodstock parents (i.e., each allele in an identified offspring matched to at least 1 allele in both parents). All parentage analyses were completed with freely available R scripts (32, 33). To account for genotyping errors, we allowed an offspring to mismatch at one allele in both parents (6, 28, 32), although 81% of assignments contained no mismatches. No F1 hatchery offspring matched to more than one broodstock pair because we had an average of 36 alleles per locus and because we knew the hatchery pairings, which substantially reduced the required number of pair-wise comparisons. We also performed a more conservative analysis in which we did not allow for any mismatching loci (see below). Broodstock that were not assigned as parents to any F1 fish were removed from further analyses. In the Hood River, very few hatchery fish residualize (i.e., remain in freshwater as residents) (28, 30), such that the estimates of broodstock reproductive success should not be influenced by this alternate life history strategy (cf. 34).
To determine the reproductive success of the F1 hatchery fish, we next used the assigned F1 hatchery fish, sorted by run-year, as putative parents. The 15 run-years of fish in our dataset allowed for 8 run-years of F1 fish to be considered as putative parents. The small number of F1 hatchery fish that were taken for use as broodstock were removed as candidate parents for F2 fish. Wild fish, sorted by brood-year, were used as putative offspring. As above, we used wild fish ± 1 brood-year as putative offspring. We used Mendelian exclusion to assign the wild fish to hatchery parents and allowed for one mismatch between parents and offspring. Of the F2 fish that were assigned to a F1 hatchery parent, the majority were assigned only to one parent because their other parent was either a wild or resident fish (27, 28). There was no relationship between F1 family size and assignment to either one or both parents. A small number of F2 fish (n = 162) were assigned to two putative parents of the same sex. To resolve these assignments, we used maximum likelihood approaches as implemented in CERVUS (35, 36) to determine the most likely parents. CERVUS assignments based on the simulation-based P values and highest log-likelihood scores were identical. As above, we also performed a more conservative assignment in which we only assigned individuals that matched at all eight loci.
F1 per Capita Reproductive Success.
We first measured the reproductive success of each broodstock fish by counting the number of assignments of F1 hatchery fish to those broodstock fish. We next calculated the per capita reproductive success for all F1 fish assigned to a given broodstock. For example, if a broodstock fish had 10 F1 offspring, of which only 1 produced a single adult F2 fish, the per capita F1 reproductive success equaled 0.1. We next plotted the reproductive success of broodstocks vs. the per capita reproductive success of their F1 offspring. We fit a GLM using a “gamma” family with a model link equal to “inverse.” We chose a gamma distribution because our response variable was not normally distributed and could not be transformed to fit normality. The mean and variance of the response variable were well-approximated with a gamma distribution, and there was no overdispersion (i.e., the mean and variance were approximately equal) (37). All analyses were implemented in the R statistical software environment (33).
We performed separate analyses for all individual F1 run-years (Fig. 2 and Table S1) and for male and female broodstock (Fig. S1). To explore alternate explanations, we additionally analyzed the data in five different ways. First, we performed analyses for each F1 run-year using only fish that were assigned at all eight loci. Second, we performed the analyses for each broodstock pair rather than for each individual broodstock fish. Third, because it was plausible that age of returning F1 fish may correlate with reproductive success, we performed a separate analysis using only age 3 fish, which was the most common age class. Lastly, we performed analyses by F1 brood-year as opposed to run-year. Repeat spawners were unlikely to influence these patterns, because there is no a priori reason to believe that fish from large families are more likely to be iteroparous. We also examined the distribution of F1 return times and found that that was no relationship between F1 family size and return date (SI Materials and Methods, Fig. S3, and Table S4). No matter how the data were analyzed, we see evidence for a strong tradeoff; larger F1 families have lower per capita reproductive success in the wild (Table S2).
We also explored the possibility that hatchery managers were inadvertently selecting on phenotypic traits of the broodstock. However, performance in the hatchery could not be explained by the phenotypic traits of broodstock. The high variance in reproductive success of broodstock fish (Fig. S2) could not be explained by broodstock length, weight, run-timing, salt water age, age, temperature regime of the eggs, or the number of eggs that successfully hatched (Table S5). Each explanatory variable was considered simultaneously within a multiple linear regression, and the response variable was log-transformed for normality when required (although there were no qualitative differences in the results either way). These results suggest that selection is occurring on F1 fish in the hatchery and not on the broodstock fish. Lastly, to explore the possibility that broodstock fish in more recent years had a greater proportion of hatchery genes, we performed parentage analysis of relevant broodstock fish and found that this was unlikely to be occurring (SI Materials and Methods).
Inbreeding.
One competing explanation for our results is that large F1 families were more likely to interbreed with each other. Because there are substantial costs associated with inbreeding, this could result in low F1 per capita reproductive success for large F1 families. To examine this possibility, we performed a series of simulated matings to determine the inbreeding-associated reduction in fitness of related hatchery fish. We considered a scenario in which F1 hatchery fish bred randomly with other F1 hatchery fish. This is a conservative scenario because we know from our pedigree records that hatchery and wild fish commonly interbreed (27, 28). Using our pedigree and database records, we split returning F1 hatchery fish into males and females by run-year. For each run-year, we randomly sampled one male and one female hatchery fish with replacement. Using our pedigree, we matched the sampled hatchery fish to their known broodstock parents and calculated the number of unique parents. If the hatchery fish we sampled had four unique parents, they were unrelated. If the hatchery fish had three unique parents, they were half-sibs, and if the hatchery fish had two unique parents, they were full-sibs. We also matched each sample with the F1 family size associated with each broodstock. We repeated the above procedure 10,000 times per family and tabulated the number of observed full-sib, half-sib, and unrelated matings in every broodstock family.
To calculate the relative reproductive success (RRS) of hatchery fish compared with a randomly mating population, we used the following equation:
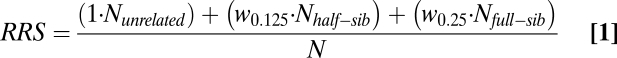 |
where N equals the total number of draws (here, 10,000) and N unrelated, half-sib, and full-sib equal the number of observed matings between unrelated individuals, half-sibs, and full-sibs, respectively. The fitness costs associated with various degrees of inbreeding were calculated from published estimates of genetic load in steelhead (∼11 lethal equivalents) (21) as w0.125 = 0.51 and w0.25 = 0.26, where w0.125 equals the relative fitness of the offspring of half-sibs and w0.25 equals the relative fitness of offspring of full-sibs. We next multiplied each F1 family's inbreeding RRS by the average per capita reproductive success of broodstock that had one returning offspring (Fig. 3A).
Sampling Variance.
Each estimate of the per capita F1 reproductive success consists of an average of reproductive success for F1 hatchery fish from the same family. For large F1 families, the estimate of the per capita F1 reproductive success has little variance. However, estimates of per capita F1 reproductive success based on smaller F1 families (e.g., 1 offspring per broodstock) have a larger associated variance. To establish that our observations were not simply attributable to increased sampling variance with smaller families, we preformed a randomization procedure. First, for each F1 run-year, we created a sample space of all observed values for F1 reproductive success (including 0's). We next randomly sampled our observed F1 reproductive success values by the total number of F1 fish used to calculate the observed mean and used the randomly sampled values to calculate an expected mean. For each F1 family size, we repeated this procedure 10,000 times and calculated the expected mean and 95% confidence intervals. To test whether our observed pattern deviated from the distribution expected under random sampling, we performed a likelihood-ratio test (G-test) (38) on the number of observed points that lay outside of the 95% confidence intervals vs. the expected number (Fig. 3B and Tables S1 and S2). The samples of F1 reproductive success were appropriately modified for the alternate approaches presented in Table S2.
Adaptation to Captivity.
To test whether hatchery fish were adapting to the captive environment, we compared the lifetime reproductive success of F1 hatchery broodstock and wild broodstock. In 5 run-years, 1995–1999, a portion of F1 hatchery fish (n = 90) were successfully used as hatchery broodstock. A roughly equal number of male and female F1 hatchery fish were used in each run-year, and each F1 fish was mated with a wild partner. We examined each sex separately. Because the fitness estimates of wild fish would be biased if they had mated with an F1 hatchery fish, we examined the fitness of wild fish that were mated only with other wild fish. We calculated the mean reproductive success for both broodstock types for each of the 5 y (sample sizes for calculating the mean were approximately equal in both groups). We next compared the mean reproductive success of wild and F1 hatchery broodstock using a paired t test, where pairings are within run-years. For male and female fish, P values for t tests equaled 0.1296 and 0.07, respectively, if run-year 1995 was included. However, we excluded run-year 1995 because the returning F1 hatchery fish came from smolt releases that were treated very differently than in the other 4 y (Table S3). In the year that these F1 fish were released into the wild, only a handful of broodstock fish were used (62% fewer than in other years) and substantially fewer smolts were released (4,600 vs. a mean of 52,700), which likely created less opportunity for selection (Tables S1 and S3). After removing run-year 1995, we find that both female (P < 0.0122) and male (P < 0. 0203) F1 hatchery broodstock had greater reproductive success than their wild counterparts. Above, we report one-sided P values because we did not expect F1 hatchery fish to perform worse than their wild counterparts, but results from a two-sided test for female (P < 0.0244) and male (P < 0.0406) fish were still significant at α = 0.05.
Supplementary Material
Acknowledgments
We thank W. Ardren, B. Cooper, V. Amarasinghe, J. Stephenson, and the Oregon State Center for Genome Research and Biotechnology for laboratory protocols and genotyping. We acknowledge all Oregon Department of Fish and Wildlife staff who collected data and acquired tissue samples for this dataset. We also thank H. Araki, S. J. Arnold, J. Hard, R. Waples, C. Searle, J. Tennessen, I. Phillipsen, and L. Curtis for insightful discussions. This research was funded by a grant from the Bonneville Power Administration (to M.S.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. F.W.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111073109/-/DCSupplemental.
References
- 1.Hedrick P. Genetics of Populations. Sudbury, UK: Jones and Bartlett; 2005. [Google Scholar]
- 2.Allendorf FW, Luikart G. Conservation and the Genetics of Populations. Blackwell, Oxford; 2007. pp. 452–481. [Google Scholar]
- 3.Seddon PJ, Armstrong DP, Maloney RF. Developing the science of reintroduction biology. Conserv Biol. 2007;21:303–312. doi: 10.1111/j.1523-1739.2006.00627.x. [DOI] [PubMed] [Google Scholar]
- 4.Fraser DJ. How well can captive breeding programs conserve biodiversity? A review of salmonids. Evol Appl. 2008;1:535–586. doi: 10.1111/j.1752-4571.2008.00036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snyder NFR, et al. Limitations of captive breeding in endangered species recovery. Conserv Biol. 1996;10:338–348. [Google Scholar]
- 6.Araki H, Cooper B, Blouin MS. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science. 2007;318:100–103. doi: 10.1126/science.1145621. [DOI] [PubMed] [Google Scholar]
- 7.Frankham R. Genetic adaptation to captivity in species conservation programs. Mol Ecol. 2008;17:325–333. doi: 10.1111/j.1365-294X.2007.03399.x. [DOI] [PubMed] [Google Scholar]
- 8.Williamson KS, Murdoch AR, Pearsons TN, Ward EJ, Ford MJ. Factors influencing the relative fitness of hatchery and wild spring Chinook salmon (Oncorhynchus tshawytscha) in the Wenatchee River, Washington, USA. Can J Fish Aquat Sci. 2010;67:1840–1851. [Google Scholar]
- 9.Lynch M, O'Hely M. Captive breeding and the genetic fitness of natural populations. Conserv Genet. 2001;2:363–378. [Google Scholar]
- 10.Araki H, Berejikian BA, Ford MJ, Blouin MS. Fitness of hatchery-reared salmonids in the wild. Evol Appl. 2008;1:342–355. doi: 10.1111/j.1752-4571.2008.00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlesworth D, Willis JH. The genetics of inbreeding depression. Nat Rev Genet. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- 12.Hansen MM, Mensberg KLD. Admixture analysis of stocked brown trout populations using mapped microsatellite DNA markers: Indigenous trout persist in introgressed populations. Biol Lett. 2009;5:656–659. doi: 10.1098/rsbl.2009.0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelletier F, Réale D, Watters J, Boakes EH, Garant D. Value of captive populations for quantitative genetics research. Trends Ecol Evol. 2009;24:263–270. doi: 10.1016/j.tree.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Neff BD, Garner SR, Heath JW, Heath DD. The MHC and non-random mating in a captive population of Chinook salmon. Heredity. 2008;101:175–185. doi: 10.1038/hdy.2008.43. [DOI] [PubMed] [Google Scholar]
- 15.Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: What have we learned about natural selection in 15 years? J Evol Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- 16.Ford MJ. Selection in captivity during supportive breeding may reduce fitness in the wild. Conserv Biol. 2002;16:815–825. [Google Scholar]
- 17.Kostow K. Factors that contribute to the ecological risks of salmon and steelhead hatchery programs and some mitigating strategies. Rev Fish Biol Fish. 2009;19:9–31. [Google Scholar]
- 18.Naish KA, et al. An evaluation of the effects of conservation and fishery enhancement hatcheries on wild populations of salmon. Adv Mar Biol. 2007;53:61–194. doi: 10.1016/S0065-2881(07)53002-6. [DOI] [PubMed] [Google Scholar]
- 19.Laikre L, Schwartz MK, Waples RS, Ryman N. GeM Working Group Compromising genetic diversity in the wild: unmonitored large-scale release of plants and animals. Trends Ecol Evol. 2010;25:520–529. doi: 10.1016/j.tree.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Good TP, Waples RS, Adams P. Updated status of federally listed ESUs of West Coast salmon and steelhead. 2005. US Department of Commerce NOAA Tech. Memo. NMFS-NWFSC-66, 598 p. Available at http://www.nwr.noaa.gov/Publications/Biological-Status-Reviews/Salmon.cfm. Accessed November 28, 2011.
- 21.Thrower FP, Hard JJ. Effects of a single event of close inbreeding on growth and survival in steelhead. Conserv Genet. 2008;10:1299–1307. [Google Scholar]
- 22.Reisenbichler R, Rubin S, Wetzel L, Phelps S. In: Stock Enhancement and Sea Ranching. Leber KM, Kitada S, Blakenship HL, Svasand T, editors. Blackwell, Oxford; 2004. pp. 371–384. [Google Scholar]
- 23.Heath DD, Heath JW, Bryden CA, Johnson RM, Fox CW. Rapid evolution of egg size in captive salmon. Science. 2003;299:1738–1740. doi: 10.1126/science.1079707. [DOI] [PubMed] [Google Scholar]
- 24.Roberge C, Normandeau E, Einum S, Guderley H, Bernatchez L. Genetic consequences of interbreeding between farmed and wild Atlantic salmon: Insights from the transcriptome. Mol Ecol. 2008;17:314–324. doi: 10.1111/j.1365-294X.2007.03438.x. [DOI] [PubMed] [Google Scholar]
- 25.Frankham R, Lobel DA. Modeling problems in conservation genetics using captive Drosophila populations: Rapid genetic adaptation to captivity. Zoo Biol. 1992;11:333–342. [Google Scholar]
- 26.Quinn TP. The Behavior and Ecology of Pacific Salmon and Trout. Seattle: University of Washington Press; 2005. [Google Scholar]
- 27.Araki H, Ardren WR, Olsen E, Cooper B, Blouin MS. Reproductive success of captive-bred steelhead trout in the wild:Evaluation of three hatchery programs in the Hood river. Conserv Biol. 2007;21:181–190. doi: 10.1111/j.1523-1739.2006.00564.x. [DOI] [PubMed] [Google Scholar]
- 28.Christie MR, Marine ML, Blouin MS. Who are the missing parents? Grandparentage analysis identifies multiple sources of gene flow into a wild population. Mol Ecol. 2011;20:1263–1276. doi: 10.1111/j.1365-294X.2010.04994.x. [DOI] [PubMed] [Google Scholar]
- 29.Olsen EA. Annual Report 2000\x{2013}01 of the Oregon Department of Fish and Wildlife. Portland, OR: Oregon Department of Fish and Wildlife; 2003. Hood River and Pelton ladder evaluation studies. [Google Scholar]
- 30.Kostow KE. Differences in juvenile phenotypes and survival between hatchery stocks and a natural population provide evidence for modified selection due to captive breeding. Can J Fish Aquat Sci. 2004;61:577–589. [Google Scholar]
- 31.Seamons TR, Dauer MB, Sneva J, Quinn TP. Use of parentage assignment and DNA genotyping to validate scale analysis for estimating steelhead age and spawning history. N Am J Fish Manage. 2009;29:396–403. [Google Scholar]
- 32.Christie MR. Parentage in natural populations: Novel methods to detect parent-offspring pairs in large data sets. Mol Ecol Resour. 2010;10:115–128. doi: 10.1111/j.1755-0998.2009.02687.x. [DOI] [PubMed] [Google Scholar]
- 33.R Development Core Team A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna) 2010. . Available at http://www.R-project.org. Accessed December 11, 2010.
- 34.Pearse DE, et al. Over the falls? Rapid evolution of ecotypic differentiation in steelhead/rainbow trout (Oncorhynchus mykiss) J Hered. 2009;100:515–525. doi: 10.1093/jhered/esp040. [DOI] [PubMed] [Google Scholar]
- 35.Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- 36.Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- 37.Zuur AF, Ieno EN, Walker N, Saveliev AA, Smith GM. Mixed Effects Models and Extensions in Ecology with R. New York: Springer; 2009. [Google Scholar]
- 38.Sokal RR, Rohlf J. Biometry. New York: Freeman; 1995. pp. 685–715. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.