Abstract
Individual microRNAs (miRNAs) are rapidly down-regulated during conditions of cellular activation and infection, but factors mediating miRNA turnover are poorly understood. Infection of mouse cells with murine cytomegalovirus (MCMV) induces the rapid down-regulation of an antiviral cellular miRNA, miR-27. Here, we identify a transcript produced by MCMV that binds to miR-27 and mediates its degradation. UV-crosslinking and high-throughput sequencing [CRAC (UV-crosslinking and analysis of cDNA)] identified MCMV RNA segments associated with the miRNA-binding protein Argonaute 2 (Ago2). A cluster of hits mapped to a predicted miR-27-binding site in the 3′UTR of the previously uncharacterized ORF, m169. The expression kinetics of the m169 transcript correlated with degradation of miR-27 during infection, and m169 expression inhibited miR-27 functional activity in a reporter assay. siRNA knockdown of m169 demonstrated its requirement for miR-27 degradation following infection and did not affect other host miRNAs. Substitution of the miR-27-binding site in m169 to create complementarity to a different cellular miRNA, miR-24, resulted in down-regulation of only miR-24 following infection. The m169 transcript is cytoplasmic, capped, polyadenylated, and interacts with miRNA-27 through seed pairing: characteristic features of the normal messenger RNA (mRNA) targets of miRNAs. This virus–host interaction reveals a mode of miRNA regulation in which a mRNA directs the degradation of a miRNA. We speculate that RNA-mediated miRNA degradation could be a more general viral strategy for manipulating host cells.
Keywords: herpesvirus, RNA crosslinking, RNA degradation, RNA turnover
Viruses devote a large portion of their genomes to strategies for manipulating host cells and evading antiviral defense mechanisms. Numerous host miRNA-binding sites have been predicted in different viral genomes, but the validity and functional relevance of most predictions remain unclear. The best studied miRNA–virus interactions demonstrate that RNA viruses can use cellular miRNAs to regulate their life cycles; for example, the interaction between hepatitis C virus and miR-122 enhances viral replication (1), whereas the interaction between HIV-1 and miR-29 mediates its localization to P bodies (2). Direct interactions between host miRNAs and viral genes can also suppress viral gene expression and replication (3–6) (reviewed in Ref. 7). However, the factors driving the evolution of these interactions remain somewhat controversial, because they may relate to viral mechanisms for persistence and latency rather than host defense (8, 9). At the same time, the expression levels of specific miRNAs can indirectly influence infections; miRNAs are key components of the innate immune response (10, 11) and exert antiviral properties by modulating host cofactors and pathways required by viruses (12–15). We previously showed that miR-27 limits the replication capacity of murine cytomegalovirus (MCMV) but is rapidly degraded during the lytic infection (16). Actinomycin D treatment upon infection prevents miR-27 down-regulation, suggesting that an RNA produced during infection might be involved (16). A small nuclear RNA (snRNA) in Herpesvirus saimiri (HVS), HSUR-1, was recently shown to bind to miR-27 and mediate its degradation (17). However, no homolog of HSUR-1 has been identified in other viral families. miRNA turnover mechanisms in animals remain poorly characterized (18). The aim of this work was to identify miR-27 interaction sites in the MCMV transcriptome and determine whether any of these could mediate miR-27 turnover. To achieve this aim, we implemented a UV-crosslinking method that precisely mapped a miR-27-binding site to the previously uncharacterized viral transcript, m169. Our findings demonstrate that, despite hundreds of predicted miR-27-binding sites, one sequence is responsible for the degradation of miR-27 upon infection.
Results
High-Throughput Identification of MCMV RNAs Associated with Mouse Argonaute 2.
The probability of finding potential miRNA-binding sites at random is high in the large (230-kb) MCMV genome. At least 134 potential miR-27–binding sites are predicted using criteria of seed pairing and a minimal free energy cutoff of −20 kcal/mol (Fig. S1). We, therefore, attempted to experimentally identify binding sites for miR-27 on the MCMV transcriptome. miRNAs function within the RNA-induced silencing complex (RISC), the effectors of which are Argonaute (Ago) proteins. We adapted the UV-crosslinking and analysis of cDNA (CRAC) technique (19) to identify RNA sequences bound to the Ago2 protein. Living cells expressing tagged Ago2 were UV-irradiated at 254 nm, and RNA was purified and reverse transcribed, and the cDNAs were sequenced on a Solexa GAII. The approach is similar to previous HITS-CLIP (high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation) analyses (20), but the Ago2 protein was N-terminally tagged with protein A and His6 (PTH-Ago2) to enable purification under highly denaturing conditions (Fig. 1 and SI Materials and Methods). Transfection with the PTH-Ago2 construct resulted in modest over-expression (1.4-fold) of the tagged protein compared with endogenous Ago2 in NIH 3T3 cells under conditions where > 70–80% of cells are transfected (Fig. S2). Transfection with PTH-Ago2 did not interfere with the viral infection, as assessed by comparing immediate early viral gene expression (ie1) and viral growth kinetics in untransfected and transfected cells (Fig. S2). For the purpose of this study, cells were mock-infected or infected with MCMV at 20 h posttransfection at a multiplicity of infection (MOI) of 5. Analyses were performed 8 h postinfection; under these infection conditions, approximately half of miR-27 is degraded (Fig. S3), indicating that the putative inhibitor is likely to be associated with the RISC at this time point.
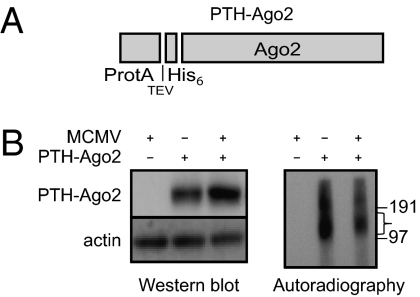
Fig. 1.
Ago2-CRAC method in MCMV-infected NIH 3T3 cells. (A) Schematic of PTH-Ago2 construct. (B) Left, Western blot showing expression of PTH-Ago2 in uninfected and infected cells at 8 h postinfection, detected using an anti-PAP antibody recognizing the protein A tag. (B) Right, nitrocellulose membrane showing 32P-labeled RNA crosslinked to mouse PTH-Ago2 following immunoprecipitation and resolution on a 4–12% Bis-Tris NuPage gel. The excised fragment used for further steps in the CRAC protocol is bracketed and protein size markers indicated.
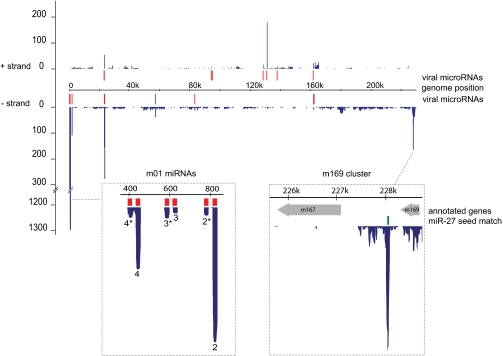
Cells were crosslinked and lysed, and the RNA–protein complexes were purified as described (19), with modifications detailed in SI Materials and Methods. RNA classes identified in uninfected and infected cells are indicated in Table S1. In this study, we focus exclusively on reads mapped to the viral genome (∼1% of total reads; Table S1). Almost all regions with high hit density represented annotated viral miRNAs; clusters of reads from 15 of the 18 previously reported pre-miRNAs (21, 22) are recovered at this early time point (Table S2). The clusters with the highest hit density mapped to miRNAs in the m01 and M23 clusters (Fig. 2 and Table 1). The data shown in Fig. 2 are based on one sample, but the rankings of viral hit densities were comparable in another sample prepared from infected cells at the same time point, where miR-27 was overexpressed by transfection of a synthetic mimic (Table S3).
Fig. 2.
Location of Ago2-binding sites in the MCMV transcriptome. Distribution of Solexa reads (blue) on the (+) and (−) strands of the MCMV genome, in comparison with location of annotated viral miRNAs (red). Dashed boxes show a zoomed in view of two regions of the genome with high hit density clusters (Table 1): the m01 miRNAs (left) and a region of the genome downstream of the annotated ORF, m169 (right). The predicted binding site for miR-27 is shown in green.
Table 1.
Top Ago2-binding sites in the MCMV transcriptome
| Rank | Start | End | Genome strand | Cluster hit density | Annotation |
| 1 | 811 | 835 | − | 1,020 | miR-m01-2 |
| 2 | 433 | 457 | − | 462 | miR-m01-4 |
| 3 | 23,665 | 23,689 | − | 237 | miR-M23-1–3p |
| 4 | 131,353 | 131,377 | + | 169 | miR-m88-1 |
| 5 | 228,044 | 228,068 | − | 146 | 3′ UTR of predicted m169 |
| 6 | 23,468 | 23,492 | − | 130 | miR-M23-2 |
| 7 | 2,008 | 2,032 | − | 95 | Unannotated miR-m01-1* |
| 8 | 574 | 598 | − | 85 | miR-m01-3* |
| 9 | 395 | 419 | − | 81 | miR-m01-4* |
| 10 | 767 | 791 | − | 59 | miR-m01-2* |
m169 RNA Is a 1,669-nt mRNA with a miR-27-Binding Site in Its 3′UTR That Cross-Links to Argonaute 2.
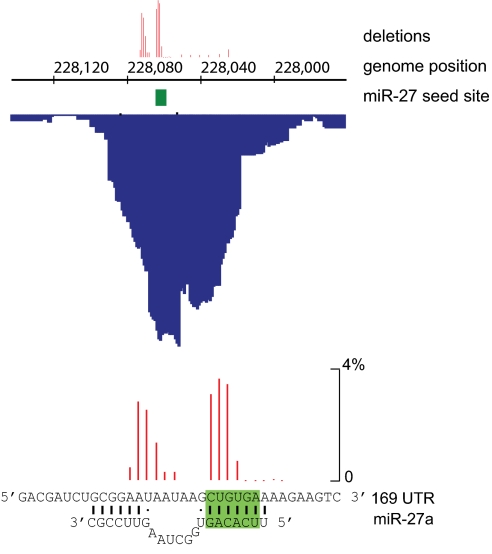
The cluster with the fifth highest peak hit density mapped to positions 228,044–228,068 in the (−) strand of the viral genome and included a predicted miR-27-binding site with complementarity to nucleotides 1–7 and 16–21 in the miRNA (Fig. 3). In CRAC and related techniques, an increased rate of microdeletions is seen in the sequence data at the site of protein crosslinking (19, 23–26). This is attributable to errors introduced by reverse transcriptase when bypassing the crosslinked site and can be used to pinpoint the precise protein-binding site. Strikingly, the highest incidence of deletions localized to the region complementary to miR-27 (Fig. 3). This miR-27-binding site is 231 nt downstream of the m169 ORF, which has predicted TATA-binding elements and a polyadenylation signal as indicated in Fig. 4A. Using 5′ RACE (27), we identified a transcript produced from this locus with a transcriptional start site at position 229,097 (including a nontemplated G at the 5′ end, consistent with the presence of a cap), and this was validated by primer extension (Fig. 4B). The 3′ end of the transcript was identified based on reverse transcription with an oligo(dT) adapter primer followed by PCR and sequencing, as described in Ref. 28. This procedure confirmed the presence of a poly(A) tail and mapped the 3′ end to position 227,429, which is 23 nt downstream of the predicted polyadenylation signal (Fig. 4). Northern blot analysis with a probe spanning the miR-27-binding site confirmed that a ∼1.7-kb RNA is produced from this locus, and no smaller processing products were observed (Fig. 4B). The expression kinetics of the m169 RNA were closely correlated with the degradation of miR-27 upon infection (Fig. 4C). As expected, treatment of cells with actinomycin D blocked m169 RNA expression (Fig. 4C) and rescued miR-27 down-regulation upon infection. Absolute quantitative (q)RT-PCR analysis of miR-27 and m169 levels in infected cells in comparison with synthetic miR-27 and in vitro-transcribed m169 standards demonstrated that similar amounts of each RNA are expressed between 12–24 h postinfection (Fig. 4D). We conclude that MCMV generates a 1,669-nt transcript that is produced during the lytic infection and contains a miR-27-binding site in its 3′UTR that precisely matches a site of Ago2 association.
Fig. 3.
Site of Ago2 crosslinking to the m169 transcript. Distribution of Solexa reads (blue) downstream of the m169 ORF (Upper) and complementarity between the viral RNA sequence and miR-27 (Lower). Location of deletions identified in Solexa reads are shown in red on a scale depicting percentage deletions at each location; the miR-27 seed site is shown in green.
Fig. 4.
m169 is a 1,669-nt transcript with expression kinetics that correlate with the degradation of miR-27. (A) Schematic of m169 gene locus based on 5′ and 3′ RACE. (B) Left, validation of m169 transcriptional start site based on primer extension using a 20-nt primer starting at position 229,039. (B) Center, PCR product of the m169 transcript using a forward primer at the transcriptional start site and reverse primer at the 3′ end; the cDNA was created with an oligo(dT) primer. (B) Right, Northern blot analysis with a 80-nt probe that spans the miR-27-binding site in m169. RNA was collected from NIH 3T3 cells mock-infected (−) or infected with MCMV at MOI = 5 (+) for 48 h and then resolved on a formaldehyde-agarose gel. (C) Left, Northern blot analysis with RNA collected from NIH 3T3 cells mock infected (−) or infected (+) with MCMV (MOI = 5) for the indicated times. (C) Right, Northern blot analysis with RNA collected from NIH 3T3 cells mock-infected or infected with MCMV (MOI = 5) for 16 h in the absence and presence of 5 μg/mL actinomycin D (ACT); 5 μg of total RNA was loaded per lane. (D) Absolute quantification of m169 and miR-27 at the indicated time points postinfection (MOI = 5) based on qRT-PCR, using synthetic miR-27 and in vitro transcribed m169 as standards.
MCMV m169 RNA Blocks miR-27 Functional Activity and Mediates miR-27 Degradation.
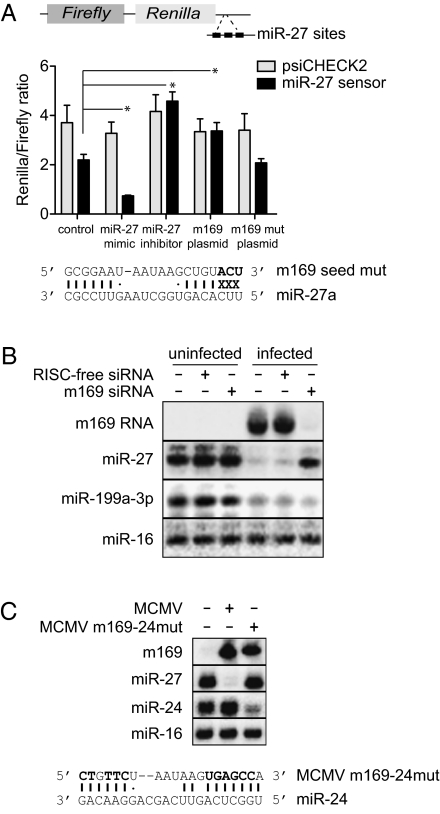
To examine the effect of m169 RNA on the functional activity of miR-27, we constructed a miR-27 sensor containing three miR-27-binding sites behind the 3′UTR of Renilla luciferase in the psiCHECK2 vector (Fig. 5A). The miR-27-binding sites are perfectly complementary to miR-27 but contain an internal loop at nucleotides 9–12 to prevent endonucleolytic cleavage, as described in Ref. 29. The miR-27 sensor displayed an approximately twofold reduction in the ratio of Renilla/Firefly expression compared with the control psiCHECK2 vector, consistent with regulation by endogenous miR-27 (Fig. 5A). miR-27-dependent repression of the Renilla luciferase was enhanced by cotransfection with a synthetic miR-27 mimic (resulting in an approximately fivefold reduction in the Renilla/Firefly ratio). In contrast, cotransfection of a synthetic miR-27 inhibitor relieved the repression, Fig. 5A. Cotransfection of the sensor with a plasmid encoding m169 produced the same result as the synthetic miR-27 inhibitor, relieving the repression of Renilla luciferase with no effects on the control psiCHECK2 vector. Mutation of nucleotides in m169 that interact with positions 1–3 in miR-27 eliminated this effect (Fig. 5A). To directly examine the impact of m169 on miR-27 during infection, we measured miR-27 levels in cells in which m169 RNA was depleted by a custom pool of 3 siRNAs targeting the viral transcript. Transfection with the m169 siRNAs resulted in >95% knockdown of m169 and resulted in 80% relief of miR-27 down-regulation at 24 h postinfection (Fig. 5B). Transfection of RISC-free siRNA (a small RNA duplex that is taken up by cells but is chemically modified to prevent incorporation into RISC) or an siRNA targeting GAPDH had no effect on miR-27 levels in uninfected or infected cells (Fig. 5B and Fig. S4). We, therefore, conclude that the rescue of miR-27 levels in these experiments is not attributable to nonspecific effects of transfection or saturation of the RISC machinery by the siRNAs. The specificity of m169 function was confirmed by comparison with two other host miRNAs: miR-16, a miRNA that does not change in expression level in response to infection and miR-199a-3p, another miRNA that is down-regulated during infection (14). Neither miRNA displayed a significant change in expression level as a result of m169 knockdown (Fig. 5B), supporting the conclusion that the m169 RNA specifically targets miR-27, rather than globally inhibiting the host miRNA machinery. To confirm that miR-27 degradation requires base pair interactions with m169, we introduced 11 mutations into the miRNA-binding site in the viral genome. The mutations eliminate pairing to miR-27 and introduce complementarity to another cellular miRNA, miR-24. miR-24 is derived from the same primary transcript as miR-27 and also displays antiviral properties against MCMV (14) but does not change in expression level during the lytic infection (16). As shown in Fig. 5C, substitution of 11 nt in m169 abolished virus-induced degradation of miR-27 and resulted in a gain-of-function ability of MCMV to down-regulate miR-24.
Fig. 5.
m169 blocks miR-27 functional activity, is responsible for miR-27 degradation and can be directed to target another cellular miRNA. (A) The miR-27 sensor contains three miR-27-binding sites spaced 11-nt apart in the 3′UTR of Renilla luciferase in the psiCHECK2 vector. The graph shows ratios of Renilla to Firefly luciferase for the psiCHECK2 vector compared with the miR-27 sensor vector following transfection of 50 ng of each into NIH 3T3 cells in combination with: 25 nM synthetic miR-27 mimic, 25 nM synthetic miR-27 inhibitor (Thermofisher), 50 ng of pCMV-m169 plasmid, 50 ng of pCMV-m169-mut plasmid, or 50 ng pCMV control plasmid (n = 4 independent experiments; error bars represent SDs; *P < 0.005). The location and identity of the seed mutations in the m169 mutant plasmid are noted in bold. (B) Northern blot analysis following transfection of NIH 3T3 cells with 25 nM m169 siRNA pool or RISC-free control siRNA before mock infection (“uninfected”) or infection with MCMV at MOI = 5 for 24 h. (C, Upper) Northern blot analysis of NIH 3T3 cells mock-infected or infected with the wild-type virus or the mutant virus (MCMV-24mut) at MOI = 5 for 24 h. (C, Lower) Schematic of the mutations in m169 (bold) that introduce complementarity to miR-24 and eliminate complementarity to miR-27.
m169 RNA Is Unrelated to the Other Known miR-27 Inhibitor, HSUR-1, and Localizes to the Cytoplasm.
The only other known miRNA inhibitor that has been described in a mammalian system is HSUR-1, a snRNA that mediates miR-27 degradation in HVS-transformed T cells (17). A comparison of HSUR-1 and m169 sequences shows identical sequence at the site of base-pairing to nucleotides 2–7 and 16–19 in miR-27. However, the length of the central bulge and extent of 3′ pairing to miR-27 differs in these two RNAs: HSUR-1 is complementary to eight consecutive nucleotides in the 3′ end of miR-27 with a 3-nt central bulge, whereas m169 is complementary to six consecutive nucleotides in the 3′ end with a 5-nt central bulge flanked by G-U wobble pairs (Fig. 6A). These RNAs differ significantly in length (143 nt versus 1,669 nt) and, apart from the miR-27-binding site, do not display obvious homology in primary sequence or predicted folding around the miR-27-binding site. Because m169 and HSUR-1 are unlikely to derive from a common ancestor, their roles in mediating miR-27 degradation may represent convergent evolution. The mechanism of miR-27 degradation induced by HSUR-1 is not yet known, but HSUR-1, like other snRNAs, associates with Sm proteins and localizes to the nucleus (30). There are also reports that some cellular miRNAs display nuclear localization (31, 32). To examine localization of m169 and miR-27, we carried out in situ hybridizations with digoxigenin (DIG)-labeled probes directed against these RNAs in uninfected and infected cells. As shown in Fig. 6B, the miR-27 signal in uninfected cells appears in the cytoplasm and displays a clear reduction in intensity at 16 h postinfection. A probe against miR-16 demonstrates that this miRNA also localizes to the cytoplasm but shows no loss of signal upon infection (Fig. 6B). A probe against m169 RNA revealed it also to be localized within the cytoplasm in infected cells, and no signal was observed using a probe against the antisense sequence of m169 (Fig. 6B). We, therefore, conclude that m169 RNA and miR-27 predominantly localize to the cytoplasm in these cells.
Fig. 6.
Sequence alignment of miR-27-binding sites in HSUR-1 and m169 and cytoplasmic localization of miR-27 and m169 in NIH 3T3 cells. (A) Complementarity of m169 and HSUR-1 to miR-27; green box indicates the seed site. (B) In situ detection of DIG-labeled probes against miR-27, miR-16, m169, and antisense m169 in NIH 3T3 cells, uninfected or at 16 h postinfection. DIG-labeled probes were detected by FITC-tyramide amplification (green) and overlaid on nuclear DAPI signal (blue). Images are representative of three independent experiments.
Discussion
The interpretation of miRNA-binding sites within animal and viral genomes has generally been based on the expectation that miRNAs exert a negative regulatory function on the genes with which they interact. The findings presented here suggest that the opposite can also occur, with the “target” mRNA actively regulating the miRNA. Specifically, we demonstrate that MCMV encodes a miRNA inhibitor that is responsible for the robust down-regulation of miR-27 observed during the lytic infection (16). We further show that the miR-27 inhibitor is capped, polyadenylated, and expressed in the cytoplasm at a comparable level to miR-27 between 12 and 24 h postinfection (Figs. 4 and 6B). Hundreds of potential miR-27 interaction sites are predicted in the MCMV transcriptome. However, these analyses demonstrate that m169 is the only inhibitor responsible for the degradation of miR-27 upon infection. This underscores the importance of biochemical approaches for identifying functional miRNA-binding sites in viruses.
Only one other miRNA inhibitor has been identified to date in a viral system: HSUR-1, the snRNA encoded by HVS, which does not share obvious features in common with m169, beyond the miR-27-binding site. HVS is a γ-herpesvirus that infects New World primates and is separated from MCMV by hundreds of millions of years of evolution (33). The existence of a miR-27 inhibitor in MCMV suggests that this class of molecule might be a more common feature in viral genomes and illuminates the potential diversity of RNAs that can mediate miRNA degradation. Our results also suggest that miRNA inhibitors might readily be mistaken for targets, because m169 appears strikingly similar to a “normal” targeted mRNA. Specifically, the miRNA-binding site is in the 3′UTR of an annotated ORF and the miR-27–m169 binding interaction involves seed pairing, a predominant recognition criteria for bona fide targets of miRNAs (34). It is, therefore, possible that other “killer targets” exist that have not been annotated as such.
RNA–RNA recognition is the central means by which small guide RNAs, including miRNAs, mediate specificity in interactions between ribonucleoprotein complexes. Our findings support the idea that this principle extends to regulation of the guide RNAs themselves, enabling specific miRNAs to be regulated without global effects on the miRNAome. This concept was first introduced in the context of “miRNA sponges,” a term coined for transgenes containing multiple miRNA-binding sites that act as competitive inhibitors of a miRNA (35). In several cases, sponges, or related constructs, have also been shown to cause a reduction in the level of the endogenous miRNA (36, 37). Ameres et al. (38) recently showed that extensive complementarity between a miRNA and an artificial target can induce miRNA degradation in flies, which was also observed in HeLa cells. This degradation was attributed to the 3′ end pairing and subsequent trimming of the miRNAs, which may relate to the observations with sponges, reviewed in Ref. 39. The m169–miR-27 binding interaction does not appear to fit the miRNA degradation model of Ameres et al. (38), because of the length of the central bulge. However, further work is required to define the criteria that determine whether a miRNA interaction partner is a target, sponge, or other type of inhibitor. This is likely to relate to the concentration of the interaction partner in relation to the miRNA, the pattern and extent of pairing between the two RNAs (beyond seed site recognition), and, potentially, the presence of motifs in the interaction partner involved in recruiting factors involved in degradation. At present, it is unclear why only miR-27 is targeted by MCMV. This miRNA is derived from a primary transcript that also encodes miR-23 and miR-24. All three miRNAs in this cluster target genes associated with cell proliferation, differentiation, and cancer and are predicted to function in a cooperative fashion (40). miR-24, in addition to miR-27, displays antiviral properties when overexpressed (14), and we show that the virus can be engineered to target miR-24 by mutation of 11 nt in the miRNA-binding site (Fig. 5C). Our findings suggest that viruses may have the capacity to target various cellular miRNAs. Further analyses will shed light on why two diverse herpesviruses have devoted genome space to specifically inhibiting miR-27.
Materials and Methods
For a full description of all methods used here, see SI Materials and Methods. All oligonucleotides used in these studies are listed in Table S4. The tagged mouse Ago2 was constructed by ligating the PTH tag sequence (amplified from the pRS415 plasmid containing the ProtA-TEV-His6-MCS construct, a generous gift from the laboratory of Markus T. Bohnsack, Institut für Molekulare Biowissenschaften, Goethe Universität Frankfurt, Frankfurt, Germany) to the N terminus of mouse Ago2, which was then cloned into the pCDNA3 vector. NIH 3T3 cells (1.2 × 107 cells per sample, distributed in four 15-cm dishes) were transfected with 60 μg of pcDNA3-PTH-Ago2 plasmid in 0.3% Lipofectamine 2000 (Invitrogen) and 20 h posttransfection infected with MCMV (Smith strain) at an MOI of 5. At 8 h postinfection, cells were washed with PBS, immediately UV-irradiated at 254 nm (400 mJ/cm2; Uvitec), and the CRAC pull-down technique was performed as described previously (19), with modifications detailed in SI Materials and Methods. For construction of the miR-27 sensor, three miR-27-binding sites spaced 11 nt apart were cloned behind the 3′UTR of Renilla luciferase in the psiCHECK2 vector (Promega) as in Ref. 29. Knockdown of m169 was carried out by transfecting a pool of custom siRNAs into NIH 3T3 cells at a final concentration of 25 nM in 0.3% DharmaFECT 1 (Thermofisher) 24 h before infection. MCMV was derived from the pSM3fr BAC (41). The MCMV-24 mutant was constructed using the adapted protocol described in Ref. 42, with modifications described in SI Materials and Methods. For in situ hybridization analyses, LNA-incorporated miRNA probes were purchased from Exiqon and the m169 probe and antisense control probe were generated using the DIG RNA labeling kit (Roche).
Supplementary Material
Acknowledgments
We thank the Genepool at the University of Edinburgh for assistance with sequencing and Anna Hoy, Nouf Laqtom, and Hannah Stevens for technical assistance and helpful discussions. This work was supported by the Biotechnology and Biological Sciences Research Council (A.H.B.) and Wellcome Trust (D.T.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114204109/-/DCSupplemental.
References
- 1.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 2.Nathans R, et al. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pedersen IM, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lecellier CH, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 5.Otsuka M, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Ahluwalia JK, et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahajan VS, Drake A, Chen J. Virus-specific host miRNAs: antiviral defenses or promoters of persistent infection? Trends Immunol. 2009;30:1–7. doi: 10.1016/j.it.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 10.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagos D, et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 12.Sung TL, Rice AP. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog. 2009;5:e1000263. doi: 10.1371/journal.ppat.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang FZ, et al. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol. 2008;82:9065–9074. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santhakumar D, et al. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc Natl Acad Sci USA. 2010;107:13830–13835. doi: 10.1073/pnas.1008861107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Triboulet R, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 16.Buck AH, et al. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA. 2010;16:307–315. doi: 10.1261/rna.1819210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci USA. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buck AH, et al. Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol. 2007;81:13761–13770. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dölken L, et al. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J Virol. 2007;81:13771–13782. doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 2011;30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 27.Scotto-Lavino E, Du G, Frohman MA. 5′ end cDNA amplification using classic RACE. Nat Protoc. 2006;1:2555–2562. doi: 10.1038/nprot.2006.480. [DOI] [PubMed] [Google Scholar]
- 28.Scotto-Lavino E, Du G, Frohman MA. 3′ end cDNA amplification using classic RACE. Nat Protoc. 2006;1:2742–2745. doi: 10.1038/nprot.2006.481. [DOI] [PubMed] [Google Scholar]
- 29.Vermeulen A, et al. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA. 2007;13:723–730. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SI, Murthy SC, Trimble JJ, Desrosiers RC, Steitz JA. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988;54:599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- 31.Castanotto D, Lingeman R, Riggs AD, Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci USA. 2009;106:21655–21659. doi: 10.1073/pnas.0912384106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 33.McGeoch DJ, Cook S, Dolan A, Jamieson FE, Telford EA. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- 34.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horie T, et al. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem Biophys Res Commun. 2009;389:315–320. doi: 10.1016/j.bbrc.2009.08.136. [DOI] [PubMed] [Google Scholar]
- 37.Sayed D, et al. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ameres SL, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a∼27a∼24-2 cluster and its implication in human diseases. Mol Cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner M, Jonjic S, Koszinowski UH, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Britt WJ, Jarvis M, Seo JY, Drummond D, Nelson J. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J Virol. 2004;78:539–543. doi: 10.1128/JVI.78.1.539-543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.