Abstract
Binding of EGF to its receptor induces dimerization of the normally monomeric receptor. Activation of its intracellular tyrosine kinase then occurs through the formation of an asymmetric kinase dimer in which one subunit, termed the “receiver” kinase, is activated by interaction with the other subunit, termed the “activator” kinase [Zhang, et al. (2006) Cell 125: 1137–1149]. Although there is significant experimental support for this model, the relationship between ligand binding and the mechanics of kinase activation are not known. Here we use luciferase fragment complementation in EGF receptor (EGFR)/ErbB2 heterodimers to probe the mechanics of ErbB kinase activation. Our data support a model in which ligand binding causes the cis-kinase (the EGFR) to adopt the receiver position in the asymmetric dimer and to be activated first. If the EGF receptor is kinase active, this results in the phosphorylation of the trans-kinase (ErbB2). However, if the EGF receptor kinase is kinase dead, the ErbB2 kinase is never activated. Thus, activation of the kinases in the EGFR/ErbB2 asymmetric dimer occurs in a specific sequence and depends on the kinase activity of the EGF receptor.
The EGF receptor is a member of the ErbB family of receptor tyrosine kinases that also includes ErbB2, ErbB3, and ErbB4 (1, 2). All ErbB receptors contain an extracellular ligand-binding domain, a single pass transmembrane domain, an intracellular tyrosine kinase, and a C-terminal tail (3). The EGF receptor, ErbB3, and ErbB4 are activated through the binding of a family of homologous ligands (4). Unique among the ErbB receptors, ErbB2 has no known ligand (5, 6).
Binding of an ErbB ligand to its receptor induces dimerization of the receptor through interaction of the extracellular domains. Essentially all combinations of ErbB receptors are possible. However, the ligandless ErbB2 appears to be the preferred heterodimerization partner among ErbB receptors (7, 8).
Ligand-induced dimerization of the extracellular domains leads to the activation of the intracellular tyrosine kinase through the formation of an asymmetric kinase dimer (9–11). In the asymmetric dimer, the C lobe of the activator kinase interacts with the N lobe of the receiver kinase in a manner similar to that in which cyclin A interacts with cyclin-dependent kinase (12). This interaction leads to the activation of the receiver kinase, which then phosphorylates the C-terminal tail of the activator kinase.
Although a substantial body of evidence supports this model for EGF receptor kinase activation (9–11, 13), it is not clear how the binding of ligand to the extracellular domain directs the assembly of the intracellular asymmetric kinase dimer. In particular, if ligand binds to one subunit in an ErbB dimer, which kinase domain adopts the activator and which adopts the receiver position in the asymmetric dimer, or is the choice made randomly? Once formed, does the asymmetric kinase dimer readily shift from one configuration to the reciprocal one, activating each kinase in turn, or is this a controlled process?
We have previously used luciferase fragment complementation imaging to monitor the dimerization of and conformational dynamics in the EGF receptor in real time in live cells (14, 15). Stimulation of full-length receptors with EGF leads to a rapid decrease in luciferase activity followed by a slower recovery back to baseline levels. The results suggest that luciferase complementation can take place in EGF receptor predimers (16–20). Upon addition of EGF, a conformational change occurs that initially separates the luciferase fragments but subsequently they are brought back into proximity. These conformational dynamics are strictly dependent on kinase activation. In the kinase-dead EGF receptor, where dimerization occurs but no phosphorylation takes place, only a monotonic rise in luciferase activity is observed (14). Thus, the pattern of luciferase complementation is distinctly different for wild-type and kinase-dead receptors.
In the work reported here, we use luciferase fragment complementation imaging to monitor the ability of the EGF receptor to interact with and activate the ligandless ErbB2. We find that EGF induces the same conformational dynamics in the kinase-active EGF receptor (EGFR)/ErbB2 heterodimer as it does in the kinase-active EGF receptor homodimer. Similarly, EGF elicited the characteristic monotonic rise in luciferase activity in the kinase-dead version of the EGF receptor/ErbB2 heterodimer. Interestingly, in “half-dead” heterodimers containing one wild-type and one kinase-dead receptor, the pattern of luciferase activity was determined by the activity status of the kinase domain of the ligand-binding EGF receptor subunit. Assays of kinase and signaling activities in the half-dead pairs were consistent with the results of the luciferase assays.
The data support a model in which ligand binding directs the EGF receptor to adopt the receiver position in the asymmetric dimer and to phosphorylate ErbB2. In the absence of this phosphorylation event, the ErbB2 kinase domain is never activated. These observations provide key mechanistic insight into the activation and functioning of the EGF receptor/ErbB2 asymmetric kinase dimer.
Results
Dimerization of the EGF Receptor and ErbB2.
Firefly luciferase can be split into an N-terminal fragment and a C-terminal fragment, neither one of which exhibits enzymatic activity on its own. However, when the fragments are brought together, they interact to form a functional luciferase enzyme (21, 22) (Fig. S1). Thus, luciferase fragment complementation can be used to assess changes in the proximity of the proteins to which the fragments have been fused.
The N-terminal fragment of firefly luciferase (NLuc) was fused to the C terminus of a truncated version of ErbB2 that contained only the extracellular and transmembrane domains of that receptor (ΔC-B2-NLuc). The C-terminal fragment of firefly luciferase (CLuc) was fused to the C terminus of a similarly truncated EGF receptor (ΔC-B1-CLuc) (see Fig. S1 for constructs used in these experiments). For sake of simplicity, the EGF receptor is referred to as “B1” in all constructs and ErbB2 is referred to as “B2”.
CHO cells do not express EGF receptors and express barely detectable levels of endogenous ErbB2. They were therefore used as the parental cell line in these experiments. CHO cells were transfected with ΔC-B1-CLuc and ΔC-B2-NLuc, and a double-stable cell line was selected. Because a ΔC-B2-NLuc must partner with a ΔC-B1-CLuc to reconstitute an active luciferase, this system only reports on heterodimer formation. However, it is possible that heterodimer complementation is occurring within the context of larger oligomers. Although EGF receptor homodimers may form, they do not produce a signal. We have previously generated the equivalent double-stable ΔC-B1-NLuc/ΔC-B1-CLuc CHO cell line for monitoring dimerization in the truncated homodimer (14) and that line was used here for comparison purposes.
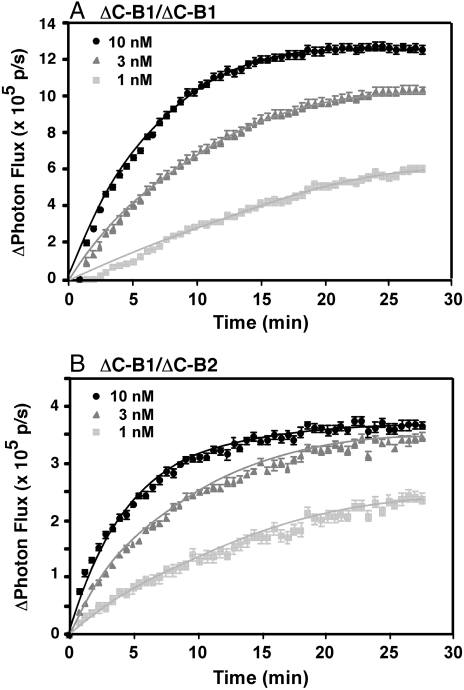
Both cell lines were assayed for EGF-stimulated luciferase activity. Addition of EGF to CHO cells expressing ΔC-B1-NLuc/ΔC-B1-CLuc homodimers led to a dose-dependent increase in luciferase activity (Fig. 1A). These data indicate that, as expected, EGF induces the formation of EGF receptor homodimers, resulting in enhanced complementation of the luciferase fragments. Similar findings were observed with the ΔC-B2-NLuc/ΔC-B1-CLuc heterodimeric pair (Fig. 1B). Both the time course and the dose response to EGF were similar in the truncated EGF receptor homodimers and the truncated EGFR/ErbB2 heterodimers.
Fig. 1.
Luciferase fragment complementation using truncated forms of the EGF receptor and ErbB2. CHO cells stably expressing ΔC-EGFR-NLuc and ΔC-EGFR-CLuc (A) or ΔC-ErbB2-NLuc and ΔC-EGFR-CLuc (B) were plated in 96-well, black-wall dishes 48 h prior to assay. For assay, cultures were incubated with 0.6 mg/mL d-luciferin for 20 min prior to the addition of the indicated concentrations of EGF. Photon flux in the absence or presence of EGF was monitored over time. The change in photon flux was calculated by subtracting the average photon flux in the absence of EGF from the photon flux observed in the presence of EGF. The data represent the average of five independent replicates and the standard error is shown.
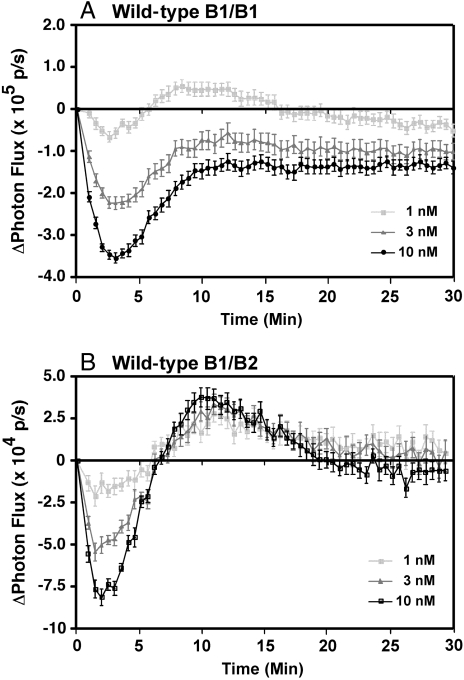
Similar experiments were performed using stable cell lines expressing full-length EGF receptors and full-length ErbB2 fused to the luciferase fragments. The results are shown in Fig. 2. As we have reported previously (14, 15), addition of EGF to cells expressing B1-NLuc/B1-CLuc homodimers resulted in a rapid dose-dependent decrease in luciferase activity followed by a slow recovery toward baseline levels (Fig. 2A). A similar pattern was seen in cells expressing the B2-NLuc/B1-CLuc heterodimers (Fig. 2B). However, luciferase activity in the heterodimers consistently recovered to a level above baseline before dropping back to basal levels of complementation. This result may be because of greater stability or more extensive dimerization of the heterodimers. As with the truncated receptors, both the kinetics and the dose response to EGF for the heterodimerization of the EGF receptor and ErbB2 were comparable to those for the homodimerization of the EGF receptor.
Fig. 2.
Luciferase fragment complementation using full-length EGF receptor and full-length ErbB2. CHO cells stably expressing EGFR-NLuc and EGFR-CLuc (A) or ErbB2-NLuc and EGFR-CLuc (B) were plated in 96-well, black-wall dishes 48 h prior to assay. For assay, cultures were incubated with 0.6 mg/mL d-luciferin for 20 min prior to the addition of the indicated concentrations of EGF. Photon flux in the absence or presence of EGF was monitored over time. The data represent the average of five independent replicates and the standard error is shown.
Complementation in Kinase-Dead and Mixed Wild-Type/Kinase-Dead Pairs.
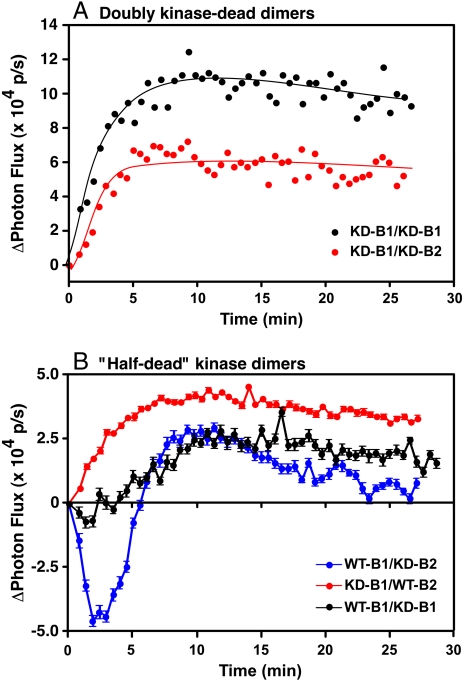
The conformational dynamics observed in the full-length homo- and heterodimers are dependent on the kinase activity of the receptors. In cells expressing full-length, kinase-dead EGF receptors fused to the luciferase fragments (KD-B1/KD-B1), addition of EGF resulted in a monotonic rise in luciferase activity (Fig. 3A), similar to what was seen when the C-terminally truncated EGF receptor constructs were used. As we have noted previously (14), this result suggests that EGF induces dimerization of the kinase-dead EGF receptors but the intracellular conformational dynamics do not occur because they require activation of the receptor tyrosine kinase. EGF stimulated a similar monotonic increase in luciferase activity in cells expressing the full-length, kinase-dead EGF receptor paired with full-length, kinase-dead ErbB2 and fused to the appropriate luciferase fragments (KD-B1/KD-B2). Thus, the kinase-dead homodimers and heterodimers both yield a pattern of luciferase complementation that is distinct from that observed for the wild-type dimers.
Fig. 3.
Luciferase fragment complementation in kinase-dead dimers and dimers containing wild-type and kinase-dead EGF receptors and ErbB2. (A) Luciferase complementation in response to 10 nM EGF in CHO cells that were transiently transfected with kinase-dead versions of the EGF receptor or ErbB2 prior to assay. Black line, cells expressing K721A-EGFR-NLuc and K721A-EGFR-CLuc. Red line, cells expressing K721A-EGFR-CLuc and K732M-ErbB2-NLuc. (B) Luciferase complementation in response to 10 nM EGF in CHO cells expressing mixed pairs of wild-type and kinase-dead receptors. Blue line, wild-type EGFR-CLuc and K732M-ErbB2-NLuc (WT-B1/KD-B2); red line, K721A-EGFR-CLuc and wild-type ErbB2-NLuc (KD-B1/WT-B2); or black line, K721A-EGFR-NLuc and wild-type EGFR-CLuc.
Several questions arise when considering the EGF-induced activation of the tyrosine kinase domains in the EGFR/ErbB2 heterodimer. First, is there a particular order of activation of the kinases or is the selection of the activator and receiver positions in the asymmetric dimer random? Second, if the order is not random, is the kinase of the ligand-binding EGF receptor subunit activated first (the cis-kinase) or is the kinase in the non-ligand-binding ErbB2 subunit (the trans-kinase) activated first?
To address these questions, we generated two double-stable CHO cell lines in which one of the heterodimeric partners was wild type, and the other partner was kinase-dead. In cells expressing wild-type EGF receptor but kinase-dead ErbB2 (WT-B1/KD-B2), addition of EGF resulted in a pattern of luciferase activity similar to that seen in dimers in which both partners had wild-type kinase activity (Fig. 3B, blue line). There was a rapid decrease in luciferase activity followed by a slow recovery back to baseline levels. Because luciferase complementation requires interaction of the NLuc and CLuc fragments, this result reflects the behavior of only the EGFR/ErbB2 heterodimers. The EGF receptor homodimers, which would be entirely wild type, do not produce a signal in this assay. Thus, the heterodimers containing wild-type EGF receptors but kinase-dead ErbB2 exhibit fully wild-type conformational dynamics.
In contrast to the behavior of the WT-B1/KD-B2 pair, in cells expressing kinase-dead EGF receptors and wild-type ErbB2 (KD-B1/WT-B2), addition of EGF led to a slow, monotonic rise in luciferase activity (Fig. 3B, red line). This pattern is characteristic of the luciferase complementation seen in dimers in which both partners were kinase-dead. Thus, the pattern of luciferase complementation in the two sets of half-dead heterodimers is different and appears to be determined by the activity status of the EGF receptor kinase domain.
The kinase-dead K721A mutation of the EGF receptor used here impairs kinase activity because it precludes the formation of a salt bridge with Glu-738 that stabilizes the α-C helix in its active conformation (9, 11, 23). This mutation may also impact the ability of the subunit to serve as a receiver kinase, enhancing its kinase-dead phenotype. However, the K721A-EGF receptor is perfectly capable of serving as an activator kinase. Thus, the failure of this kinase-dead receptor to activate the ErbB2 kinase in the half-dead heterodimer is not because of its inability to form an activating asymmetric dimer with ErbB2 in the receiver position.
One difference between the EGF receptor and ErbB2 is that the EGF receptor binds ligand whereas ErbB2 does not. Therefore, the difference in luciferase complementation observed between the WT-B1/KD-B2 and KD-B1/WT-B2 pairings could be related to whether the ligand-binding subunit is kinase-active or kinase-dead. To further explore the relationship between ligand binding and the pattern of luciferase complementation, a cell line coexpressing wild-type EGFR-NLuc and kinase-dead EGFR-CLuc (WT-B1/KD-B1) was generated. This pairing differs from the half-dead heterodimers because, in the half-dead EGF receptor homodimer, both subunits can bind ligand. In the half-dead heterodimers, only one subunit can bind ligand. Addition of EGF to this half-dead homodimer cell line (Fig. 3B, black line) resulted in a phenotype that was distinct from that observed for either of the half-dead EGFR/ErbB2 heterodimers. The initial rapid decrease in luciferase activity was largely absent, but a slow rise in activity occurred over a time course similar to that of the recovery of activity in the WT-B1/KD-B2 pairing. This phenotype can be viewed as intermediate between the fully wild-type and the fully kinase-dead patterns. Importantly, this phenotype is different from that observed for either of the half-dead heterodimers, indicating that the ability of both subunits of the dimer to bind ligand gives rise to a different pattern of luciferase complementation. Only in the KD-B1/WT-B2 pairing, in which ligand binding is restricted to the kinase-dead subunit, is the fully kinase-dead phenotype in luciferase activity observed.
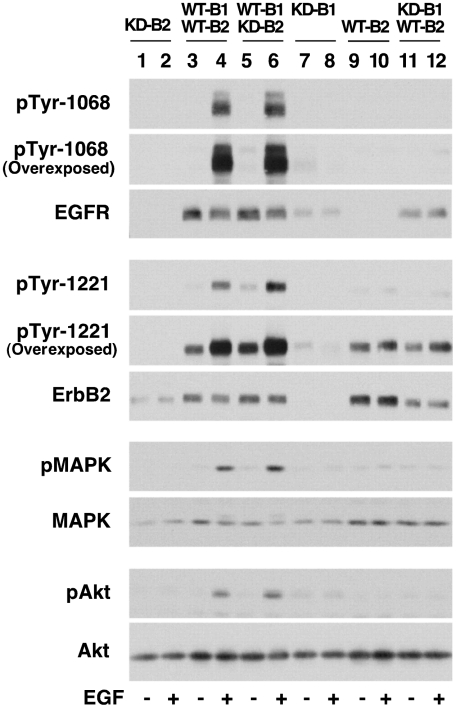
Based on the patterns of EGF-stimulated luciferase activity, our data would suggest that the WT-B1/KD-B2 pair should be kinase-active, and the KD-B1/WT-B2 pair should be kinase-dead. To test this prediction, EGF-stimulated tyrosine phosphorylation of the EGF receptor and ErbB2 in the half-dead heterodimers was determined along with the ability of these pairings to mediate the activation of MAP kinase and Akt. The results are shown in Fig. 4.
Fig. 4.
Signaling in cells expressing mixed wild-type and kinase-dead EGFR/ErbB2 heterodimers. CHO cells expressing the indicated pairs of EGF receptors and/or ErbB2 were treated without or with 10 nM EGF for 2 min. Lysates were prepared and proteins separated by SDS-polyacrylamide gel electrophoresis followed by Western blotting with the indicated antibodies. Because the NLuc fragment interferes with recognition of ErbB2 on Western blots, these experiments were carried out in cells stably expressing receptors that lacked the luciferase fragments.
Expression of kinase-dead ErbB2 by itself in CHO cells resulted in no phosphorylation of ErbB2 and no activation of MAP kinase or Akt (Fig. 4, lanes 1 and 2). Thus, kinase-dead ErbB2 is completely inactive. As expected, when wild-type EGF receptor and wild-type ErbB2 were coexpressed in CHO cells, both receptors were tyrosine phosphorylated in response to EGF and there was strong activation of MAP kinase and Akt (Fig. 4, lanes 3 and 4). Coexpression of wild-type EGF receptor with kinase-dead ErbB2 also resulted in strong EGF-dependent tyrosine phosphorylation of both the EGF receptor and ErbB2 (Fig. 4, lanes 5 and 6). In addition, the activity of both MAP kinase and Akt was stimulated in response to EGF.
In this half-dead heterodimer, phosphorylation of the kinase-dead ErbB2 presumably occurs as a result of heterodimer formation with the wild-type EGF receptor. This observation indicates that ErbB2 does not need to be kinase active to serve as a substrate for the EGF receptor. Autophosphorylation of the EGF receptor would presumably take place in the context of homodimers of wild-type EGF receptors. Thus, the fact that the level of EGF receptor phosphorylation was high and that MAP kinase and Akt were strongly activated in this half-dead heterodimer does not provide diagnostic information on the function of the heterodimer. However, the finding that ErbB2 is phosphorylated normally is consistent with the results of the luciferase assay that show wild-type conformational dynamics in this pairing.
In cells expressing only the kinase-dead EGF receptor (Fig. 4, lanes 7 and 8), addition of EGF failed to induce receptor autophosphorylation, even after very long exposures of the blots. Furthermore, there was no activation of either MAP kinase or Akt, indicating that this mutant receptor is inactive. Expression of wild-type ErbB2 alone (Fig. 4, lanes 9 and 10) resulted in very weak phosphorylation of ErbB2 that could only be visualized by long-term exposure of the blots, and there was no activation of MAP kinase or Akt. Neither receptor autophosphorylation nor downstream signaling was affected by the addition of EGF, indicating that the EGF receptor must be present to stimulate signaling via ErbB2. Surprisingly, despite the presence of an EGF receptor to bind ligand and a wild-type ErbB2 kinase domain to phosphorylate substrates, no EGF-stimulated receptor phosphorylation occurred when wild-type ErbB2 was coexpressed with kinase-dead EGF receptors (Fig. 4, lanes 11 and 12). The EGF receptor, which could potentially be phosphorylated by the wild-type ErbB2 in the heterodimer, was not phosphorylated in this half-dead pairing. Likewise, the level of ErbB2 phosphorylation was very low when compared to what was seen in either the fully wild-type heterodimer (Fig. 4, lanes 3 and 4) or the half-dead heterodimer containing wild-type EGF receptor but kinase-dead ErbB2 (Fig. 4, lanes 5 and 6). In fact, the level of ErbB2 phosphorylation was similar to what was seen when ErbB2 was expressed by itself in cells (Fig. 4, lanes 10 and 11) and was not stimulated by the addition of EGF. In keeping with the low levels of receptor phosphorylation seen in this half-dead heterodimer, addition of EGF failed to stimulate the activity of either MAP kinase or Akt. This lack of receptor autophosphorylation and downstream signaling is consistent with the kinase-dead phenotype observed for this pairing in the luciferase assay.
Discussion
ErbB receptors have been shown to be activated through the formation of an asymmetric kinase dimer (9–11). However, the mechanics of asymmetric dimer formation and its relationship to the ligand-binding event are open questions. In this work, we used luciferase fragment complementation to address these issues. Two features of this work enable an unequivocal interpretation of the effects of ligand binding on kinase activation. First, we studied the activation of the kinase domains within the EGF receptor/ErbB2 heterodimer in which only one subunit bound ligand. Thus, in the half-dead heterodimers, it is clear whether it is the kinase-active or kinase-dead subunit that is binding ligand. Second, the analyses were done using a luciferase fragment complementation assay in which a signal is generated only when the NLuc and CLuc fragments come into proximity. This approach permits the assessment of the behavior of only the EGF receptor/ErbB2 heterodimers, even in the presence of EGF receptor homodimers.
The results reported here demonstrate that EGF receptor/ErbB2 heterodimers behave very similarly to EGF receptor homodimers in terms of dimerization and kinase activation, consistent with the notion that the EGF receptor and ErbB2 are largely functionally interchangeable. Both wild-type homodimers and the wild-type EGFR/ErbB2 heterodimers exhibited a rapid decrease followed by a slow recovery in luciferase activity following stimulation by EGF. We interpret this observation as indicating the presence of complementation within inactive predimers that is disrupted following agonist-stimulated kinase activation. Phosphorylation of the C-terminal tail of the activator kinase apparently leads to a conformational change that separates the luciferase fragments, leading to a reduction in luciferase complementation. The data presented here do not address the molecular basis of the recovery of luciferase activity after its initial decline. The recovery may relate to the phosphorylation status of the receptor because mutants that exhibit enhanced or prolonged phosphorylation have a delayed or absent recovery phase (14, 15).
When both partners in the homodimers or heterodimers were mutated to their kinase-dead versions, stimulation with EGF resulted in a monotonic rise in luciferase activity. These data suggest that EGF induces dimerization of the kinase-dead subunits but that, in the absence of a phosphorylation event, the receptors are trapped in a nonfunctional complex and the steps that lead to the repositioning of the C-terminal tails do not occur.
We took advantage of these easily distinguishable patterns of behavior to explore the relationship between ligand binding and the mechanics of kinase activation using half-dead heterodimers. Although coexpression of wild-type EGF receptor and kinase-dead ErbB2 resulted in entirely wild-type conformational dynamics in the luciferase assay, heterodimers in which a kinase-dead EGF receptor was paired with a wild-type ErbB2 exhibited a monotonic rise in luciferase activity characteristic of pairings when both partners were kinase dead, suggesting that EGF can induce the dimerization of the EGF receptor and ErbB2 subunits, but that the presence of a kinase-dead EGF receptor renders the entire heterodimer effectively kinase dead. These findings imply that, in an EGFR/ErbB2 heterodimer, ligand-binding signals the EGF receptor kinase to adopt the receiver position in the asymmetric dimer and to be activated first. The data also suggest that the EGF receptor must phosphorylate the C-terminal tail of ErbB2 before the ErbB2 kinase domain can be activated. Receptor phosphorylation and the activation of downstream signaling pathways were not different in cells expressing WT-EGFR/WT-ErbB2 and cells expressing WT-EGFR but kinase-dead ErbB2, which can be ascribed to the activity of wild-type EGF receptor homodimers. However, it remains a formal possibility that the ErbB2 kinase domain is never actually activated in a wild-type EGFR/ErbB2 heterodimer and that all the phosphorylation is carried out by the EGF receptor kinase.
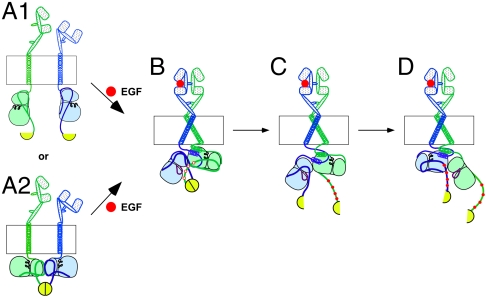
Together our findings suggest the model for the mechanics of kinase activation in the EGFR/ErbB2 heterodimer that is shown in Fig. 5. In the basal state, receptors exist either as monomers in which no luciferase complementation occurs (Fig. 5A1) or as inactive predimers in which complementation of the luciferase fragments can occur (Fig. 5A2). Binding of ligand to the EGF receptor induces dimerization of the receptor with ErbB2 and directs the kinase domain of the EGF receptor (the cis-kinase) to adopt the receiver position in the asymmetric dimer. Thus, the EGF receptor kinase is activated first (Fig. 5B). If the cis-kinase domain is catalytically competent, it phosphorylates the C-terminal tail of ErbB2, the trans-kinase. This transphosphorylation leads to a change in the position of the C-terminal tail of ErbB2, resulting in the separation of the luciferase fragments and a decrease in luciferase activity (Fig. 5C). This phosphorylation-induced conformational change must occur prior to the formation of the reciprocal asymmetric dimer in which the kinase domain of ErbB2 is activated (Fig. 5D). If the cis-kinase cannot phosphorylate its partner, the conformational change cannot occur and the trans-kinase domain is never activated.
Fig. 5.
Model for the activation of the kinase domains in the EGF receptor/ErbB2 heterodimer. The EGF receptor is shown as the blue subunit, and ErbB2 is shown as the green subunit. The red circles represent the EGF ligand. The C-terminal luciferase fragments are shown as yellow semicircles. See text for complete description of the model.
Variations on this mechanism are likely utilized by other ErbB receptor dimers. For example, results from our recent 125I-EGF ligand-binding studies are consistent with the interpretation that, in the EGF receptor homodimer, the binding of ligand to the second site on the dimer provides the final driving force for the shift to the reciprocal asymmetric dimer and the activation of the second kinase domain (24). If this model applies across all heterodimers, activation of the ErbB2/ErbB3 heterodimer by ErbB3 ligands would appear to be problematic. The ligand-binding ErbB3 subunit has been reported to be kinase dead (25), so it would be unable to carry out the initial transphosphorylation of ErbB2, which would result in the formation of a nonproductive complex with ErbB3 in the receiver position. However, recent work has demonstrated that ErbB3 does in fact possess a weak kinase activity that is capable of trans-phosphorylation within heterodimers (26). This activity may be sufficient to permit the shift to the reciprocal asymmetric dimer in which ErbB2 occupies the receiver position. Alternatively, in the ErbB2/ErbB3 heterodimer, ligand binding could direct ErbB3 into the activator position, obviating the problems associated with its lack of kinase activity.
These results have been interpreted based on the most parsimonious model of the luciferase fragments complementing each other within the context of a dimer. Given that there is evidence for the formation of higher order oligomers (16, 20, 27, 28), it is possible that the complementation that we are measuring occurs, not within a dimer, but between two dimers in a tetramer or other oligomer. Although this feature would make the physical model more complex, it does not change the overall interpretation of the data. Regardless of whether the receptors are in a simple dimer or a dimer of dimers, the kinase domain of the ligand-binding subunit must be active to enable wild-type conformational dynamics.
In summary, our results indicate that activation of the kinase domains in the EGFR/ErbB2 heterodimer occurs in a defined sequence. Ligand binding directs the EGF receptor kinase domain to adopt the receiver position and to be the first kinase activated in the heterodimer. If the EGF receptor is kinase active, this activation is followed by the phosphorylation of the ErbB2 subunit. However, if the EGFR kinase is kinase dead, phosphorylation of ErbB2 does not occur and the ErbB2 kinase domain is never activated. These findings provide insight into the mechanics of activation of ErbB kinases and may be valuable in the design of inhibitors of these validated drug targets.
Materials and Methods
DNA Constructs.
The ErbB2-NLuc construct was generated by engineering a BsiWI site at the 3′ end of the wild-type ErbB2 sequence in pcDNA3. This plasmid was digested with NheI and BsiWI, and the fragment containing ErbB2 was isolated. The EGFR-NLuc pBI-Tet vector (14) was digested with the same restriction enzymes and the NheI-BsiWI ErbB2 fragment ligated into the plasmid in place of the EGF receptor to generate the ErbB2-NLuc pBI-Tet vector. A similar approach was used to generate the ΔC-ErbB2-NLuc plasmid from the ΔC-EGFR-NLuc plasmid (14). Both were expressed from the pBi-Tet vector. The ΔC-EGFR-CLuc was on pcDNA6. The kinase-dead K732M-ErbB2-NLuc pBI-Tet construct was generated from the ErbB2-NLuc pBI-Tet plasmid using Quik Change site-directed mutagenesis (Stratagene).
Cell Lines.
For the wild-type and half-dead pairs, double-stable cell lines were generated. CHO-K1 Tet-on cells (Clontech) were transfected with either wild-type EGFR-NLuc, wild-type ErbB2-NLuc, or kinase-dead-ErbB2-NLuc in pBI-Tet plus pTK-Hyg. Clones were selected by growth in 400 μg/mL hygromycin (Invitrogen). Single stable cell lines were then transfected with wild-type or kinase-dead EGF receptor-CLuc on pcDNA3.1(+)/Zeo and stable lines selected by growth in 500 μg/mL Zeocin.
Transient transfections were used for the double kinase-dead experiments. The K721A-EGFR-NLuc and the K732M-ErbB2-NLuc were in the pBI-Tet vector. The K721A-EGFR-CLuc was in pcDNA 3.1. The plasmids were transiently transfected into CHO-K1 cells as described previously (14). Transfection efficiency was monitored by cotransfection with and assay for Renilla luciferase (14).
Luciferase Assays.
Cells were plated in 96-well black-walled dishes 48 h prior to use. Cultures were transferred to Dulbecco’s modified Eagle’s medium without serum or phenol red but with 1 mg/mL bovine serum albumin and incubated with 0.6 mg/mL d-luciferin for 20 min at 37 °C. EGF was then added at the concentration indicated and cell radiance (photons per second per square centimeter per steradian) was measured at 30-s intervals over a 25 min time course using a cooled CCD camera and an in vivo imaging system (IVIS 50). Points were done in quintuplicate and the data analyzed as described previously (14).
Kinase Activation and Downstream Signaling.
Cells stably expressing the indicated EGF receptor and/or ErbB2 constructs were grown to confluence in 35-mm dishes. Cultures were treated without or with 10 nM EGF for 5 min at 37 °C. Radioimmunoprecipitation assay lysates were prepared and equal amounts of protein separated by SDS polyacrylamide gel electrophoresis. Proteins were transferred to PVDF membranes and probed by Western blotting with the indicated antibodies.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health (NIH) Grant R01GM082824 (to L.J.P.) and NIH Grant P50 CA94056 (to D.P.-W.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111316109/-/DCSupplemental.
References
- 1.Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res. 2009;315:638–648. doi: 10.1016/j.yexcr.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hynes NE, Lane HA. ErbB receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson KM. Structure-based view of epidermal growth factor receptor regulation. Annu Rev Biophys. 2008;37:353–373. doi: 10.1146/annurev.biophys.37.032807.125829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson KJ, et al. Functional selectivity of EGF family peptide growth factors: Implications for cancer. Pharmacol Ther. 2009;122:1–8. doi: 10.1016/j.pharmthera.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho H-S, et al. Structure of the extracellular region of HER2 alone and in complex with the herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 6.Klapper LN. The ErbB2/HER2 oncoprotein of human carcinomas may function solely as a shared coreceptor for multiple stroma-derive growth factors. Proc Natl Acad Sci USA. 1999;96:4995–5000. doi: 10.1073/pnas.96.9.4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graus Porta D, Beerli RR, Daly JM, Hynes NE. ErbB2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzahar E, et al. A Hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brewer MR, et al. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jura N, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, et al. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Jeffrey PD, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 13.Monsey J, Shen W, Schlesinger P, Bose R. Her4 and Her2/neu tyrosine kinase domains dimerize and activate in a reconstituted in vitro system. J Biol Chem. 2010;285:7035–7044. doi: 10.1074/jbc.M109.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang KS, Ilagan MXG, Piwnica-Worms D, Pike LJ. Luciferase fragment complementation imaging of conformational changes in the EGF receptor. J Biol Chem. 2009;284:7474–7482. doi: 10.1074/jbc.M808041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang KS, Macdonald-Obermann JL, Piwnica-Worms D, Pike LJ. Asp-960/Glu-961 Control the movement of the C-terminal tail of the EGF receptor to regulate asymmetric dimer formation. J Biol Chem. 2010;285:24014–24022. doi: 10.1074/jbc.M110.103317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clayton AHA, et al. Ligand-induced dimer-tetramer transition during the activation of cell surface epidermal growth factor receptor. A Multidimensional microscopy analysis. J Biol Chem. 2005;280:30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Fernandez M, et al. Preformed oligomeric epidermal growth factor receptors undergo and ectodomain structure change during signaling. Biophys J. 2002;82:2415–2427. doi: 10.1016/S0006-3495(02)75585-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sako Y, Minoguchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 19.Tao R-H, Maruyama IN. All EGF (ErbB) receptors have preformed homo- and heterodimeric structures in living cells. J Cell Sci. 2008;121:3207–3217. doi: 10.1242/jcs.033399. [DOI] [PubMed] [Google Scholar]
- 20.Yu X, et al. Ligand-independent dimer formation of epidermal growth factor receptor (EGFR) is a step separable from ligand-induced EGFR signaling. Mol Biol Cell. 2002;13:2547–2557. doi: 10.1091/mbc.01-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luker KE, et al. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci USA. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villalobos VM, Naik S, Piwnica-Worms D. Current state of imaging protein-protein interactions in vivo with genetically encoded reporters. Annu Rev Biomed Eng. 2007;9:321–349. doi: 10.1146/annurev.bioeng.9.060906.152044. [DOI] [PubMed] [Google Scholar]
- 23.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 24.Adak S, Yang KS, Macdonald-Obermann JL, Pike LJ. The membrane-proximal intracellular domain of the EGF receptor underlies negative cooperativity in ligand binding. J Biol Chem. 2011 doi: 10.1074/jbc.M111.274175. 10.1074/jbc.M111.274175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guy PM, et al. Insect cell-expressed p180ErbB3 possesses an impaired tyrosine kinase activity. Proc Natl Acad Sci USA. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi F, et al. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci USA. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriki T, Maruyama H, Maruyama IN. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J Mol Biol. 2001;311:1011–1026. doi: 10.1006/jmbi.2001.4923. [DOI] [PubMed] [Google Scholar]
- 28.Saffarian S, Li Y, Elson EL, Pike LJ. Oligomerization of the EGF receptor investigated by live cell fluorescence intensity distribution analysis. Biophys J. 2007;93:1021–1031. doi: 10.1529/biophysj.107.105494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.