Abstract
Checkpoint kinase 1 (Chk1) is a key regulator of checkpoint signaling in both the unperturbed cell cycle and DNA damage response. Under these conditions, Chk1 becomes active to prevent premature CDK1 activation and mitotic entry until DNA is properly replicated or repaired. It is unclear how Chk1 activity is controlled in the unperturbed cell cycle. During DNA damage, Chk1 is activated by ataxia telangiectasia and Rad3 related (ATR)-mediated phosphorylation; however, it is not entirely clear how this phosphorylation results in Chk1 activation. Here we report an N-terminally truncated alternative splice variant of Chk1, Chk1-S. Importantly, we show that Chk1-S is an endogenous repressor and regulator of Chk1. In the unperturbed cell cycle, Chk1-S interacts with and antagonizes Chk1 to promote the S-to-G2/M phase transition. During DNA damage, Chk1 is phosphorylated, which disrupts the Chk1–Chk1-S interaction, resulting in free, active Chk1 to arrest the cell cycle and facilitate DNA repair. Higher levels of Chk1-S are expressed, along with Chk1, in fetal and cancer tissues than in normal tissues. However, forced overexpression of Chk1-S in cultured cells and tumor xenografts induces premature mitotic entry, mitotic catastrophe, and reduction of tumor growth. The identification of Chk1-S as a unique splice variant and key regulator of Chk1 provides insights into cell cycle regulation and DNA damage response.
The cell cycle involves orderly transitions from G1, S, and G2 to M phase, resulting in cell division and proliferation. These transitions are under the vigilant surveillance of checkpoint pathways, which are activated to prevent entry into the next cell cycle phase until the current phase is properly completed. Checkpoint signaling is also crucial to the DNA damage response, where it induces cell cycle arrest and activates the process of DNA repair (1–4). Checkpoint kinase 1 (Chk1) is a serine/threonine protein kinase originally identified as the key regulator of the DNA damage checkpoint in yeast and mammalian cells (5, 6). It is now recognized that Chk1 also has an essential role in normal cell cycle checkpoints, cell proliferation, and viability in all eukaryotes (7–12). In response to DNA damage, Chk1 is phosphorylated and activated by ataxia telangiectasia and Rad3 related (ATR) (7, 13, 14) and, upon activation, Chk1 phosphorylates cdc25 and Wee1 family proteins, resulting in the inactivation of CDK1 and delay of mitotic entry to facilitate DNA repair (5, 15–20). In the unperturbed cell cycle, Chk1 regulates DNA replication in S phase, G2/M transition or mitotic entry, and the completion of mitosis (7, 15, 21–27). Despite these remarkable roles, it is unknown how Chk1 activity is controlled in various phases of the cell cycle. During DNA damage, Chk1 is activated by phosphorylation in its C-terminal domain, but it is unclear how the C-terminal phosphorylation leads to the activation of the N-terminal kinase domain (28–31). Earlier work suggested that the C-terminal domain may antagonize the N-terminal kinase domain via an intramolecular interaction that can be disrupted by phosphorylation, leading to Chk1 activation (31). This “autoinhibition” model, although supported by some observations (28), has been seriously challenged (30). Alternatively, Chk1 activity may be governed by a repressing factor(s), the dissociation of which from Chk1 leads to Chk1 activation (29). However, the identity of such a factor is unknown. In this study, we have identified an alternative splice variant of Chk1, Chk1-S. Chk1-S interacts with Chk1 and acts as an endogenous inhibitor of Chk1. Working together, Chk1 and Chk1-S regulate cell cycle (S-to-G2/M phase) and DNA damage checkpoints.
Results and Discussion
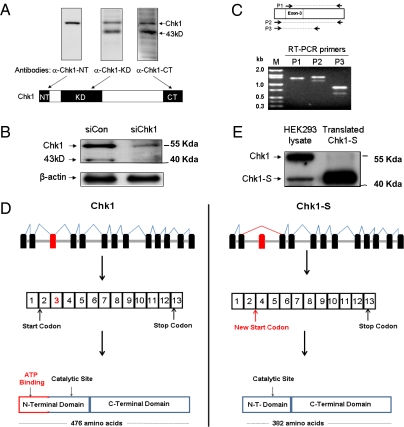
During our study of DNA damage signaling (32, 33), we observed two prominent bands in Chk1 immunoblots: the Chk1 band at 56 kDa and a faster-migrating band of 43 kDa. The faster-migrating band was detected by Chk1 antibodies that were reactive to the internal kinase domain or the C-terminal sequence, but not by Chk1 antibodies recognizing the N terminus (Fig. 1A). This band was not recognized by the G4 monoclonal antibody from Santa Cruz Biotechnology that is commonly used for immunoblot analysis of Chk1 (24, 29). Knockdown of Chk1 via siRNA led to the disappearance of both Chk1 and the 43-kDa band, further confirming their relevance (Fig. 1B). The 43-kDa protein was not affected by proteasome and protease inhibitors (SI Appendix, Fig. 1), raising the possibility that it could be an alternative splice variant of Chk1. The National Center for Biotechnology Information database currently lists three alternatively spliced human Chk1 mRNAs that have varying untranslated regions but encode the same full-length Chk1 protein of 476 amino acids (http://www.ncbi.nlm.nih.gov/gene/1111). However, our analysis using an EST-based alternative splicing predictive database (http://genome.ewha.ac.kr/ECgene) suggested the possibility of a unique splice variant of Chk1 in which exon 3 is alternatively spliced or deleted. To directly test this possibility, we performed RT-PCR using primers within the Chk1 coding sequence (Fig. 1C). RT-PCR using the primer set P1, in which the forward primer was designed within exon 3, generated a single amplicon of the expected size, whereas RT-PCR with the P2 or P3 primer set (both based on sequences flanking exon 3) generated two amplicons, one with the expected size of Chk1 and the other ∼200 bp shorter (Fig. 1C). Sequencing confirmed that the longer amplicon was indeed Chk1 and, notably, the shorter amplicon was an alternative splice variant of Chk1 lacking exon 3 (Fig. 1D and SI Appendix, Fig. 2A). This splice variant was predicted to translate into an N-terminally truncated form of Chk1 consisting of 382 amino acids that we named Chk1-short, or Chk1-S (Fig. 1D and SI Appendix, Fig. 2). In gel electrophoresis, in vitro translated Chk1-S migrated similarly as the 43-kDa protein from HEK293 cells (Fig. 1E). We further immunoprecipitated the 43-kDa protein from HEK293 cell lysate for mass spectrometry and confirmed its identity as Chk1-S. N-terminal truncation in Chk1-S is consistent with the immunoblot results showing that it was not recognized by the antibodies reactive to the N-terminal sequence of Chk1 (Fig. 1A). Real-time and RT-PCR analysis detected Chk1-S mRNA expression in multiple human tissues, and the expression is generally higher in fetal tissues (SI Appendix, Fig. 3 A and D). Immunoblot analysis further detected Chk1-S protein expression in human, mouse, and rat cell lines, and also in human fetal tissues (SI Appendix, Fig. 3B) and mouse primary renal tubular cells (SI Appendix, Fig. 3C). The full-length cDNA of Chk1-S was also cloned from three normal human tissues (thymus, colon, fetal liver) and sequenced to confirm that it encodes an N-terminally truncated splice variant of Chk1. Together, these experiments have identified a unique splice variant of Chk1 that is N-terminally truncated and widely expressed in mammalian cells and tissues.
Fig. 1.
Identification of Chk1-S as a unique, N-terminally truncated splice variant of Chk1. (A) HEK293 cell lysate was analyzed by immunoblotting using antibodies specifically recognizing the N terminus (α-Chk1-NT), kinase domain (α-Chk1-KD), or C terminus of Chk1 (α-Chk1-CT). In addition to Chk1, a 43-kDa protein was revealed by α-Chk1-KD and α-Chk1-CT, but not α-Chk1-NT. (B) HEK293 cells were transfected with Chk1 siRNA (siChk1) or a scrambled sequence (siCon) for 48 h to collect whole-cell lysate for immunoblot analysis using α-Chk1-KD. siChk1 diminished the expression of both Chk1 and the 43-kDa protein. (C) RNA was isolated from HEK293 cells for RT-PCR using three different sets of primers for Chk1: P1, P2, and P3 (relative sequence locations are shown in the diagram). Two amplicons were detected by RT-PCR using the primer sets flanking exon 3 (P2 and P3), whereas only one amplicon was amplified using the primer set P1, of which the forward primer was within exon 3 of Chk1. (D) Schematic representation of alternative splicing of Chk1 resulting in an N-terminally truncated protein, Chk1-S. (E) Chk1-S was cloned for in vitro translation, and the translated protein along with HEK293 cell lysate were analyzed by immunoblot analysis. In vitro translated Chk1 migrated similarly to the 43-kDa band in HEK293 cells.
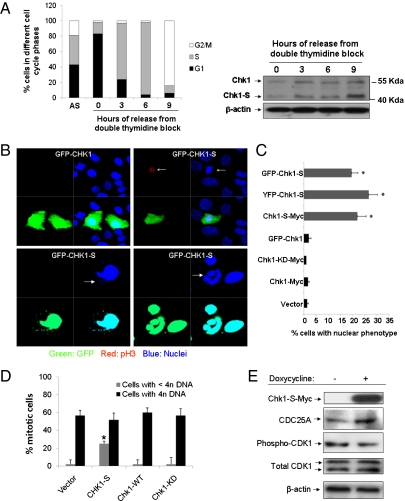
What is the function of Chk1-S? Does it regulate cell cycle checkpoints like Chk1? With these questions, we first determined the temporal and spatial expression patterns of Chk1-S in the cell cycle. HEK293 cells were synchronized at G1, G1/S, or G2/M phases by serum starvation, double thymidine block, or nocodazole, respectively. Chk1-S was low in G1-phase cells and higher in G1/S and G2/M cells (SI Appendix, Fig. 4 A and B). Moreover, when cells were synchronized at G1/S phase by double thymidine block and then released, both Chk1 and Chk1-S expression increased after entering S phase (Fig. 2A). A distinct difference between Chk1 and Chk1-S expression was that whereas Chk1 expression peaked at mid-S phase, Chk1-S expression kept increasing from the S to M phase (Fig. 2A). Interestingly, high levels of Chk1-S expression were observed at 7 h after cell cycle release from double thymidine block, a time point when cells were yet to be mitotic (SI Appendix, Fig. 4C). We further quantified the Chk1-S:Chk1 ratio in asynchronous and S or G2/M synchronized cells. Despite the variations of Chk1-S expression in HEK293, U2OS, and primary renal tubular cells, the Chk1-S:Chk1 ratio was above 1 in all cells at G2/M phase (SI Appendix, Fig. 5). In asynchronous cells, Chk1-S and Chk1 were mainly localized in the nucleus (SI Appendix, Fig. 6A). However, in G2/M-phase cells, some Chk1-S and Chk1 accumulated in centrosomes (SI Appendix, Fig. 6 B and C). The localization of Chk1 in centrosomes is critical for its G2/M checkpoint function, where it prevents premature activation of Cdk1 and mitotic entry (34, 35). Our results indicate that Chk1-S can also localize to centrosomes, providing a spatial and temporal regulation of Chk1 to initiate mitotic entry. Next, we determined the effects of Chk1 or Chk1-S overexpression in HEK293 and U2OS cells. Compared with GFP-Chk1, GFP-Chk1-S induced a distinguished nuclear phenotype that was characterized by nuclear condensation and formation of micronuclei and multiple or multilobed nuclei (Fig. 2B). At earlier time points, the transfected cells showed nuclear/chromatin condensation marked by faint and punctate histone-H3 phosphorylation (Fig. 2B), indicative of premature mitotic entry. Later, these cells developed micronuclei or multilobed nuclei, characteristics of mitotic catastrophe (Fig. 2B). Cell counting showed that GFP-Chk1-S induced the nuclear phenotype in ∼20% of U2OS cells (Fig. 2C). Similar effects were shown by YFP-Chk1-S and Chk1-S-Myc (Fig. 2C), indicating that the observed nuclear phenotype was induced by Chk1-S and not by the fusion protein tags. In contrast, premature mitotic entry was not induced by Chk1 or its kinase-dead mutant Chk1-KD (Fig. 2C). Chk1-S also induced the striking nuclear phenotype in other cell types (SI Appendix, Fig. 7). Notably, a similar nuclear phenotype was reported in cells and murine models lacking one or both Chk1 alleles (7, 36, 37), suggesting that Chk1-S may act as an endogenous inhibitor of Chk1. To further test this possibility, we generated Tet-on U2OS cell lines that can be induced to express Chk1, Chk1-KD, or Chk1-S. The cells were synchronized at G1/S phase by double thymidine block, induced by doxycycline, and then released into nocodazole-containing medium. About 80% of Chk1-S-expressing cells entered mitosis [phospho-histone H3 (pH3)-positive], whereas ∼60% of Chk1- or Chk1-KD-expressing cells did (Fig. 2D). Importantly, whereas most mitotic cells in the Chk1- and Chk1-KD-expressing groups had 4n DNA, ∼25% of mitotic cells from the Chk1-S-expressing group had less than 4n DNA content (Fig. 2D), indicative of aberrant mitotic entry in these cells without completion of DNA replication. In addition, Chk1-S overexpression resulted in earlier entry into mitosis, as indicated by the appearance of pH3-positive cells (SI Appendix, Fig. 8). Chk1 is a key regulator of mitotic entry or the S-to-G2/M transition in the cell cycle. By phosphorylating CDC25A (inducing its degradation) and Wee1, Chk1 prevents CDK1 activation and mitotic entry (5, 15–20). We showed that induction of Chk1-S by doxycycline in the Tet-on U2OS cells resulted in higher levels of CDC25A and lower levels of phospho-CDK1, suggesting that Chk1-S antagonized Chk1 to induce mitotic entry (Fig. 2E). Specific knockdown of Chk1-S via RNAi was not successful because the Chk1-S sequence is contained in Chk1 mRNA. However, an antisense oligonucleotide complementary to the unique exon2–exon4 junction in Chk1-S could specifically reduce Chk1-S expression (SI Appendix, Fig. 9A). Chk1-S down-regulation markedly reduced cell proliferation (SI Appendix, Fig. 9B). Interestingly, Chk1-S down-regulation did not significantly change the cell cycle profile (SI Appendix, Fig. 9C). Because Chk1 functions at several cell cycle checkpoints (e.g., G2/M, spindle, intra-S), Chk1-S may antagonize Chk1 to facilitate cell cycle progression at multiple sites. As a result, inhibition of Chk1-S may slow down the cell cycle at various phases and result in the suppression of proliferation without major changes of cell cycle distribution.
Fig. 2.
Regulation of the cell cycle by Chk1-S. (A) HEK293 cells were synchronized by double thymidine block and then released into nocodazole-containing medium. (Left) Cell cycle profile analyzed by propidium iodide (PI) staining and FACS analysis. (Right) Immunoblot analysis of Chk1 and Chk1-S in the cell lysate collected at indicated time points after the release from thymidine block. AS, asynchronous cells (B) U2OS cells were transfected with GFP-Chk1 or GFP-Chk1-S (green), and then fixed for immunofluorescence of phospho-histone H3 (red) and nuclear staining with Hoechst33342 (blue). (Upper) Chk1-S, but not Chk1, induced premature chromatin condensation and weak pH3 staining (arrows). (Lower) At late stage, Chk1-S-transfected cells showed the characteristics of mitotic catastrophe including micronuclei and multilobed nuclei (arrows). (C) U2OS cells were transfected with the indicated genes, and cells with the nuclear phenotype of premature chromatin condensation and mitotic catastrophe were counted. Data indicate mean ± SD; *P < 0.05 versus vector group. The results show that Chk1-S overexpression specifically led to the nuclear phenotype. (D) U2OS cells were transfected with the indicated genes, synchronized by double thymidine block, and released for 7 h. The cells were then fixed for pH3 immunofluorescence and PI staining and analyzed by FACS. Data indicate mean ± SD; *P < 0.05 versus vector group. The results show that Chk1-S specifically induced premature mitotic entry without completion of DNA replication (cells with <4n DNA). (E) Tet-on U2OS cells were induced with or without doxycycline. The cells were then synchronized at S phase by double thymidine block and released for 7 h. Whole-cell lysate was collected for immunoblot analysis of the indicated proteins. The results show that induced Chk1-S expression led to an increase of CDC25A and decrease of phospho-CDK1, contributing to premature mitotic entry.
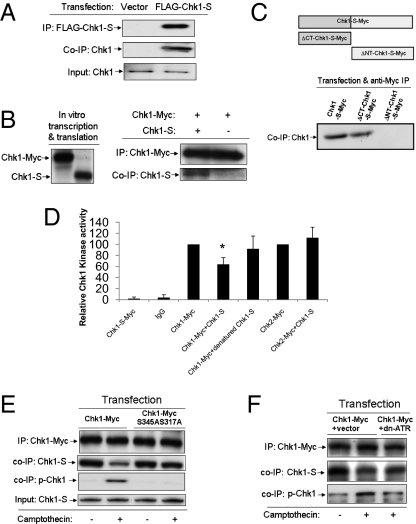
How does Chk1-S antagonize Chk1? We hypothesized that Chk1-S might interact with Chk1 to block its kinase activity. Consistently, FLAG-Chk1-S expressed in HEK293 cells coimmunoprecipitated with endogenous Chk1 (Fig. 3A). Moreover, in vitro translated Myc-Chk1 and Chk1-S coimmunoprecipitated (Fig. 3B), suggesting a direct Chk1–Chk1-S interaction. We further demonstrated the coimmunoprecipitation of Chk1 with the N-terminal domain, but not the C-terminal domain, of Chk1-S (Fig. 3C), suggesting the requirement of the N-terminal sequence in Chk1-S for its interaction with Chk1. Functionally, we determined the effects of Chk1-S on Chk1 kinase activity. Chk1-Myc was expressed and immunoprecipitated for in vitro kinase assay in the absence or presence of purified Chk1-S. As shown in Fig. 3D, Chk1 kinase activity was partially yet significantly suppressed by Chk1-S, but not by heat-denatured Chk1-S. In contrast, Chk1-S did not diminish Chk2 activity, which has similar substrate preferences as Chk1. Compared with Chk1, Chk1-S lacks part of the kinase domain including the ATP-binding site (Fig. 1D) and, as expected, did not show significant protein kinase activity (Fig. 3D). Chk1-S can also suppress the kinase activity of N-terminally tagged Chk1 (FLAG-Chk1) in an in vitro kinase assay (SI Appendix, Fig. 10). These results suggest that Chk1-S may inhibit Chk1 via molecular interaction. A recent study showed that washing of Chk1 immunoprecipitates with a more stringent Radio Immuno-Precipitation Assay (RIPA) buffer can markedly increase Chk1 kinase activity, suggesting that Chk1 is normally inhibited by repressing factors (29); however, the identity of the repressing factors is unknown. We confirmed the effect of RIPA buffer washing and, notably, we further showed that addition of exogenous Chk1-S could reverse the effect of RIPA buffer washing on Chk1 kinase activity (SI Appendix, Fig. 11), suggesting that Chk1-S may be one of the key endogenous repressing factors for Chk1.
Fig. 3.
Chk1-S interacts with Chk1 to suppress its kinase activity. (A) HEK293 cells were transfected with FLAG-Chk1-S or empty vector to collect lysate for immunoprecipitation (IP) using anti-FLAG antibodies. The immunoprecipitates were analyzed for Chk1 and FLAG-Chk1-S by immunoblotting using anti-FLAG and anti-N terminus Chk1 antibodies, respectively. The results show the co-IP of FLAG-Chk1-S with endogenous Chk1. (B) Chk1-Myc and Chk1-S were produced using the TnT in vitro transcription/translation kit (Promega). (Left) In vitro produced Chk1-Myc and Chk1-S shown by immunoblotting. (Right) Chk1-Myc was immunoprecipitated using anti-Myc antibodies after incubation with or without Chk1-S. The immunoprecipitates were analyzed for Chk1-S and Chk1-Myc by immunoblotting. The results indicate a direct interaction between Chk1 and Chk1-S. (C) HEK293 cells were transfected with Myc-tagged Chk1-S or its C- or N-terminal deletion mutants to collect lysate for immunoprecipitation using anti-Myc antibodies. The immunoprecipitates were examined for the presence of Chk1. The results demonstrate the coimmunoprecipitation of Chk1 with Chk1-S and its C-terminal deletion mutant, but not with its N-terminal deletion mutant. (D) HEK293 cells were transfected with Myc-tagged Chk1-S, Chk1, or Chk2 to collect lysate for immunoprecipitation using anti-Myc antibodies. The immunoprecipitates, containing Chk1-S-Myc, Chk1-Myc, or Chk2-Myc, were incubated with or without Chk1-S for 1 h and then added to the kinase activity assay using Chktide as substrate. Denatured Chk1-S was prepared by boiling. Data indicate mean ± SD; *P < 0.05 versus Chk1-Myc group. The results show that native Chk1-S can specifically inhibit Chk1. (E) HEK293 cells were transfected with Chk1-Myc or Chk1-Myc(S317A/S345A) mutant. The cells were then untreated or treated with 100 nM camptothecin for 2 h to collect lysate for immunoprecipitation with anti-Myc antibodies. The immunoprecipitates were analyzed for Myc-Chk1, phosphorylated (serine 345) Chk1, and Chk1-S by immunoblotting. Chk1 input was also verified in the samples. The results show that Chk1-S coimmunoprecipitated or associated with Chk1 in normal cells and that the association was diminished during camptothecin-induced DNA damage. However, the association between Chk1-S and the Chk1(S345A/S317A) mutant was not disrupted during DNA damage, suggesting that the phosphorylation of Chk1 at S345 and S317 may be required for the dissociation of Chk1-S from Chk1. (F) HEK293 cells were cotransfected with either Chk1-Myc + empty vector or Chk1-Myc + dn-ATR, followed by treatment with 100 nM camptothecin for 2 h. The cellular lysate was collected for immunoprecipitation with anti-Myc antibodies, followed by immunoblot analysis of the indicated proteins. The results show that camptothecin-induced disruption of Chk1–Chk1-S dissociation depends on ATR and Chk1 phosphorylation.
In DNA damage response (DDR), Chk1 is markedly activated via phosphorylation at S345 and S317 residues (7, 13, 38, 39). Despite recognition of the importance of S345/S317 phosphorylation, it remains unclear how this phosphorylation activates Chk1 (28–30). We showed that Chk1-S expression did not change appreciably in DDR (SI Appendix, Fig. 12). However, the Chk1–Chk1-S interaction was attenuated in the DDR induced by camptothecin, a DNA topoisomerase I inhibitor and DNA-damaging agent (Fig. 3E). Notably, the dissociation of Chk1 from Chk1-S in DDR appeared dependent on Chk1 phosphorylation at S345/S317, because the Chk1(S345A/S317A) mutant did not dissociate from Chk1-S during camptothecin treatment (Fig. 3E). In DDR, Chk1 activation depends on ATR-mediated phosphorylation (7, 13, 14). We showed that inhibition of ATR via a dominant-negative mutant (dn-ATR) not only suppressed camptothecin-induced Chk1-S phosphorylation but also prevented Chk1–Chk1-S dissociation (Fig. 3F). Similarly, Chk1 dissociated from Chk1-S during cisplatin-induced DDR, and the dissociation was prevented by dn-ATR (SI Appendix, Fig. 13). Together, the results suggest that phosphorylation of Chk1 may disrupt its interaction with the endogenous inhibitor Chk1-S, leading to Chk1 activation in DDR.
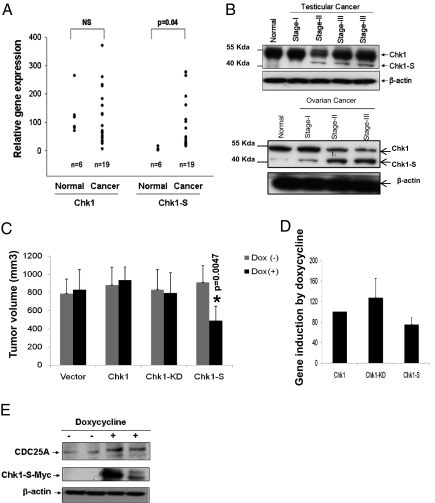
The identification of Chk1-S as a unique regulator of cell cycle progression and cellular proliferation prompted us to examine its expression in cancer tissues. At the mRNA level, most cancer tissues expressed higher levels of Chk1 and Chk1-S than normal tissues (SI Appendix, Fig. 14). Interestingly, testicular carcinomas showed a marked up-regulation of Chk1-S but not Chk1 (Fig. 4A). Specific up-regulation of Chk1-S was further confirmed by immunoblot analysis in testicular carcinoma tissues, especially in late-stage cancer samples (Fig. 4B). As reported previously (5), both normal and malignant testicular tissues had high levels of Chk1 expression. Increased expression of Chk1-S was also detected during the progression of ovarian cancer (Fig. 4B). The relatively high level of Chk1 and Chk1-S expression in both fetal (SI Appendix, Fig. 3) and cancer (Fig. 4) tissues suggests that Chk1-S may accelerate cell cycle progression, promoting cell proliferation under these conditions.
Fig. 4.
Chk1-S regulation in cancer. (A) Real-time PCR analysis of Chk1 and Chk1-S mRNA expression in normal testicular tissues and testicular carcinoma samples, showing up-regulation of Chk1-S in testicular carcinomas. (B) Immunoblot analysis of Chk1 and Chk1-S in human normal testicular and testicular cancer tissues, showing increased Chk1-S expression in late-stage cancer tissues. (C) Nude mice were injected with 10 × 106 Tet-on MDA-MB-231 cells that were doxycycline-inducible to express Chk1, Chk1-KD, or Chk1-S, respectively. After tumor establishment to ∼100 mm3, the mice were maintained on drinking water with or without doxycycline. Tumor volume was measured weekly (shown for fourth-week values, n = 8). Data indicate mean ± SD. The results show that induced expression of Chk1-S, but not Chk1 or Chk-KD, inhibited tumor growth. (D) Densitometry of immunoblot results of doxycycline-induced Chk1-Myc, Chk1-KD-Myc, and Chk1-S-Myc expression in excised tumors (n = 3 for each group). The signals were normalized with Chk1-Myc (arbitrarily set as 100). Data indicate mean ± SD. The results show that doxycycline induced similar levels of expression of Chk1-Myc, Chk1-KD-Myc, and Chk1-S-Myc in the tumors. (E) Nude mice were injected with Chk1-S-inducible cells to establish tumors and then maintained on drinking water with or without doxycycline for 4 wk. Tumor tissues were collected for immunoblot analysis. The results show higher CDC25A expression in the tumor xenografts with doxycycline-induced Chk1-S expression.
We further examined the effect of ectopic Chk1-S expression on xenograft tumor growth in mice. Tumor xenografts were established in nude mice using Tet-on MDA-MB-231 breast cancer cell lines that can be induced by doxycycline to express Chk1, Chk1-S, or Chk1-KD. Induction of Chk1 or Chk1-KD by doxycycline did not affect tumor growth; however, induction of Chk1-S resulted in a 40% reduction in tumor volume (Fig. 4C). Chk1-S–induced tumor tissues also showed cdc25A accumulation, indicating the inhibition of Chk1 (Fig. 4E). Although the results may imply a unique anticancer strategy, the physiological relevance of forced overexpression of ectopic Chk1-S remains unclear. These results, however, provide a proof of principle that tilting the Chk1–Chk1-S balance may lead to mitotic catastrophe and reduction of cell proliferation.
The observation that Chk1-S overexpression reduced xenograft tumor growth (Fig. 4 C–E) seemed contradictory to the observation that the tumor samples from human patients expressed relatively high levels of Chk1-S for cell proliferation (Fig. 4 A and B and SI Appendix, Fig. 14). To reconcile these data, it is important to recognize the differences between the experimental conditions. Relatively high Chk1-S expression in human tumors can be attributed to the presence of proliferative cells in these tissues. Consistently, Chk1-S is also high in fetal tissues that are highly proliferative (SI Appendix, Fig. 3A). Importantly, Chk1-S expression in these proliferative tissues is temporally restricted to the late S to M phase (Fig. 2 and SI Appendix, Fig. 4). This temporal regulation may ensure that Chk1 activity is inhibited after (and only after) the completion of DNA replication in S phase for cell cycle progression into G2/M phase. However, when ectopic Chk1-S is forced to overexpress in cultured cells or xenograft tumors, the temporal restriction of Chk1-S expression is disrupted; in other words, Chk1-S is high throughout the cell cycle including S phase, resulting in constant blockade of Chk-1 and premature mitotic entry, leading to mitotic catastrophe and reduced cell proliferation.
In conclusion, this study has identified a splice variant of Chk1, Chk1-S, that is a key regulator of Chk1 in the normal cell cycle and during DDR (SI Appendix, Fig. 15). In the unperturbed cell cycle, Chk1 expression increases at S phase and some Chk1 molecules might be phosphorylated by ATR preventing Chk1-S binding, resulting in high Chk1 activity and S-phase maintenance until the completion of DNA replication. In G2 phase, Chk1-S expression increases and, as reported (29), Chk1 phosphorylation decreases, promoting a Chk1–Chk1-S interaction to suppress Chk1 activity. The decrease in Chk1 activity at G2/M phase is critical to mitotic entry. Under conditions of Chk1-S induction or overexpression, excessive amounts of Chk1-S sequester Chk1 and diminish its kinase activity during S phase, resulting in premature mitotic entry without completion of DNA replication, leading to mitotic catastrophe. During DNA damage, Chk1 is phosphorylated, resulting in decreased Chk1-S binding, increased Chk1 activity, and G2/M arrest (SI Appendix, Fig. 11). Our findings support the “de-repression” mechanism of Chk1 activation (29). Importantly, Chk1-S appears to be one of the key repressing factors of Chk1 activity. By antagonizing Chk1, Chk1-S may accelerate the cell cycle, leading to increased proliferation in fetal and cancerous tissues. High levels of Chk1 in proliferating cells coordinate S phase and mitosis, whereas high levels of Chk1-S may provide a powerful switch for the S-to-G2/M phase transition. In contrast, forced overexpression of Chk1-S at excessive levels, unbalanced by Chk1, induces premature mitotic entry and cell death (Fig. 2) and suppresses tumor growth in tumor xenograft models (Fig. 4). Chk1 has been suggested to be an effective therapeutic target in cancer therapy, and Chk1 inhibitors are being evaluated in clinical trials (40–43). The identification of Chk1-S as an endogenous inhibitor of Chk1 may open new areas of research in cell cycle regulation, DNA damage response, and cancer therapy.
Materials and Methods
Alternative splicing of Chk1 was analyzed by immunoblotting, RT-PCR, and sequencing. Chk1 and Chk1-S expression in various cell types and tissues was determined by real-time PCR and immunoblot analysis. The role of Chk1-S in cell cycle regulation was examined in asynchronous or cell cycle synchronized cells. Aberrant mitotic entry in cells overexpressing Chk1-S protein was shown by microscopic, biochemical, and FACS analyses. The interaction between Chk1 and Chk1-S was verified by coimmunoprecipitation analysis of cellular lysates and in vitro translated proteins. Tumor xenograft models inducibly expressing Chk1 or Chk1-S were used to determine the effect of Chk1-S overexpression in tumors. More details are included in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Piwnica-Worms at Washington University School of Medicine for the pcDNA3-Chk1-Myc and pcDNA3-Chk1-KD-Myc plasmids, Dr. Hopfer at Case Western Reserve University for the RPTC cell line, and Dr. Lieberthal and Dr. Shwartz at Boston University for the BUMPT cell line. The study was supported by grants from the National Institutes of Health and Veterans Administration (Z.D.) and a predoctoral fellowship from the American Heart Association (K.B.). Z.D. is a Veterans Administration Research Career Scientist.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence of Chk1-S reported in this paper has been deposited in the GenBank database (accession no. JF289264).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104767109/-/DCSupplemental.
References
- 1.Nurse P. A long twentieth century of the cell cycle and beyond. Cell. 2000;100(1):71–78. doi: 10.1016/s0092-8674(00)81684-0. [DOI] [PubMed] [Google Scholar]
- 2.Paulovich AG, Toczyski DP, Hartwell LH. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 3.Zhou BB, Elledge SJ. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 4.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez Y, et al. Conservation of the Chk1 checkpoint pathway in mammals: Linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 6.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 7.Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 8.Takai H, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 9.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 10.Walworth NC. Cell-cycle checkpoint kinases: Checking in on the cell cycle. Curr Opin Cell Biol. 2000;12:697–704. doi: 10.1016/s0955-0674(00)00154-x. [DOI] [PubMed] [Google Scholar]
- 11.Zachos G, Gillespie DA. Exercising restraints: Role of Chk1 in regulating the onset and progression of unperturbed mitosis in vertebrate cells. Cell Cycle. 2007;6:810–813. doi: 10.4161/cc.6.7.4048. [DOI] [PubMed] [Google Scholar]
- 12.Zaugg K, et al. Cross-talk between Chk1 and Chk2 in double-mutant thymocytes. Proc Natl Acad Sci USA. 2007;104:3805–3810. doi: 10.1073/pnas.0611584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng CY, et al. Mitotic and G2 checkpoint control: Regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 17.O'Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Kumagai A, Dunphy WG. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Mol Biol Cell. 2001;12:551–563. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397(6715):172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 20.Mailand N, et al. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 21.Feijoo C, et al. Activation of mammalian Chk1 during DNA replication arrest: A role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sørensen CS, et al. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–258. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 23.Petermann E, et al. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26:3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zachos G, et al. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–260. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang J, Erikson RL, Liu X. Checkpoint kinase 1 (Chk1) is required for mitotic progression through negative regulation of polo-like kinase 1 (Plk1) Proc Natl Acad Sci USA. 2006;103:11964–11969. doi: 10.1073/pnas.0604987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilsker D, Petermann E, Helleday T, Bunz F. Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc Natl Acad Sci USA. 2008;105:20752–20757. doi: 10.1073/pnas.0806917106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci USA. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsuragi Y, Sagata N. Regulation of Chk1 kinase by autoinhibition and ATR-mediated phosphorylation. Mol Biol Cell. 2004;15:1680–1689. doi: 10.1091/mbc.E03-12-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker M, Black EJ, Oehler V, Gillespie DA, Scott MT. Chk1 C-terminal regulatory phosphorylation mediates checkpoint activation by de-repression of Chk1 catalytic activity. Oncogene. 2009;28:2314–2323. doi: 10.1038/onc.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tapia-Alveal C, Calonge TM, O'Connell MJ. Regulation of Chk1. Cell Div. 2009;4:8. doi: 10.1186/1747-1028-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen P, et al. The 1.7 Å crystal structure of human cell cycle checkpoint kinase Chk1: Implications for Chk1 regulation. Cell. 2000;100:681–692. doi: 10.1016/s0092-8674(00)80704-7. [DOI] [PubMed] [Google Scholar]
- 32.Pabla N, Huang S, Mi QS, Daniel R, Dong Z. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J Biol Chem. 2008;283:6572–6583. doi: 10.1074/jbc.M707568200. [DOI] [PubMed] [Google Scholar]
- 33.Pabla N, Ma Z, McIlhatton MA, Fishel R, Dong Z. hMSH2 recruits ATR to DNA damage sites for activation during DNA damage-induced apoptosis. J Biol Chem. 2011;286:10411–10418. doi: 10.1074/jbc.M110.210989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krämer A, Lukas J, Bartek J. Checking out the centrosome. Cell Cycle. 2004;3:1390–1393. doi: 10.4161/cc.3.11.1252. [DOI] [PubMed] [Google Scholar]
- 35.Krämer A, et al. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 36.Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6(1):45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 37.Fishler T, et al. Genetic instability and mammary tumor formation in mice carrying mammary-specific disruption of Chk1 and p53. Oncogene. 2010;29:4007–4017. doi: 10.1038/onc.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capasso H, et al. Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J Cell Sci. 2002;115:4555–4564. doi: 10.1242/jcs.00133. [DOI] [PubMed] [Google Scholar]
- 39.Latif C, den Elzen NR, O'Connell MJ. DNA damage checkpoint maintenance through sustained Chk1 activity. J Cell Sci. 2004;117:3489–3498. doi: 10.1242/jcs.01204. [DOI] [PubMed] [Google Scholar]
- 40.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–383. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merry C, Fu K, Wang J, Yeh IJ, Zhang Y. Targeting the checkpoint kinase Chk1 in cancer therapy. Cell Cycle. 2010;9:279–283. doi: 10.4161/cc.9.2.10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole KA, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci USA. 2011;108:3336–3341. doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17(2):88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.