Abstract
We report the three-dimensional structure of a β-catenin armadillo repeat in complex with the liver receptor homolog-1 (LRH-1) ligand binding domain at 2.8 Å resolution as the first structure of β-catenin in complex with any nuclear receptor. The surface of β-catenin that binds LRH-1 partly overlaps defined contact sites for peptide segments of β-catenin partners, including T-cell factor-4. The surface of LRH-1 that engages β-catenin is comprised of helices 1, 9, and 10 and is distinct from known interaction surfaces of LRH-1, including corepressor and coactivator binding sites. Targeted mutagenesis of amino acids forming both sides of the LRH-1/β-catenin interface reveals that they are essential for stable interactions between these proteins in solution. The LRH-1 binding site in β-catenin is also required for association with androgen receptor, providing evidence that the observed LRH-1/β-catenin interaction may be prototypic.
Keywords: crystal structure, crystallography, protein–protein interaction
Liver receptor homolog-1 (LRH-1, NR5A2) is a member of the nuclear hormone receptor (NR) family of transcription factors with essential roles in development, metabolism, and cancer (1). LRH-1 is critical in multiple stages in early development (2, 3) and maintenance of mouse embryonic stem (ES) cell pluripotency (3). In this context, LRH-1 is an upstream regulator of Oct4, a transcription factor that is essential for embryonic stem cell identity (3), and can substitute for Oct4 in generation of induced pluripotent stem cells (4). LRH-1 is also implicated in pancreas development, differentiation, and function (5) and regulates steroid synthesis and cholesterol and bile acid homeostasis in several tissues (1, 6–9) and can convert mesenchymal stem cells into steroidogenic cells (10). Additionally, LRH-1 promotes aberrant growth in multiple cancers, including gastrointestinal tumors (11) and some breast cancers (12). A genome-wide association study identified five of the eight highly significant SNPs for pancreatic cancer in the LRH-1 gene 5′ region (13). A recent report showed that LRH-1 regulates pancreatic cancer cell growth and proliferation (14).
Like other NRs, LRH-1 and its close homolog steroidogenic factor 1 [SF-1; also Ad4 binding protein (Ad4BP) or NR5A1] are multidomain proteins. LRH-1 and SF-1 are comprised of a C-terminal ligand binding domain (LBD), and an N-terminal domain linking a DNA binding domain (DBD) comprised of two zinc fingers and a Fushi tarazu-F1 motif that allows the receptor to bind to DNA as monomers (15, 16) connected by a long hinge that bridges the DBD and LBD and is the target of posttranslational modification (17). Three-dimensional structures of human and mouse LRH-1 LBDs reveal a canonical NR fold with an extra helical element stabilizing the receptor in the absence of hormone and accounting for constitutive activity (18–21). The hLRH-1 LBD structures revealed bacterial phosphatidylglycerol and phosphatidylethanolamine in the well-formed hormone binding pocket suggesting this class of ligands are potential physiological agonists (19, 20). In addition, Lee et al. recently reported that dilauroyl phosphatidylcholine is an LRH-1 agonist ligand in vitro (22).
LRH-1 modulates transcription by directing transcription factor assembly at appropriate response elements on DNA. Two special NRs of the NR0B subfamily, small heterodimer partner (SHP) and dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1 (Dax-1) (23–25), act as repressors of constitutive LRH-1 activity and bind the LBD at the activation function-2 (AF-2) site where classic NR coactivators steroid receptor coactivators (SRCs) also interact (26). LRH-1 also binds the fundamentally distinct coactivator β-catenin, which accumulates in the nucleus upon Wnt activation (27). LRH-1/β-catenin interactions are implicated in induction of cyclin D1 and E1 and LRH-1-dependent cell proliferation (27). SF-1 (28–31) and nuclear hormone receptor (NHR) 25 (32), the Caenorhabditis elegans SF-1 ortholog, also bind β-catenin. Moreover, unrelated NRs, including androgen receptor (AR) (33) and others (34) modulate transcription through β-catenin association. Presently, NR/β-catenin contact modes are not well understood at an atomic level.
As the principal agent of Wnt-dependent effects on cell adhesion, differentiation, and cancer, β-catenin engages in multiple protein–protein contacts (35, 36), some of which are understood in atomic detail (37, 38). β-catenin is comprised of a central association region, the armadillo-repeat region (ARM) (39), and N- and C-terminal transactivation domains. Whereas the N- and C-terminal regions are intrinsically unstructured (40), the ARM is comprised of 12 repeated three-helix structures (ARM-1 to -12), which adopt a superhelical organization. X-ray structures of the ARM have been determined with interacting fragments of TCF-4 (41, 42), inhibitor of beta-catenin and TCF-4 (ICAT) (43), Axin (44), B-cell chronic lymphocytic leukemia/lymphoma 9 (BCL9) (45), and others (37, 38). In most cases, α-helical or unstructured polypeptide segments from the partner dock into a positively charged groove formed by linked ARMs (38). BCL9 binds the N-terminal tip of the ARM and can form a trimeric complex with β-catenin and TCF-4 (45).
NR/β-catenin contact modes are complex, but there is evidence that NR LBDs are important for contact in some cases (34). Although phosphorylation-dependent interactions between the retinoid orphan receptor α N-terminal domain and β-catenin are important for attenuation of Wnt/β-catenin signaling (46), biochemical and genetic evidence reveals that LRH-1 and AR LBDs bind the ARM (27, 33, 34), with AR exhibiting strong dependence on the ARM-5 and ARM-6 (33). β-catenin synergizes with SRC-2 to hyperactivate AR and the isolated AR LBD (47) suggesting that the β-catenin binding surface differs from AF-2 (47).
Here, we report the crystal structure of a complex of the LRH-1 LBD and the β-catenin at 2.8 Å resolution. The LRH-1 LBD utilizes a novel interaction surface to dock into the positively charged groove at a site that partially overlaps the binding surface for TCF-4. Mutational analysis verified the observed interaction and raises the possibility that LRH-1/β-catenin interactions may be prototypic for other NRs.
Results
The LRH-1 LBD/β-Catenin Complex.
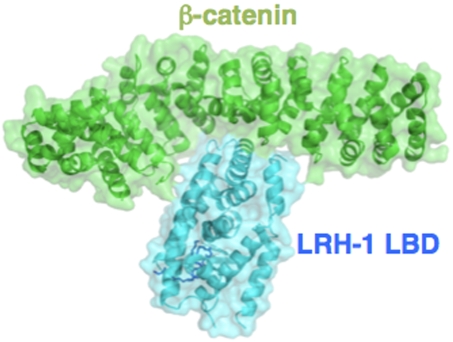
We expressed, purified, and cocrystallized protein fragments corresponding to the β-catenin armadillo repeat (amino acids 138–663) and the hLRH-1 hinge LBD (amino acids 191–541) (SI Materials and Methods). Biochemical measurements suggested that the complex was specific. Crystals of the complex grew readily. The structure of the complex was solved from the P21 crystal form (Fig. 1). The final model is refined to 2.8 Å with Rfree/Rwork values of 24.26/19.88. The polypeptide chains, forming the complex, were well resolved except for a loop region in ARM-10 of β-catenin and the loop between helix (H) 2 and H3, H3 and H4, and H11 and H12 in LRH-1 LBD and includes residues L148–Q548 and Q559–R661 for β-catenin and A299–A331, K338–S363, Q374–V461, and Q468–A538 for LRH-1 LBD (Fig. 1). The LRH-1 hinge (aa191–298), which was included in the expression vector, was invisible in the electron density map and was not included in the structure. Details of structure determination and refinement are summarized in Table S1.
Fig. 1.
Structure of the LRH-1/β-catenin complex. β-catenin is represented in green, and hLRH-1 is in cyan. The ligand of the LRH-1 is in blue (ball and stick).
Visual inspection of both components of the complex reveals characteristic overall folds observed in prior X-ray structures (37–39, 48). There are no significant changes in the LRH-1 LBD relative to previous hLRH-1 structures (19, 20) and the ligand binding pocket holds a phosphatidyl lipid of bacterial origin (stick representation, Fig. 1). However, comparisons of α-carbon positions with previous β-catenin [Protein Data Bank (PDB): 1JDH] yielded root mean squared deviations of 1.75 Å, suggesting that β-catenin undergoes significant conformational changes on binding to LRH-1. To understand the mechanisms of β-catenin conformational change, we compared β-catenin structures in complex with LRH-1 LBD (this study) and TCF-4 (1JDH). DynDom analysis (49) of LRH-1 and TCF-4 complexes identified two domains in β-catenin. Domains 1 and 2 consist of amino acid residues 150–388, 394–409, and 414–419 and 389–393, 410–413, and 420–659, respectively (Table S2). Rmsd of domain 1 and 2 were 1.18 Å and 0.7 Å, respectively. An angle relating the principal axes of these two was also defined (49). The rotation angle is 12.1°, translation is -0.1 Å, and the closure is 24.4%. The analysis also identified amino acid residues 388–396, 409–414, and 419–420 (these residues are in ARM-6 and -7) as the hinge region for the conformational change. Fig. S1 shows that domain 1- or 2-superposed structures of β-catenin complexed with TCF-4 (1JDH) to LRH-1/β-catenin complex. The hinge regions allowed the conformational changes of β-catenin to accommodate and bind to LRH-1. A green arrow points to a part of β-catenin in the LRH-1 complex that is closer to the partner, LRH-1, than when β-catenin binds TCF-4. Thus, β-catenin closes more with LRH-1 than TCF-4 suggesting that β-catenin has the capacity to adapt to different interacting proteins.
LRH-1/β-Catenin Interactions.
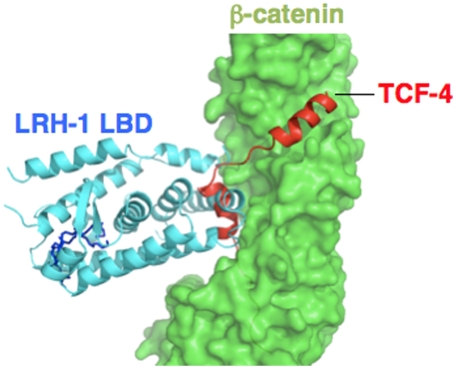
The LRH-1 binding region in β-catenin overlaps that for previously defined β-catenin interacting proteins, including TCF-4, ICAT, and others, but the binding mode of the LRH-1 LBD to β-catenin is unique. Like many other β-catenin partners, the LRH-1 LBD docks into the positively charged armadillo-repeat groove. The binding site partly overlaps the main contact surface for TCF-4 (Fig. 2); LRH-1 occupies a location that binds one of two TCF-4 α-helical segments that contact the groove. Analysis of ratios of buried surface area (BSA) in each ARM versus total surface area buried by LRH-1 and by TCF-4 confirms that binding surfaces only partly overlap (Fig. S2). Whereas ARM-4 (aa 277–318) makes most contacts with LRH-1 and TCF-4 (Fig. S2), LRH-1 contacts are mostly restricted to ARM-3 to -7 and the TCF-4 interface is more widely distributed throughout ARM-3 to -9. Unlike other β-catenin partners, LRH-1 does not utilize unstructured peptide or long α-helical segments to bind β-catenin and retains its classic NR α-helical sandwich configuration.
Fig. 2.
Superposition of the β-catenin structures of TCF-4 complex (PDB: 1JDH) and of LRH-1 LBD complex (this work). The β-catenin surface is shown in green, and the TCF-4 peptide is colored red. LRH-1 LBD is colored in cyan.
LRH-1 engages β-catenin quite differently from other coregulators such as peroxisome proliferator-activated receptor gamma coactivator 1-apha, SRC-2, and Dax-1. The LBD surface that binds β-catenin has never previously been implicated in cofactor contact; it is comprised of predominantly polar surface exposed residues from H1, H9, H10, and the intervening loop between the latter helices (Fig. S3). The surface appears well conserved in LRH-1 sequences from other species and mostly conserved in SF-1 (Fig. S4). The LRH-1 contact surface with β-catenin is larger than AF-2; calculated BSA for the LRH-1/β-catenin complex is 1,591 Å2 compared to 1,162 Å2 for the LRH-1/SRC-2 NR box 3 peptide (50) (Figs. S3 and S5, Left). Moreover, it is widely separated from AF-2, by 30–40 Å and is on the opposite side of the LBD from H7-H11, which contacts one component of the Dax-1 dimer in a heterotrimeric LRH-1/(Dax-1)2 complex (25) (Fig. S5, Right). Thus, LRH-1 could retain the capacity to bind other coregulators when assembled with β-catenin.
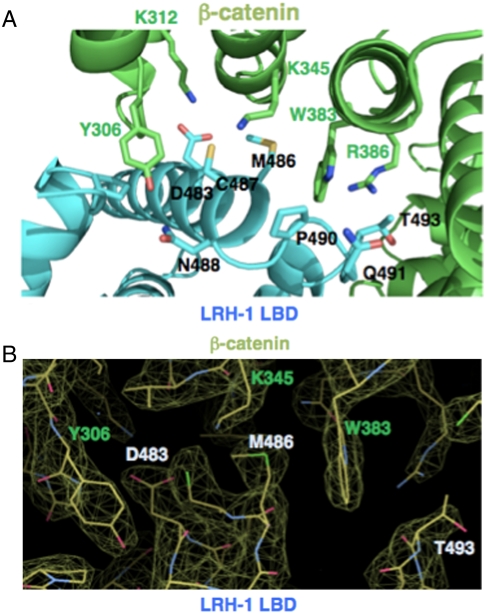
We analyzed specific amino acids involved in LRH-1/β-catenin contacts. The interface is formed by 29 side chains from β-catenin and 28 side chains from LRH-1. β-catenin Y306 presents the largest interface (128 Å2 BSA) for LRH-1 and forms hydrogen bonds with LRH-1 S300, D483, and N488 side chains (Fig. 3 A and B). The β-catenin W383 indole ring presents additional 98 Å2 BSA, forming a hydrogen bond with LRH-1 P490. β-catenin residues K312 and R386 make hydrogen bonds with LRH-1 D483 and T493, respectively. Although many β-catenin amino acids that bind LRH-1 are also implicated in contacts with TCF-4 and other proteins, there are clear differences in their binding modes. For example, β-catenin W383 contacts LRH-1 but not TCF-4 (PDB: 1JDH); another key interface residue of β-catenin, K345, presents 49 Å2 BSA for LRH-1 compared to only 25 Å2 BSA for TCF-4. On the LRH-1 surface, M486 also has significant contact with β-catenin, 106 Å2 BSA, and forms a hydrogen bond with N387.
Fig. 3.
(A) Interfacial residues of the complex of β-catenin and LRH-1 LBD. β-catenin is colored in green and Y306, K345, and W383 in the β-catenin are shown in ball and stick model. LRH-1 is colored in cyan and D483, M486, and T493 in the LRH-1 are shown ball and stick. (B) 2Fo-Fc omit map of LRH-1 LBD and β-catenin interface. The map is contoured at 1.77σ, visualized with model in Coot (Fig. S4).
Mutational Analysis of the LRH-1/β-Catenin Interface.
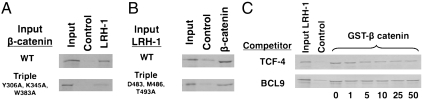
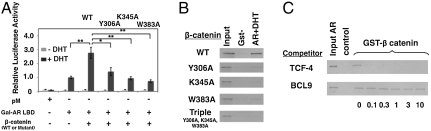
To verify the observed protein–protein interactions, we examined the effects of mutations on either side of the interface using solution GST-pulldown and competing peptide binding assays. Mutation of key β-catenin amino acids Y306, K345, and W383 or LRH-1 residues D483, M486, and T493 to Ala inhibited formation of the LRH-1/β-catenin complex (Fig. 4 A and B). In addition, a TCF-4 peptide inhibited LRH-1 binding to their partially overlapping site on the β-catenin surface, whereas a BCL9 peptide, which binds at a distinct location, had no effect (Fig. 4C).
Fig. 4.
Point mutations diminish the LRH-1/β-catenin LBD interaction. (A) 35S-labeled β-catenin or β-catenin mutants pulled down by bacterially expressed GST-LRH-1 LBD. In this figure, and similar figures below, input refers to 10% of the total amount of radiolabeled input protein included in the initial binding reaction and control refers to the amount of material retained on beads that contain GST protein without an in frame fusion. (B)35S-labeled full length LRH-1 or LRH-1 mutants pulled down by bacterially expressed GST-β-catenin armadillo repeat (138–663). (C) 35S-labeled full length LRH-1 pulled down by bacterially expressed GST-β-catenin armadillo repeat +/- increasing doses of TCF-4 or BCL9 derived peptide.
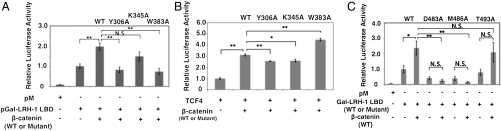
Effects of β-catenin and LRH-1 mutations were also analyzed in transfection assays that utilized expression vectors for a fusion protein composed of the yeast GAL4 DNA binding domain and the LRH-1 LBD, β-catenin, and a GAL4 responsive reporter in cultured CV-1 cells. As previously shown by Botrugno et al. (27) we found that β-catenin is a relatively weak LRH-1 coactivator, yielding twofold potentiation of reporter activity in the presence of GAL-LRH-1 at low doses (10 ng) of transfected β-catenin vector (Fig. 5A), and auto-inhibition of LRH-1 activity at higher doses. Key β-catenin mutations Y306A and W383A blocked the ability of β-catenin to coactivate GAL-LRH-1 (Fig. 5A). Similar effects were not seen at a TCF-4 responsive reporter, where the β-catenin Y306A and K345A mutants exhibited partially reduced coactivation relative to wild-type β-catenin (Fig. 5B) and W383A mutant showed partially enhanced coactivation. This finding implies that β-catenin contact surfaces for LRH-1 are overlapping, but nonidentical, and that β-catenin mutants that fail to coactivate LRH-1 (Y306A and W383A) are expressed in functional form. Likewise, LRH-1 point mutations that affect the β-catenin binding interface (D483A, M486A) reduced transcriptional activity of the GAL-LRH-1 LBD fusion and blocked its capacity for coactivation by β-catenin (Fig. 5C).
Fig. 5.
(A) Results of luciferase assays performed on extracts of cells transfected with a GAL-responsive reporter and expression vectors for a GAL-LRH-1 fusion protein (pM is the empty vector) and wild type or mutant β-catenin. Number of the data point, n = 6. (B) Luciferase assays performed on cells transfected with a TCF-4 driven expression vector and expression vectors for TCF-4 +/- wild-type or mutant β-catenin. n = 3 (C) As in Fig. 5A except that experiment utilized wild type or mutant versions of GAL-LRH-1 LBD expression vector +/- wild-type β-catenin. n = 5. This is another independent experiment from the one in Fig. 5A. (A–C) Bar represents mean + /-SEM *P < 0.05; **P < 0.01. N.S., not significant.
Might these points of contact play a role in associations with other NRs? β-catenin mutations that inhibit binding to LRH-1 also block interactions of β-catenin with AR. β-catenin is an effective coactivator of a GAL-AR LBD fusion protein in the presence of hormone (dihydrotestosterone, DHT). This activating effect was reduced by mutating key β-catenin residues that participate in LRH-1 binding (Fig. 6A). Likewise, the same mutations inhibited β-catenin interactions with a bacterially expressed AR LBD-DHT complex in GST-pulldown assays (Fig. 6B). Complementing these data, the TCF-4 peptide, but not the BCL9 peptide, inhibited AR/β-catenin binding interactions (Fig. 6C). Finally, our bacterially expressed LRH-1 LBD preparations competed for AR binding to GST-β-catenin (Fig. S6A). Based on these results, we hypothesize that the observed LRH-1 interactions with β-catenin may be prototypic for AR LBD, and possibly other NR LBDs.
Fig. 6.
Mutations of β-catenin amino acids that bind LRH-1 also disrupt AR interactions. (A) Results of luciferase assays using GAL-AR LBD +/- DHT and wild type or mutant β-catenin expression vector, as in Fig. 6D. n = 4. Bar represents mean + /-SEM *P < 0.05; **P < 0.01 (B) 35S-labeled β-catenin or β-catenin mutants pulled down by bacterially expressed GST-AR LBD. (C) 35S-labeled full length AR pulled down by bacterially expressed GST-β-catenin armadillo repeat +/- TCF-4 or BCL9 derived peptide.
Discussion
The LRH-1/β-Catenin Interface.
The structure of a complex of a NR with β-catenin reveals surprising analogies between interaction modes of the LRH-1 LBD and other β-catenin partners. Whereas the α-helical globular organization of the LRH-1 LBD and its molecular surface differs greatly from the contact surfaces of other β-catenin partners, which present as unstructured peptides or short α-helices linked by coiled regions (37, 38), LRH-1 nevertheless docks into the charged groove formed by the β-catenin armadillo repeats at a position that overlaps previously defined binding sites for TCF-4 and other partners. In support of the validity of the observed interaction, targeted β-catenin mutations and a TCF-4 peptide disrupt LRH-1/β-catenin complex formation in solution, whereas a BCL9 peptide, previously shown to bind to β-catenin at a distinct location from TCF-4, does not. A clear example of induced fit, the structure shows that β-catenin adapts to fit the bulky NR by a distributed conformational change, with bending residues found in ARM-6 and ARM-7 (Fig. S1, Table S2).
In contrast, LRH-1 utilizes a region of the LBD surface not previously implicated in protein–protein interactions to contact β-catenin. This region is comprised of predominantly polar residues presented by helices H1, H9, H10, and the loop connecting H9 and H10. This surface is distinct from the LRH-1 AF-2, which contacts SRCs and NR0B family corepressors such as Dax-1 and SHP, and lies on the opposite side of the LBD from the H7–H11 region implicated in contacts with one of the two Dax-1 subunits in the LRH-1/(Dax-1)2 heterotrimeric complex (25).
The observed binding mode may edit assemblies of LRH-1. Part of the LRH-1/β-catenin binding surface overlaps a segment of the LBD (H10 with its preceding loop) that mediates dimer or heterodimer contacts in other NRs, including thyroid hormone receptors, retinoic acid receptors, and peroxisome proliferator-activated receptors (51–53). We have proposed that surface-exposed acidic residues in H10 of LRH-1 and SF-1 result in repulsive interactions that prevent formation of canonical homodimers and retinoid X receptor (RXR)/heterodimers by these receptors (18) and these sequence alterations could also help to create the β-catenin interaction surface described here (18, 54).
How General Is the Observed Association?
The LRH-1/β-catenin structure may provide clues to possible interaction modes of other NRs with β-catenin. It is likely that other NR5A subfamily members share an analogous β-catenin binding surface; LRH-1 residues that interact with β-catenin are highly conserved among different species and are well conserved in the close homolog SF-1 (NR5A1, Fig. S4). Previously published reports of mutational studies proposed that helix H1 of SF-1 is needed for the receptor binding to β-catenin (30, 31, 55). Although our structure confirms that H1 plays a role in β-catenin contacts, the critical binding segment defined in these mutational studies is positioned at the C terminus of H1 and does not contact β-catenin in our structure (31). However, it is possible that multiple mutations of SF-1 H1 introduce local changes to the protein fold and could diminish β-catenin binding via indirect effects on the receptor fold.
In addition to the NR5A subfamily, correlations between LRH-1 and AR interactions with β-catenin are apparent. ARM-5 and -6 are essential for β-catenin interaction with AR in yeast two-hybrid assays (33). This region of β-catenin (Fig. S6B), colored in blue) constitutes part of its binding interface with LRH-1 (Fig. S2). We find that multiple mutations that disrupt LRH-1/β-catenin binding also disrupt AR/β-catenin interactions (Fig. 6), and that the TCF-4 peptide competes highly efficiently for AR binding to β-catenin. Two additional studies support the idea that AR binds to the same region of β-catenin. First, mutations of K312A (ARM-4) and K386A (ARM-6) in β-catenin were shown to abolish the interaction of β-catenin with liganded AR LBD (56). Second, p300-dependent acetylation of β-catenin K345 was shown to inhibit AR/β-catenin association (57), and we verified the importance of this residue for AR interactions with β-catenin in this study (Fig. 6). Thus, LRH-1 and AR dock onto a similar β-catenin surface.
There may be analogies between β-catenin docking surface for LRH-1 and AR, the nature of other NR LBD β-catenin binding interfaces remains a mystery. Outside of the NR5A family, we do not observe any significant conservation of amino acids homologous to those contacting β-catenin in the LRH-1/β-catenin structure. Part of the observed LRH-1 surface (H9–H10) contacting β-catenin will be in competition with homo- or heterodimer interactions in most NRs. For AR and other steroid hormone receptors, which do not form RXR-type LBD-LBD dimers, this region might be occluded by the extended F domain at the C terminus of H12 (58). This suggests that NRs that are not obligate monomers or contain an F-domain extension must either contain unique β-catenin interaction surfaces that differ from the one seen in LRH-1 or that other NRs rearrange to bind to β-catenin. The structure of β-catenin might be flexible enough to accommodate binding of various protein partners by adjusting its conformation (Fig. S1, Table S2). Ultimately, the nature of β-catenin interactions with other NR will require further investigation.
Implications for LRH-1 and Wnt/β-Catenin Signaling.
Botrugno et al. suggested that LRH-1 can interact with β-catenin in two ways (27). In the first, LRH-1 recruits β-catenin as a classic coactivator. In the second, it is independent of direct LRH-1/DNA contacts; LRH-1 interacts with a TCF-4/β-catenin complex, potentiating its activity. Our data, however, suggest that simultaneous association of LRH-1 and TCF-4 with β-catenin in the manner observed in respective X-ray structures with the two factors is sterically impossible. Indeed, overexpression of TCF-4 inhibits β-catenin coactivation of NR (59) supporting the notion that NR and TCF-4 might compete for binding to β-catenin. How then could LRH-1 enhance cyclin D1 expression by acting as a coactivator of the TCF-4/β-catenin complex? One possibility is that LRH-1 utilizes a distinct β-catenin contact surface to form the proposed assembly. We favor another model, in which LRH-1 might displace one of the two TCF-4 α-helices contacting β-catenin for the two proteins to interact jointly with β-catenin. Clearly, this mechanism will require further investigation.
Finally, the structure suggests interesting mechanisms for assembly of multifactor transcription complexes. NR LBDs are known to engage in multiple protein–protein contacts with coregulators (60), and β-catenin is shown to bind many proteins including a wide variety of transcription factors (34). Because the LRH-1/β-catenin structure reveals a unique protein interaction interface that is distinct from AF-2, the LRH-1 LBD may have the capacity to accept β-catenin along with other interacting partners, nucleating formation of multicomponent transcription complexes. We and others have suggested that it may be possible to develop new compounds that inhibit NR action by binding directly to key protein interaction surfaces. Thus, it is plausible that β-catenin binding site of LRH-1 and other NRs could be a target for drug discovery. Further focused screening efforts are needed to investigate this possibility.
Materials and Methods
A complementary DNA (cDNA) fragment encoding human LRH-1 residues 191–541 corresponding to hLRH-1 LBD with the preceding hinge region and a cDNA fragment encoding human β-catenin residues 138–663 were cloned into pET32 Xa/LIC and pCDF-2 Ek/ligation independent cloning vectors, respectively, with introduced tobacco etch virus protease cleavable sites, coexpressed in BL21Star (DE3) cells (Invitrogen), and purified using Ni-nitrilotriacetate beads (Qiagen). The proteins were further purified with Superdex 200 10/30 (GE Healthcare), concentrated to 15 mg/mL, and crystallized in 20 mM Tris·HCl (pH 8.5) and 40% PEG200 at 20 °C. The diffraction data was obtained at the Advanced Light Source (Lawerence Berkeley National Laboratory) Beamline (BL) 8.3.1. The space group of the crystal was P21 with cell dimensions of a = 49.8 Å, b = 151.6 Å, c = 76.1 Å, and β = 96.96°. The structure was solved by molecular replacement using the program Phaser (61) in Collaborative Computation Project 4 suite (62) using a β-catenin fragment (PDB ID: 2Z6H (40)) and the LRH-1 LBD (PDB ID: 1YOK (19)) structures as the search models. The structure was refined to 2.8 Å with Rfree/Rwork values of 24.26/19.88 with programs Coot (63) and Phenix.refine (64). Details of the materials, methods, and associated references are in the SI Materials and Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Drs. James Holton and George Meigs at the Advanced Light Source BL 8.3.1 (Lawerence Berkeley National Laboratory) for assistance with data collection. We are grateful to Drs. Holly A Ingraham and Miyuki Suzawa for the cDNA of human LRH-1, to Dr. Pascal Egre for advice on the protein crystallography, and to Dr. James Bayrer for critical reading of the manuscript. The work was supported by National Institutes of Health Grants R01 DK078075 and R21 DK084504 (R.F.) and DK51281 and DK41482 (to J.D.B.). F.Y. was supported by the Uehara Memorial Foundation Postdoctoral Fellowship (Tokyo, Japan), Hubert Boyer Postdoctoral Fellowship [Department of Biophysics and Biochemistry, University of California, San Francisco (UCSF)], and Program for Breakthrough Biomedical Research Postdoctoral Research Fellowship in UCSF.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3TX7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117036108/-/DCSupplemental.
References
- 1.Fayard E, Auwerx J, Schoonjans K. LRH-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Paré JF, et al. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J Biol Chem. 2004;279:21206–21216. doi: 10.1074/jbc.M401523200. [DOI] [PubMed] [Google Scholar]
- 3.Gu P, et al. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol. 2005;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heng JC, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Annicotte JS, et al. Pancreatic-duodenal homeobox 1 regulates expression of liver receptor homolog 1 during pancreas development. Mol Cell Biol. 2003;23:6713–6724. doi: 10.1128/MCB.23.19.6713-6724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M, et al. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. J Biol Chem. 1998;273:29022–29031. doi: 10.1074/jbc.273.44.29022. [DOI] [PubMed] [Google Scholar]
- 7.Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: An orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci USA. 1999;96:6660–6665. doi: 10.1073/pnas.96.12.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueller M, et al. The nuclear receptor LRH-1 critically regulates extra-adrenal glucocorticoid synthesis in the intestine. J Exp Med. 2006;203:2057–2062. doi: 10.1084/jem.20060357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boerboom D, Pilon N, Behdjani R, Silversides DW, Sirois J. Expression and regulation of transcripts encoding two members of the NR5A nuclear receptor subfamily of orphan nuclear receptors, steroidogenic factor-1 and NR5A2, in equine ovarian cells during the ovulatory process. Endocrinology. 2000;141:4647–4656. doi: 10.1210/endo.141.12.7808. [DOI] [PubMed] [Google Scholar]
- 10.Yazawa T, et al. Liver receptor homolog-1 regulates the transcription of steroidogenic enzymes and induces the differentiation of mesenchymal stem cells into steroidogenic cells. Endocrinology. 2009;150:3885–3893. doi: 10.1210/en.2008-1310. [DOI] [PubMed] [Google Scholar]
- 11.Schoonjans K, et al. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc Natl Acad Sci USA. 2005;102:2058–2062. doi: 10.1073/pnas.0409756102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Annicotte JS, et al. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene. 2005;24:8167–8175. doi: 10.1038/sj.onc.1208950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen GM, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15. Nat Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benod C, et al. Nuclear receptor liver receptor homologue 1 (LRH-1) regulates pancreatic cancer cell growth and proliferation. Proc Natl Acad Sci USA. 108:16927–16931. doi: 10.1073/pnas.1112047108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lala DS, Rice DA, Parker KL. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol Endocrinol. 1992;6:1249–1258. doi: 10.1210/mend.6.8.1406703. [DOI] [PubMed] [Google Scholar]
- 16.Morohashi K, Honda S, Inomata Y, Handa H, Omura T. A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J Biol Chem. 1992;267:17913–17919. [PubMed] [Google Scholar]
- 17.Lee YK, Choi YH, Chua S, Park YJ, Moore DD. Phosphorylation of the hinge domain of the nuclear hormone receptor LRH-1 stimulates transactivation. J Biol Chem. 2006;281:7850–7855. doi: 10.1074/jbc.M509115200. [DOI] [PubMed] [Google Scholar]
- 18.Sablin EP, Krylova IN, Fletterick RJ, Ingraham HA. Structural basis for ligand-independent activation of the orphan nuclear receptor LRH-1. Mol Cell. 2003;11:1575–1585. doi: 10.1016/s1097-2765(03)00236-3. [DOI] [PubMed] [Google Scholar]
- 19.Krylova IN, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 20.Ortlund EA, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol. 2005;12:357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, et al. Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner. Proc Natl Acad Sci USA. 2005;102:9505–9510. doi: 10.1073/pnas.0501204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JM, et al. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 2011;474:506–510. doi: 10.1038/nature10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodwin B, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki T, Kasahara M, Yoshioka H, Morohashi K, Umesono K. LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1. Mol Cell Biol. 2003;23:238–249. doi: 10.1128/MCB.23.1.238-249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sablin EP, et al. The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proc Natl Acad Sci USA. 2008;105:18390–18395. doi: 10.1073/pnas.0808936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu PL, et al. Molecular mechanism for the potentiation of the transcriptional activity of human liver receptor homolog 1 by steroid receptor coactivator-1. Mol Endocrinol. 2004;18:1887–1905. doi: 10.1210/me.2003-0334. [DOI] [PubMed] [Google Scholar]
- 27.Botrugno OA, et al. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol Cell. 2004;15:499–509. doi: 10.1016/j.molcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Gummow BM, Winnay JN, Hammer GD. Convergence of Wnt signaling and steroidogenic factor-1 (SF-1) on transcription of the rat inhibin alpha gene. J Biol Chem. 2003;278:26572–26579. doi: 10.1074/jbc.M212677200. [DOI] [PubMed] [Google Scholar]
- 29.Hossain A, Saunders GF. Synergistic cooperation between the beta-catenin signaling pathway and steroidogenic factor 1 in the activation of the Müllerian inhibiting substance type II receptor. J Biol Chem. 2003;278:26511–26516. doi: 10.1074/jbc.M300804200. [DOI] [PubMed] [Google Scholar]
- 30.Jordan BK, Shen JH, Olaso R, Ingraham HA, Vilain E. Wnt4 overexpression disrupts normal testicular vasculature and inhibits testosterone synthesis by repressing steroidogenic factor 1/beta-catenin synergy. Proc Natl Acad Sci USA. 2003;100:10866–10871. doi: 10.1073/pnas.1834480100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizusaki H, et al. -1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1) gene transcription is regulated by wnt4 in the female developing gonad. Mol Endocrinol. 2003;17:507–519. doi: 10.1210/me.2002-0362. [DOI] [PubMed] [Google Scholar]
- 32.Asahina M, Valenta T, Silhankova M, Korinek V, Jindra M. Crosstalk between a nuclear receptor and beta-catenin signaling decides cell fates in the C. elegans somatic gonad. Dev Cell. 2006;11:203–211. doi: 10.1016/j.devcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Yang F, et al. Linking beta-catenin to androgen-signaling pathway. J Biol Chem. 2002;277:11336–11344. doi: 10.1074/jbc.M111962200. [DOI] [PubMed] [Google Scholar]
- 34.Mulholland DJ, Dedhar S, Coetzee GA, Nelson CC. Interaction of nuclear receptors with the Wnt/beta-catenin/Tcf signaling axis: Wnt you like to know? Endocr Rev. 2005;26:898–915. doi: 10.1210/er.2003-0034. [DOI] [PubMed] [Google Scholar]
- 35.van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 36.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 37.Shapiro L, Weis WI. Structure and biochemistry of cadherins and catenins. Cold Spring Harb Perspect Biol. 2009;1:a003503. doi: 10.1101/cshperspect.a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu W, Kimelman D. Mechanistic insights from structural studies of beta-catenin and its binding partners. J Cell Sci. 2007;120:3337–3344. doi: 10.1242/jcs.013771. [DOI] [PubMed] [Google Scholar]
- 39.Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 40.Xing Y, et al. Crystal structure of a full-length beta-catenin. Structure. 2008;16:478–487. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham TA, Ferkey DM, Mao F, Kimelman D, Xu W. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat Struct Biol. 2001;8:1048–1052. doi: 10.1038/nsb718. [DOI] [PubMed] [Google Scholar]
- 42.Poy F, Lepourcelet M, Shivdasani RA, Eck MJ. Structure of a human Tcf4-beta-catenin complex. Nat Struct Biol. 2001;8:1053–1057. doi: 10.1038/nsb720. [DOI] [PubMed] [Google Scholar]
- 43.Daniels DL, Weis WI. ICAT inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol Cell. 2002;10:573–584. doi: 10.1016/s1097-2765(02)00631-7. [DOI] [PubMed] [Google Scholar]
- 44.Xing Y, Clements WK, Kimelman D, Xu W. Crystal structure of a beta-catenin/axin complex suggests a mechanism for the beta-catenin destruction complex. Genes Dev. 2003;17:2753–2764. doi: 10.1101/gad.1142603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sampietro J, et al. Crystal structure of a beta-catenin/BCL9/Tcf4 complex. Mol Cell. 2006;24:293–300. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Lee JM, et al. RORα attenuates Wnt/β-catenin signaling by PKCα-dependent phosphorylation in colon cancer. Mol Cell. 2010;37:183–195. doi: 10.1016/j.molcel.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 47.Li H, Kim JH, Koh SS, Stallcup MR. Synergistic effects of coactivators GRIP1 and beta-catenin on gene activation: Cross-talk between androgen receptor and Wnt signaling pathways. J Biol Chem. 2004;279:4212–4220. doi: 10.1074/jbc.M311374200. [DOI] [PubMed] [Google Scholar]
- 48.Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: Insights into physiology and therapeutics. Annu Rev Physiol. 2010;72:247–272. doi: 10.1146/annurev-physiol-021909-135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poornam G, Matsumoto A, Ishida H, Hayward S. A method for the analysis of domain movements in large biomolecular complexes. Proteins. 2009;76:201–21. doi: 10.1002/prot.22339. [DOI] [PubMed] [Google Scholar]
- 50.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 51.Wagner RL, et al. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 52.Bourguet W, et al. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000;5:289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 53.Chandra V, et al. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solomon IH, et al. Crystal structure of the human LRH-1 DBD-DNA complex reveals Ftz-F1 domain positioning is required for receptor activity. J Mol Biol. 2005;354:1091–1102. doi: 10.1016/j.jmb.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 55.Salisbury TB, Binder AK, Grammer JC, Nilson JH. Maximal activity of the luteinizing hormone beta-subunit gene requires beta-catenin. Mol Endocrinol. 2007;21:963–971. doi: 10.1210/me.2006-0383. [DOI] [PubMed] [Google Scholar]
- 56.Song LN, Gelmann EP. Interaction of beta-catenin and TIF2/GRIP1 in transcriptional activation by the androgen receptor. J Biol Chem. 2005;280:37853–37867. doi: 10.1074/jbc.M503850200. [DOI] [PubMed] [Google Scholar]
- 57.Lévy L, et al. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol Cell Biol. 2004;24:3404–3414. doi: 10.1128/MCB.24.8.3404-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Centenera M, Harris JM, Tilley WD, Butler LM. The contribution of different androgen receptor domains to receptor dimerization and signaling. Mol Endocrinol. 2008;22:2373–2382. doi: 10.1210/me.2008-0017. [DOI] [PubMed] [Google Scholar]
- 59.Mulholland DJ, Read JT, Rennie PS, Cox ME, Nelson CC. Functional localization and competition between the androgen receptor and T-cell factor for nuclear beta-catenin: A means for inhibition of the Tcf signaling axis. Oncogene. 2003;22:5602–5613. doi: 10.1038/sj.onc.1206802. [DOI] [PubMed] [Google Scholar]
- 60.Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 61.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.