Abstract
Proteins segregate into discrete subcellular compartments via a variety of mechanisms, including motor protein transport, local binding, and diffusion barriers. This physical separation of cell functions serves, in part, as a mechanism for controlling compartment activity by allowing regulation of local protein concentrations. In this study we explored how soluble protein size impacts access to the confined space within the retinal photoreceptor outer segment signaling compartment and discovered a strikingly steep relationship. We find that GFP monomers, dimers, and trimers expressed transgenically in frog rods are present in the outer segment at 1.8-, 2.9-, and 6.8-fold lower abundances, relative to the cell body, than the small soluble fluorescent marker, calcein. Theoretical analysis, based on statistical–mechanical models of molecular access to polymer meshes, shows that these observations can be explained by the steric hindrance of molecules occupying the highly constrained spaces between outer segment disc membranes. This mechanism may answer a long-standing question of how the soluble regulatory protein, arrestin, is effectively excluded from the outer segments of dark-adapted rods and cones. Generally, our results suggest an alternate mode for the control of protein access to cell domains based on dynamic, size-dependent compartmental partitioning that does not require diffusion barriers, active transport, or large numbers of immobile binding sites.
Keywords: signaling, structure, phototransduction, vision, transducin
Many cellular functions take place in highly spatially constrained compartments such as primary cilia, microvilli, endoplasmic reticulum (ER)/Golgi, filopodia, or dendritic spines. Delivery and retention of membrane-associated proteins to these areas are mediated by several complementary mechanisms, including intraflagellar transport (1), local binding to relatively immobile structures (2), and barriers to diffusion between the specialized compartment and the rest of the cell (3–4). Although it is generally recognized that the activities of these compartments are regulated by soluble proteins, especially in the case of cell signaling (5), specific rules governing soluble protein access to the compartments are not well defined. We addressed this problem in rod photoreceptors of Xenopus laevis where visual signal transduction occurs within the narrow confines of a sensory cilium.
Photoreceptor outer segments are ciliary organelles containing a stack of up to ∼2,000 flattened membrane vesicles called lamellar discs (5). The main protein component of these membranes is the visual pigment rhodopsin. Most other signaling proteins acting downstream of rhodopsin are tethered to the discs by posttranslational lipidation or single-pass transmembrane anchorage (6). An exception is arrestin, a soluble protein involved in the termination of the rhodopsin-mediated light response (7). An important feature of photoresponse regulation by arrestin is that arrestin levels in the outer segment are modulated in a light-dependent manner. Arrestin is nearly undetectable in the outer segment of dark-adapted photoreceptors, where it is found primarily in the cell body. By contrast nearly all arrestins are found in the outer segment of light-adapted photoreceptors. At a concentration of ∼3 mM (8), arrestin is one of the most abundant proteins in photoreceptors, and thus its light-dependent subcellular localization represents a major problem for which the identity of underlying mechanisms remains highly controversial (5).
Here we examine the possibility that soluble protein access to spatially constrained subcellular signaling compartments is governed by the size and shape of the molecules and the geometry of the compartment. We conducted a systematic, quantitative analysis of the subcellular distribution patterns of benchmark soluble fluorescent molecules of varying size that were introduced into the rod cytoplasm. We then analyzed the patterns using a statistical–mechanical approach (9–11) that takes into account the molecule size and the outer segment cytoplasmic structure. This analysis revealed an unintuitively steep dependence between the molecular size and the amount of soluble marker found within the outer segment interdiscal spaces, which, however, can be accurately predicted on the basis of the interplay between the Stokes radii of the markers and the highly constrained geometry of the outer segment cytoplasm. These results reveal a general cellular mechanism, differential steric volume exclusion, where even small changes in molecular size or shape, or compartment dimensions, have profound consequences for soluble protein access to specialized organelles.
Results
Densities of Structures in the Rod Cytoplasm Are Dramatically Heterogeneous.
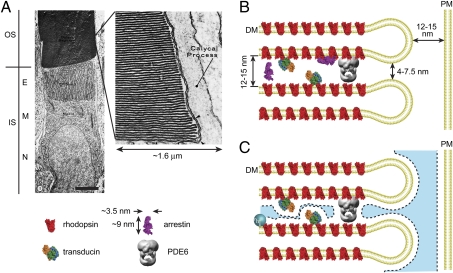
Examination of the ultrastructure of rod cells reveals that the two major subcellular compartments, the cell body and the ciliary outer segment, contain structures in dramatically heterogeneous spatial densities (Fig. 1A). The outer segment is densely packed with membranous discs that occupy roughly half of the outer segment volume and that have a highly uniform spacing of 12–15 nm (12–13). Remarkably, this spacing is on the order of the size of soluble arrestin (Fig. 1B), an approximately spherocylindrical molecule ∼9 nm long and ∼3.5 nm in diameter (14). In addition to the discs, the high, millimolar concentrations of signal transduction proteins further reduce the outer segment volume by 6–8% (Fig. 1B and Table S1). These structural components, however, reduce the effective cytoplasmic volume available to soluble molecules more than predicted by the geometric volume they occupy alone. An additional volume is unavailable to solutes due to the steric hindrance of the approach of their centers of mass to the surfaces of the structures (Fig. 1C). Together, the steric volume exclusion due to membranes and proteins within the space between the closely juxtaposed discs will result in a significant decrease in the volume available to soluble macromolecules in the rod outer segment. Importantly, this decrease will depend on the size of the molecules.
Fig. 1.
The density of structures in rod photoreceptor compartments is heterogeneous. (A) Transmission EM of an amphibian rod. E, ellipsoid; IS, inner segment (cell body); M, myoid; N, nucleus; OS, outer segment. The OS is densely packed with lamellar disc membranes (Inset) whereas the IS myoid is relatively sparsely populated with membranous vesicles and organelles (© Townes-Anderson et al., 1985. Originally published in Journal of Cell Biology, 100:175–188). (Scale bar, 5 μm.) (B) The geometry of the outer segment with disc membranes (DM), the major transduction proteins (pymol representations), and the plasma membrane (PM), drawn to scale. Rhodopsins, present at an average spatial density of ∼6 nm in the disc membrane, possess cytoplasmic loops that extend ∼2 nm into the interdiscal space. Transducins, tethered to the disc membranes by acylation of the α-subunits and farnesylation of the γ-subunits, are approximately spherical with radius 2.9 nm (34) and are present at ∼1:10 with respect to rhodopsin. PDE6, the largest protein in the interdiscal space spanning almost the entire gap, is ∼1:200 per rhodopsin. (C) The blue shaded region represents the volume of the outer segment aqueous cytoplasm accessible to the center of mass of a spherical solute with rh ∼ 3 nm.
The rod cell body appears significantly less densely populated with structural elements (Fig. 1A). We reasoned that this difference in density of structures, and the resulting differences in volume available to solutes due to steric constraints, would produce a shift in the mass of soluble proteins toward the less spatially constrained cell body. A prediction of differential steric volume exclusion is that the proportion of molecules present in the more structurally constrained outer segment will progressively decrease, relative to the cell body, as the size of the molecules increases.
Access of Soluble Molecules to the Rod Outer Segment Is Steeply Molecular Size Dependent.
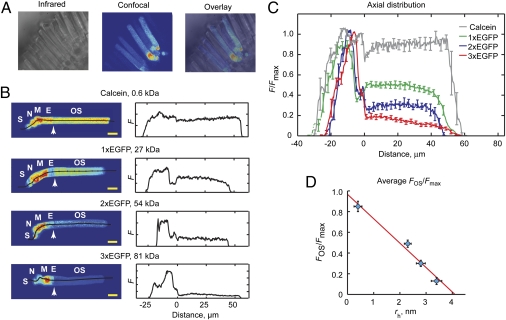
We tested this prediction by examining the steady-state distribution patterns of obligate soluble molecules of broadly different sizes, from 0.6 to 81 kDa, introduced into the cytoplasm of X. laevis rod photoreceptors (Fig. 2). Confocal images of live retinal tissue slices loaded with calcein, a polar fluorescene derivative with virtually no tendency to bind cell structures or membranes (15), or transgenic frogs expressing GFP monomers or concatemers in rods, revealed that the relative concentration of soluble proteins in the outer segments was indeed molecular size dependent (Fig. 2 B and C). The myoid region of the cell body contains the highest density of all size probes whereas the average fluorescence in the outer segment relative to the maximal fluorescence (FOS/Fmax) declined steeply and approximately linearly as a function of increasing molecular size with complete exclusion projected for molecules with hydrodynamic radii (rh) greater than ∼4 nm (Fig. 2D). The small proximal to distal negative slope of the fluorescence for 3× GFP in the outer segment may indicate a small, gradual decrement in spacing between the discs (SI Text). Interestingly, the fluorescence levels in the nucleus also appear to decline in a molecular size-dependent manner (Fig. 2B).
Fig. 2.
Soluble molecule levels in the rod outer segment depend on molecular size. (A) Images of live retinal slices. (B) Axial distributions of fluorescent molecules in rods. (Left) Central confocal z-sections; lines indicate positions from which fluorescence was read. S, presynaptic spherule. (Scale bars, 10 μm.) (Right) Fluorescence intensities plotted as a function of distance from the IS-OS interface (Left arrows). (C) Intensity distributions normalized to maximum fluorescence and averaged across multiple cells. (D) Fraction of fluorescence in the OS as a function of estimated hydrodynamic radii. Vertical error bars, F/Fmax SEM. n = 5, 14, 7, and 7 for calcein and 1×, 2×, and 3× GFP. Horizontal error bars represent SEM of rh approximation. Line was found from linear regression of the data.
The differential steric volume exclusion mechanism relies on macromolecular solutes being freely exchangeable between cytoplasmic compartments. To confirm this we examined the mobility of the smallest and largest GFP size variants in and between the major rod compartments, using photoactivatable GFP (PAGFP) and the FRAPa method (16). Upon cell body photoactivation, monomer and 3× PAGFP began entering the outer segment immediately and eventually reached distribution patterns that were essentially identical to those of the prephotoconverted molecules. Importantly, flux through the ciliary transition zone was not significantly different, and thus no diffusion barrier for soluble proteins up to ∼80 kDa exists within this structure (Fig. S1). Also of note is that 3× PAGFP entry into the outer segment appears to be channeled along the periphery, between the discs and the plasma membrane. This channeling is likely due to the narrowing of the entryway into the interdiscal space caused by disc end loops and may facilitate more rapid and uniform filling of the outer segment with regulatory proteins such as arrestin (SI Text).
Access of Soluble Molecules to the Outer Segment Interdiscal Spaces Depends on the Size of the Molecules and the Spacing of the Discs.
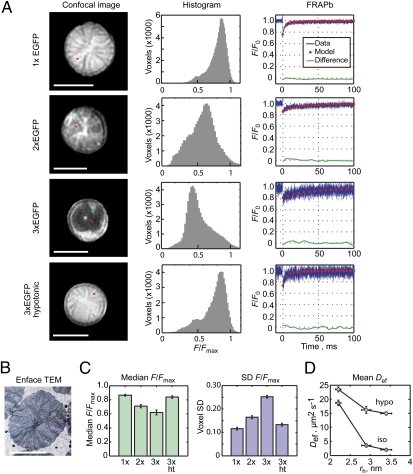
To further test the hypothesis that the close juxtaposition of disc membranes is responsible for the size-dependent variation in the concentration of soluble macromolecules in the outer segment, we examined the distributions of molecules within the interdiscal spaces under conditions that result in different spacing of the discs. Confocal images of outer segments arranged perpendicular to the scanning plane, the “end-on” imaging orientation, were obtained (Fig. 3A, Left). Upon averaging several images at a given z level, fine structures of the discs were revealed. In 1× GFP expressers, thin lines of reduced fluorescence indicated the positions of disc incisures, clefts that extend from the periphery toward the center of the disc membranes (13, 17) (Fig. 3B). Histograms of the end-on images show that the fluorescence intensity of 1× GFP was fairly uniform (Fig. 3A), with median F/Fmax = 0.87 and SD = 0.12 (Fig. 3C).
Fig. 3.
Soluble protein access to outer segment interdiscal spaces is molecular size dependent. (A) End-on imaging of the outer segment in live rods expressing GFP variants. (Left) Red dots in confocal images indicate position of FRAPa experiments shown at Right. F0, fluorescence before photobleach. (B) EM of a Rana catesbeiana rod showing incisures.[Reprinted from Experimental Eye Research, 45/1, Yoshihiko Tsukamoto; The number, depth and elongation of disc incisures in the retinal rod of Rana catesbeiana, 105–116, Copyright 1987, with permission from Elsevier (ref. 17)]. (C) Median voxel intensities and their SDs, averaged from 10, 4, 5, and 6 1×, 2×, 3×, and 3× hypotonic (ht) rods, respectively. (D) Diffusion coefficients in isotonic and hypotonic rods plotted as a function of hydrodynamic radii. n = 7, 7, and 6 for 1×, 2×, and 3× isotonic and 9, 7, and 6 for hypotonic. (A–D) Error bars, SEM. Horizontal error bars are as in Fig. 2. (Scale bars, 5 μm.) Hypotonic medium was 0.25 isotonic.
The fluorescence distributions of 2× and 3× GFP were significantly less homogeneous, with lower median voxel intensities that were spread over a broader range (Fig. 3 A and C). The radial incisure lines were less apparent or absent and the brightest voxels did not appear at the center of the 3× GFP expressers and were found instead at the periphery. The molecular size-dependent differences disappeared upon hypotonic swelling of the cells, which resulted in 15–20% lengthening of the outer segment, previously shown to mechanically increase the spacing of discs (18). Thus, the amount of soluble proteins within the interdiscal spaces was significantly dependent on molecule size and the spacing of the discs, consistent with the steric volume exclusion model.
To show that the interdiscal distribution patterns of the GFP variants were not due to immobile proteins, the mobility of the GFP concatemers between discs was examined using the multiphoton FRAPb method (16) (Fig. 3A, Right). Although the size-dependent access to the interdiscal space was accompanied by reduced mobility (Fig. 3D), this reduction was likely due to the highly constrained diffusion paths between the narrowly spaced discs and would not, in itself, lead to steady-state differences in molecular distributions (SI Text). Indeed, cell swelling resulted in an increase in the effective diffusion coefficients, Def, for all of the GFPs and a decrease in the differences between Def of 1× GFP and those of GFP concatemers (Fig. 3D).
Quantification of Relative Steric Volume Exclusion.
To quantify the impact of steric volume exclusion on the distribution of soluble macromolecules of a given size in rods, we reasoned that the ratio of fluorescence (F) recorded from any two positions in the cell represents the ratio of the fraction of volumes accessible to molecules of a given size (fac),
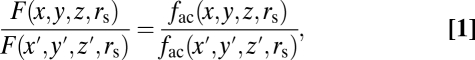 |
where rs is the hydrated radius of a spherical solute (Figs. S2 and S3 and SI Text). We estimated fac(rs) for the outer segment (fac,OS(rs)) from the well-established geometry of the disc membranes and the known numbers and volumes of the major proteins in this compartment as follows.
For idealized, spherical solutes, the ratio of the accessible volume to the geometric volume, as a function of hydrated radius, rs, may be written
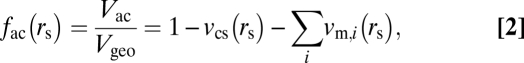 |
where vcs is the geometric volume fraction excluded by cytosolic structures, and vm,i is the fraction excluded by the ith endogenous macromolecule. The major cytosolic structures in the outer segment are the disc membranes, which are uniformly spaced and parallel. Thus, the volume fraction excluded to solutes by the discs, as a function of solute size, is
 |
where L is the distance between discs. This equation states that the volume excluded to spherical solutes between parallel membranes is linearly related to the solute radius and inversely related to the distance between membranes.
The two most abundant proteins in the interdiscal spaces of dark-adapted photoreceptors are transducin and rhodopsin (Fig. 1 and Table S1). The fraction of the interdisc volume excluded to solutes of various sizes by the portion of transducin that is >rs from the membrane surface is
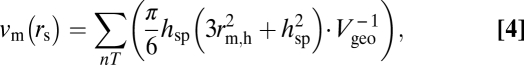 |
where hsp is the height of the portion of transducin that is >rs from the membrane surface, 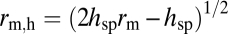 is the radius of transducin at hsp, Vgeo is the average geometric volume of the cytoplasmic space between a pair of discs, and nT is the number of transducins in the interdiscal space. The calculation of the volume excluded by the cytoplasmic loops of rhodopsin molecules is described in SI Text.
is the radius of transducin at hsp, Vgeo is the average geometric volume of the cytoplasmic space between a pair of discs, and nT is the number of transducins in the interdiscal space. The calculation of the volume excluded by the cytoplasmic loops of rhodopsin molecules is described in SI Text.
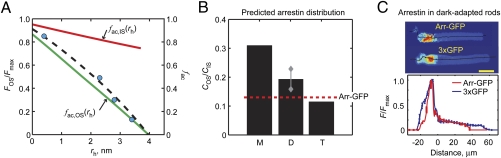
fac,OS(rs) predicted by this analysis is slightly steeper and lies below the measured size dependence of the FOS/Fmax ratio (Fig. 4A). However, the rod cell cytoplasm is a closed, self-contained system and thus the relative steric volume exclusion in neighboring compartments will impact the levels of macromolecules found in each (Fig. S2C). It is thus necessary to estimate the size dependence of the accessible volume fraction within the inner segment to fully account for the observed molecular distributions.
Fig. 4.
The steric volume exclusion model for regulation of soluble protein concentrations in spatially constrained cellular compartments. (A) FOS/Fmax vs. rh from Fig. 2D (blue circles). Lines are referenced to the right ordinate: green, calculated fac,OS(rh); red, estimated fac,IS(rh); dashed line, fac,OS(rh)/fac,IS(rh). (B) Predicted OS/IS concentration ratios for monomer (M), dimer (D), and tetramer (T) arrestin in dark-adapted rods. Gray diamonds represent maximum and minimum values calculated for dimers of various shapes (text). Dashed line indicates arrestin–GFP ratio (from C). (C) Arrestin–GFP distribution in dark adapted rods is similar to that of 3× GFP (cf. ref. 32).
Estimation of fac(rs) in the Inner Segment Compartment.
The structures and protein complement of the rod inner segment are less well characterized. However, having calculated fac,OS(rs) for the outer segment, and having measured the size-dependent fluorescent intensity ratios (FOS/FIS), the fraction of volume accessible to solutes as a function of their size in the inner segment, fac,IS(rs), or virtually any other cell compartment, may be estimated by finding the relation that produces an fac,OS(rs)/fac,IS(rs) ratio that fits the measured FOS/FIS ratio (Fig. 4A). This analysis leads directly to the size dependence of the proportional concentrations of approximately spherical macromolecules, relative to the accessible cytoplasmic spaces, in the two regions. Importantly, the analysis reveals that the outer segment discs play an overwhelmingly dominant role in setting the soluble protein distribution in the cytoplasm of rod photoreceptors.
Prediction of Arrestin Distribution in Dark-Adapted Rods.
The above analysis is relevant only to approximately spherical solutes. Arrestin, however, is nonspherical (Fig. 1). Moreover, arrestin has been shown to form dimers and tetramers at physiological concentrations (19, 20). Thus, to analyze the impact of the structural heterogeneity of rod compartments on soluble arrestin distribution, we used statistical mechanical approaches to estimating accessible volumes that allow extension of the steric volume exclusion analysis to molecules of virtually any shape (9, 11).
For simplicity, we assume that arrestin cannot fit between rhodopsin molecules on the disc surface and, thus, that the interdiscal space is reduced by ∼4 nm. Additionally, on the basis of the analysis outlined above, we determined that transducin represents a small fraction of the total volume reduction in the outer segment and ignore its contribution. With these assumptions, the effective fraction of the geometrical volume that is accessible to the center of mass of the spherocylindrical, monomer arrestin is
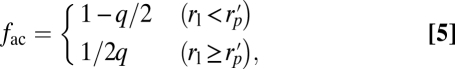 |
where rl is half the length of the cylinder, 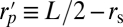 , rs is the radius of the spherical ends of the spherocylinder, and
, rs is the radius of the spherical ends of the spherocylinder, and 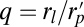 .
.
Arrestin tetramers are predicted to be approximately flattened, diamond shaped with overlapping C- and N-terminal lobes (19). We thus treated tetramer arrestin as a spherodisc with 9 nm diameter and 7 nm thickness. The effective fraction of the geometrical interdisc volume that is accessible to the center of mass of the spherodiscoidal, tetramer arrestin is
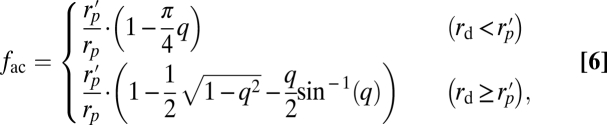 |
where here 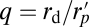 and rd is the disc radius.
and rd is the disc radius.
The structure of dimer arrestin is not known; however, limits on its shape-dependent accessible volume fraction may be examined. We chose two shapes that represent the extremes in estimated excluded volume fraction: bound end to end, thus forming an 18-nm long by 3.5-nm diameter spherocylinder, and side by side, which we treated as a 9 × 5.3-nm spherocylinder.
The results of this analysis are presented in Fig. 4B, where they are compared with the distribution pattern of a transgenically expressed arrestin–GFP fusion protein in dark-adapted rods (Fig. 4C). The concentration distributions of 3× GFP and arrestin–GFP in dark-adapted Xenopus rods are remarkably similar, as would be predicted if arrestins were mostly tetrameric with a roughly spherodiscoidal shape (19) (Fig. 4 B and C). The spacing of the outer segment discs appears tuned for regulating the outer segment concentrations of soluble proteins the size and shape of arrestin and its multimers. Monomer arrestin is expected to favor the cell body 3-fold over the outer segment, dimer arrestin 4- to 6-fold, and arrestin tetramers ∼10-fold (Fig. 4B).
Discussion
Much of cell signaling takes place in highly spatially constrained microcompartments including primary cilia, microvilli, ER/Golgi, and filopodia. Steric volume exclusion-mediated control of soluble protein concentrations is, thus, likely to be a general mechanism for setting sensitivity to environmental stimuli. Moreover, size-dependent access to the nucleus suggests that steric interactions with chromatin may be important for setting the local concentration of transcriptional regulators. Protein concentrations may be regulated by protein–protein association or by changes in compartment morphology, as has been demonstrated in, for example, dendritic spines. Whereas the potential implications of steric volume exclusion have been recognized (10–11), its impact on cell signaling has been inaccessible to direct study due primarily to the fact that most of these compartments are smaller than can be resolved with current live cell fluorescence imaging technologies. The light-controlled modulation of arrestin levels in the ciliary rod outer segment, combined with the fact that this signaling compartment is large enough to allow quantitative fluorescence imaging, make photoreceptors an unparalleled model system for studying these mechanisms.
Binding of arrestin to light-activated rhodopsin is the likeliest mechanism for arrestin localization to the outer segment of rods adapted to moderately bright light (5, 21). However, the mechanism by which arrestin is excluded from the dark-adapted outer segment has eluded explanation for decades. Essentially two mechanisms have been postulated: a diffusion barrier in the ciliary transition zone that would prevent arrestin from entering the outer segment without motor protein transport (22, 23) and a binding partner, other than rhodopsin, that would localize arrestin to the inner segment (24). Our recent study, bolstered by results presented here, effectively ruled out the diffusion barrier hypothesis (25). Three binding partners have been identified for arrestin in dark-adapted rods, α-tubulin (24), enolase 1 (26), and NSF (27). However, at ∼50 μm in neural cells (28) and ∼5 μm in rods (26), respectively, α-tubulin and enolase 1 are present in at least two orders of magnitude lower abundance than the greater than millimolar levels of arrestin (8) and NSF is not codistributed with arrestin in dark- or light-adapted rods, being found predominantly within the synaptic spherule (27). Thus, these proteins fail to meet two important criteria required to account for arrestin localization: capacity and codistribution.
Differential steric volume exclusion provides an elegant solution to this problem. The steep relationship between size and shape of soluble proteins and the levels of protein found in the ciliary rod outer segment shows that dynamic arrestin self-association and steric volume exclusion, alone, may be sufficient to explain the heavily cell body-weighted distribution of arrestin in dark-adapted rods.
The G protein, transducin, also undergoes redistribution from the outer segment to the cell body in response to light via unknown mechanisms (5). Recently it has been demonstrated that transducin subunits associate with soluble proteins, the α-subunit with UNC119 (29) and the βγ-subunits with phosducin (30, 31), that shield membrane anchoring lipid moieties and increase their masses. Because arrestin moves to the outer segment at dimmer light levels than those that drive transducin movement (5) and is 10-fold more abundant than transducin, our model calculations project that the light-dependent redistribution of transducin may also be explained by the steric volume exclusion effect.
Materials and Methods
Transgenic X. laevis expressing the constructs illustrated in Fig. S4 or arrestin–GFP (32) were generated by the REMI method (33). Animal experiments were approved by the State University of New York Upstate Committee for the Humane Use of Animals and carried out in accordance with the Guide for the Care and Use of Laboratory Animals (1996; National Academy of Sciences, Washington, DC). Quantitative imaging and multiphoton FRAPb and FRAPa experiments were conducted using a custom-built confocal and multiphoton microscope described previously (16, 25). Detailed materials and methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Edward N. Pugh, Jr., Eduardo Solessio, David R. Mitchell, and Arkadii M. Perzov for comments on an earlier version of the manuscript and Tina Gill, Maria Goralski, and Julia Bedy for technical assistance. This work was supported by National Eye Institute Grant R01EY018421 and by the Karl Kirchgessner Foundation (P.D.C.). P.D.C. is the recipient of a Career Development Award from Research to Prevent Blindness. State University of New York Upstate Ophthalmology Department is the recipient of an unrestricted grant from Research to Prevent Blindness and support from Lions District 20-Y1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.N.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115109109/-/DCSupplemental.
References
- 1.Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 2.Francis SS, Sfakianos J, Lo B, Mellman I. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J Cell Biol. 2011;193:219–233. doi: 10.1083/jcb.201009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dishinger JF, et al. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Q, et al. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329:436–439. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Jr, Arshavsky VY. Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 2006;16:560–568. doi: 10.1016/j.tcb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Wensel TG. Signal transducing membrane complexes of photoreceptor outer segments. Vision Res. 2008;48:2052–2061. doi: 10.1016/j.visres.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross OP, Burns ME. Control of rhodopsin's active lifetime by arrestin-1 expression in mammalian rods. J Neurosci. 2010;30:3450–3457. doi: 10.1523/JNEUROSCI.5391-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giddings JC, Kucera E, Russell CP, Myers MN. Statistical theory for the equilibrium distribution of rigid molecules. J Phys Chem. 1968;72:4397–4408. [Google Scholar]
- 10.Janson LW, Ragsdale K, Luby-Phelps K. Mechanism and size cutoff for steric exclusion from actin-rich cytoplasmic domains. Biophys J. 1996;71:1228–1234. doi: 10.1016/S0006-3495(96)79367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minton AP. Confinement as a determinant of macromolecular structure and reactivity. Biophys J. 1992;63:1090–1100. doi: 10.1016/S0006-3495(92)81663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nickell S, Park PS, Baumeister W, Palczewski K. Three-dimensional architecture of murine rod outer segments determined by cryoelectron tomography. J Cell Biol. 2007;177:917–925. doi: 10.1083/jcb.200612010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenkranz J. New aspects of the ultrastructure of frog rod outer segments. Int Rev Cytol. 1977;50:25–158. doi: 10.1016/s0074-7696(08)60098-4. [DOI] [PubMed] [Google Scholar]
- 14.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. A model for arrestin's regulation: The 2.8 Å crystal structure of visual arrestin. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 15.Papadopoulos MC, Kim JK, Verkman AS. Extracellular space diffusion in central nervous system: Anisotropic diffusion measured by elliptical surface photobleaching. Biophys J. 2005;89:3660–3668. doi: 10.1529/biophysj.105.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calvert PD, Peet JA, Bragin A, Schiesser WE, Pugh EN., Jr Fluorescence relaxation in 3D from diffraction-limited sources of PAGFP or sinks of EGFP created by multiphoton photoconversion. J Microsc. 2007;225:49–71. doi: 10.1111/j.1365-2818.2007.01715.x. [DOI] [PubMed] [Google Scholar]
- 17.Tsukamoto Y. The number, depth and elongation of disc incisures in the retinal rod of Rana catesbeiana. Exp Eye Res. 1987;45:105–116. doi: 10.1016/s0014-4835(87)80082-9. [DOI] [PubMed] [Google Scholar]
- 18.Heller J, Ostwald TJ, Bok D. The osmotic behavior of rod photoreceptor outer segment discs. J Cell Biol. 1971;48:633–649. doi: 10.1083/jcb.48.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson SM, et al. A model for the solution structure of the rod arrestin tetramer. Structure. 2008;16:924–934. doi: 10.1016/j.str.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamoto Y, Tamura C, Kamikubo H, Kataoka M. Concentration-dependent tetramerization of bovine visual arrestin. Biophys J. 2003;85:1186–1195. doi: 10.1016/S0006-3495(03)74554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson SM, et al. Each rhodopsin molecule binds its own arrestin. Proc Natl Acad Sci USA. 2007;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marszalek JR, et al. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 23.Peterson JJ, et al. A role for cytoskeletal elements in the light-driven translocation of proteins in rod photoreceptors. Invest Ophthalmol Vis Sci. 2005;46:3988–3998. doi: 10.1167/iovs.05-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nair KS, et al. Direct binding of visual arrestin to microtubules determines the differential subcellular localization of its splice variants in rod photoreceptors. J Biol Chem. 2004;279:41240–41248. doi: 10.1074/jbc.M406768200. [DOI] [PubMed] [Google Scholar]
- 25.Calvert PD, Schiesser WE, Pugh EN., Jr Diffusion of a soluble protein, photoactivatable GFP, through a sensory cilium. J Gen Physiol. 2010;135:173–196. doi: 10.1085/jgp.200910322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith WC, et al. Interaction of arrestin with enolase1 in photoreceptors. Invest Ophthalmol Vis Sci. 2011;52:1832–1840. doi: 10.1167/iovs.10-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang SP, Brown BM, Craft CM. Visual arrestin 1 acts as a modulator for N-ethylmaleimide-sensitive factor in the photoreceptor synapse. J Neurosci. 2010;30:9381–9391. doi: 10.1523/JNEUROSCI.1207-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiller G, Weber K. Radioimmunoassay for tubulin: A quantitative comparison of the tubulin content of different established tissue culture cells and tissues. Cell. 1978;14:795–804. doi: 10.1016/0092-8674(78)90335-5. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, et al. UNC119 is required for G protein trafficking in sensory neurons. Nat Neurosci. 2011;14:874–880. doi: 10.1038/nn.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukov GL, et al. Role of the isoprenyl pocket of the G protein beta gamma subunit complex in the binding of phosducin and phosducin-like protein. Biochemistry. 2004;43:5651–5660. doi: 10.1021/bi035903u. [DOI] [PubMed] [Google Scholar]
- 31.Sokolov M, et al. Phosducin facilitates light-driven transducin translocation in rod photoreceptors. Evidence from the phosducin knockout mouse. J Biol Chem. 2004;279:19149–19156. doi: 10.1074/jbc.M311058200. [DOI] [PubMed] [Google Scholar]
- 32.Peet JA, et al. Quantification of the cytoplasmic spaces of living cells with EGFP reveals arrestin-EGFP to be in disequilibrium in dark adapted rod photoreceptors. J Cell Sci. 2004;117:3049–3059. doi: 10.1242/jcs.01167. [DOI] [PubMed] [Google Scholar]
- 33.Knox BE, Schlueter C, Sanger BM, Green CB, Besharse JC. Transgene expression in Xenopus rods. FEBS Lett. 1998;423:117–121. doi: 10.1016/s0014-5793(98)00018-0. [DOI] [PubMed] [Google Scholar]
- 34.Lambright DG, et al. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.