Abstract
Oxidation of genomic DNA forms the guanine lesion 7,8-dihydro-8-oxoguanine (8-oxoG). When in the template base position during DNA synthesis the 8-oxoG lesion has dual coding potential by virtue of its anti- and syn-conformations, base pairing with cytosine and adenine, respectively. This impacts mutagenesis, because insertion of adenine opposite template 8-oxoG can result in a G to T transversion. DNA polymerases vary by orders of magnitude in their preferences for mutagenic vs. error-free 8-oxoG lesion bypass. Yet, the structural basis for lesion bypass specificity is not well understood. The DNA base excision repair enzyme DNA polymerase (pol) β is presented with gap-filling synthesis opposite 8-oxoG during repair and has similar insertion efficiencies for dCTP and dATP. We report the structure of pol β in binary complex with template 8-oxoG in a base excision repair substrate. The structure reveals both the syn- and anti-conformations of template 8-oxoG in the confines of the polymerase active site, consistent with the dual coding observed kinetically for this enzyme. A ternary complex structure of pol β with the syn-8-oxoG:anti-A Hoogsteen base pair in the closed fully assembled preinsertion active site is also reported. The syn-conformation of 8-oxoG is stabilized by minor groove hydrogen bonding between the side chain of Arg283 and O8 of 8-oxoG. An adjustment in the position of the phosphodiester backbone 5′-phosphate enables 8-oxoG to adopt the syn-conformation.
Keywords: oxidative DNA lesion, X-ray crystallography
Reactive oxygen species (ROS) are produced during aerobic respiration throughout nature and during inflammatory responses in higher organisms. To balance ROS-induced stress, a reducing environment is maintained within cells through several enzymatic systems, including superoxide dismutase, catalase, and glutathione reductase, among others. Yet, long term exposure to oxidative stress, ultraviolet light and ionizing radiation can eventually result in genomic DNA oxidation (1) and adverse effects (2, 3). The modified guanine base 8-oxoG is a well-recognized DNA lesion secondary to oxidative stress (4), and this lesion is associated with a signature type of mutagenesis in genomic DNA, the G to T transversion (5). The predominant tautomeric form of 8-oxoG at physiological pH has a carbonyl group at C8 and is protonated at N7 (4–7), defining its Hoogsteen base pair hydrogen bonding capacity with adenine; in addition, 8-oxoG forms the Watson–Crick base pair with cytosine (4) (Fig. S1A). Thus, during DNA synthesis, template 8-oxoG has dual coding potential, adopting the anti-conformation when base paired with incoming dCTP and the syn-conformation when base paired with incoming dATP.
Many DNA polymerases insert dATP opposite template 8-oxoG, presumably involving the Hoogsteen base pair in the preinsertion complex. Yet, the DNA polymerase preference for insertion of incoming dCTP vs. dATP opposite template 8-oxoG varies by orders of magnitude (Table S1). In general, however, once the template 8-oxoG:A base pair is incorporated into DNA and is at the template-primer terminus during DNA synthesis, it is well-extended by DNA polymerases facilitating G to T mutagenesis. The structural basis for DNA polymerase preferences in mutagenic bypass of the 8-oxoG lesion is emerging across the DNA polymerase families (8–12) and is the subject of the present report.
Repair of the 8-oxoG-DNA lesion in mammalian cells proceeds by several DNA repair pathways (13–15), but the major pathway is considered to be a form of the base excision repair (BER) pathway referred to as the “GO system” (16). An early step in this repair system involves DNA glycosylase (MYH) removal of A from the 8-oxoG:A base pair that was introduced during DNA replication or repair (Fig. S1B). Action of this DNA glycosylase results in a single-nucleotide gapped BER intermediate with 8-oxoG in the template position in a single-nucleotide gap. In subsequent BER steps, 8-oxoG serves as the template base during gap-filling DNA synthesis by pol β. This can lead to error-prone lesion bypass of 8-oxoG, because pol β inserts dATP and dCTP opposite template 8-oxoG with similar efficiencies and within about fivefold of that for dCTP insertion opposite unmodified dG (Fig. S1B and Table S1).
A variety of studies of 8-oxoG-DNA have supported the hypothesis that chronic inflammation and attendant oxidative stress is a contributing risk factor in chronic disease, such as cancer (2, 17, 18). First, 8-oxoG-DNA is linked to mutagenesis in genomic DNA (4, 5, 19), and this is a likely component of carcinogenesis. Second, in studies of mouse models, deficiency in 8-oxoG-DNA repair was linked to increased tumor formation and to the G to T mutation in an oncogene (20). In clinical studies, a form of inherited colon cancer, MYH-associated polyposis, is associated with deficiency in the DNA glycosylase specific for A removal from the 8-oxoG:A base pair (21). And, increases in G to T mutations were found in lung tumor genomic DNA through large-scale sequencing (22, 23). An underlying contributing factor in these examples appears to be 8-oxoG-induced mutations in genes critical for growth control. In light of the role of base excision repair in protection against 8-oxoG-DNA accumulation, it is important to gain a deeper understanding of how the BER enzymes respond to this lesion.
In addition to 8-oxoG-DNA, oxidative stress generates oxidized nucleotides in the dNTP pool including 8-oxodGTP (24), and this nucleotide can be incorporated into DNA opposite template A. To prevent this type of mutagenic DNA synthesis, 8-oxodGTPases are expressed in a number of biological systems eliminating virtually all 8-oxodGTP from dNTP pools. Nevertheless, 8-oxodGTP can accumulate under oxidative stress conditions and become incorporated into DNA, eventually leading to the A to C transversion (25). We recently reported the crystal structure of a preinsertion ternary complex of pol β with template A Hoogsteen base paired with 8-oxodGTP in a single-nucleotide gapped BER intermediate (26, 27). The structure revealed that the syn-conformation of incoming 8-oxodGTP is stabilized in the preinsertion active site through a hydrogen bond between O8 and Nδ of Asn279 and an intramolecular hydrogen bond (N2 and a nonbridging oxygen on Pα). This type of structural insight on the preinsertion complex is invaluable in understanding DNA polymerase contributions to 8-oxoG-induced mutagenesis.
Previous work to obtain a pol β structure with template 8-oxoG in the syn-conformation opposite incoming dATP resulted in an “open” conformation of the polymerase (28). In this structure, the template 8-oxoG base was in anti-conformation and staggered register with the base of the incoming nucleotide, dATP, resulting in the inactive conformation of pol β’s N subdomain (29). In addition to the aberrant base position, electron density was not observed for the β- and γ-phosphates of the incoming dATP, indicating hydrolysis of the dNTP. This structure was obtained with a dideoxy-terminated primer used to freeze the polymerase reaction. In light of the unusual features of the active site, this structure is of limited value in understanding how the syn-conformation of 8-oxoG is stabilized in the preinsertion nascent base pair binding site.
The goal of the present study was to obtain pol β crystal structures that would inform both insertion opposite the template 8-oxoG lesion and extension off a template-primer terminus containing 8-oxoG. We now report several crystal structures with this mutagenic base lesion in the nascent base pair binding pocket and positioned at the template-primer terminus. Information from these pol β structures provides insight on the fidelity of 8-oxoG lesion bypass by virtue of various strategies of stabilizing the syn- and anti-conformations of the 8-oxoG base.
Results
Binary Complex Structure with 8-oxoG in the Template Base Position.
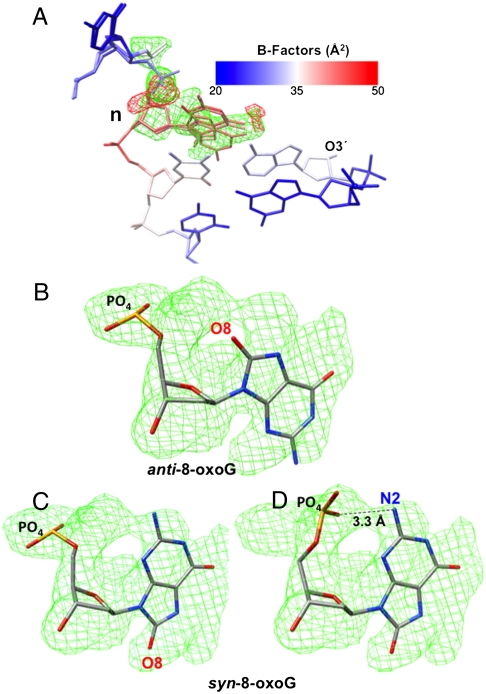
A binary complex crystal structure was obtained with 8-oxoG in the template position of the single-nucleotide gapped DNA substrate representing a base excision repair intermediate. The crystal diffracted to 2.1 Å resolution, and the structure was solved by molecular replacement. The crystallographic data are summarized in Table S2. The enzyme is in the open conformation, as expected in the absence of an incoming nucleotide, and is very similar to the enzyme in a reference binary complex structure [Protein Data Bank (PDB) ID code 3ISB] with unmodified gapped DNA (30). The rmsd for all 326 Cα is 0.2 Å when compared with the reference structure (Fig. S2A). We found that the 8-oxoG base could be modeled in the electron density in both syn- and anti-conformations (Fig. 1). The B-factor for the 8-oxoG base is higher than those of the neighboring bases (Fig. 1A); however, when 8-oxoG is in a ternary complex base paired with incoming nucleotide, or when 8-oxoG is base paired at the template-primer terminus, the B-factors for 8-oxoG are low and similar to those of the neighboring unmodified base pairs (see below). The electron density corresponding to the 5′-phosphate of the phosphodiester backbone of 8-oxoG is indicative of three main conformations (Fig. 1 B–D). As it is difficult to calculate the absolute occupancies for the different conformations, the occupancy values were assigned based on the refinement that showed the lowest rfree value. All of the electron density of the omit map can be accounted for by these multiple conformations of 8-oxoG with no significant residual difference (Fo - Fc) density. Also, no negative electron density was observed in the vicinity of the modeled 8-oxoG. As discussed below, two of these phosphodiester backbone 5′-phosphate conformations correspond to those in ternary complexes with incoming dCTP opposite anti-8-oxoG and incoming dATP opposite syn-8-oxoG. One of the conformations seen with syn-8-oxoG reveals a hydrogen bond between N2 and a 5′-phosphate oxygen (Fig. 1D). The global structure of the single-nucleotide gapped DNA is similar to that in the reference structure, including the sharp 90° bend at the 5′-phosphodiester bond of the template nucleotide, 8-oxoG (Fig. S2B). We conclude from this pol β binary complex structure that unpaired 8-oxoG is accommodated in both its syn- and anti-conformations.
Fig. 1.
Alternate confirmations of 8-oxoG in a binary pol β complex crystal structure. (A) The 8-oxoG base can be modeled into the omit map in both syn- and anti-conformations and the 5′-phosphate can be modeled in three positions. The B-Factor color scale used is shown. The templating nucleotide, 8-oxoG, is labeled n. The O3′ of the primer terminus is shown. The simulated annealing Fo - Fc electron density omit map (cyan) and difference density (Fo - Fc) map (red) are contoured at 3σ. No negative density was observed in the vicinity of O8 in the anti-conformation. (B) The anti-conformation of the 8-oxoG base modeled into the omit map. The 5′-phosphate and O8 are shown. The omit map is contoured at 3σ. (C) The syn-conformation of the 8-oxoG base modeled into the omit map, and the 5′-phosphate is modeled in one position, as illustrated. The omit map is contoured at 3σ. (D) The syn-conformation of the 8-oxoG base modeled into the omit map, and the 5′-phosphate is modeled in the alternate position, as illustrated. The omit map is contoured at 3σ. The 5′-phosphate N2 H-bond is shown.
Template 8-oxoG Conformations in Ternary Complex Crystal Structures.
To evaluate the 8-oxoG nucleotide conformations observed in the binary complex structure, we obtained preinsertion ternary complex structures of pol β with 8-oxoG in the template position of a single-nucleotide gapped DNA substrate. This was achieved by soaking binary complex crystals with nonhydrolyzable nucleotide analogues, dAMP(CH2)PP (referred to as dATP or dAMPCPP) or dCMP(CF2)PP (referred to as dCTP), and using a primer terminus with an O3′. For the ternary complex with incoming dATP, the resulting crystals diffracted to 2.3 Å resolution, and the structure was solved by molecular replacement. The crystallographic data are summarized in Table S2. This ternary complex structure reveals that the enzyme is in the “closed” conformation (29) and very similar to the enzyme in a reference structure (PDB ID code 2FMS) with an unmodified base pair in the fully assembled preinsertion active site (30). The rmsd for all 326 Cα is 0.3 Å when compared with the reference structure (Fig. S3A). The structure of the single-nucleotide gapped DNA is similar to that in the reference structure.
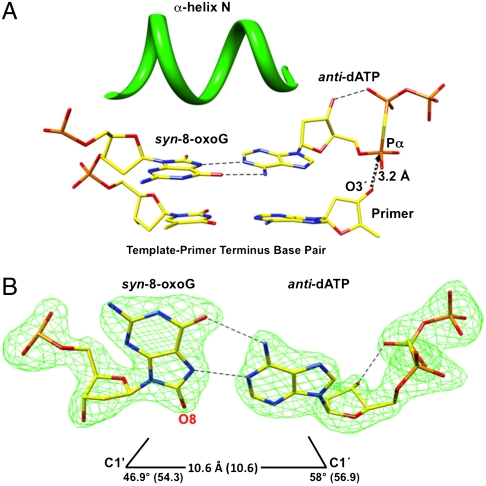
The syn-8-oxoG:anti-dATP Hoogsteen base pair is in the nascent base pair binding pocket (Fig. 2). The planar Hoogsteen base pair is tightly sandwiched between the template-primer terminus base pair and α-helix N (Fig. 2A). One of the positions of the phosphodiester backbone 5′-phosphate of the templating syn-8-oxoG seen in the binary complex structure (Fig. 1C) is similar to that observed in this ternary complex structure (Fig. S3B). Most of the atoms in the active site are in similar positions to those found in the reference structure. Yet, the distance between O3′ and Pα is 3.2 Å (Fig. 2A) vs. 3.4 Å in the reference structure. The C1′–C1′ distance of the syn-8-oxoG:anti-dATP base pair is similar to that of a Watson–Crick base pair; the λ1 angle for syn-8-oxoG is reduced (46.9°), as expected (Fig. 2B) (26, 31).
Fig. 2.
Ternary complex crystal structure with syn-8-oxoG in the template position. (A) Image of detailed active site features of the structure of human pol β in the fully assembled preinsertion ternary complex. The nascent base pair binding pocket with template syn-8-oxoG and incoming anti-dAMPCPP (dATP) is illustrated, along with the base pair at the template-primer terminus. The perspective is from the major groove. The image shows that the template base 8-oxoG and incoming base of dATP are planar, and the nascent base pair is sandwiched between the template-primer terminus nucleotides and closed conformation of α-helix N (green). O3′ of the primer is 3.2 Å from Pα of the incoming dATP, as illustrated by the arrow. H-bonds are shown as dashed lines. (B) Image illustrating the Hoogsteen base pair with the syn-conformation of template 8-oxoG and anti-conformation of incoming dATP. The C1′ to C1′ distance for the base pair is indicated, and the λ1 and λ2 angles are shown., the values in parentheses are from the reference structure (30) with template A and incoming dUMPNPP (PDB ID code 2FMS). The omit map (green) is contoured at 4σ. For the incoming dATP, intramolecular hydrogen bonding between O3′ and a nonbridging oxygen on Pβ is shown. H-bonds are shown as dashed lines.
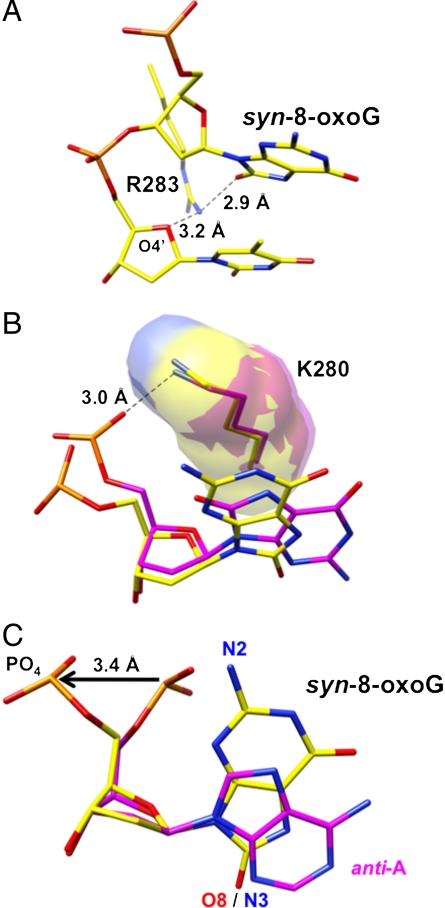
Regarding interactions that stabilize the syn-conformation of 8-oxoG, the side chain of Arg283 in α-helix N occupies the minor groove and forms a hydrogen bond with 8-oxoG (O8) and the sugar ring (O4′) of the upstream template nucleotide (Fig. 3A). In addition, a stacking interaction between the base of 8-oxoG and the methylene side chain of Lys280 is observed (Fig. 3B). Finally, adjustment of the phosphodiester backbone 5′-phosphate enables 8-oxoG to adopt the syn-conformation, but at the cost of a potential H-bond with N2 (Fig. 3C). The phosphodiester backbone torsion angle (O3′-P-O5′-C5′) is -142°, resulting in displacement of the backbone phosphate by approximately 3.4 Å (Fig. 3C). This position of the 5′-phosphate in the 8-oxoG syn-conformation corresponds with one of the 5′-phosphate positions of the syn-conformation in the binary complex structure (Fig. 1 C and D).
Fig. 3.
Detailed illustrations of features of the ternary complex crystal structure with syn-8-oxoG:dATP in the nascent base pair binding pocket. (A) A close up illustration of the distances between Nη1 of Arg283 and O8 of 8-oxoG (2.9 Å) and the O4′ of the sugar ring of the template strand nucleotide at the template-primer terminus (3.2 Å). Arg283 of α-helix N stabilizes the syn-conformation of 8-oxoG through O8 and Nη1 hydrogen bonding, as shown. (B) Superimposed structures illustrating the relative positions of Lys280 and 8-oxoG in the complexes with the syn-8-oxoG and anti-dATP base pair (yellow) and the anti-8-oxoG and anti-dCTP base pair (magenta). The side chain of Lys280 (K280) is similar in the two structures and maintains stacking interaction over the purine ring of syn-8-oxoG. Yet, the hydrogen bond between Nζ of Lys280 and the 5′-phosphoryl oxygen is seen in the anti-8-oxoG structure, but is not observed in the syn-8-oxoG structure. The hydrogen bond is illustrated (3.0 Å). (C) Illustration of the phosphodiester backbone adjustment accommodating template 8-oxoG in syn-conformation (yellow). The image of 5′-phosphate groups and template bases is taken from superimposed ternary complex structures with the syn-8-oxoG:anti-dATP base pair in the active site (yellow) and the reference pol β structure with A in the template base position and incoming dUMPNPP (PDB ID code 2FMS) (magenta). To accommodate 8-oxoG in syn-conformation, the O3′-P-O5′-C5′ torsion angle is rotated approximately 200° and displaced by 3.4 Å, as illustrated by arrow. N2 and O8 of 8-oxoG and N3 of A are indicated, along with the 5′-phosphate groups.
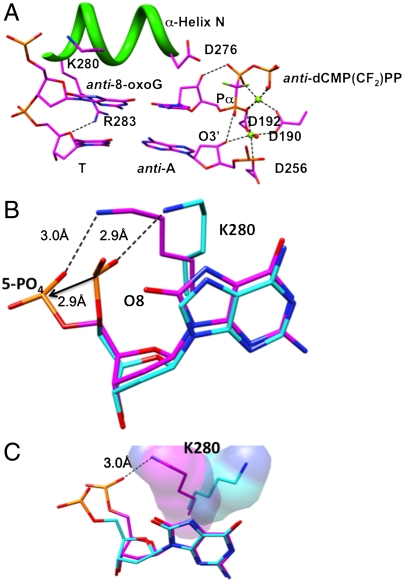
To further evaluate the alternate 8-oxoG conformations observed in the binary complex structure, another structure was obtained of a preinsertion ternary complex with incoming dCTP (Table S2). The anti-8-oxoG:anti-dCTP base pair is in the nascent base pair binding pocket (Fig. 4), and the enzyme is in the closed conformation. The position of the backbone 5′-phosphate (Fig. 4B) is similar to that observed for the anti-conformation in the binary complex (Fig. 1B). In addition, in this case with the 8-oxoG in anti-conformation, Lys280 stacks over the base of 8-oxoG and its Nζ is in position to hydrogen bond with the phosphate oxygen of 8-oxoG. This hydrogen bond interaction is reminiscent of, but not identical to, that seen in ternary complex structures with unmodified template nucleotides (Fig. 4B). Finally, the position of Lys280 in this structure is quite different from that in our earlier structure (28) with 8-oxoG in the anti-conformation (Fig. 4C). A survey of the various ternary complex structures of pol β revealed that the Lys280 side chain is flexible and can adopt multiple conformations (Fig. S4).
Fig. 4.
Ternary complex of human pol β with the 8-oxoG:C base pair in the nascent base pair binding pocket. (A) A detailed view of the active site with template anti-8-oxoG and incoming anti-dCMP(CF2)PP is shown. The perspective is from the major groove. The unmodified T:A base pair at the template-primer terminus in the closed ternary complex is shown. The side chain of Arg283 (R283), that hydrogen bonds to O8 and stabilizes the syn-conformation of 8-oxoG when it is positioned both as the template and at the template-primer terminus, is too far away to interact with O8 of 8-oxoG in anti-conformation. Helix N and its Lys280 (K280) and Asp276 (D276) side chains are illustrated, as is the hydrogen bond between Nη1 of Arg283 (R283) and the sugar oxygen of the template nucleotide at the template-primer terminus. Two active site water molecules (red spheres) and two magnesium ions (green spheres) are illustrated. The O3′ to Pα distance is 3.5 Å. (B) A superposition illustrating of the hydrogen bond between the 5′-phosphoryl oxygen and Nζ of the Lys280 (K280) side chain in two structures: The reference structure with unmodified template dG (blue) and the structure in panel A with template anti-8-oxoG (magenta). The 5′-phosphate is repositioned 2.9 Å to make room for O8 of anti-8-oxoG, but an approximately 2.9 Å hydrogen bond between Nζ of Lys280 (K280) and the 5′-phosphate is maintained. (C) A superposition illustrating key differences between the current ternary complex structure with anti-8-oxoG in the template and an earlier 8-oxoG:dCTP structure (6). The orientation of the Lys280 (K280) side chain in the current structure provides for stacking interactions with the purine base and hydrogen bonding with the 5′-phosphate group, as illustrated here and in (B).
Primer Extension from the Mutagenic Template-Primer Terminus.
Another important feature in 8-oxoG-induced mutagenesis is extension off the syn-8-oxoG:anti-A Hoogsteen base pair and anti-8-oxoG:anti-C base pair at the template-primer terminus. Kinetic characterization of extension of 8-oxoG-containing base pairs is summarized in Table S3. Extension off the template-primer terminus combinations of 8-oxoG/A and 8-oxoG/C was similar to that for unmodified G/C and nearly 1,000 times more efficient than extension off the mismatched G/A terminus (Table S3), indicating that the mutagenic base pair (8-oxoG/A) is well accommodated at the template-primer terminus. To gain insight on the structural basis for the efficient pol β extension off a template-primer terminus with the 8-oxoG:A base pair, we obtained the corresponding binary and ternary complex structures (Table S2). First, the binary complex crystal structure reveals that the enzyme is in the open conformation, as expected, and the syn-8-oxoG:anti-A base pair is at the template-primer terminus (Fig. S5A). At this 8-oxoG position in the DNA strand, repositioning of the phosphodiester backbone is not required to accommodate the syn-conformation. Next, the ternary complex crystal structure with the 8-oxoG:A base pair at the template-primer terminus was examined (Table S2 and Fig. S5B). The structure reveals the syn-8-oxoG:anti-A base pair at the template-primer terminus, and Arg283 stabilizes the syn-conformation of 8-oxoG through a hydrogen bond (3.27 Å) with O8. The positions of O3′ and key active site atoms are very similar to those in the reference structure with an unmodified template-primer terminus base pair (Fig. S5B). Taken together, these results indicate that pol β accommodates the syn-8-oxoG:anti-A base pair at the template-primer terminus very well, and the structural results are consistent with the kinetic data in Table S3. Similarly, the anti-8-oxoG:anti-C base pair at the template-primer terminus is not perturbed as shown in Fig. S6. The crystallographic data for these structures are presented in Table S4.
Discussion
An underlying molecular mechanism in etiology of some cancers appears to be 8-oxoG-induced mutations in growth control genes, among others, and in light of the central role of base excision repair in protection against 8-oxoG-induced mutagenesis, it is important to gain a better understanding of how this repair pathway responds to the lesion. We have reported here on features of how the gap-filling base excision repair enzyme pol β accommodates the mutagenic syn-conformation of template 8-oxoG in a Hoogsteen base pair with incoming dATP in the anti-conformation. The Hoogsteen base pairing itself stabilizes the syn-conformation of 8-oxoG, but the enzyme also stabilizes the syn-conformation using hydrogen bonding between O8 of 8-oxoG and a side chain (Arg283), along with an adjustment in the phosphodiester backbone of the template strand. This structural information is consistent with kinetic studies showing that pol β is capable of relatively strong promutagenic insertion of dATP opposite 8-oxoG in a BER substrate (Table S1). In addition, and as illustrated in Fig. 1, the 8-oxoG base in syn- and anti-conformations in the binary complex facilitates either dATP or dCTP insertion opposite template 8-oxoG, and there are corresponding differences in the positions of the 5′-phosphate of the 8-oxoG nucleotide.
Any potential clash between the 5′-phosphate and the 8-oxoG base appears to be minimized by several intrinsic features of the polymerase–DNA complex. The template strand has a 90° bend at the position of the template base, but this does not preclude the local rotation of backbone phosphate. A similar observation regarding a backbone bend at the template 8-oxoG base in anti-conformation was made for a ternary complex of the RB69 polymerase (32). In addition, the pol β active site has only one side chain in the immediate vicinity, Lys280, and this side chain does not restrict rotation of the phosphodiester backbone in the case of template 8-oxoG; in structures with unmodified substrates, the Nζ of Lys280 hydrogen bonds with the backbone 5′-phosphate of the template nucleotide at the approximately 90° bend. This interaction is preserved in the case of anti-8-oxoG (Fig. 4B), but is lost in the case of syn-8-oxoG. In the latter, the methylene side chain of Lys280 moves to stack with and stabilize the purine base, and the phosphodiester backbone 5′-phosphate moves away from the base (Fig. 3B). The stacking interaction of Lys280 over the template anti-8-oxoG base is reminiscent of the role proposed for Met135 in the human pol κ structure (33, 34). The flexibility of the phosphodiester backbone in the vicinity of the template base in human pol β also is reminiscent of the flexibility observed with the Y567A mutant of the RB69 polymerase (35, 36). Like pol β, this mutant enzyme has similar catalytic efficiencies for error-free and error-prone insertion opposite 8-oxoG (35), but the potential contribution of syn-, anti-conformational differences in the binary complex is unknown for this mutant enzyme, because the binary complex structure has not been reported.
To confirm the relevance of the positions suggested in the binary 8-oxoG complex conformations, we obtained preinsertion ternary complex structures corresponding to the syn- and anti-conformations of 8-oxoG. As noted above, previous efforts to obtain a pol β structure with the combination of template 8-oxoG and incoming dATP in the active site (28) resulted in an open conformation of the polymerase and an unusual arrangement in the active site including a hydrolyzed incoming nucleotide. This structure contained a dideoxy-terminated primer. In light of this experience, we used DNA with a primer O3′, and to freeze the reaction, we employed nonhydrolyzable incoming dNTPs. The recent use of nonhydrolyzable incoming dNTPs has enhanced pol β crystallography and has been found to be nondistorting for assembly of the preinsertion active site (31). The approach was successful here in that we obtained a high-resolution structure with the closed fully assembled preinsertion ternary complex containing the Hoogsteen base pair in the confines of the active site (Fig. 2). The key findings regarding stabilization of the mutagenic syn-8-oxoG:anti-A base pair may be summarized as follows: (i) there is a hydrogen bond between Arg283 and O8 of 8-oxoG in the minor groove, and (ii) there is a phosphodiester backbone adjustment to make room for 8-oxoG in the syn-conformation. This backbone adjustment at the 5′-phosphodiester bond of 8-oxoG is greater than that predicted from previous modeling of syn-8-oxoG into the pol β active site (37). The modeling predicted that N2 of 8-oxoG would be within 3.0 Å of its phosphoryl oxygen in the phosphodiester backbone, and that this could stabilize the syn-conformation without altering the position of phosphodiester backbone. This predicted N2 and 5′-phosphoryl oxygen proximity is similar to that seen in one of the conformations in the binary complex structure (Fig. 1D). However, the corresponding distance in the ternary complex structure is 5.8 Å.
Interestingly, the syn-8-oxoG:anti-A base pair is maintained when it is positioned at the template-primer terminus in the structures of binary and ternary complexes reported here (Fig. S5). A phosphodiester backbone adjustment is not observed in these cases. In the ternary complex with the syn-8-oxoG:anti-A base pair in the template-primer position, a hydrogen bond is observed between O8 of 8-oxoG and the side chain of Arg283 (Fig. S5B). Taken together, it appears that stabilization of the syn-conformation of 8-oxoG at the template-primer terminus is multifaceted. It is well-recognized from structural studies of duplex DNA that base pairing with A stabilizes the syn-conformation of 8-oxoG (27) and that local sequence context can influence 8-oxoG conformations (38). With the structures of Dpo4 and BF polymerases in binary complex, a similar situation was found regarding the syn-8-oxoG:anti-A base pair at the template-primer terminus. These structures, like the pol β structure, exhibit a lack of distortion in the enzyme and in the template and primer strands (11, 39). Thus, the syn-8-oxoG:anti-A base pair at the template-primer terminus is well accommodated by these three DNA polymerases. Another interesting feature regarding the syn-8-oxoG:anti-A base pair at the template-primer terminus is seen in kinetic studies of replicative polymerases with associated proofreading activity, such as T7 DNA polymerase, mammalian DNA polymerase δ, and RB69 DNA polymerase. These enzymes are more efficient at extending the syn-8-oxoG:anti-A base pair than the anti-8-oxoG:anti-C base pair (32, 40, 41). Also, it appears that the syn-8-oxoG:anti-A base pair is not detected as a mismatch in the proofreading activity of these enzymes (42). As shown here (Table S3), pol β has equal catalytic efficiencies for extending template-primers with the anti-8-oxoG:anti-C and syn-8-oxoG:anti-A base pairs. In contrast, Y-family DNA polymerases preferentially extend from anti-8-oxoG:anti-C than syn-8-oxoG:anti-A base pairs.
For repair of 8-oxoG-DNA, organisms use overlapping repair pathways (13, 15, 20), including DNA glycosylases and other BER enzymes, in successfully limiting accumulation of the oxidized base. The action of the DNA glycosylases may be coordinated with other components of the BER system, but nevertheless, glycosylase action results in potentially toxic single-strand breaks or abasic sites that are intermediates in the BER pathway. To avoid toxicity, it is likely that these intermediates are quickly processed to fully repaired DNA, including the gap-filling step by pol β. A block in the gap-filling step could trigger accumulation of toxic intermediates. Insertion of dCTP with unmodified template G is approximately 10-fold stronger than with template 8-oxoG. It remains to be seen whether this lower efficiency of gap-filling synthesis with template 8-oxoG could cause accumulation of sufficient BER intermediate to trigger the cell killing observed after exposure to oxidative stress (43). Also, an explanation for the lower insertion efficiency opposite 8-oxoG is not apparent from the present work.
Finally, the structural analyses of pol β in complex with 8-oxoG-containing substrates reported here and elsewhere (26) indicate that active site residues Arg283 and Asn279 form hydrogen bonds with O8 of the 8-oxoG base to help stabilize the mutagenic conformation when it is in the template position and the incoming nucleotide position, respectively. In both cases, the base combination of syn-8-oxoG and anti-adenine forms a Hoogsteen base pair that is compatible with significant pol β gap-filling synthesis. It is interesting that these two active site amino acids also contribute key roles in DNA synthesis with unmodified substrates. In addition, the sharp 90° bend in the architecture of the template strand in the pol β-DNA complex and the absence of protein interactions are associated with an intrinsic flexibility in the template strand phosphodiester backbone when 8-oxoG is the template base. Thus, the ability to accommodate the mutagenic syn-conformation of the 8-oxoG base appears to be an intrinsic feature of pol β. That this feature coevolved along with the ability of the enzyme to conduct the nucleotidyl transfer reaction with normal unmodified substrates suggests that the cell has potent repair strategies capable of limiting the deleterious effects of 8-oxoG (Fig. S1).
Materials and Methods
Human pol β was overexpressed in E. coli and purified (44). Binary complex crystals with 8-oxoG as the template base in a 1-nucleotide gapped DNA were grown as previously described (30). The sequence of the template strand (16-mer) with 8-oxoG (underlined) in the template (coding) position was 5′- CCG ACG TCG CAT CAG C- 3′. The primer strand (10-mer) sequence was 5′- GCT GAT GCG A -3′. The downstream oligonucleotide (5-mer) was phosphorylated, and the sequence was 5′- GTC CC—3′. The binary complex crystals were then soaked in artificial mother liquor (50 mM imidazole, pH 7.5, 20% PEG3350, 90 mM sodium acetate, 200 mM MgCl2 with nonhydrolyzable dNTP analogs [5 mM dAMP(CH2)PP or dCMP(CF2)PP] and 12% ethylene glycol as cryoprotectant. This resulted in ternary complex crystals. Further information regarding materials and methods can be found in SI Text.
Supplementary Material
Acknowledgments.
We thank Thomas Upton for synthesis of dCMP(CF2)PP, Lars Pedersen and Bret Freudenthal for comments on the manuscript, and Bonnie Mesmer for editorial assistance. We thank Rajen Prasad and Esther Hou for purifying the enzyme. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (Z01-ES050158) and in association with NIH Grant 1U19CA105010.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 3RJE—3RJK).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112235108/-/DCSupplemental.
References
- 1.Dizdaroglu M. Chemical determination of free radical-induced damage to DNA. Free Radic Biol Med. 1991;10:225–242. doi: 10.1016/0891-5849(91)90080-m. [DOI] [PubMed] [Google Scholar]
- 2.Ames BN, Gold LS. Endogenous mutagens and the causes of aging and cancer. Mutat Res. 1991;250:3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 3.Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: Formation, measurement, and biochemical features. Mutat Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Nishimura S. 8-Hydroxyguanine: From its discovery in 1983 to the present status. Proc Jpn Acad Ser B Phys Biol Sci. 2006;82:127–141. doi: 10.2183/pjab.82.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: An enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 6.Culp SJ, Cho BP, Kadlubar FF, Evans FE. Structural and conformational analyses of 8-hydroxy-2′-deoxyguanosine. Chem Res Toxicol. 1989;2:416–422. doi: 10.1021/tx00012a010. [DOI] [PubMed] [Google Scholar]
- 7.Uesugi S, Ikehara M. Carbon-13 magnetic resonance spectra of 8-substituted purine nucleosides. Characteristic shifts for the syn conformation. J Am Chem Soc. 1977;99:3250–3253. doi: 10.1021/ja00452a008. [DOI] [PubMed] [Google Scholar]
- 8.Broyde S, et al. Lesion processing: High-fidelity versus lesion-bypass DNA polymerases. Trends Biochem Sci. 2008;33:209–219. doi: 10.1016/j.tibs.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vega M, Salas M. A highly conserved tyrosine residue of family B DNA polymerases contributes to dictate translesion synthesis past 8-oxo-7,8-dihydro-2′-deoxyguanosine. Nucleic Acids Res. 2007;35:5096–5107. doi: 10.1093/nar/gkm545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eoff RL, Angel KC, Egli M, Guengerich FP. Molecular basis of selectivity of nucleoside triphosphate incorporation opposite O6-benzylguanine by sulfolobus solfataricus DNA polymerase Dpo4: steady-state and pre-steady-state kinetics and X-ray crystallography of correct and incorrect pairing. J Biol Chem. 2007;282:13573–13584. doi: 10.1074/jbc.M700656200. [DOI] [PubMed] [Google Scholar]
- 11.Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 12.Rechkoblit O, et al. Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases. Structure. 2009;17:725–736. doi: 10.1016/j.str.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boiteux S, le Page F. Repair of 8-oxoguanine and Ogg1-incised apurinic sites in a CHO cell line. Prog Nucleic Acid Res Mol Biol. 2001;68:95–105. doi: 10.1016/s0079-6603(01)68092-9. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura S. Mammalian Ogg1/Mmh gene plays a major role in repair of the 8-hydroxyguanine lesion in DNA. Prog Nucleic Acid Res Mol Biol. 2001;68:107–123. doi: 10.1016/s0079-6603(01)68093-0. [DOI] [PubMed] [Google Scholar]
- 15.Russo MT, et al. Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Ogg1 DNA glycosylases. Cancer Res. 2004;64:4411–4414. doi: 10.1158/0008-5472.CAN-04-0355. [DOI] [PubMed] [Google Scholar]
- 16.Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feig DI, Reid TM, Loeb LA. Reactive oxygen species in tumorigenesis. Cancer Res. 1994;54:1890s–1894s. [PubMed] [Google Scholar]
- 18.Meira LB, et al. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J Clin Invest. 2008;118:2516–2525. doi: 10.1172/JCI35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan X, Grollman AP, Shibutani S. Comparison of the mutagenic properties of 8-oxo-7,8-dihydro-2′-deoxyadenosine and 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA lesions in mammalian cells. Carcinogenesis. 1999;20:2287–2292. doi: 10.1093/carcin/20.12.2287. [DOI] [PubMed] [Google Scholar]
- 20.Xie Y, et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 21.Al-Tassan N, et al. Inherited variants of MYH associated with somatic G : C → T : A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 22.Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 23.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 25.Nakabeppu Y. Regulation of intracellular localization of human MTH1, OGG1, and MYH proteins for repair of oxidative DNA damage. Prog Nucleic Acid Res Mol Biol. 2001;68:75–94. doi: 10.1016/s0079-6603(01)68091-7. [DOI] [PubMed] [Google Scholar]
- 26.Batra VK, et al. Mutagenic conformation of 8-oxo-7,8-dihydro-2′-dGTP in the confines of a DNA polymerase active site. Nat Struct Mol Biol. 2010;17:889–890. doi: 10.1038/nsmb.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beard WA, Batra VK, Wilson SH. DNA polymerase structure-based insight on the mutagenic properties of 8-oxoguanine. Mutat Res. 2010;703:18–23. doi: 10.1016/j.mrgentox.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krahn JM, et al. Structure of DNA polymerase β with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 29.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase β. Chem Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 30.Batra VK, et al. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14:757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batra VK, et al. Structures of DNA polymerase β with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol Cell. 2008;30:315–324. doi: 10.1016/j.molcel.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freisinger E, Grollman AP, Miller H, Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irimia A, Eoff RL, Guengerich FP, Egli M. Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase κ opposite the 7,8-dihydro-8-oxo-2′-deoxyguanosine adduct. J Biol Chem. 2009;284:22467–22480. doi: 10.1074/jbc.M109.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasquez-Del Carpio R, et al. Structure of human DNA polymerase κ inserting dATP opposite an 8-oxoG DNA lesion. PLoS ONE. 2009;4:e5766. doi: 10.1371/journal.pone.0005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckman J, et al. Substitution of ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine. Biochemistry. 2010;49:4116–4125. doi: 10.1021/bi100102s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hogg M, et al. Kinetics of mismatch formation opposite lesions by the replicative DNA polymerase from bacteriophage RB69. Biochemistry. 2010;49:2317–2325. doi: 10.1021/bi901488d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krahn JM, Beard WA, Wilson SH. Structural insights into DNA polymerase β deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure. 2004;12:1823–1832. doi: 10.1016/j.str.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Cheng X, et al. Dynamic behavior of DNA base pairs containing 8-oxoguanine. J Am Chem Soc. 2005;127:13906–13918. doi: 10.1021/ja052542s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oda Y, et al. NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 1991;19:1407–1412. doi: 10.1093/nar/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Einolf HJ, Schnetz-Boutaud N, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of 8-oxo-7,8-dihydroguanosine triphosphate incorporation and extension by replicative and repair DNA polymerases. Biochemistry. 1998;37:13300–13312. doi: 10.1021/bi981346d. [DOI] [PubMed] [Google Scholar]
- 41.Furge LL, Guengerich FP. Pre-steady-state kinetics of nucleotide insertion following 8-oxo-7,8-dihydroguanine base pair mismatches by bacteriophage T7 DNA polymerase exo. Biochemistry. 1998;37:3567–3574. doi: 10.1021/bi9722094. [DOI] [PubMed] [Google Scholar]
- 42.Brieba LG, et al. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakabeppu Y, et al. Programmed cell death triggered by nucleotide pool damage and its prevention by MutT homolog-1 (MTH1) with oxidized purine nucleoside triphosphatase. Mutat Res. 2010;703:51–58. doi: 10.1016/j.mrgentox.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Beard WA, Wilson SH. Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol. 1995;262:98–107. doi: 10.1016/0076-6879(95)62013-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.