Abstract
Adenosine-5′-phosphosulfate (APS) kinase (APSK) catalyzes the phosphorylation of APS to 3′-phospho-APS (PAPS). In Arabidopsis thaliana, APSK is essential for reproductive viability and competes with APS reductase to partition sulfate between the primary and secondary branches of the sulfur assimilatory pathway; however, the biochemical regulation of APSK is poorly understood. The 1.8-Å resolution crystal structure of APSR from A. thaliana (AtAPSK) in complex with β,γ-imidoadenosine-5′-triphosphate, Mg2+, and APS provides a view of the Michaelis complex for this enzyme and reveals the presence of an intersubunit disulfide bond between Cys86 and Cys119. Functional analysis of AtAPSK demonstrates that reduction of Cys86-Cys119 resulted in a 17-fold higher kcat/Km and a 15-fold increase in Ki for substrate inhibition by APS compared with the oxidized enzyme. The C86A/C119A mutant was kinetically similar to the reduced WT enzyme. Gel- and activity-based titrations indicate that the midpoint potential of the disulfide in AtAPSK is comparable to that observed in APS reductase. Both cysteines are invariant among the APSK from plants, but not other organisms, which suggests redox-control as a unique regulatory feature of the plant APSK. Based on structural, functional, and sequence analyses, we propose that the redox-sensitive APSK evolved after bifurcation of the sulfur assimilatory pathway in the green plant lineage and that changes in redox environment resulting from oxidative stresses may affect partitioning of APS into the primary and secondary thiol metabolic routes by having opposing effects on APSK and APS reductase in plants.
Keywords: enzyme kinetics, sulfur metabolism, X-ray crystallography, metabolic evolution
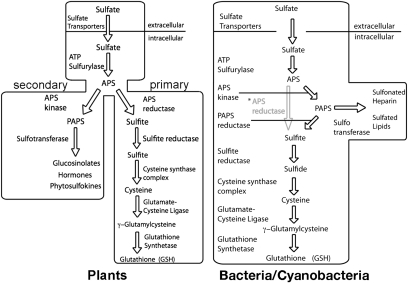
Sulfur is an essential element for all living organisms and is required for the biosynthesis of a diverse array of metabolites and macromolecules (1–4). Plants and prokaryotes are the primary assimilatory organisms that convert inorganic sulfate (SO42−), the predominant form of environmental sulfur, into physiologically useful forms of sulfur (5, 6). The metabolic organization of sulfur assimilation varies among plants and microbes (Fig. 1) (1–8). In yeast, fungi, and enterobacteria, including Escherichia coli, sulfate is incorporated into adenosine-5′-phosphate (APS), then converted to 3′-phospho-APS (PAPS) as a biologically “activated” compound that is reduced to sulfide (6, 7). In other sulfate-assimilating bacteria, such as Pseudomonas aeruginosa, APS can be used for reduction of sulfide (8). Plants possess bifurcated thiol metabolism pathways, which may reflect the metabolic needs of sessile organisms adapted to a range of environmental stresses and nutrient fluctuations (Fig. 1) (5). These pathways branch after formation of APS. The primary sulfur metabolic route in plants uses APS as the activated high-energy compound for sulfur reduction and the production of cysteine, which is crucial for the synthesis of proteins, methionine, iron–sulfur clusters, vitamin cofactors, and compounds that protect against oxidative stresses, including glutathione and phytochelatin peptides (9–12). Alternatively, APS can be converted into PAPS to provide a sulfate donor for the modification of multiple natural products, brassinosteroid and jasmonate hormones, phytosulfokines, and other sulfonated molecules (13–15). Partitioning of sulfate at the branch point between primary (i.e., reductive) and secondary metabolic pathways in plants is controlled by APS reductase (APSR) and APS kinase (APSK), respectively (16, 17).
Fig. 1.
Sulfur assimilatory pathways in plants (Left) and bacteria/cyanobacteria (Right). Various bacteria and cyanobacteria use APS kinase and PAPS reductase (black arrows) or APS reductase (gray arrow and text marked with an asterisk) for formation of sulfite.
APSR catalyzes the glutathione-dependent reduction of APS to sulfite (SO32−) and AMP. Extensive studies of APSR in plants demonstrate a critical role for this enzyme in regulating flux through the primary sulfur assimilatory pathway (18–21). Modulation of activity by redox changes in the APSR from Arabidopsis thaliana (AtAPSR) occurs through a regulatory disulfide bond, which upon reduction attenuates activity (18). Additional evidence suggests that APSR is important for supplying reduced sulfur under stress conditions, such as nutrient deprivation and chilling (22). Conversely, application of cysteine and glutathione to Arabidopsis results in decreased APSR transcript levels and activity (23). In contrast to APSR, the role of APSK in plant thiol metabolism has only begun to be examined.
Recent studies in Arabidopsis revealed that APSK is essential for plant reproduction and viability (17, 24–27). Use of T-DNA knockout lines targeting each of the four AtAPSK isoforms established a connection between this branch of sulfur metabolism and glucosinolate biosynthesis (24, 27). Subsequent analysis of triple AtAPSK knockout lines demonstrated that AtAPSK1 alone could maintain wild-type levels of growth and development in Arabidopsis (26). Moreover, measurements of flux through the primary sulfur assimilation pathway was increased when APSK activity was reduced, which implies that alterations in either APSR or APSK activity aids in partitioning of sulfur between the two branches of thiol metabolism (17).
In plants, the biochemical regulation of APSK is not well understood, and we are aware of no study to date that has linked stress conditions to the control of its activity. APSK catalyzes the transfer of the γ-phosphate from ATP to the 3′-hydroxyl group of the APS adenine ring to yield PAPS and ADP. The reaction sequence suggested by kinetic studies of the APSK from Penicillium chrysogenum, E. coli, and A. thaliana follows an ordered mechanism with substrate inhibition by APS (28–30). Although no structural information for a plant APSK is available, X-ray crystal structures of the P. chrysogenum APSK and bifunctional ATP sulfurylase-APSK from human, Aquifex aeolicus, and Thiobacillus denitrificans have been determined and reveal a canonical α/β purine nucleotide binding fold (31–36). For the bifunctional human enzyme, the substrate inhibition by APS on APSK activity is linked to an approximately 20-aa N-terminal loop of low sequence homology (36) (Fig. S1); however, the general role of this region across different species remains unclear. In addition, earlier studies of AtAPSK suggest that oxidation reduces specific activity (29).
To better understand the molecular function of the plant APSK, we determined the crystal structure of AtAPSK isoform 1, lacking its chloroplast localization leader, in complex with β,γ-imidoadenosine-5′-triphosphate (AMP-PNP), Mg2+, and APS. Although the overall structure is similar to that of the APSK from fungi and humans, the AtAPSK⋅AMP-PNP⋅Mg2+⋅APS complex new insight on the reaction sequence of this enzyme and reveals an intersubunit disulfide bond formed between Cys86 in the N-terminal loop of one subunit and Cys119 in the adjacent monomer. Kinetic analysis and redox titrations of WT and the C86A/C119A mutant of AtAPSK indicate that the disulfide bond forms at a physiologically relevant redox potential and affects catalytic efficiency and substrate inhibition by APS. Because sequence comparisons suggest a unique regulatory element of the plant APSK, we also functionally compare AtAPSK and the APSK from the cyanobacterium Synechocystis sp. PCC 6803, which is evolutionarily related to plant chloroplasts (37), to demonstrate that a homologue lacking the cysteines is not redox-sensitive. Based on structural, functional, and sequence analyses, we propose that evolution of a thiol-based redox switch in the plant APSK evolved with bifurcation of the sulfur assimilatory pathway later in the plant lineage, and that changes in redox environment resulting from oxidative stresses may coordinate flux between the primary and secondary thiol metabolic routes by modulating APSR and APSK activity in plants.
Results
Overall Structure.
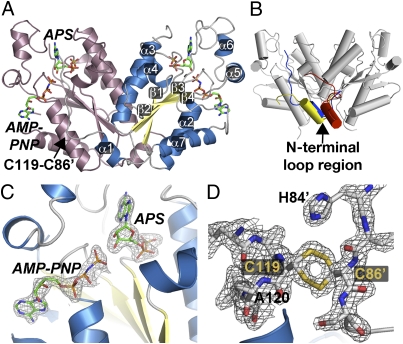
AtAPSK isoform 1 (residues 78–276) (38) lacking the 77-aa N-terminal chloroplast localization sequence was used for protein crystallography. The structure of AtAPSK in complex with AMP-PNP, Mg2+, and APS was solved by molecular replacement using the P. chrysogenum APSK (32) as a search model with three monomers in the asymmetric unit (Fig. 2 and Table S1). Chain A forms a crystallographic dimer, and chains B and C yield a second noncrystallographic dimer.
Fig. 2.
Structure of AtAPSK. (A) Ribbon diagram of the dimer. Each monomer is colored in pink and blue, respectively. The positions of AMP-PNP and APS are shown as stick molecules. The position of the C86⋅C119 disulfide bond is indicated in the left monomer. Secondary structure features are labeled in the right monomer. (B) Structural comparison of APSK structures. The overall AtAPSK structure is shown in gray. The position of the N-terminal region of the AtAPSK⋅AMP-PNP⋅Mg2+⋅APS complex is in blue. The N-terminal of the APSK domain from the human PAPS synthetase⋅ADP⋅PAPS complex is nearly identical to that of AtAPSK. The N-terminal of P. chrysogenum APSK apoenzyme is in yellow and the position in the P. chrysogenum APSK⋅ADP⋅APS complex is in red. (C) The 2Fo-Fc omit map (1.5 σ) for AMP-PNP and APS in the AtAPSK active site. (D) The 2Fo-Fc omit map (1.5 σ) for Cys86-Cys119 disulfide bond. Note that the cysteine residues are found in alternate conformations.
The overall fold of each monomer in the dimer consists of a variable N-terminal region (residues 80–98), which includes α1 and entwines the adjacent monomer, a canonical α/β-purine nucleotide-binding domain (β1-α2-β2-α3-β3-α4-β4-α7), and a small domain, which includes α5 and α6, that caps the nucleotide binding sites (Fig. 2A). Although AtAPSK shares a common fold with the bifunctional PAPS synthetase from human (1.2 Å rmsd for 187 Cα atoms; 51% sequence identity) (35) and the P. chrysogenum APSK (1.3 Å rmsd. for 176 Cα atoms; 55% sequence identity) (32), the positioning of the N-terminal region differs in these structures (Fig. 2B). This region is characterized by an unstructured loop followed by a approximately 7-aa α-helix (α1) in various APSK structures (32–36) and displays sequence divergence between the APSK homologues from bacteria, cyanobacteria, green algae, fungi, plants, and human (Fig. S1A).
In the initial 2Fo-Fc maps of the AtAPSK structure, clear electron density for AMP-PNP, APS, and Mg2+ was observed (Fig. 2C). Additional contiguous density was observed between Cys86 in monomer B and Cys119 of α2 in monomer C (Fig. 2D), Cys86 in monomer C and Cys119 in monomer B, and Cys86 of monomer A and Cys119 in the crystallographic symmetry-mate of monomer A. The distance between the sulfur atoms of each cysteine falls within the range of expected bond length (∼2.05 Å) for a disulfide linkage. The Cys86(A)-Cys119(A) and Cys86(B)-Cys119(C) disulfides were fully oxidized and the Cys86(C)-Cys119(B) disulfide was partially oxidized. The cysteines forming the disulfide are invariant across the APSK from plants and mosses, but are generally missing in the homologues from other organisms (Fig. S1).
Structure of Active Site and N-Terminal Loop.
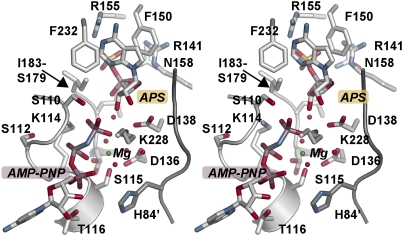
The structure of AtAPSK with a nonhydrolyzable ATP analogue (i.e., AMP-PNP), Mg2+, and APS bound in the active site provides a view of the Michaelis complex in the APSK reaction sequence (Fig. 3). APS is locked in the active site through multiple binding contacts (Fig. 3). The adenine ring is stacked between Phe150 and Phe232. Arg141 and Asn158 provide bridging interactions between the sulfate and phosphate groups of the substrate. Arg155 and the backbone nitrogen of Ile181 also interact with the sulfate and phosphate groups, respectively. Asp138 forms a bidentate interaction with the ribose hydroxyl groups to position the 3′-OH proximal to the γ-phosphate group of AMP-PNP for the ensuing phosphoryl transfer reaction.
Fig. 3.
Stereoview of the AtAPSK active site. The N-terminal loop of the adjacent monomer is shown as the dark gray loop, which includes His84.
Binding of AMP-PNP is mediated almost exclusively through the phosphate moieties of the ligand to residues in the active site and an extensive water network centered on the bound Mg2+ ion (Fig. 3). The adenine ring is positioned between Pro257 and Arg215. Extending into the active site, phosphate groups of AMP-PNP form multiple contacts with main-chain atoms of residues 110 to 116 in the P-loop (39). The α-phosphate group is bound by interactions with the side-chain hydroxyl group and backbone nitrogen of Thr116 and the backbone nitrogen of Gly113. Lys114 contacts both the β- and γ-phosphates with Ser110 and Lys228, forming additional interactions to the γ-phosphate. The β- and γ-phosphates of AMP-PNP, three water molecules, the hydroxyl-groups of Ser115 and Ser179, and the carboxylate of Asp136 octahedrally coordinate the Mg2+ ion. Asp136 and Asp138 form part of the DGDN-loop that bridges the APS and ATP⋅Mg2+ binding sites and are critical for catalytic activity in APSK (31, 35, 40).
Although not immediately in contact with ligands in the AtAPSK active site, part of the N-terminal loop (residues 80–84) of the adjacent monomer abuts the DGDN-loop and the side-chain of His84 hydrogen bonds to a water molecule in the network that coordinates the Mg2+ ion (Fig. 3). The structure of the AtAPSK N-terminal region is nearly identical to that of the APSK domain from human PAPS synthetase in complex with ADP and PAPS, but differs from the P. chrysogenum APSK apoenzyme and ADP⋅APS complex (Fig. 2B). Changes in the N-terminal region of PAPS synthetase were suggested to promote conformational changes that stabilize interactions between the DGDN-loop and substrates during the catalytic cycle (33). Interestingly, the average B-factors for the N-terminal loop residues (i.e., 80–90) in the monomers containing the fully oxidized Cys86(A)-Cys119(A) and Cys86(B)-Cys119(C) disulfides were 39 Å2 and 47 Å2, respectively, which were lower than the average B-factor of 56 Å2 in the monomer with the partially oxidized Cys86(C)-Cys119(B) disulfide bond. Crystallographic analysis of the Cys86-Cys119 disulfide bond in AtAPSK suggests that redox changes may alter the positioning and/or mobility of the N-terminal loop near the active site and potentially affect enzymatic activity.
Cys86-Cys119 Disulfide and Redox Regulation.
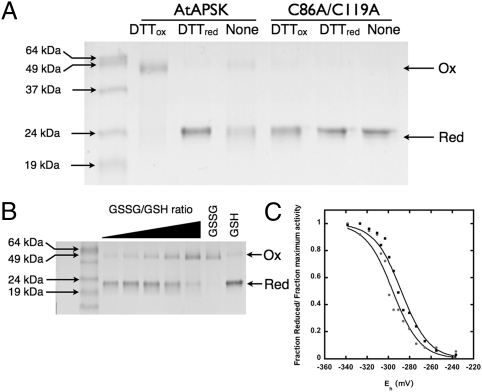
Formation of the Cys86-Cys119 disulfide linkage in the N-terminal region of AtAPSK may have a functional role in modulating enzymatic activity. Nonreducing SDS/PAGE analysis of AtAPSK incubated with either reduced DTT (DTTred) or trans-4,5-dihydroxy-1,2-dithiane (DTTox) showed the enzyme as reduced monomeric (22 kDa) and oxidized dimeric (44 kDa) forms, respectively (Fig. 4A). The same analysis using the C86A/C119A mutant confirmed that mutation of the cysteines prevented cross-linking of the two monomers (Fig. 4A). Both the WT and C86A/C119A mutant AtAPSK migrated as dimers upon size-exclusion chromatography in the presence or absence of reducing agents.
Fig. 4.
Functional analysis of redox activity. (A) SDS/PAGE of WT and C86A/C119A AtAPSK incubated in the absence or presence of 5 mM DTTred or DTTox. Protein (0.3–1 μg) was stained with Coomassie blue. Arrows on the left side of the gel indicate positions of the molecular weight markers. Arrows on the right indicate positions of the oxidized dimeric and reduced monomeric forms. (B) Gel-based redox titration of AtAPSK. Protein was incubated in increasing ratios of oxidized:reduced glutathione (GSSG:GSH; 20 mM total; Eh, −240 to −340 mV). (C) Comparison of gel-based (squares) and activity-based (circles) redox titrations. Solid lines represent fits to the Nernst equation.
Kinetic analysis of AtAPSK incubated in the presence of DTTred or DTTox demonstrated that reduction of the disulfide bond resulted in enhanced catalytic efficiency (i.e., kcat/Km) and a decreased effect of substrate inhibition by APS (Table 1 and Fig. S2). Reduced AtAPSK was 17-fold more efficient and had a Ki value for APS 15-fold higher compared with the oxidized protein (Table 1). Moreover, the Ki values of the reduced protein increased dramatically with decreasing ATP concentrations (Fig. S2D); however, substrate inhibition by APS was comparable at all ATP concentrations for the oxidized enzyme (Fig. S2E). Steady-state kinetic analysis of the C86A/C119A mutant showed similar kinetic parameters for both forms of the enzyme that were comparable to those of the reduced AtAPSK (Table 1). The C86A/C119A mutant treated with DTTred showed little difference in kinetic parameters with respect to AtAPSKred. Similarly, treatment of the mutant with DTTox caused a modest 1.5-fold decrease in activity compared with DTTred, but had little effect on either Km or Ki. These results suggest that redox environment may have a role in regulating the activity of AtAPSK.
Table 1.
Steady-state kinetic parameters
| Parameter | kcat, s−1 | Km, μM | Ki, μM | kcat/Km, M−1 s−1 × 108 |
| AtAPSKred | 272 ± 39 | 0.48 ± 0.41 | 37.5 ± 6.9 | 5.67 |
| AtAPSKox | 14.1 ± 2.3 | 0.43 ± 0.26 | 2.51 ± 0.84 | 0.328 |
| C86A/C119Ared | 239 ± 44 | 1.90 ± 0.53 | 34.8 ± 2.7 | 1.25 |
| C86A/C119Aox | 203 ± 31 | 0.87 ± 0.49 | 41.6 ± 7.1 | 2.33 |
| SynAPSKred | 18.6 ± 4.5 | 0.33 ± 0.17 | 27.6 ± 3.5 | 0.56 |
| SynAPSKox | 15.9 ± 2.2 | 0.57 ± 0.26 | 21.6 ± 3.7 | 0.28 |
Average values ± SE (n = 3) are shown.
To determine the redox midpoint potential (Em) for the Cys86-Cys119 disulfide, titrations using a gel-based system and activity assays were performed (Fig. 4 B and C). AtAPSK was incubated in solutions containing varied ratios of oxidized:reduced glutathione (GSSG:GSH; total concentration 20 mM). Aliquots from incubations (Eh of −340 to −240 mV) were analyzed by nonreducing SDS/PAGE and activity assays. Triplicate titrations of AtAPSK at pH 7.5 yielded average Em values of −295 ± 12 mV and −286 ± 18 mV in the gel- and activity-based assays, respectively. At pH 7, the titrations yielded average Em values of −249 ± 9 mV and −260 ± 17 mV in the gel- and activity-based assays, respectively. Compared with the Em values of other redox-regulated plant enzymes, which range from −390 to −237 mV (18, 41–45), these values suggest that changes in redox environment may modulate APSK activity in vivo, which could provide a strategy for modulating flux through the primary and secondary sulfur metabolism pathways in plants.
Characterization of APSK from Synechocystis sp. PC 6803.
Comparison of APSK sequences indicates that the residues corresponding to Cys86 and Cys119 are invariant in the enzymes from monocot and dicot plants and the mosses Selaginella moellendorffii and Physcomitrella patens (Fig. S1B). Cys119 is conserved in the APSK from green alga Chlorella variabilis and Chlamydomonas reinhardtii, but is not found in the enzyme from cyanobacteria Synechocystis and Cyanothece (Fig. S1A). These sequence alignments suggest that redox-regulation of APSK appeared later in the lineage of photosynthetic organisms. To confirm that an evolutionarily earlier homologue is not redox-sensitive, the APSK from Synechocystis sp. PC 6803 was cloned, expressed, purified, and assayed. Steady-state kinetic analysis of SynAPSK in the presence of DTTred or DTTox indicates that its activity does not change with redox environment (Table 1).
Discussion
The sulfur assimilation pathway in plants supports sulfur reduction and the synthesis of cysteine in the primary metabolic branch and the generation of sulfonated molecules for specialized metabolism in the secondary branch. Partitioning of sulfate at the branchpoint between primary and secondary metabolic pathways in plants is balanced by the activities of APSR and APSK (Fig. 1) (16, 17). Recent work reveals an essential role of APSK for the reproductive viability of Arabidopsis (17, 24–27), but the biochemical regulation of this enzyme in plants is not well understood. Structural and functional studies of AtAPSK provide insights on the molecular basis for PAPS formation and redox-control of this important metabolic branchpoint enzyme.
The reaction sequence of APSK is proposed to follow an ordered mechanism with ATP⋅Mg2+ binding first, followed by APS, catalysis, release of PAPS, and release of ADP⋅Mg2+ (46). Structures of the P. chrysogenum APSK apoenzyme (32), the P. chrysogenum APSK⋅ADP⋅Mg2+⋅APS dead-end complex (33), the ADP⋅Mg2+⋅PAPS product complex of the APSK domain from human PAPS synthetase (35), and AtAPSK⋅AMP-PNP⋅Mg2+⋅APS Michaelis complex (Figs. 2 and 3) clearly define the active site changes accompanying ATP-dependent phosphorylation of APS to PAPS. Shifts in the side chains of residues in the P-loop (Ser110-Thr116 in AtAPSK) position the phosphate backbone of ADP or AMP-PNP into the active site with additional conformational changes in residues of the APS site allowing for binding of this ligand. These alterations are accompanied by ordering of the “lid”-domain, which in AtAPSK contains α5 and α6, over the active site to constrain the flexibility of the structure.
These crystal structures suggest critical roles for the Mg2+ ion in catalysis and organization of the active site architecture. The interactions centered on the Mg2+ ion intricately link the ATP binding site, the active site water network, and residues in the catalytically essential DGDN-loop, which includes Asp136 and Asp138 of AtAPSK. Sekulic et al. (35) noted that Mg2+ binding induces the DGDN-loop to switch from an inactive to active conformation. Contacts from Asp136 to the Mg2+ and active site water molecules position Asp138 in proximity to the ribose hydroxyl groups of APS. In the AtAPSK structure, the carboxylate oxygens of Asp138 are 2.67 Å and 3.15 Å from the 3′-OH and 2'-OH groups of the APS ribose, respectively (Fig. 3). This suggests that the negatively charged oxygen of Asp138 serves as a general base in the catalytic mechanism to abstract a proton from the 3′-hydroxyl group of APS and promotes its nucleophilic attack on the γ-phosphate group of ATP.
Phosphorylation of APS to yield PAPS requires an orchestrated series of structural changes to organize the APSK active site for catalysis. Comparisons of various APSK in complex with different ligands (Fig. 2B) suggest that the conformational flexibility of the N-terminal loop region likely affects these structural changes. Within the active site, this loop sterically contacts the DGDN-loop (Fig. 3) and is proposed to position catalytically essential residues to promote ligand binding and phosphoryl transfer (35, 36). Deletion of the N-terminal region from the APSK domain of human PAPS synthetase indicates that this region, which is not in direct contact with ligands in the active site, is responsible for substrate inhibition by APS (36). The AtAPSK structure reveals a regulatory control feature in the N-terminal loop, which is unique to plants.
Formation of a disulfide bond between Cys86 and Cys119 in the N-terminal loop region cross-links subunits of the AtAPSK dimer (Figs. 2D and 4), decreases catalytic efficiency, and enhances substrate inhibition by APS (Table 1). Comparisons of the kinetic properties of the reduced and oxidized forms of AtAPSK indicate that reduction of the disulfide yields a more active enzyme. Mutagenesis of the two cysteines in AtAPSK also mimics reduction of the disulfide, as demonstrated by activity assays (Table 1) and gel-based analysis (Fig. 4A). Moreover, redox titrations of the disulfide bond using gel-based and activity-based assays indicate that the Em is within a physiological range (Fig. 4 B and C) (18, 41–45, 47). The redox Em of the Cys86-Cys119 disulfide in AtAPSK is comparable to that determined for AtAPSR (Em of −255 at pH 7). Interestingly, both AtAPSK and AtAPSR are closer to the redox midpoint for GSH/GSSG than to the reported values associated with plant thioredoxins (48–50). Structurally, changes in the oxidation state of Cys86 and Cys119 in AtAPSK likely alter the dynamic movements of the N-terminal loop to affect catalytic efficiency and substrate inhibition. Physiologically, the Cys86-Cys119 disulfide bond may act as a regulatory switch to coordinate flux between the primary and secondary branches of sulfur assimilation in Arabidopsis and other plants.
Several plant thiol metabolism enzymes [i.e., APSK, APSR, and glutamate-cysteine ligase (GCL); Fig. 1] possess cysteines that form disulfide bonds to regulate enzymatic activity in response to changes in redox environment (18, 43–45). This suggests that oxidative stresses may simultaneously affect multiple proteins across sulfur assimilation and metabolism in plants. Because APSR and APSK share similar redox Em values and partition the flow of APS into different branches of sulfur metabolism (16, 17), coordinated biochemical regulation of their activities may be important to meet metabolic demands for sulfur reduction and/or production of PAPS for specialized sulfonation reactions. Redox regulation of these branchpoint proteins would provide a mechanism for controlling sulfur allocation between the two metabolic routes. For example, under oxidative stress conditions that increase demand for sulfur reduction to support glutathione synthesis for maintaining redox state, formation of the disulfides in APSR and APSK would increase and decrease their activities, respectively, and result in direction of APS into cysteine and glutathione synthesis and away from synthesis of PAPS. Moreover, previous studies demonstrate that formation of a key disulfide bond in the plant GCL under multiple oxidative stress conditions activates this enzyme to increase glutathione synthesis (42–45). Thus, both APSR and GCL in the primary thiol metabolic pathway would be active in response to stress and APSK activity attenuated in the secondary pathway. Further experimental studies in plants that analyze metabolite distribution and sulfur pathway flux in response to biotic and abiotic stresses that alter oxidation state is required to explore this potential biochemical regulatory mechanism.
Sequence comparisons of the APSK from plants, mosses, fungi, human, green algae, and cyanobacteria show that active site residues are highly conserved (24–29, 31–36) and that the N-terminal regions are divergent (Fig. S1), which may reflect the need for specialization of APSK regulation in organisms with differing metabolic demands for sulfur assimilation. Residues corresponding to Cys86 and Cys119 of AtAPSK are nearly invariant across the homologues from plants and mosses with some unicellular alga and yeast retaining Cys119; however, in the APSK from organisms lacking plastids, neither cysteine is found (Fig. S1B). In addition, of the four APSK isoforms in Arabidopsis (22), the disulfide cysteines are found in the three plastid-localized isoforms, but only Cys119 is conserved in the cytosolic isoform. Given that the chloroplast is a highly redox-active organelle (9, 10, 12), this difference between AtAPSK isoforms suggests the evolution of a specialized regulatory control in the enzymes of this organelle. Through the lineage from cyanobacteria to green algae to mosses and “higher” plants, there is also a shift in the organization of the sulfur assimilation pathway from a linear one in cyanobacteria to a branched pathway in the later evolving mosses and plants (Fig. 1) (5, 37). Thus, the redox-sensitive disulfide observed in the plant and moss APSK may be a later adaptation to the development of chloroplasts and branched sulfur metabolism pathways in these multicellular photosynthetic organisms, and reflects a greater need for regulatory control in response to more specialized metabolic demands for coordination of flux between the primary and secondary sulfur pathways by reciprocally modulating APSR and APSK activity in plants.
Materials and Methods
Protein Expression and Purification.
Generation of the pET-28a-AtAPSKΔ77 bacterial expression vector, which encodes AtAPSK isoform 1 lacking the plastid localization sequence and with an N-terminal hexahistidine tag, was previously described (38). Transformed E. coli BL21(DE3) cells were grown at 37 °C in Terrific broth containing 50 μg mL−1 kanamycin until an absorbance at 600 nm of approximately 0.8. After induction with 1 mM isopropyl 1-thio-β-d-galactopyranoside, the cultures were grown at 20 °C overnight. Cells were pelleted by centrifugation and resuspended in 50 mM Tris (pH 8.0), 500 mM NaCl, 20 mM imidazole, 1 mM β-mercaptoethanol (βME), 10% (vol/vol) glycerol, and 1% Tween-20. After sonication and centrifugation, the supernatant was passed over a Ni2+-nitriloacetic acid (Qiagen) column. The column was washed with buffer minus Tween-20 and the bound protein eluted by using 250 mM imidazole in wash buffer. The eluent was dialyzed overnight at 4 °C against 25 mM Hepes (pH 7.5), 200 mM KCl, 5% glycerol, and 5 mM βME, and then loaded onto a Superdex-200 26/60 HiLoad FPLC size-exclusion column equilibrated in the same buffer. Fractions corresponding to the major protein peak were pooled, and judged to be more than 95% pure by SDS/PAGE. Protein was concentrated (Amicon) to 10 mg mL−1 with protein concentration determined by using a molar extinction coefficient (ε280nm of 22,430 M−1⋅cm−1) calculated in ProtParam (http://web.expasy.org/protparam). Protein was dialyzed against 25 mM Hepes (pH 7.5), 200 mM KCl, 10% glycerol, and 5 mM βME, flash-frozen in liquid nitrogen, and stored at −80 °C. For crystallization and redox titrations, βME was removed by buffer exchange.
Crystallography.
Crystals of the AtAPSK⋅AMP-PNP⋅Mg2+⋅APS complex grew at 4 °C in hanging drops from a 1:1 ratio of protein preincubated with 5 mM APS, 5 mM AMP-PNP, and 10 mM MgCl2 and crystallization buffer (100 mM Hepes, pH 7.5, 200 mM MgCl2, and 15–17.5% PEG 2000). For data collection, crystals were transferred to cryoprotectant (mother liquor containing ligands and 20% glycerol) and then frozen in liquid nitrogen. Diffraction data (100 K) were collected at beam 19-ID of the Advanced Photon Source/Argonne National Laboratory. Diffraction intensities were integrated, merged, and scaled by using the HKL3000 software suite (51). The structure of AtAPSK was determined by molecular replacement with PHASER (52) by using the structure of APSK from P. chrysogenum (PDB ID 1M7G) (32) as the search model. After iterative rounds of model building in COOT (53) and refinement in PHENIX (54), the R-factors converged to those reported in Table S1. Coordinates and structure factors for the AtAPSK⋅AMP-PNP⋅Mg2+⋅APS complex have been deposited in the Protein Data Bank.
Mutagenesis.
The AtAPSK C86A/C119A mutant was generated using the QuikChange PCR method (Stratagene) with pET-28a-AtAPSKΔ77 as the template. Two pairs of oligonucleotides (Table S2) were used to sequentially generate the double-mutant. Mutations were confirmed by sequencing (Washington University DNA Sequencing Facility). Protein expression and purification were performed as described for WT protein.
Enzyme Assays and Redox Titrations.
Enzymatic activity of purified WT and mutant AtAPSK was determined spectrophotometrically by using a coupled assay (30). Steady-state kinetic parameters were determined by initial velocity experiments with data fitted to a general substrate inhibition model:
The effect of redox potential on AtAPSK activity was examined with assays containing defined ratios of reduced (i.e., GSH) and oxidized (i.e., GSSG) glutathione (Sigma-Aldrich). Protein (5 μM) was equilibrated in degassed 100 mM Hepes (pH 7.5 or 7.0), 200 mM KCl, 5% glycerol, and 20 mM GSH/GSSG for 60 min at 25 °C. Aliquots were removed and transferred to the coupled assay (final protein concentration of 10–30 nM), and initial velocities measured. Values for Em were determined by fitting titration data to the Nernst equation by using Kaleidagraph (Synergy Software):
with an RT/F of 25.7 mV (44, 47) and n = 2. The effect of redox potental on AtAPSK was also evaluated by nonreducing SDS/PAGE. Protein was equilibrated with varying ratios of GSSG/GSH, DTTred, or DTTox, and then separated into reduced and oxidized forms by SDS/PAGE. The fraction of reduced vs. oxidized AtAPSK was calculated by quantifying the fractions of oxidized cross-linked (44 kDa) and reduced monomeric (22 kDa) species by densitometry (Quantity One software; BioRad).
Cloning, Expression, Purification, and Analysis of APSK from Synechocystis sp. PCC 6803.
The cDNA encoding APSK from Synechocystis sp PCC. 6803 was PCR-amplified by using PfuUltraHF polymerase and gene-specific oligonucleotides (Table S2). The PCR product was digested and cloned into the NheI and EcoRI sites of pET28a. Protein expression, purification, and activity assays were performed as described for AtAPSK.
Supplementary Material
Acknowledgments
We thank the Pakrasi laboratory for providing Synechocystis genomic DNA. This work was supported by National Science Foundation Grant MCB-0904215. Portions of this research were carried out at the Argonne National Laboratory Structural Biology Center of the Advanced Photon Source, a national user facility operated by the University of Chicago for the Department of Energy Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3UIE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115772108/-/DCSupplemental.
References
- 1.Leustek T, Martin MN, Bick JA, Davies JP. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:141–165. doi: 10.1146/annurev.arplant.51.1.141. [DOI] [PubMed] [Google Scholar]
- 2.Yi H, Galant A, Ravilious GE, Preuss ML, Jez JM. Sensing sulfur conditions: Simple to complex protein regulatory mechanisms in plant thiol metabolism. Mol Plant. 2010;3:269–279. doi: 10.1093/mp/ssp112. [DOI] [PubMed] [Google Scholar]
- 3.Yi H, Ravilious GE, Galant A, Krishnan HB, Jez JM. Thiol metabolism in soybean: sulfur to homoglutathione. Amino Acids. 2010;39:963–978. doi: 10.1007/s00726-010-0572-9. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol. 2011;62:157–184. doi: 10.1146/annurev-arplant-042110-103921. [DOI] [PubMed] [Google Scholar]
- 5.Patron NJ, Durnford DG, Kopriva S. Sulfate assimilation in eukaryotes: Fusions, relocations and lateral transfers. BMC Evol Biol. 2008;8:39. doi: 10.1186/1471-2148-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marzluf GA. Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu Rev Microbiol. 1997;51:73–96. doi: 10.1146/annurev.micro.51.1.73. [DOI] [PubMed] [Google Scholar]
- 7.Sekowska A, Kung HF, Danchin A. Sulfur metabolism in Escherichia coli and related bacteria: Facts and fiction. J Mol Microbiol Biotechnol. 2000;2:145–177. [PubMed] [Google Scholar]
- 8.Bick JA, Dennis JJ, Zylstra GJ, Nowack J, Leustek T. Identification of a new class of 5′-adenylylsulfate (APS) reductases from sulfate-assimilating bacteria. J Bacteriol. 2000;182:135–142. doi: 10.1128/jb.182.1.135-142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noctor G, Foyer CH. Ascorbate and glutathione: Keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 10.Meyer AJ. The integration of glutathione homeostasis and redox signaling. J Plant Physiol. 2008;165:1390–1403. doi: 10.1016/j.jplph.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Cobbett C, Goldsbrough P. Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- 12.Galant A, Preuss ML, Cameron JC, Jez JM. Plant glutathione biosynthesis: diversity in biochemical regulation and reaction products. Front Plant Sci. 2011;2:45. doi: 10.3389/fpls.2011.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein M, Papenbrock J. The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J Exp Bot. 2004;55:1809–1820. doi: 10.1093/jxb/erh183. [DOI] [PubMed] [Google Scholar]
- 14.Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- 15.Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:18333–18338. doi: 10.1073/pnas.0706403104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rausch T, Wachter A. Sulfur metabolism: A versatile platform for launching defence operations. Trends Plant Sci. 2005;10:503–509. doi: 10.1016/j.tplants.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Mugford SG, Lee BR, Koprivova A, Matthewman C, Kopriva S. Control of sulfur partitioning between primary and secondary metabolism. Plant J. 2011;65:96–105. doi: 10.1111/j.1365-313X.2010.04410.x. [DOI] [PubMed] [Google Scholar]
- 18.Bick JA, et al. Regulation of the plant-type 5′-adenylyl sulfate reductase by oxidative stress. Biochemistry. 2001;40:9040–9048. doi: 10.1021/bi010518v. [DOI] [PubMed] [Google Scholar]
- 19.Martin MN, Tarczynski MC, Shen B, Leustek T. The role of 5′-adenylylsulfate reductase in controlling sulfate reduction in plants. Photosynth Res. 2005;86:309–323. doi: 10.1007/s11120-005-9006-z. [DOI] [PubMed] [Google Scholar]
- 20.Loudet O, et al. Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat Genet. 2007;39:896–900. doi: 10.1038/ng2050. [DOI] [PubMed] [Google Scholar]
- 21.Scheerer U, et al. Sulphur flux through the sulphate assimilation pathway is differently controlled by adenosine 5′-phosphosulphate reductase under stress and in transgenic poplar plants overexpressing gamma-ECS, SO, or APR. J Exp Bot. 2010;61:609–622. doi: 10.1093/jxb/erp327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phartiyal P, Kim WS, Cahoon RE, Jez JM, Krishnan HB. The role of 5′-adenylylsulfate reductase in the sulfur assimilation pathway of soybean: Molecular cloning, kinetic characterization, and gene expression. Phytochemistry. 2008;69:356–364. doi: 10.1016/j.phytochem.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Vauclare P, et al. Flux control of sulphate assimilation in Arabidopsis thaliana: Adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J. 2002;31:729–740. doi: 10.1046/j.1365-313x.2002.01391.x. [DOI] [PubMed] [Google Scholar]
- 24.Mugford SG, et al. Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell. 2009;21:910–927. doi: 10.1105/tpc.109.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopriva S, Mugford SG, Matthewman C, Koprivova A. Plant sulfate assimilation genes: Redundancy versus specialization. Plant Cell Rep. 2009;28:1769–1780. doi: 10.1007/s00299-009-0793-0. [DOI] [PubMed] [Google Scholar]
- 26.Mugford SG, Matthewman CA, Hill L, Kopriva S. Adenosine-5′-phosphosulfate kinase is essential for Arabidopsis viability. FEBS Lett. 2010;584:119–123. doi: 10.1016/j.febslet.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Yatusevich R, et al. Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J. 2010;62:1–11. doi: 10.1111/j.1365-313X.2009.04118.x. [DOI] [PubMed] [Google Scholar]
- 28.Satishchandran C, Markham GD. Adenosine-5′-phosphosulfate kinase from Escherichia coli K12. Purification, characterization, and identification of a phosphorylated enzyme intermediate. J Biol Chem. 1989;264:15012–15021. [PubMed] [Google Scholar]
- 29.Lillig CH, et al. Molecular and catalytic properties of Arabidopsis thaliana adenylyl sulfate (APS)-kinase. Arch Biochem Biophys. 2001;392:303–310. doi: 10.1006/abbi.2001.2453. [DOI] [PubMed] [Google Scholar]
- 30.MacRae IJ, Segel IH. Adenosine 5′-phosphosulfate (APS) kinase: Diagnosing the mechanism of substrate inhibition. Arch Biochem Biophys. 1999;361:277–282. doi: 10.1006/abbi.1998.0993. [DOI] [PubMed] [Google Scholar]
- 31.MacRae IJ, Segel IH, Fisher AJ. Crystal structure of adenosine 5′-phosphosulfate kinase from Penicillium chrysogenum. Biochemistry. 2000;39:1613–1621. doi: 10.1021/bi9924157. [DOI] [PubMed] [Google Scholar]
- 32.Lansdon EB, Segel IH, Fisher AJ. Ligand-induced structural changes in adenosine 5′-phosphosulfate kinase from Penicillium chrysogenum. Biochemistry. 2002;41:13672–13680. doi: 10.1021/bi026556b. [DOI] [PubMed] [Google Scholar]
- 33.Yu Z, Lansdon EB, Segel IH, Fisher AJ. Crystal structure of the bifunctional ATP sulfurylase-APS kinase from the chemolithotrophic thermophile Aquifex aeolicus. J Mol Biol. 2007;365:732–743. doi: 10.1016/j.jmb.2006.10.035. [DOI] [PubMed] [Google Scholar]
- 34.Gay SC, Segel IH, Fisher AJ. Structure of the two-domain hexameric APS kinase from Thiobacillus denitrificans: Structural basis for the absence of ATP sulfurylase activity. Acta Crystallogr D Biol Crystallogr. 2009;65:1021–1031. doi: 10.1107/S0907444909026547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekulic N, et al. Elucidation of the active conformation of the APS-kinase domain of human PAPS synthetase 1. J Mol Biol. 2007;367:488–500. doi: 10.1016/j.jmb.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekulic N, Konrad M, Lavie A. Structural mechanism for substrate inhibition of the adenosine 5′-phosphosulfate kinase domain of human 3′-phosphoadenosine 5′-phosphosulfate synthetase 1 and its ramifications for enzyme regulation. J Biol Chem. 2007;282:22112–22121. doi: 10.1074/jbc.M701713200. [DOI] [PubMed] [Google Scholar]
- 37.Raven JA, Allen JF. Genomics and chloroplast evolution: What did cyanobacteria do for plants? Genome Biol. 2003;4:209. doi: 10.1186/gb-2003-4-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phartiyal P, Kim WS, Cahoon RE, Jez JM, Krishnan HB. Soybean ATP sulfurylase, a homodimeric enzyme involved in sulfur assimilation, is abundantly expressed in roots and induced by cold treatment. Arch Biochem Biophys. 2006;450:20–29. doi: 10.1016/j.abb.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 39.Saraste M, Sibbald PR, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 40.Singh B, Schwartz NB. Identification and functional characterization of the novel BM-motif in the murine phosphoadenosine phosphosulfate (PAPS) synthetase. J Biol Chem. 2003;278:71–75. doi: 10.1074/jbc.M206688200. [DOI] [PubMed] [Google Scholar]
- 41.Bick JA, Aslund F, Chen Y, Leustek T. Glutaredoxin function for the carboxyl-terminal domain of the plant-type 5′-adenylylsulfate reductase. Proc Natl Acad Sci USA. 1998;95:8404–8409. doi: 10.1073/pnas.95.14.8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jez JM, Cahoon RE, Chen S. Arabidopsis thaliana glutamate-cysteine ligase: Functional properties, kinetic mechanism, and regulation of activity. J Biol Chem. 2004;279:33463–33470. doi: 10.1074/jbc.M405127200. [DOI] [PubMed] [Google Scholar]
- 43.Hothorn M, et al. Structural basis for the redox control of plant glutamate cysteine ligase. J Biol Chem. 2006;281:27557–27565. doi: 10.1074/jbc.M602770200. [DOI] [PubMed] [Google Scholar]
- 44.Hicks LM, et al. Thiol-based regulation of redox-active glutamate-cysteine ligase from Arabidopsis thaliana. Plant Cell. 2007;19:2653–2661. doi: 10.1105/tpc.107.052597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gromes R, et al. The redox switch of γ-glutamylcysteine ligase via a reversible monomer-dimer transition is a mechanism unique to plants. Plant J. 2008;54:1063–1075. doi: 10.1111/j.1365-313X.2008.03477.x. [DOI] [PubMed] [Google Scholar]
- 46.Renosto F, Seubert PA, Segel IH. Adenosine 5′-phosphosulfate kinase from Penicillium chrysogenum. Purification and kinetic characterization. J Biol Chem. 1984;259:2113–2123. [PubMed] [Google Scholar]
- 47.Hutchison RS, Ort DR. Measurement of equilibrium midpoint potentials of thiol/disulfide regulatory groups on thioredoxin-activated chloroplast enzymes. Methods Enzymol. 1995;252:220–228. doi: 10.1016/0076-6879(95)52025-2. [DOI] [PubMed] [Google Scholar]
- 48.Dangoor I, Peled-Zehavi H, Levitan A, Pasand O, Danon A. A small family of chloroplast atypical thioredoxins. Plant Physiol. 2009;149:1240–1250. doi: 10.1104/pp.108.128314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirasawa M, et al. Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry. 1999;38:5200–5205. doi: 10.1021/bi982783v. [DOI] [PubMed] [Google Scholar]
- 50.Collin V, et al. The Arabidopsis plastidial thioredoxins: New functions and new insights into specificity. J Biol Chem. 2003;278:23747–23752. doi: 10.1074/jbc.M302077200. [DOI] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 52.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.