Abstract
The circadian clock coordinates daily oscillations of essential physiological and behavioral processes. Conversely, aberrant clocks with damped amplitude and/or abnormal period have been associated with chronic diseases and aging. To search for small molecules that perturb or enhance circadian rhythms, we conducted a high-throughput screen of approximately 200,000 synthetic compounds using Per2∷lucSV reporter fibroblast cells and validated 11 independent classes of molecules with Bmal1:luciferase reporter cells as well as with suprachiasmatic nucleus and peripheral tissue explants. Four compounds were found to lengthen the period in both central and peripheral clocks, including three compounds that inhibited casein kinase Iε in vitro and a unique benzodiazepine derivative acting through a non-GABAA receptor target. In addition, two compounds acutely induced Per2∷lucSV reporter bioluminescence, delayed the rhythm, and increased intracellular cAMP levels, but caused rhythm damping. Importantly, five compounds shortened the period of peripheral clocks; among them, four compounds also enhanced the amplitude of central and/or peripheral reporter rhythms. Taken together, these studies highlight diverse activities of drug-like small molecules in manipulating the central and peripheral clocks. These small molecules constitute a toolbox for probing clock regulatory mechanisms and may provide putative lead compounds for treatment of clock-associated diseases.
The mammalian circadian clock is an essential timing system driving daily oscillations of physiology and behavior, including sleep/wake cycles, cell division cycles, metabolism, cardiovascular functions, hormone secretion, and mood balance (1–6). In mammals, the central pacemaker of the hierarchical clock system resides in the hypothalamic suprachiasmatic nucleus (SCN), functioning to respond to photic input signals and to regulate output pathways through neuronal and hormonal signals (7, 8). The peripheral, non-SCN clocks are subsequently synchronized by SCN-derived signals to perform tissue-specific functions (9, 10). At the molecular level, the cellular oscillator is similar in both SCN and peripheral tissues, containing interlocked negative feedback loops (11, 12). In the primary clock feedback loop, heterodimeric transcription factors (CLOCK/BMAL1 and NPAS2/BMAL1) drive expression of the Period1/2 and Cryptochrome1/2 genes. The encoded PER1/2 and CRY1/2 proteins in turn heterodimerize and repress CLOCK/BMAL and NPAS2/BMAL1 activity to inhibit their own expression (13, 14). In addition, a secondary feedback loop consisting of the nuclear hormone receptors (REV–ERBs and RORs) directly regulates Bmal1 gene transcription, thus modulating the transcriptional output of the primary loop (15, 16).
In addition to the core transcriptional feedback loops, signaling and regulatory steps are also key. For example, the cAMP-dependent signaling pathway has been shown to be an integral component of the clock network, transmitting external signaling to the core oscillator to trigger rapid adaptive responses (17, 18). Furthermore, posttranscriptional control mechanisms such as phosphorylation and ubiquitination have also been shown to play important roles in clock regulation (19–22). The PER proteins, for example, are regulated by casein kinase I (CKI)-directed phosphorylation, which consequently leads to polyubiquitination by the E3 ligase β-TRCP for proteosome-dependent degradation, a process central to molecular oscillation of the clock (23, 24). Overall, complex clock regulatory pathways appear to act in concert to render precision and robustness of the clock.
Chemical screening has emerged as a powerful tool to investigate clock regulatory mechanisms (25) and other processes (26). Several recent studies have employed cell-based chemical screening strategy to identify small molecules with clock-altering activities. For example, screening of off-patent drug collections has revealed multiple casein kinase I inhibitors that markedly lengthened the circadian period (27) and a period-shortening compound that acts on glycogen synthase kinase-3β (GSK-3β) (28). More recently, screening of a large collection (170,000) of uncharacterized compounds identified another period-lengthening compound that interacts with multiple kinases, including CKIα previously unknown to be involved in clock regulation (29). These studies provide proof-of-principle evidence for the chemical screening approach.
The above screens have mainly identified period-altering compounds, with the great majority lengthening the period. On the other hand, circadian amplitude, a measure of robustness, is technically more difficult to quantify and can be influenced by a number of downstream factors (30, 31). Circadian reporter assays based upon the cycling core clock elements are ideal for screens because they are close to the mechanism, can be measured in real time, and can be used to estimate rhythm amplitude. In the current study, we employed a cell-based circadian reporter assay in a high-throughput chemical screen of 200,000 synthetic small molecules. In addition to unique CKI inhibitors, a benzodiazepine derivative was also found to significantly lengthen the period. Interestingly, we identified previously uncharacterized compounds that shortened the period of central and/or peripheral clocks. Several of these period-shortening compounds also appeared to enhance the amplitude of reporter rhythms in wild-type and ClockΔ19/+ cells and tissue explants. Furthermore, two compounds were found to enhance intracellular cAMP levels, and caused acute reporter induction followed by significant phase delay of the reporter rhythm. These small molecules may be utilized in the future as probes of clock regulatory mechanisms as well as potential candidates for therapeutic development.
Results
High-Throughput Screen for Chemical Modulators of the Mammalian Circadian Clock.
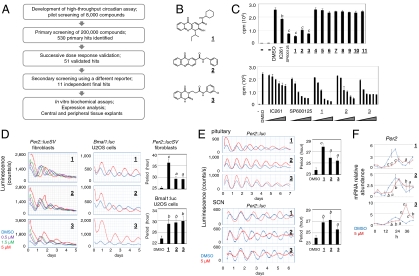
We first developed a high-throughput assay to allow continuous monitoring of circadian cycles in adult mouse ear fibroblast cells expressing a PER2-luciferase fusion reporter from the endogenous Per2 locus (Fig. S1). These cells were derived from Per2∷lucSV knockin mice analogous to the published Per2∷luc knockin mice (32, 33). Screening of 200,000 commercial compounds (University of Texas Southwestern) was carried out over a 10 month period (Fig. 1A). A number of period-lengthening compounds were found to severely reduce the amplitude of reporter rhythms. Because cell toxicity could underlie such effects, these compounds were not analyzed further in the current study. Subsequently, 530 primary hits underwent consecutive dose response validation, first in 384-well plates followed by 35 mm dishes (Fig. S2), with the latter step using chemicals reordered from commercial suppliers. Fifty one validated hits were subjected to a secondary screen using Bmal1:luc U2OS osterosarcoma cells (34) containing a luciferase reporter driven by the Bmal1 promoter (Fig. S3) that is regulated by a distinct feedback loop compared with Per2 (10). The final 11 structurally distinct, independent classes of compounds (Fig. S4), exhibiting minimal cytotoxicity in fibroblast cells (Fig. S5), were classified into three groups, including period-lengthening (Fig. S4A, compounds 1–4), cAMP enhancing (Fig. S4B, compounds 5 and 6), and period-shortening (Fig. S4C, compounds 7–11). A number of compounds also enhanced the amplitude of reporter bioluminescence in Per2∷lucSV cells (Fig. S2). All the hit compounds caused greater than 2x SD changes in period and/or amplitude as analyzed by the MultiCycle data analysis software (Fig. S1B, Actimetrics) in Per2∷lucSV cells at 1.5 μM.
Fig. 1.
Casein kinase I inhibitors. (A) Flowchart of the screen and characterization. (B) Chemical structures of compounds 1–3. (C) In vitro CKIε inhibition assay. (Top) The first two lanes correspond to no enzyme and no substrate controls. IC261 and SP600125 serve as positive controls. The compounds and controls were added to the final concentration of 20 μM. Data was analyzed by one-way ANOVA, Dunnett’s test. Compared with the DMSO control: b, p < 0.01; c, p < 0.001. (Bottom) Dose-dependent inhibition of three CKI inhibitors, compounds 1–3. IC261 and SP600125 serve as positive controls. The concentration gradient for each compound was 0.6, 2, 6, 20, 40, and 60 μM. (D) Effects of compounds 1–3 at the indicated concentrations on reporter rhythms in Per2∷lucSV and Bmal1:luc U2OS cells. Data was analyzed by one-way ANOVA, Dunnett’s test. Compared with the DMSO control: a, p < 0.05; b, p < 0.01; c, p < 0.001. (E) Period-lengthening effects of compounds in pituitary and SCN tissues from Per2∷luc mice. The graphs to the right show quantitation. Data was analyzed by one-way ANOVA, Dunnett’s test. Compared with the DMSO control: a, p < 0.05; b, p < 0.01; c, p < 0.001. (F) Real-time qPCR analysis of Per2 mRNA expression in wild-type MEF cells without reporter. Cells were treated with compounds or DMSO at time 0, and collected every 4 h up to 44 h (12 samples per set). Data was analyzed by two-way ANOVA, Bonferroni’s test. Compared with the DMSO control: a, p < 0.05; b, p < 0.01; c, p < 0.001.
Period Lengthening by Compounds Acting as CKI Inhibitors.
We first examined the mechanisms of the period-lengthening compounds (Fig. S4A). Given the clock-lengthening effects of CKI inhibitors (27, 29, 35), we performed in vitro casein kinase Iε inhibition assays. Three compounds (1–3, Fig. 1B) were found to be potent inhibitors of CKI, displaying more potent activities than the classical CKI inhibitor IC261 (Fig. 1C). Consistent with their robust inhibition of CKIε activity, compounds 1–3 were able to markedly lengthen the period in both peripheral (fibroblast/U2OS cells and pituitary tissues) and central SCN clocks (Fig. 1 D and E). Furthermore, compound 1 showed the most robust inhibition of CKIε activity, and correspondingly elicited the most significant period-lengthening effects. As expected, real-time quantitative PCR (qPCR) analysis revealed that Per2 mRNA rhythms exhibited longer periods and delayed phases in parallel with the reporter rhythm (Fig. 1F, Fig. S6). These results underscore the prominent role of CKI in the regulation of clock period.
Dual Action by a Period-Lengthening Benzodiazepine Compound.
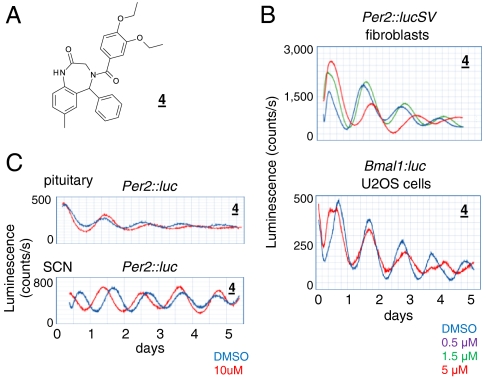
The other period-lengthening compound, compound 4, showed no activity in the CKI assay (Fig. 1C). This compound contains a benzodiazepine scaffold (Fig. 2A) found in classical agonists of the inhibitory GABAA receptors. Benzodiazepine agonists augment GABA-induced neuronal inhibition and have been used extensively as sedative drugs in humans (36). More recently, benzodiazepines have been utilized as a versatile scaffold for synthesis of derivatives with unique biological activities (37). Compound 4 showed broad period-lengthening activities in peripheral and SCN tissues (Fig. 2 B and C). An analogue, 4–1, lengthened the periods of both SCN and peripheral clocks, albeit to lesser degrees relative to compound 4 (Fig. S7 A–C); whereas, the classical benzodiazepine drug diazepam caused modest period lengthening in SCN slices, and showed no activity in fibroblast cells or peripheral tissues (Fig. S7 A–C).
Fig. 2.
A dual-acting benzodiazepine derivative. (A) Structure of compound 4. (B) Effects of compound 4 on reporter rhythms in Per2∷lucSV and Bmal1:luc U2OS cells. (C) Period-lengthening effects of compound 4 on central and peripheral tissues.
In whole-cell voltage-clamp recording using rat hypothalamic brain slices (38), diazepam and compound 4, but not 4–1 and two other inactive analogues, increased the peak amplitude and decay times of the GABAA-dependent inhibitory postsynaptic currents (IPSCs) (Fig. S7 D and E). These observations suggest a dual function of compound 4 requiring an additional pharmacophore other than the benzodiazepine scaffold. In central neurons, although induction of GABAA receptor by compound 4 may contribute moderately to period lengthening similar to diazepam, the majority of its activity is likely mediated via an additional target. In peripheral, nonneuronal cells where GABAA receptors are not abundantly expressed, compound 4 likely acts on a non-GABAA receptor target.
Complex Circadian Effects by cAMP-Inducing Compounds.
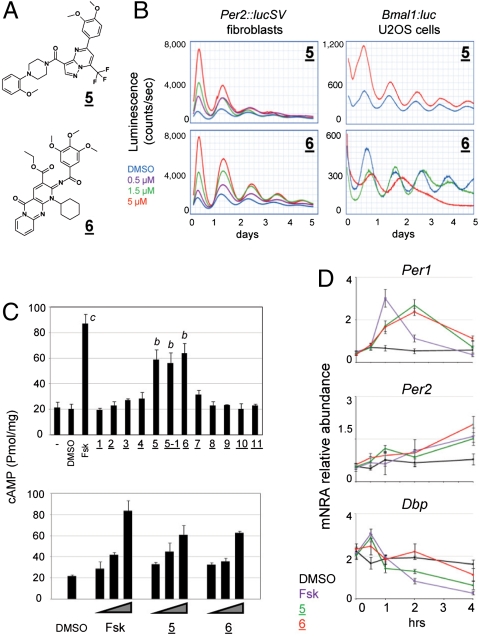
Compounds 5 and 6 (Fig. 3A) caused acute induction of reporter bioluminescence, followed by significant phase delay of reporter rhythms in fibroblast cells (Fig. 3B). In addition, compound 5, as well as an analogue 5–1, also caused rapid damping of Per2∷lucSV rhythms (Fig. 3B, Fig. S8A). These observations are reminiscent of the effects seen previously in SCN slices treated with forskolin (17). We therefore conducted cAMP ELISA assays and showed that treatment of compounds 5, 5–1, and 6 dose-dependently increased intracellular cAMP levels (Fig. 3C). Furthermore, a well-established inhibitor of the cAMP-catabolizing enzyme phosphodiesterase 4, rolipram, also caused acute bioluminescence induction and subsequent phase delay (Fig. S8B). Previously, forskolin has been shown to acutely induce Per1 mRNA levels, but not those of Per2 and the clock output gene Dbp (39, 40). As shown in Fig. 3D, the effects of compounds 5 and 6 on Per1, Per2, and Dbp gene expression were highly concordant with those of forskolin, except that the acute induction of Per1 by compounds 5 and 6 appeared weaker yet more persistent. Taken together, these results indicate that compounds 5 and 6 induce intracellular cAMP levels.
Fig. 3.
Compounds that enhance intracellular cAMP levels. (A) Structures of compounds 5 and 6. (B) Effects of compound 5 on Per2∷lucSV and Bmal1:luc U2OS cells. (C) Intracellular cAMP measurement. Wild-type MEF cells were treated with the compounds at 5 μM, and cAMP levels in the lysates were quantified via absorbance measurement. Forskolin (5 μM) serves as a positive control. Data was analyzed by one-way ANOVA, Dunnett’s test. Compared with the DMSO control: b, p < 0.01; c, p < 0.001. In the dose response experiments, compounds were added to 0.5, 1.5, and 5 μM. (D) Acute effects of the compounds on clock gene expression. MEF cells were treated with DMSO (0.1%) and compounds (forskolin, compounds 5 and 6 in 0.1% DMSO final concentration) at time 0, and cells were collected at 0, 0.5, 1, 2, and 4 h for real-time qPCR analysis of Per1, Per2, and Dbp mRNA levels.
The effects of these molecules on circadian rhythm appeared highly complex. In the primary screen, these compounds caused significant initial induction of the reporter bioluminescence and a concomitant phase delay in the following cycles (Fig. 3B, Left). The dose response patterns in other cells, however, are highly variable (Fig. 3B, Right; Fig. S8 A and C), suggesting the potential of a complex web of intracellular targets, perhaps corresponding to the various downstream effectors of the cAMP signaling pathway. In pituitary explants, both 5 and 5–1, but not 6, shortened the period (Fig. S8D). No significant effects were observed in the SCN tissue explant (Fig. S8E), perhaps partly due to poor solubility of these compounds.
Period-Shortening Compounds.
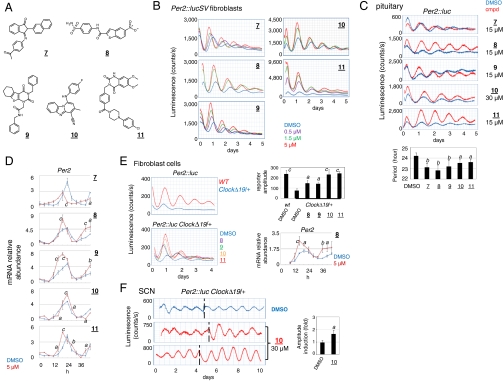
Compounds 7–11 were found to shorten the period of reporter bioluminescence in fibroblast cells (Fig. 4 A and B), reducing the period by 1.5–2.5 h at 5 μM concentration (Table S1). Several active analoguess were also found to exhibit robust period-shortening effects, whereas an inactive analogue of compound 10 showed insignificant clock-altering activities (Fig. S9). Compounds 7–11 are not structurally related with previously identified GSK-3β inhibitors (28), and did not exhibit inhibitory activities in GSK-3β kinase inhibition assay (Fig. S10). When administered to peripheral tissue explants (pituitary and lung) (Fig. 4C, Fig. S11A, respectively), compounds 7–11 consistently shortened the periods of reporter rhythms, and the extent of shortening appeared to be tissue-specific. Real-time qPCR analysis revealed phase advancement of Per2 mRNA accumulation in accordance with the observed period-shortening effects (Fig. 4D). Thus, compounds 7–11 strongly shortened period in peripheral clocks, but were less effective on the SCN (Fig. S11B), perhaps due to the known robustness of the SCN network to perturbation (41, 42).
Fig. 4.
Period-shortening compounds. (A) Structures of compounds 7–11. (B) Effects of compounds 7–11 on Per2∷lucSV cells. (C) Period shortening in pituitary explants. Compared with the control: *, p < 0.05. Data was analyzed by one-way ANOVA, Dunnett’s test. Compared with the DMSO control: a, p < 0.05; b, p < 0.01. (D) Real-time qPCR analysis of Per2 mRNA expression in wild-type MEF cells without reporter. Cells were treated with compounds or DMSO at time 0, and collected every 4 h up to 44 h (12 samples per set). RNA and cDNA samples were prepared for qPCR analysis. Data was analyzed by two-way ANOVA, Bonferroni’s test. Compared with the DMSO control: a, p < 0.05; b, p < 0.01; c, p < 0.001. (E) Amplitude enhancement of reporter oscillation in Clock∆19/+ mutant reporter cells. (Upper Left) Per2∷luc reporter rhythms in wild-type and Clock∆19/+ hetetozygous fibroblast cells. (Lower Left) Induction of reporter rhythms by compounds 8–11 in Clock∆19/+ hetetozygous fibroblast cells. (Upper Right) Comparison of bioluminescence amplitude in wild-type and treated Clock∆19/+ hetetozygous fibroblast cells. Luminescence amplitude for the first two cycles was measured using LumiCycle software and values are presented as mean ± SD from 3–5 independent samples. Data was analyzed by one-way ANOVA, Dunnett’s test. Compared with the DMSO Clock∆19/+ control: a, p < 0.05. (Lower Right) Real-time qPCR analysis of Per2 mRNA expression in Clock∆19/+ MEF cells. Cells were treated with compound 8 or DMSO at time 0, and collected every 4 h from 8 h post-treatment up to 44 h (10 samples per set). Data was analyzed by two-way ANOVA, Bonferroni’s test. Compared with the DMSO control: a, p < 0.05; b, p < 0.01; c, p < 0.001. (F) Amplitude enhancing effects of compound 10 in SCN explants from Clock∆19/+ mice. Time of compound or DMSO (0.1% final concentration) administration is indicated by dotted lines. Two representative compound 10 treated samples are shown. The graph on the right indicates fold induction of reporter rhythm amplitude compared with pretreatment baselines. Two-tailed t test: a, p < 0.05 versus the DMSO control.
In addition to period shortening, these compounds had distinct effects on the amplitude of oscillations. Compound 7 caused only modest enhancement of the reporter bioluminescence amplitude in cells (< 50%, Fig. 4B) and no effects in tissue explants (Fig. 4C). In contrast, compounds 8–11 showed dose-dependent amplitude enhancement in fibroblast cells (2.1–3.5-fold, Fig. 4B, Table S1). Moreover, in pituitary explants, compounds 8–11 also enhanced reporter bioluminescence amplitude in conjunction with their period-shortening effects (Fig. 4C). Expression of Per2 and two CLOCK output genes, Dbp (43) and Rev-erbα (44), was also induced to varying extents in a time- and compound-dependent manner in mouse embryonic fibroblast (MEF) cells (Fig. 4D, Fig. S11C), further supportive of the amplitude enhancing effects by compounds 8–11. Interestingly, the most significant enhancement of the SCN rhythm amplitude was observed with compound 10 (Fig. S11B).
Previous studies have demonstrated that a number of pathological conditions are associated with damped clocks (31, 45). Next we investigated whether compounds 8–11 can restore the amplitude of damped clocks. ClockΔ19 mutant mice are known to exhibit damped amplitude of circadian rhythms, accompanied by various physiological and behavioral deficiencies (31, 46). ClockΔ19/+ heterozygous cells displayed approximately threefold reduction in amplitude relative to wild-type cells (Fig. 4E, Upper Left). Treatment of the ClockΔ19/+ cells with compounds 8–11 enhanced the amplitude of reporter oscillation in the first two cycles by two- to threefold (Fig. 4E, Upper Right), with the most pronounced effects observed for compounds 10 and 11. Therefore, these compounds appeared to be capable of rescuing the amplitude of the mutant peripheral clock to near normal levels. In accordance, Per2 mRNA levels were also enhanced to varying degrees in ClockΔ19/+ MEF cells treated with compounds 8 and 11 compared with the control (Fig. 4E, Lower Right and Fig. S11D). Next, compounds 8–11 were found to elicit more robust reporter rhythms in ClockΔ19/+ pituitary explants relative to the DMSO control, most significantly compound 11 (Fig. S11E). Moreover, compound 10 also enhanced reporter amplitude in ClockΔ19/+ SCN explants (Fig. 4F). Overall, these observations highlight potential therapeutic effects of these small molecules in restoring normal oscillation amplitude.
Discussion
Diverse Clock-Manipulating Activities of Small Chemicals.
High-throughput screening of synthetic compounds offers an approach to identify distinct chemical modulators of biological processes. Previous circadian chemical screens have identified several compounds that inhibited the CKI family of enzymes and caused dramatic period lengthening (27, 29). In the current study, we report screening of a 200,000 chemical collection and identification of 11 small molecules that exhibited wide-ranging effects on circadian rhythms and are structurally distinct from previously identified hit molecules (27–29).
Candidate approaches aiming at well-established clock regulatory pathways have revealed putative modes of action for two groups of compounds. Three period-lengthening compounds showed robust CKI inhibitory activities. Previously identified CKI inhibitors have been shown to have promiscuous effects on paralogous CKI kinases (12, 27, 29, 47); it is likely that the compounds identified in this study act similarly. It is worth noting that unlike compounds 1 and 2, compound 3 significantly induced the levels of Per2 mRNAs (Fig. 1F), suggesting a unique mechanism for this CKI inhibitor. In addition, a group of compounds were found to induce intracellular cAMP levels, causing acute bioluminescence induction followed by significant phase delay and rapid damping of the reporter rhythms. However, the circadian effects of these compounds appear context-dependent, as evidenced by complex dose response patterns and distinct effects in cell culture versus tissue explants. The complex effects of these compounds perhaps reflect distinct effector pathways associated with cAMP induction.
Central Versus Peripheral Clocks.
The central SCN clock in the mammalian clock system has been shown to be particularly robust due to intercellular coupling (42), thus resistant to genetic perturbation (41). Among different tissues, there is also considerable variation in clock-regulated gene expression and physiological functions (6). The current screen was initially conducted using fibroblast cells, which can be regarded as a peripheral clock. Further examination using pituitary tissues revealed largely concordant effects of the hit compounds, although the relative efficacy appears to vary among cells and various tissue explants. Such tissue-specific circadian responses to chemicals may correspond to distinct expression and/or activity of intracellular targets in various tissues, which could have important implications on therapeutic efficacy and toxicity for tissue-specific clocks.
Given the tissue-specific nature and the intercellular coupling efficiency in the SCN, several hit molecules did not show significant activities in SCN tissues. Nonetheless, a number of compounds were found to function in both central and peripheral tissues. The CKI inhibitors act via a universal clock regulatory pathway. On the other hand, compound 4 showed strong GABAA agonist activity, likely corresponding to the benzodiazepine moiety. However, compound 4 also appears to contain additional structural groups that confer its function in peripheral cells and tissues where GABAA receptors are not known to be fully active. A peripheral benzodiazepine target, translocator protein 18 kDa (TSPO), has been previously identified (48). However, in a large-scale siRNA screen, knockdown of the TSPO gene showed no effects on circadian rhythms (49). Identification of the additional pharmacophore on compound 4 will require significant medicinal chemistry efforts to broadly interrogate functional group substitutions. Regardless, these observations highlight the complexity of circadian regulation, and also reveal unexpected versatility of small molecules.
Enhancement of Reporter Rhythms by Small Molecules.
A number of compounds identified in the current screen were found to increase reporter bioluminescence either acutely or persistently. The molecular mechanisms underlying such effects, however, may be complex. In the case of the cAMP-inducing compounds 5 and 6, acute induction of PER2-luciferase reporter bioluminescence contrasts with their lack of effects on Per2 transcript levels (Fig. 3D, Fig. S6). In comparison, compounds 8–11 displayed dose-dependent period-shortening effects in conjunction with enhancement of reporter rhythm amplitude over several cycles. These compounds were also found to moderately elevate Per2 mRNA levels, although the fold induction of Per2 mRNA expression is less pronounced than that of reporter bioluminescence. These compounds also showed distinct enhancing effects on mRNA expression of two other CLOCK target genes Dbp and Rev-erbα, consistent with their putative clock-enhancing functions and also suggestive of divergent mechanisms of action. Furthermore, whereas compounds 8–11 potently enhanced the reporter rhythm in peripheral clocks, only compound 10 appeared to be effective in SCN explants. Future studies will investigate the effects of these various chemicals on single-cell bioluminescence and molecular clock functions, which may also shed light on the observed correlation between period shortening and amplitude enhancement by these molecules.
The Clock∆19 mutant mice have been shown to exhibit pronounced circadian damping and altered phase resetting, accompanied by far-ranging physiological and behavioral defects (31, 46). Interestingly, we found that compounds 8–11 were able to restore the amplitude of damped clocks associated with the Clock∆19 mutation to wild-type levels. These small molecules are thus ideally suited for future investigation on whether clock-associated physiology can be enhanced by small molecules.
The current high-throughput screen has led to identification of potent chemical molecules capable of manipulating peripheral and central clocks. These molecules display diverse clock-modulatory activities, potentially functioning as versatile probes of circadian biology. Future studies will focus on understanding their mechanisms of action using unbiased approaches, and investigating the potential beneficial effects of the compounds in physiological settings.
Materials and Methods
Adult mouse ear fibroblast cells from Per2∷lucSV reporter mice were immortalized (50) and used for the high-throughput primary screen. The cell culture conditions were based on the procedure previously described (33). Briefly, cells were grown to full confluency in 384-well plates, and treated with 5 μM forskolin for 1–2 h. After compounds were added to the plates with robot (Beckman), the plates were tightly sealed with membranes (GeneMate) and subjected to continuous monitoring by an EnVision microplate reader (Perkin Elmer). By using MultiCycle software (Actimetrics), the data were detrended (first-order polynomial) and then best-fit to a sine wave estimated by a Levenberg–Marquardt algorithm for measurement of period, phase, amplitude, and damping rate. The primary hits were cherry-picked and subjected to two rounds of dose response validation. Hits were subsequently tested in the U2OS cell line containing a Bmal1 promoter-driven luciferase reporter. Eleven compounds were found to exhibit robust clock-altering effects in cells, and were considered as final hits (Table S1). Sequences of primers used for real-time qPCR analysis are listed in Table S2.
The compound library at University of Texas Southwestern High-Throughput Screening Core Facility consists of highly filtered commercial collections. Please refer to the web site http://www.utsouthwestern.edu/utsw/cda/dept24734/files/342125.html for more information regarding the chemical library.
Please refer to SI Materials and Methods for additional information.
Supplementary Material
Acknowledgments.
The primary screen and initial validation in this study was conducted in Dr. Steven McKnight’s lab at University of Texas Southwestern Medical Center. We thank Dr. McKnight for conceiving the initial idea and providing generous scientific and financial support throughout the project. We thank University of Texas Southwestern High-Throughput Screening Core Laboratory, particularly M. Roth and W. Hao, for help with the screen. We also thank J. Hogenesch and J. Baggs for providing Bmal1:luc U2OS cells, D. Ferster for help with MultiCycle analysis, K. Prewit, E. Song, B. He, I. Kornblum, and K. Kim for technical assistance, H. Lin, M. Izumo, and X. Qi for discussion, and C. C. Lee and V. Kumar for critical reading of the manuscript. This work was supported in part by a Silvio O. Conte Center for Neuroscience award (J.S.T.), Welch Foundation Grant AU-1731 (to Z.C.), and National Institutes of Health Grant R01HL077400 (to H.P.). J.S.T. is an Investigator for the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118034108/-/DCSupplemental.
References
- 1.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 2.Sack RL, et al. Circadian rhythm sleep disorders: Part I, basic principles, shift work, and jet lag disorders. An American Academy of Sleep Medicine review. Sleep. 2007;30:1460–1483. doi: 10.1093/sleep/30.11.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Z, McKnight SL. A conserved DNA damage response pathway responsible for coupling the cell division cycle to the circadian and metabolic cycles. Cell Cycle. 2007;6:2906–2912. doi: 10.4161/cc.6.23.5041. [DOI] [PubMed] [Google Scholar]
- 4.Paschos GK, FitzGerald GA. Circadian clocks and vascular function. Circ Res. 2010;106:833–841. doi: 10.1161/CIRCRESAHA.109.211706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones SH. Circadian rhythms, multilevel models of emotion, and bipolar disorder—an initial step towards integration? Clin Psychol Rev. 2001;21:1193–1209. doi: 10.1016/s0272-7358(01)00111-8. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: Cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 10.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 11.Mourino M, et al. Osmolarity modulates the expression of the Hha protein from Escherichia coli. FEMS Microbiol Lett. 1998;160:225–229. doi: 10.1111/j.1574-6968.1998.tb12915.x. [DOI] [PubMed] [Google Scholar]
- 12.Yagita K, Tamanini F, van der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 13.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 14.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: An analogue of clock operative in the mammalian forebrain. Science. 2001;293:506–509. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 15.Duez H, Staels B. The nuclear receptors Rev-erbs and RORs integrate circadian rhythms and metabolism. Diab Vasc Dis Res. 2008;5:82–88. doi: 10.3132/dvdr.2008.0014. [DOI] [PubMed] [Google Scholar]
- 16.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang EE, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, et al. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol. 2008;28:6056–6065. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H, Chen R, Lee Y, Yoo S, Lee C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc Natl Acad Sci USA. 2009;106:21359–21364. doi: 10.1073/pnas.0906651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallego M, Kang H, Virshup DM. Protein phosphatase 1 regulates the stability of the circadian protein PER2. Biochem J. 2006;399:169–175. doi: 10.1042/BJ20060678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eide EJ, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maier B, et al. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23:708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirota T, Kay SA. High-throughput screening and chemical biology: New approaches for understanding circadian clock mechanisms. Chem Biol. 2009;16:921–927. doi: 10.1016/j.chembiol.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pieper AA, et al. Discovery of a proneurogenic, neuroprotective chemical. Cell. 2010;142:39–51. doi: 10.1016/j.cell.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isojima Y, et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci USA. 2009;106:15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirota T, et al. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci USA. 2008;105:20746–20751. doi: 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirota T, et al. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIalpha as a clock regulatory kinase. PLoS Biol. 2010;8:e1000559. doi: 10.1371/journal.pbio.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi JS, Zatz M. Regulation of circadian rhythmicity. Science. 1982;217:1104–1111. doi: 10.1126/science.6287576. [DOI] [PubMed] [Google Scholar]
- 31.Vitaterna MH, et al. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci USA. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo SH, et al. PERIOD2∷LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baggs JE, et al. Network features of the mammalian circadian clock. PLoS Biol. 2009;7:e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Badura L, et al. An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J Pharmacol Exp Ther. 2007;322:730–738. doi: 10.1124/jpet.107.122846. [DOI] [PubMed] [Google Scholar]
- 36.Rogers CJ, Twyman RE, Macdonald RL. Benzodiazepine and beta-carboline regulation of single GABAA receptor channels of mouse spinal neurones in culture. J Physiol. 1994;475:69–82. doi: 10.1113/jphysiol.1994.sp020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tardibono LP, Miller MJ. Synthesis and anticancer activity of new hydroxamic acid containing 1,4-benzodiazepines. Org Lett. 2009;11:1575–1578. doi: 10.1021/ol900210h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahner MR, Li DP, Pan HL. Benzodiazepine inhibits hypothalamic presympathetic neurons by potentiation of GABAergic synaptic input. Neuropharmacology. 2007;52:467–475. doi: 10.1016/j.neuropharm.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 39.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 40.Yagita K, Okamura H. Forskolin induces circadian gene expression of rPer1, rPer2, and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 2000;465:79–82. doi: 10.1016/s0014-5793(99)01724-x. [DOI] [PubMed] [Google Scholar]
- 41.Liu AC, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- 44.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 45.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walton KM, et al. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J Pharmacol Exp Ther. 2009;330:430–439. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- 48.Chen MK, Guilarte TR. Translocator protein 18 kDa (TSPO): Molecular sensor of brain injury and repair. Pharmacol Ther. 2008;118:1–17. doi: 10.1016/j.pharmthera.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang EE, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ossovskaya VS, et al. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc Natl Acad Sci USA. 1996;93:10309–10314. doi: 10.1073/pnas.93.19.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.