Abstract
Photochemical radical initiation is a powerful tool for studying radical initiation and transport in biology. Ribonucleotide reductases (RNRs), which catalyze the conversion of nucleotides to deoxynucleotides in all organisms, are an exemplar of radical mediated transformations in biology. Class Ia RNRs are composed of two subunits: α2 and β2. As a method to initiate radical formation photochemically within β2, a single surface-exposed cysteine of the β2 subunit of Escherichia coli Class Ia RNR has been labeled (98%) with a photooxidant ([Re ] = tricarbonyl(1,10-phenanthroline)(methylpyridyl)rhenium(I)). The labeling was achieved by incubation of S355C-β2 with the 4-(bromomethyl)pyridyl derivative of [Re] to yield the labeled species, [Re]-S355C-β2. Steady-state and time-resolved emission experiments reveal that the metal-to-ligand charge transfer (MLCT) excited-state 3[Re ]∗ is not significantly perturbed after bioconjugation and is available as a phototrigger of tyrosine radical at position 356 in the β2 subunit; transient absorption spectroscopy reveals that the radical lives for microseconds. The work described herein provides a platform for photochemical radical initiation and study of proton-coupled electron transfer (PCET) in the β2 subunit of RNR, from which radical initiation and transport for this enzyme originates.
Keywords: proton-coupled electron transfer, radical generation, radical transport
The initiation and transport of many amino acid radicals occurs by proton-coupled electron transfer (PCET) (1–4). Accordingly, the importance of radicals in biological function (5) provides an imperative for the description of PCET in natural systems whose functions derive from radical-based chemistry (6). The PCET activity of amino acid radicals originates from the dependence of the reduction potential on the pKa of the amino acid in its oxidized and reduced states as well as the intrinsic redox potentials of the amino acid in its protonated and deprotonated states, and its association with hydrogen bonded partners as has been shown in proteins (7,8), β-hairpin peptides with interstrand dipolar contacts (9,10), and other de novo designed protein maquettes (11).
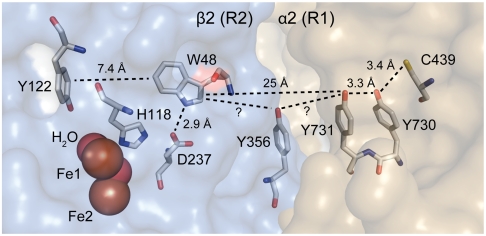
Ribonucleotide reductases (RNRs) are essential enzymes of all organisms (12) that demonstrate exquisite control of radical transport for their function (13). RNRs catalyze the conversion of nucleoside diphosphates (NDPs) to deoxynucleoside diphosphates (dNDPs) and are therefore largely responsible for maintaining the cellular pool of monomeric DNA precursors. E. coli class Ia RNR consists of two homodimeric subunits, α2 and β2 (14,15). The α2 subunit contains the enzyme active site as well as two allosteric regulation sites, whereas β2 contains a diferric tyrosyl cofactor (•Y122), which is where the radical resides in the resting state of RNR. Nucleoside reduction requires formation of an α2∶β2 complex. Substrate turnover occurs by a radical mechanism mediated by an active site cysteine thiyl radical in α2 (•C439). A docking model (Fig. S1) based on crystal structures of the individual subunits suggests that C439 (α2) and Y122 (β2) are separated by over 35 Å (16), a distance supported by pulsed electron double resonance (PELDOR) studies (17). A single-step electron tunneling mechanism at such a distance (predicted kET ∼ 10-4–10-9s-1 for β = 1.2 Å-1) is inconsistent with the observed rate of turnover (k ∼ 2–10 s-1) (13). To account for this disparity, a multistep mechanism has been proposed involving intermediate amino acid radicals comprising a highly conserved pathway between both subunits (Fig. 1): 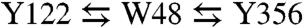 in β2 and Y731⇆Y730⇆C439 in α2.
in β2 and Y731⇆Y730⇆C439 in α2.
Fig. 1.
Key amino acids along a putative radical transport pathway in class Ia RNR. The pathway is ascertained from docking models of the α2 (gold) and β2 (blue) subunits. Substrate turnover requires charge transfer across α2 and β2 subunits over a distance of 35 Å. Radical hopping via the indicated redox-active amino acids is proposed to account for the observed rate of turnover. Distances are known for proposed hopping steps within individual subunits; distances involving Y356 have not been determined. Graphics were generated from Protein Data Bank entries 1RLR (α2) (16) and 1RIB (β2) (18).
To examine radical transport in RNR, we have created “photoRNRs” with an emphasis on the α2 subunit (photoRNR-α2). A short peptide (Y-βC19) has been employed in place of full-length β2 (19–21). This peptide contains the 20 C-terminal residues of β2, including both the binding determinant for β2 to α2 and a key tyrosine residue at the N terminus of the peptide. This tyrosine residue is in a position analogous to Y356 in β2, which facilitates radical transport at the α2∶β2 interface (22). By appending a photooxidant (PO) to this peptide (PO-Y-βC19), the equivalent of •Y356 (•Y-βC19) can be photochemically generated. Equilibrium constant measurements have shown that PO-Y-βC19 is bound to α2 with a KD = 9 μM (23). Photoinitiated substrate turnover can be observed for the PO-Y-βC19∶α2 construct upon illumination (20, 21). In these studies, the dynamics of bound βC19 are an essential factor to understand radical injection into α2 owing to peptide dissociation from α2 or conformational flexibility in the α2-βC19 system. Indeed, biexponential emission decay kinetics are observed for the bound peptide, consistent with multiple peptide conformations when bound to α2 (24). Of these conformations, only a small subset is likely to be productive toward radical injection and subsequent nucleoside diphosphate reduction, limiting yield in single turnover experiments.
Direct covalent attachment of a PO to the β2 subunit can significantly advance the study of radical transport in RNRs. PhotoRNR-β2s would: (i) enhance binding between the site of radical generation, Y356, and the α2 subunit; (ii) permit PCET to be examined in the β2 subunit; (iii) provide a platform for the first photoinitiated measurement of PCET in the α2∶β2 holoenzyme; and in doing so (iv) minimize complications arising from conformational flexibility. We now describe the creation of photoRNR-β2s by developing POs bearing an electrophilic carbon capable of alkylating surface-accessible protein thiolates. The native β2 subunit has two solvent-accessible cysteine residues C268 and C305, both of which can be mutated to serine without loss of activity (23). In the present report, we mutated these two residues to serine while also changing the serine at position 355 in the native protein to cysteine (S355C-β2). This permits a PO to be placed adjacent to Y356. A new PO, a rhenium(I) tricarbonyl phenanthroline complex (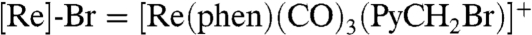 ), has been synthesized and characterized. By using the bromobenzyl derivative, the single surface cysteine variant of β2 can be labeled selectively to yield [Re]-S355C-β2. By using the flash-quench technique and transient absorption spectroscopy, we show that the production of •Y356 may be phototriggered in β2 and the radical is long lived, thus constituting a photoRNR-β2.
), has been synthesized and characterized. By using the bromobenzyl derivative, the single surface cysteine variant of β2 can be labeled selectively to yield [Re]-S355C-β2. By using the flash-quench technique and transient absorption spectroscopy, we show that the production of •Y356 may be phototriggered in β2 and the radical is long lived, thus constituting a photoRNR-β2.
Results and Discussion
[Re]-Br Photooxidant.
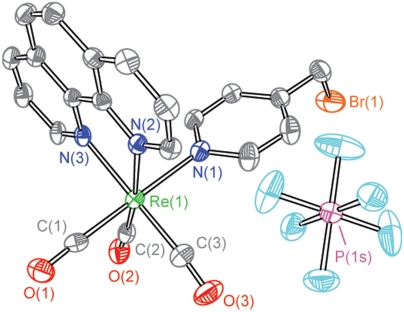
The selection of an appropriate photooxidant (PO) is essential to the construction of a photoRNR. The PO must (i) exhibit spectral separation from the protein envelope for its excitation, (ii) possess an excited-state lifetime and redox potential that are sufficient to enable amino acid oxidation, and (iii) be photostable. We have found that rhenium polypyridyl complexes, such as ReI(NN)(CO)3L (NN = 1,10-phenanthroline (phen); 2,2′-bipyridine (bpy) or derivatives; L = Cl, SCN, CN; or phosphine and pyridine derivatives) (25, 26), are superior phototriggers of amino acid radicals, especially tyrosine. They absorb well outside the absorption envelope of the protein and they are strong photooxidants. For this study, the rhenium polypyridyl complex needs to be conjugated with the protein. To accomplish this, we prepared [Re(phen)(CO)3(PyCH2Br)]PF6 according to the reaction sequence,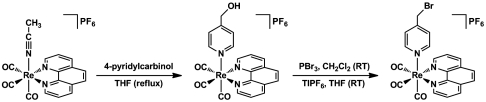 The presence of the primary bromide on the pyridyl ring provides a site for facile bioconjugation of the complex to the protein. Details of the preparation of the PO are given in SI Text. Briefly, 4-pyridylcarbinol displaces the bound MeCN ligand of [Re(phen)(CO)3(MeCN)]PF6 to yield the alcohol [Re(phen)(CO)3(PyCH2OH)]PF6, which is subsequently brominated with PBr3 to yield the protein-reactive [Re(phen)(CO)3(PyCH2Br)]PF6. The initial reaction yields a mixture of bromide and hexafluorophosphate salts, which is otherwise highly pure and suitable for use in protein labeling experiments. Of note, the treatment of the initial bromination product with TlPF6 allowed us to isolate the pure hexafluorophosphate salt. The final product was obtained in analytically pure form and 46% overall yield. The structure of [Re(phen)(CO)3(PyCH2Br)]PF6 was determined by X-ray crystallography (Fig. 2). Details of the solution of the X-ray structure (Table S1) and associated structural metrics (Table S2) are presented in SI Text. The structural parameters are consistent with an approximately octahedral geometry about rhenium with an attenuated N-Re-N angle as is typical of a phenanthroline complex. Spectroscopic studies of ultraviolet-visible absorption and steady-state emission (Fig. 3) show the characteristic features of an electronic structure derived from metal-to-ligand charge transfer (27).
The presence of the primary bromide on the pyridyl ring provides a site for facile bioconjugation of the complex to the protein. Details of the preparation of the PO are given in SI Text. Briefly, 4-pyridylcarbinol displaces the bound MeCN ligand of [Re(phen)(CO)3(MeCN)]PF6 to yield the alcohol [Re(phen)(CO)3(PyCH2OH)]PF6, which is subsequently brominated with PBr3 to yield the protein-reactive [Re(phen)(CO)3(PyCH2Br)]PF6. The initial reaction yields a mixture of bromide and hexafluorophosphate salts, which is otherwise highly pure and suitable for use in protein labeling experiments. Of note, the treatment of the initial bromination product with TlPF6 allowed us to isolate the pure hexafluorophosphate salt. The final product was obtained in analytically pure form and 46% overall yield. The structure of [Re(phen)(CO)3(PyCH2Br)]PF6 was determined by X-ray crystallography (Fig. 2). Details of the solution of the X-ray structure (Table S1) and associated structural metrics (Table S2) are presented in SI Text. The structural parameters are consistent with an approximately octahedral geometry about rhenium with an attenuated N-Re-N angle as is typical of a phenanthroline complex. Spectroscopic studies of ultraviolet-visible absorption and steady-state emission (Fig. 3) show the characteristic features of an electronic structure derived from metal-to-ligand charge transfer (27).
Fig. 2.
X-ray crystal structure of [Re]-Br. Thermal ellipsoids are reported at a 50% probability level. Hydrogen atoms, solvent molecules, and an additional molecule of [Re]-Br are omitted for clarity. Selected metric parameters are reported in Table S2.
Fig. 3.
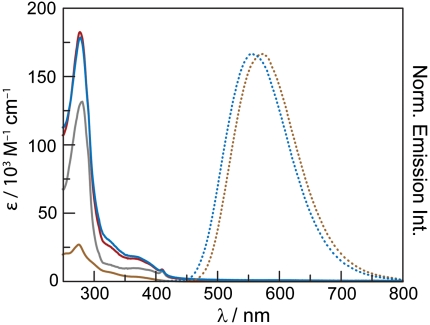
Spectroscopic comparison of [Re]-Br and [Re]-S355C-β2. The UV-vis absorption (brown) and emission spectra (λex = 355 nm) (brown) (50 μM) of [Re]-Br (MeCN solution) and the UV-vis absorption (blue) and emission (blue) (10 μM) spectra of [Re]-S355C-β2 (50 mM HEPES, 1 mM EDTA, pH 8.0) are shown. The absorption spectrum of S355C-β2 (gray) (50 mM HEPES, 1 mM EDTA, pH 8.0) is also included for reference. The simulated absorption spectrum (red) of [Re]-S355C-β2 by summing the absorption spectrum of twice the absorption spectrum of [Re]-Br and S355C-β2. The similarity of the actual (blue) and simulated (red) [Re]-S355C-β2 UV-vis absorption traces suggests that binding has no significant impact on the spectroscopic properties of [Re].
S355C-β2 Labeling with [Re] Photooxidant.
The choice of site, residue 355, for labeling β2 was guided by our previous work using PO-Y-βC19 peptides in place of β2. This peptide, representing the C terminus (355–375 of β2), contains the elements in large part responsible not only for subunit interactions but also activity; Y356 mediates radical transport between α2 and β2 (28, 29). The S355 residue was thus targeted as the site of labeling because it is directly adjacent to Y356 and it occupies a position analogous to that of the photooxidant in PO-Y-βC19, a site that has been shown to allow •Y356 generation, radical injection into α2, and subsequent substrate turnover (19–21).
To attach the PO to a single site, a β2 needed to be prepared with a single, surface-accessible cysteine at residue 355 (S355C-β2). Previous studies had shown that wt-β2 contained two such cysteines, C268 and C305, and that their mutation to serine gave C268S,C305S-β2 (23) with activity identical to wt-β2. For rapid affinity purification, all β2 constructs were constructed with an N-terminal (His)6 tag. Site-directed mutagenesis was performed with (His)6-C268S,C305S-β2 to give (His)6-C268S,C305S,S355C-β2. The protein was expressed in media containing 1,10-phenanthroline, a chelating agent, in order to express (His)6-C268S,C305S,S355C-β2 in the absence of FeII to directly obtain apo-S355C-β2 in large amounts. An alternative protocol, where expression occurs in the presence of FeII, requires a denaturing chelation step prior to cofactor reconstitution that is not conducive to large scale isolation (30). Final purification resulted in homogenous apo-S355C-β2 in a yield of 39 mg/g of cell paste. Reconstitution of the diferric tyrosyl cofactor by established methods (23) gave protein with 0.96 tyrosyl radicals/β2, demonstrating that the mutation does not disrupt cofactor assembly. Experimental details for expression and reconstitution of S355C-β2 are given in SI Text.
Prior to labeling with photooxidant, the concentration of solvent-accessible thiolate groups of S355C-β2, taken to indicate the number of surface-exposed cysteine residues, was established by reaction with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB). As expected, the results of the assay revealed 2.0 reactive thiols per β2. Sequential incubation of S355C-β2 with dithiothreitol (DTT) and [Re]-Br at pH 8 for 2 h resulted in covalent labeling of C355 via the methylene linker of the pyridyl ligand (i.e., ReI(phen)(CO)3(PyCH2 - )). Complete labeling was confirmed by a second DTNB assay, which indicated less than 500 nM free thiolate, consistent with labeling of > 98% of solution-accessible cysteine residues. Labeling was further confirmed by ESI-MS (Fig. S2); the ESI-MS (Fig. S2) shows peaks corresponding only to labeled species.
Time-Resolved Spectroscopy of Radical Initiation.
Electronic absorption and emission spectra (Fig. 3) of [Re]-S355C-β2 show the expected signatures of the [Re] complex. The MLCT transition is observed as a shoulder at λabs, max = 330 nm, and a yellow-green emission of the triplet excited state occurs at λem, max = 560 nm; the emission spectrum shows only a small blue shift relative to [Re]-Br in acetonitrile. The rising absorption envelope of the protein occurs well into the UV spectral region. The absorption spectrum for [Re]-S355C-β2 is modeled well as the sum of the unlabeled S355C-β2 spectrum and twice the spectrum of [Re]-Br. This similarity is as expected for a [Re]-S355C-β2 construct where each β monomer has been labeled with [Re], yielding a final [Re ]∶β2 ratio of 2∶1, which is reflected in the absorption spectrum, indicating that the environment of [Re] when bound to protein is similar to that of free [Re] in solution.
Nanosecond time scale transient absorption (TA) and time-resolved emission experiments were performed on [Re]-S355C-β2 to probe the effect of labeling on the spectroscopic properties of the 3[Re ]∗ excited state. TA spectra (Fig. S3) show two growth features at 300 and 450 nm; these growths are due to MLCT excited-state absorptions (31). Time-resolved emission data (Fig. S3) reveal an excited-state lifetime of 311 ns, in agreement with the excited-state lifetime derived from transient absorption kinetics measured at 450 nm, which reveal a lifetime of 328 ns. In comparison, time-resolved spectroscopic data for a small molecule analogue [Re(phen)(CO)3(PyCH3)]PF6 (Fig. S4) show almost identical transient absorption features and exhibit an emission lifetime of 228 ns in acetonitrile (in the presence of ambient O2, as present in protein experiments) and a corresponding lifetime of 237 ns monitored by transient absorption measured at 450 nm. The longer lifetime observed for [Re] bound to β2 in comparison to free [Re] in solution indicates that the modified cysteine thioether does not quench the [ReI]∗ excited state, making it available for amino acid oxidation and radical initiation.
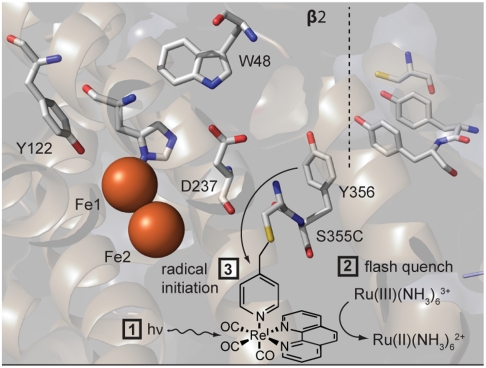
While the excited-state reduction potential for [Re(Phen)(CO)3(Py)]+(E0(ReI∗/0) = 1.7 V vs. NHE, calculated from reduction potentials and excited-state energies given for [Re(Phen)(CO)3(4-EtPy)]PF6 in ref. 32) is sufficient to oxidize tyrosine under these conditions (E0 = 0.77 V vs. NHE at pH 8.0) (11), rapid back electron transfer or significant transient absorption signals due to residual the [ReI]∗ excited state may preclude detection of a transient tyrosine radical. In order to circumvent this issue, we employed the flash-quench technique. In the presence of a sacrificial oxidant, (RuIII(NH3)6Cl3), the [ReI]∗ is rapidly quenched to yield the oxidized ground state [ReII], which also has an adequate potential for tyrosine oxidation (E0(ReII/I) = 2.0 V vs. NHE) (32), precludes any rapid back reaction and lacks significant absorption features in the spectral region of interest. As summarized in Fig. 4, the sequence of events is: (i) Laser excitation generates [ReI]∗ excited state, which is (ii) rapidly oxidized to the [ReII] ground state; (iii) the [ReII] ground state oxidizes the adjacent tyrosine residue, Y356, to yield •Y356. Presuming that Y356 is largely solvated and therefore that the pKa of Y356 and protonation state at pH 8.0 are not largely perturbed from the aqueous value of 10, under the experimental conditions, tyrosine oxidation necessarily occurs by a PCET mechanism, as Y356 is largely protonated at pH 8.0.
Fig. 4.
Strategy for the construction of a photoRNR-β2. The attachment of the PO, [Re ] = [Re(phen)(CO)3(PyCH2Br)] via the methylene group of the pyridyl ligand of the rhenium center is shown schematically. Radical initiation at Y356: (1) a 355 nm laser pulse generates the [Re]* excited state, which (2) is quenched to produce (3) the [ReII] species, which oxidizes Y356, to regenerate the [ReI] ground state and the radical. (4) The photogenerated Y356 radical is observed and monitored by transient absorption spectroscopy. Graphics were generated from Protein Data Bank entry 1MXR (β2) (15).
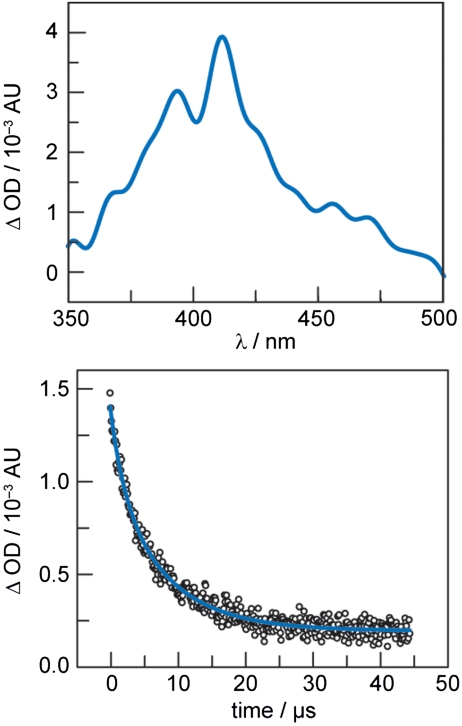
In the presence of RuIII(NH3)6Cl3 (500 eq./ Re -β2), transient absorption features due to the 3[Re ]∗ MLCT are significantly quenched upon laser excitation. After 1 μs, the only apparent features in the transient absorption spectrum are due to tyrosyl radical, •Y356 (Fig. 5). In order to maximize signal-to-noise, a total of 1,250 averages (5,000 component spectra) were collected on each of two independent samples and averaged to give a total of 2,500 averages. The transient radical vanishes with biexponential kinetics and exhibits lifetimes of 8.1 ± 1.1 and 2.0 ± 0.8 μs, with the longer component having approximately twice the amplitude of the shorter component (Fig. 5). The data presented are an average of 5,000 kinetic traces taken on a single sample and averaged.
Fig. 5.
Photochemically generated •Y356. Time-resolved spectroscopic data are recorded after excitation (λex = 355 nm) of met-[Re]-S355C-β2. Top: Transient absorption spectrum of [Re]-S355C-β2 collected 1 μs after excitation (65 μM in 50 mM HEPES, 32.5 mM RuIII(NH3)6Cl3, 1 mM EDTA, pH 8.0). The spectrum shown is obtained from 2,500 four-spectrum sequences taken on two samples (1,250 four-spectrum sequences each), averaged, and smoothed using a low-pass filter on the basis of a fast Fourier transform (FFT). Bottom: Transient absorption kinetics for transient •Y356 (λobs = 408 nm) and a biexponential fit (blue) (50 μM in 69% 50 mM HEPES, 25 mM RuIII(NH3)6Cl3, 1 mM EDTA, pH 8.0) (τ1 = 8.1 ± 1.1 μs, τ2 = 2.0 ± 0.8 μs 31%). The trace shown is obtained from 5,000 sweeps (averages) taken on a single sample.
The presence of high concentrations of RuIII(NH3)6Cl3 results in shortened apparent lifetimes; the intrinsic chemical lifetime of this species is likely to be significantly longer than that observed here. Therefore, the given lifetimes are valid only for the specific conditions given. In view of previously reported lifetimes of tyrosine radicals (77 μs) that are photoinitiated by direct irradiation (33), the observed lifetimes reported here are short. But the experiment reported here produces •Y in a more complex environment. RuII is generated stoichiometrically with the flash-quench reaction between RuIII(NH3)6Cl3 and 3[Re ]∗ excited state. Whereas large concentrations of flash-quench reagent maximize the observed signal for tyrosyl radical, the corresponding increase in RuII increases the rate of •Y reduction, thereby causing a decrease in the observed lifetime. In the absence of flash quencher, large transient absorption signatures due to 3[Re ]∗ show no significant features due to •Y or a charge-separated state. The maximum amplitude of the •Y TA spectrum (λmax = 412 nm, ΔOD412 = 3.9 mAU) allows us to estimate the overall quantum yield of •Y to be of Φ•Y ∼ 0.02 (see SI Text). In addition to the reaction of •Y with photoproduced RuII, the Φ•Y is inherently limited by: (i) the emission quantum yield of the [Re]* excited state (Φem = 0.262) (34) and (ii) incomplete quenching of the 3[Re ]∗ excited state. Notwithstanding, the concentration of photoriggered •Y is sufficient to be observed in the TA spectrum.
The observed multiexponential decay kinetics for •Y356 may indicate distinct conformations of the flexible C-terminal tail of Re-β2. In previous work, we have observed multiple peptide conformations for Re-Y-βC19 bound to the α2 subunit, (24) resulting in biexponential [Re] emission kinetics, where the PO can be closely associated with the subunit or may be largely exposed to solution. Similar dynamics of the C-terminal tail in the intact β2 subunit may result in the observed multiexponential kinetics for Re-β2.
Conclusions
[Re(phen)(CO)3(PyCH2Br)]PF6, a photooxidant bearing an electrophilic group for the selective alkylation of solution-accessible cysteine residues on proteins, has been synthesized and characterized. Its steady-state emission and electronic absorption spectra are consistent with an emissive excited state competent for photochemical tyrosine oxidation. [Re(phen)(CO)3(PyCH2Br)]PF6 has been shown to selectively and effectively alkylate surface-exposed residues on the ribonucleotide reductase mutant holo-S355C-β2 to yield a complex [Re]-S355C-β2, as confirmed by mass spectrometry. Both steady-state and time-resolved spectroscopic properties for [Re]-S355C-β2 are consistent with those reported for related compounds, indicating that the excited-state properties of [Re] are not significantly perturbed by conjugation to β2.
The successful labeling of S355C-β2 leads to the creation of a photoRNR-β2. Nanosecond flash photolysis, employing the flash-quench technique, affords •Y356, which was spectroscopically observed. The photoRNR-β2 construct is a powerful tool for the examination of PCET. Future studies will use the photoRNR-β2 to trigger radical initiation and transport in RNR. Moreover, the generation of •Y356 in an intact α2∶β2 construct provides a platform for time-resolved photochemical study of PCET in RNR. The expected enhancements in binding and minimization of detrimental conformational dynamics due to the use of an intact β2 subunit in addition to increased fidelity to the natural system will be a great asset to these studies, which are currently underway.
Experimental Methods
C355 Surface Accessibility Assay.
The solution accessibility of S355C-β2 thiolates was assessed by reaction with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB). Immediately prior to assay, S355C-β2 was pooled with dithiothreitol (DTT) (20 mM, 30 min). Excess DTT was then removed using a Sephadex G-25 column (15 mL) equilibrated in 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), 1 mM EDTA, pH 8.0. The pooled S355C-β2 (10 μM) was incubated with DTNB (50 μM total concentration in the assay buffer) and the reaction was monitored by UV-vis spectroscopy (2-min intervals, 10 min total). The concentration of free 5-thio-2-nitrobenzoic acid (TNB) was calculated (ε410,TNB = 13,600 M-1 cm-1) and corrected for background DTNB reaction by comparison with a sample prepared under identical conditions in the absence of protein. The assay was repeated in triplicate. The concentration of reactive thiolate groups was found to be 19.8 μM (1.98 per holo-S355C-β2), as expected.
Labeling of S355C-β2 with [Re(phen)(CO)3(PyCH2Br)]+.
For labeling with [Re]-Br, holo-S355C-β2 (1 mL, 211 μM) was treated with 10 mM DTT for 30 min to reduce disulfide bonds and excess DTT was removed using Sephadex G-25 column (15 mL). Labeling was performed immediately after the reduction reaction and accomplished by incubating S355C-β2 with 5 equiv of [Re(phen)(CO)3(PyCH2Br)]PF6 from a concentrated stock (50 mM in DMF) and gently shaking for 2 h at room temperature. Rhenium labeled protein ([Re]-S355C-β2) was isolated from remaining [Re(phen)(CO)3(PyCH2Br)]PF6 and DMF using another Sephadex G-25 column (15 mL). The resulting product ([Re]-S355C-β2) was subsequently concentrated to 130 μM as determined by a Bradford assay and labeling verified by ESI-MS. Wild-type β2 was used as a standard for the Bradford assay. UV-vis absorption and steady-state emission spectra were also recorded for this construct. For experiments using the reduced (met) form of [Re]-S355C-β2, reduction of the radical was performed prior to labeling by incubating with 30 mM hydroxyurea for 30 min, desalting on a Sephadex G-25 column, and labeling performed as described above.
Time-Resolved Methods.
Nanosecond time-resolved emission and transient absorption (TA) experiments were performed using a previously reported system (35) with a number of significant modifications. A description of the nanosecond flash laser apparatus is provided in SI Text.
For all protein experiments, 750-μL samples were recirculated to address sample decomposition. For small molecule experiments, samples were flowed irreversibly. Samples were held in a quartz flow-cell (585.3-Q-10/Z15, Starna) with a 1-cm path length and 3-mm diameter windows; the total sample bore was 70 μL. Fresh samples were used for all experiments as indicated. When observing the unquenched excited state of Re-β2, spectra reported are an average of 1,250 four-spectrum sequences, transient absorption kinetics are an average of 5,000 sweeps, and emission kinetics are an average of 1,000 sweeps. For experiments using the flash-quench technique to observe tyrosyl radical, the reported spectrum is an average of 2,500 four-spectrum sequences; the reported single wavelength kinetics are an average of two 5,000 sweep series (10,000 sweeps total), each of which was taken on fresh sample. The transient absorption spectrum shown has been smoothed using a low pass filter to remove high-frequency noise components due to instrumentation; the cutoff frequency was selected on the basis of a fast Fourier transform. The reported errors are the standard error of the fit, as obtained from least-squares analysis. Additionally, an inline syringe filter (0.2 μm Acrodisc, Supor membrane, 13 mm, Pall Corporation) was used for all flash-quench experiments to collect precipitate resulting from flash-quench by products. For small molecule experiments, spectra reported are an average of 2,500 four-spectrum sequences.
All experiments were conducted in buffer containing 50 mM HEPES and 1 mM EDTA at pH 8.0. Experiments in the absence of oxidative quencher were done using 10 μM [Re]-S355C-β2 (Fig. S3). Flash-quench experiments were performed in the presence of 500 eq. RuIII(NH3)6Cl3 per [Re]-S355C-β2. The reported spectrum of tyrosyl radical was collected 1 μs after excitation of a sample containing 65 μM met-[Re]-S355C-β2 and 32.5 mM RuIII(NH3)6Cl3 in buffer (Fig. 5, top); single wavelength kinetics were recorded using two samples of 50 μM met-[Re]-S355C-β2 and 25 mM in buffer (Fig. 5, bottom).
Supplementary Material
Acknowledgments.
We are grateful to Jacqueline Setas for helpful discussions. The National Institutes of Health (NIH) (GM47274) provided funding for this research. D.A.L. gratefully acknowledges the Jane Coffin Childs Memorial Fund for Medicinal Research for a postdoctoral fellowship. P.G.H. acknowledges the NIH for a postdoctoral fellowship (GM087034, Kirschstein National Research Service Award Fellowship). T.S.T. acknowledges the Fannie and John Hertz Foundation for a predoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115778108/-/DCSupplemental.
References
- 1.Reece SY, Nocera DG. Proton-coupled electron transfer in biology: Results from synergistic studies in natural and model systems. Annu Rev Biochem. 2009;78:673–699. doi: 10.1146/annurev.biochem.78.080207.092132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reece SY, Nocera DG. In: Quantum Tunnelling in Enzyme-Catalysed Reactions. Scrutton NS, Allemann RK, editors. London: Royal Society of Chemistry Press; 2009. pp. 345–377. [Google Scholar]
- 3.Hammes-Schiffer S. Hydrogen tunneling and protein motion in enzyme reactions. Acc Chem Res. 2006;39:93–100. doi: 10.1021/ar040199a. [DOI] [PubMed] [Google Scholar]
- 4.Liang Z, Klinman JP. Structural bases of hydrogen tunneling in enzymes: progress and puzzles. Curr Opin Struct Biol. 2004;14:648–655. doi: 10.1016/j.sbi.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Van der Donk WA, Stubbe J. Protein radicals in enzyme catalysis. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 6.Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. Proton-coupled electron transfer: the mechanistic underpinning for radical transport and catalysis in biology. Philos Trans R Soc London B Biol Sci. 2006;361:1351–1364. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seyedsayamdost MR, Yee CS, Reece SY, Nocera DG, Stubbe J. pH rate profiles of FnY356-R2s (n = 2, 3, 4) in Escherichia coli ribonucleotide reductase: Evidence that Y356 is a redox-active amino acid along the radical propagation pathway. J Am Chem Soc. 2006;128:1562–1568. doi: 10.1021/ja055927j. [DOI] [PubMed] [Google Scholar]
- 8.Chang CJ, Chang MCY, Damrauer NH, Nocera DG. Proton-coupled electron transfer: A unifying mechanism for biological charge transport, amino acid radical initiation and propagation, and bond making/breaking reactions of water and oxygen. Biochim Biophys Acta. 2004;1655:13–28. doi: 10.1016/j.bbabio.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Sibert RS, et al. Proton-coupled electron transfer in a biomimetic peptide as a model of enzyme regulatory mechanisms. J Am Chem Soc. 2007;129:4393–4400. doi: 10.1021/ja068805f. [DOI] [PubMed] [Google Scholar]
- 10.Sibert RS, Josowicz M, Barry BA. Control of proton and electron transfer in de novo designed, biomimetic β hairpins. ACS Chem Biol. 2010;5:1157–1168. doi: 10.1021/cb100138m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tommos C, Skalicky JJ, Pilloud DL, Wand AJ, Dutton PL. De novo proteins as models of radical enzymes. Biochemistry. 1999;38:9495–9507. doi: 10.1021/bi990609g. [DOI] [PubMed] [Google Scholar]
- 12.Nordlund N, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 13.Stubbe J, Nocera DG, Yee CS, Chang MCY. Radical initiation in the class I ribonucleotide reductase: Long-range proton-coupled electron transfer? Chem Rev. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson M, et al. Binding of allosteric effectors to ribonucleotide reductase protein R1: Reduction of active-site cysteines promotes substrate binding. Structure. 1997;5:1077–1092. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 15.Högbom M, et al. Displacement of the tyrosyl radical cofactor in ribonucleotide reductase obtained by single-crystal high-field EPR and 14-Å X-ray data. Proc Natl Acad Sci USA. 2003;100:3209–3214. doi: 10.1073/pnas.0536684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 17.Bennati M, et al. EPR distance measurements support a model for long-range radical initiation in E.coli ribonucleotide reductase. J Am Chem Soc. 2005;127:15014–15015. doi: 10.1021/ja054991y. [DOI] [PubMed] [Google Scholar]
- 18.Nordlund P, Eklund H. Structure and function of the Escherichia coli ribonucleotide reductase protein R2. J Mol Biol. 1993;232:123–164. doi: 10.1006/jmbi.1993.1374. [DOI] [PubMed] [Google Scholar]
- 19.Chang MCY, Yee CS, Stubbe J, Nocera DG. Turning on ribonucleotide reductase by light-initiated amino acid radical generation. Proc Natl Acad Sci USA. 2004;101:6882–6887. doi: 10.1073/pnas.0401718101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reece SY, Seyedsayamdost MR, Stubbe J, Nocera DG. Direct observation of a transient tyrosine radical competent for initiating turnover in a photochemical ribonucleotide reductase. J Am Chem Soc. 2007;129:13828–13830. doi: 10.1021/ja074452o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reece SY, Seyedsayamdost MR, Stubbe J, Nocera DG. Photoactive peptides for light-initiated tyrosyl radical generation and transport into ribonucleotide reductase. J Am Chem Soc. 2007;129:8500–8509. doi: 10.1021/ja0704434. [DOI] [PubMed] [Google Scholar]
- 22.Sayedsayamdost MR, Stubbe J. Site-specific replacement of Y356 with 3,4-dihydroxyphenylalanine in the β2 subunit of E. coli ribonucleotide reductase. J Am Chem Soc. 2006;128:2522–2523. doi: 10.1021/ja057776q. [DOI] [PubMed] [Google Scholar]
- 23.Hassan AQ, Wang Y, Plate L, Stubbe J. Methodology to probe subunit interactions in ribonucleotide reductases. Biochemistry. 2008;47:13046–13055. doi: 10.1021/bi8012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reece SY, Lutterman DA, Seyedsayamdost MR, Stubbe J, Nocera DG. Re(bpy)(CO)3CN as a probe of conformational flexibility in a photochemical ribonucleotide reductase. Biochemistry. 2009;48:5832–5838. doi: 10.1021/bi9005804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reece SY, Seyedsayamdost MR, Stubbe J, Nocera DG. Electron transfer reactions of fluorotyrosyl radicals. J Am Chem Soc. 2006;128:13654–13655. doi: 10.1021/ja0636688. [DOI] [PubMed] [Google Scholar]
- 26.Reece SY, Nocera DG. Direct tyrosine oxidation using the MLCT excited states of rhenium polypyridyl complexes. J Am Chem Soc. 2005;127:9448–9458. doi: 10.1021/ja0510360. [DOI] [PubMed] [Google Scholar]
- 27.Wallace L, Rillema DP. Photophysical properties of rhenium(I) tricarbonyl complexes containing alkyl- and aryl-substituted phenanthrolines as ligands. Inorg Chem. 1993;32:3836–3843. [Google Scholar]
- 28.Yee CS, Chang MCY, Ge J, Nocera DG, Stubbe J. 2,3-Difluorotyrosine at position 356 of ribonucleotide reductase R2: A proble of long-range proton-coupled electron transfer. J Am Chem Soc. 2003;125:10506–10507. doi: 10.1021/ja036242r. [DOI] [PubMed] [Google Scholar]
- 29.Yokoyama K, Uhlin U, Stubbe J. Site-specific incorporation of 3-nitrotyrosine as a probe of pKa perturbation of redox-active tyrosines in ribonucleotide reductase. J Am Chem Soc. 2010;132:8385–8397. doi: 10.1021/ja101097p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkin SE, et al. Electron injection through a specific pathway determines the outcome of oxygen activation at the diiron cluster in the F208Y mutant of Escherichia coli ribonucleotide reductase protein R2. Biochemistry. 1998;37:1124–1130. doi: 10.1021/bi9723717. [DOI] [PubMed] [Google Scholar]
- 31.Kalyanasurndaram K. Luminescence and redox reactions of the metal-to-ligand charge-transfer excited state of tricarbonylchloro(polypyridyl)rhenium(I) complexes. J Chem Soc Faraday Trans. 1986;82:2401–2415. [Google Scholar]
- 32.Dattelbaum DM, Omberg KM, Schoonover JR, Martin RL, Meyer TJ. Application of time-resolved infrared spectroscopy to electronic structure in metal-to-ligand charge-transfer excited states. Inorg Chem. 2002;41:6071–6079. doi: 10.1021/ic020400i. [DOI] [PubMed] [Google Scholar]
- 33.Cappuccio JA, et al. Modeling the active site of cytochrome oxidase: Synthesis and characterization of a cross-linked histidine-phenol. J Am Chem Soc. 2002;124:1750–1760. doi: 10.1021/ja011852h. [DOI] [PubMed] [Google Scholar]
- 34.Cleland DM, Irwin G, Wagner P, Officer DL, Gordon KC. Linker conjugation effects in rhenium(I) bifunctional hole-transport/emitter molecules. Chem Eur J. 2009;15:3682–3690. doi: 10.1002/chem.200802373. [DOI] [PubMed] [Google Scholar]
- 35.Loh Z-H, Miller SE, Chang CJ, Carpenter SD, Nocera DG. Excited-state dynamics of cofacial pacman porphyrins. J Phys Chem A. 2002;106:11700–11708. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.