Abstract
The spindle checkpoint delays the onset of anaphase until all of the chromosomes properly achieve bipolar attachment to the spindle. It has been shown that unattached kinetochores are the site that emits a signal for activation of the checkpoint. Although the components of the checkpoint such as Bub1, Mad1 and Mad2 selectively accumulate at unattached kinetochores, the answer to how they recognize unattached kinetochores has remained elusive. Mps1 pombe homolog (Mph1) kinase has been shown to function upstream of most of the components of the checkpoint and thus it is thought to recognize unattached kinetochores by itself and recruit other components. In this study we have expressed a fusion protein of Mph1 and Ndc80 (a kinetochore protein of the outer plate) and shown that the fusion protein arrests cell cycle progression in a spindle-checkpoint\x{2013}dependent manner in fission yeast. When expression of Mad2 is turned off, the cells grow normally with Mph1 constitutively localized at centromeres/kinetochores. Under this condition, Bub1 can be found with Mph1 throughout the cell cycle, indicating that localization of Mph1 at centromeres/kinetochores is sufficient to recruit Bub1. In contrast, Mad1 is found to transiently localize at kinetochores, which are presumably unattached to the spindle, but soon it dissociates from kinetochores. We propose that Mph1 is a sufficient marker for recruitment of Bub1. Mad1, in contrast, requires an additional condition/component for stable association with kinetochores.
The spindle checkpoint delays the onset of anaphase until all of the chromosomes properly achieve bipolar attachment to the spindle microtubules (reviewed in ref. 1). It has been proposed from early observation by micromanipulation experiments that an unattached kinetochore and/or a kinetochore under low tension produces a “wait anaphase” signal. In the spermatocytes of mantids, the presence of a single improperly attached chromosome is sufficient to inhibit the onset of anaphase. When this chromosome was pulled with a microneedle to place it under tension, the cell cycle arrest was released (2). Another study reported that laser ablation of the last unaligned kinetochore allows the cells to progress to anaphase in mammalian PtK1 cells (3).
The components of the spindle checkpoint have been identified through genetic screens in the budding yeast Saccharomyces cerevisiae. They include mitotic-arrest deficient (Mad) 1, 2, and 3 and budding uninhibited by benzimidazole (Bub) 1 and 3, all of which are evolutionally conserved among eukaryotes (4, 5). In addition, a dual-specificity kinase, Mps1, which was originally identified as a factor required for the duplication of spindle pole body (SPB) in S. cerevisiae, plays an important role in the checkpoint (6–8). In the presence of unaligned kinetochores, the checkpoint inhibits the activity of anaphase promoting complex/cyclosome (APC/C) to polyubiquitinate the substrates such as securin and cyclin B (9). The spindle checkpoint proteins Mad2 and Mad3/BubR1 directly bind to Slp1/Cdc20, an activator of the APC/C, thereby preventing polyubiquitination by Cdc20-APC/C (10–13).
Since the first demonstration that Mad2 accumulates at unattached kinetochores (14), subsequent studies have shown that other components of the spindle checkpoint localize at unattached kinetochores as well (15, 16). It has generally been believed that the unattached kinetochore plays a role as a factory to assemble a protein complex termed mitotic checkpoint complex (MCC), consisting of Mad2, Mad3/BubR1, Bub3, and Slp1/Cdc20, which has an inhibitory activity against APC/C (17, 18).
Overexpression of Mps1 in the budding yeast S. cerevisiae and the Mps1 pombe homolog (Mph1) kinase in the fission yeast Schizosaccharomyces pombe can induce a cell cycle arrest in a spindle-checkpoint–dependent manner without disrupting the structure of the mitotic spindle (19, 20). Because the arrest induced by overexpression of Mps1 requires other functional components of the checkpoint, it is thought that Mps1 causes the arrest by activating the Mad and Bub pathways. Other studies in higher eukaryotes have indicated that Mps1 is required for recruitment of Mad2 to unattached kinetochores (6, 21, 22). Recent studies using chemical inhibitors of Mps1 kinase have shown that Mps1 plays a critical role in regulation of other components of the spindle checkpoint at unattached kinetochores. Upon inhibition of Mps1, chromosomes cannot be properly aligned largely due to lack of the ability to correct syntelic attachments (23–25). It has also been shown that the Mps1 activity is required for activation of Mad2 by converting its conformation, and that Mps1 can dimerize and transphosphorylate, which presumably results in release from kinetochores and thereby facilitates the checkpoint signaling in the cytosol (25). The role of Mps1 kinase in the cytosol is also proposed to promote the assembly of the inhibitory complex of Cdc20-APC/C (24).
In this study, we have investigated the role of Mph1 kinase in recruiting other components of the checkpoint to the kinetochore. Our results have indicated that Bub1, which is normally found at kinetochores in mitosis, can be recruited to the centromere/kinetochore throughout the cell cycle when the Mph1 is forced to localize at centromeres/kinetochores. Mad1 is, on the other hand, found only transiently at kinetochores under the same experimental condition. It appears that Mph1 can function as a sufficient marker of unattached kinetochores for Bub1 recruitment, but that Mad1 recognizes unattached kinetochores independently from Mph1.
Results and Discussion
Requirement for Mitotic Arrest Induced by Overexpression of Mph1.
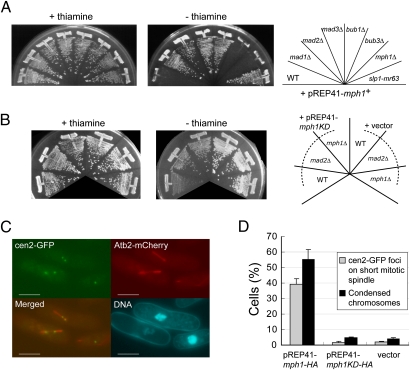
A previous study showed overexpression of Mph1 induces a mitotic arrest (20). In this study Mph1 was overexpressed in various genetic backgrounds in fission yeast. As shown in Fig. 1A and Fig. S1A, Mph1, when expressed from pREP41 in the wild-type strain, could cause a growth arrest. The effects on growth were significantly different depending on the genetic background. Overexpression of Mph1 could only affect growth of the wild-type strain or strains lacking mph1+ or bub3+. In contrast, strains expressing Slp1 defective in binding Mad2 (Slp1-mr63) or lacking one of the following components: mad1+, mad2+, mad3+, and bub1+, were resistant to overexpression of Mph1 from pREP41. These results implied that (i) a growth arrest caused by overexpression of Mph1 from pREP41 was likely due to activation of the spindle checkpoint and (ii) Mph1 activates the Mad and Bub components except for Bub3. Although Bub3 was originally identified as a component of the checkpoint in budding yeast (4), it has been reported that its homolog in fission yeast might not be required for activation of the checkpoint (26–28).
Fig. 1.
Cell cycle arrest caused by overexpression of Mph1. (A) Mph1 was overexpressed from pREP41-mph1+ in the wild-type cells or the indicated spindle checkpoint mutants. Gene expression is repressed in the media with thiamine (+thiamine) and derepressed in the media without thiamine (−thiamine). The plates were incubated at 32 °C for 3 d. (B) Mph1 kinase-dead (KD) mutant was overexpressed in the wild-type cells, mad2Δ, or mph1Δ from pREP41 as in A. (C) Centromere II's and microtubules were visualized by cen2-GFP and Atb2-mCherry, respectively. DNA was visualized by staining with Hoechst 33342. (Scale bars, 5 μm.) (D) Indexes of metaphase arrest: cen2 on the spindle (gray bars) and chromosome condensation (black bars) were determined in cells overexpressing Mph1-HA or Mph1KD-HA from pREP41, respectively. The error bars represent SEM of the three individual experiments.
We also constructed a kinase-dead (KD) mutant of Mph1 by introducing a mutation (459D to A) known to abrogate the kinase activity of Mps1 in other organisms (6, 29, 30) and tested it for the ability to activate the spindle checkpoint. As shown in Fig. 1B, when Mph1-KD was expressed from pREP41, it caused a weak growth inhibition, which was partially relieved by deletion of mad2+ or mph1+, indicating that expression of Mph1-KD from pREP41 caused a weak delay in mitotic progression as well as a growth defect for a reason unrelated to the checkpoint activation. We speculate that partially degraded Mph1-KD proteins (Fig. S2B) might be toxic to some extent.
Finally, we found that tagging green fluorescent protein (GFP) to the C terminus of Mph1 did not affect the ability to cause a growth arrest when expressed from pREP41 (Fig. S1A). Expression of the GPF-tagged Mph1 from pREP81, that had a promoter less active than that of pREP41, did not induce a growth arrest (Fig. S1A). In addition, HA epitope-tagged Mph1 from pREP41 functioned similarly to GFP-tagged Mph1 (Fig. S1A).
To confirm that overexpression of Mph1 caused a mitotic arrest, we monitored localization of centromere II (cen2-GFP) (31) and morphology of microtubules as well as chromatin. As shown in Fig. 1 C and D, 18 h after induction of Mph1-HA from pREP41, centromere II's, which were occasionally visualized as two separate dots likely due to tension between bioriented sister kinetochores, were localized on short and thick mitotic spindles in ∼40% of cells. Chromatin was tightly condensed in these cells. These phenotypes were typical of cells arrested at metaphase. We also examined localization of Mad2. Mad2 remained on kinetochores in more than 80% of the cells, indicating that the spindle checkpoint was kept active (Fig. S1 B and C). We also found that a weak growth inhibition caused by Mph1-KD expressed from pREP41 (Fig. 1B) was not due to a tight arrest in mitosis. In the cells overexpressing Mph1-KD, the three indexes for mitotic arrest (chromosome condensation, centromere II's on the spindle, and Mad2 on kinetochores), which would be clearly seen only in cells tightly arrested at mitosis, were very low (Fig 1D and Fig. S1C).
On the basis of these results, we concluded that although the interaction between the spindle and kinetochore was not interfered with, overexpression of Mph1 from pREP41 could cause a mitotic arrest by maintaining the spindle checkpoint active. Expression from pREP81, a construct with a weaker promoter, most likely could not do so because the level of Mph1 was insufficient to overcome a checkpoint silencing activity.
Localization of Mph1.
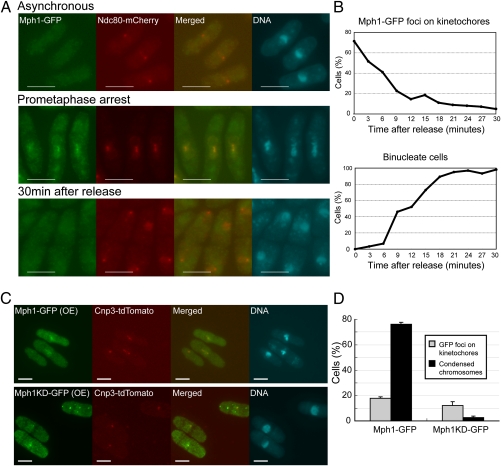
It was previously shown that Mps1 kinase, a homolog of fission yeast Mph1, was localized at the kinetochore when the spindle checkpoint was active in budding yeast (32) and higher eukaryotes as well (6, 7, 22). We tested whether this was also the case in fission yeast. Localization of Mph1-GFP expressed from the native locus was determined in a cold-sensitive mutant (nda3-KM311) that could not assemble the normal mitotic spindle at the restrictive temperature due to the mutation in the β-tubulin gene (33). In exponentially growing cells at the permissive temperature, Mph1-GFP was not localized at any particular sites. After 8 h from the shift to the restrictive temperature, Mph1-GFP was colocalized with Ndc80, a marker for the centromere/kinetochore (Fig. 2A). Upon the shift back to the permissive temperature, the cell cycle arrest was released and Mph1-GFP rapidly disappeared from kinetochores (Fig. 2 A and B), indicating a tight correlation between the arrest imposed by the spindle checkpoint and localization of Mph1 at kinetochores.
Fig. 2.
Localization of Mph1. (A) Localization of Mph1-GFP (green) was determined in the cold-sensitive mutant nda3-KM311. The cells were first precultured at 32 °C (asynchronous) and shifted down to the restrictive temperature, 20 °C for 8 h (prometaphase arrest). They were then shifted back to 32 °C to release from the arrest (30 min after release). Ndc80-mCherry (red) was used as a marker of kinetochores. DNA (blue) was visualized by staining with DAPI (4′-6-diamino-2-phenylindole). (Scale bars, 5 μm.) (B) The samples were prepared as in A and the percentages of the cells with Mph1 foci at kinetochores (Upper) and binucleate cells (Lower) were determined after release from the prometaphase arrest. (C) Localization of Mph1-GFP or Mph1-KD-GFP (green) expressed from pREP41 for 18 h at 32 °C was determined with Cnp3-tdTomato (red) as a marker of kinetochores. (Scale bars, 5 μm.) (D) The samples were prepared as in C and the percentages of cells with Mph1 foci on kinetochores (gray bars) and cells with condensed chromosomes (black bars) were determined.
We next examined localization of Mph1 when the spindle checkpoint was maintained active due to overexpression of Mph1. As shown in Fig. 2 C and D, 18 h after induction of Mph1-GFP from pREP41, Mph1-GFP was colocalized with a centromere/kinetochore marker Cnp3 (fission yeast homolog of CENP-C) in cells arrested at mitosis with condensed chromosomes. Interestingly we found that Mph1-KD-GFP overexpressed in the wild-type background was also localized with Cnp3, although it did not cause a cell cycle arrest in mitosis (Fig. 2 C and D).
Persistent Activation of the Spindle Checkpoint by Expression of Mph1-Ndc80.
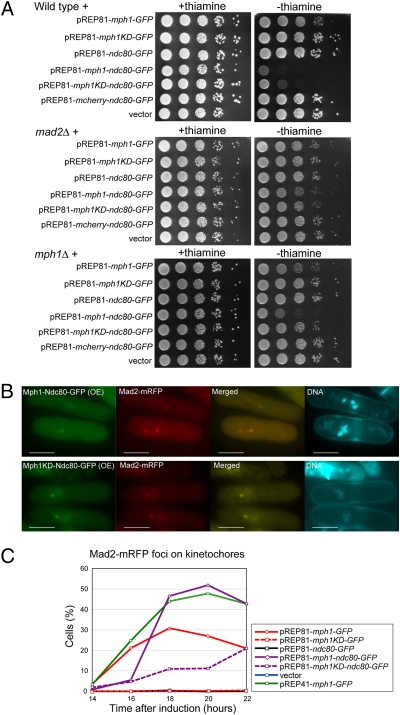
Because the above results demonstrated a strong correlation between localization of Mph1 at kinetochores and persistent activation of the spindle checkpoint, we thought that the Mph1 kinase could maintain the checkpoint active more efficiently if it was forced to localize at the kinetochore. To test this possibility, the Mph1 kinase was fused with the full length of Ndc80 tagged with GFP (Mph1-Ndc80-GFP) and expressed from pREP81, the vector that failed to induce the mitotic arrest by expressing Mph1 alone (Fig. S1A). As shown in Fig. 3A, expression of Mph1-Ndc80-GFP from pREP81 caused an arrest in the wild-type background. It failed to cause an arrest in a strain lacking mad2+, indicating that the arrest was due to activation of the spindle checkpoint. The result also indicated that expression of Mph1 fused with Ndc80, a protein at the outer plate of the kinetochore (34), did not interfere with the function of the kinetochore. Inactivation of the spindle checkpoint would cause a dramatic decrease in the cell viability if Mph1-Ndc80-GFP interfered with the function of the kinetochore.
Fig. 3.
Effect of kinetochore-tethered Mph1 expressed from pREP81. (A) Indicated fusion proteins were overexpressed from pREP81 in wild type (Top), mad2Δ (Middle), or mph1Δ cells (Bottom). The plates were incubated at 32 °C for 3 d. (B) Localization of Mph1-Ndc80-GFP or Mph1-KD-Ndc80-GFP (green) expressed from pREP81 for 22 h at 32 °C was determined with Mad2-mRFP (red). DNA was visualized by staining with Hoechst 33342. (Scale bars, 5 μm.) (C) The percentage of the cells with Mad2-mRFP foci on kinetochores was determined for cells overexpressing various fusion proteins (indicated in the box) from pREP81.
Analysis by immunoblotting indicated that the level of expression of Mph1-Ndc80-GFP was comparable with that of Mph1-GFP expressed from pREP81 (Fig. S2B), indicating that the arrest caused by Mph1-Ndc80-GFP was not due to an increased stability of Mph1 fused with Ndc80, but due to forced recruitment of Mph1 to kinetochores. Fluorescent microscopy revealed that Mph1-Ndc80-GFP formed sharp foci at kinetochores that were colocalized with Cnp3 (Fig. S2A) and Mad2 (Fig. 3B). During the course of this study, it was demonstrated that expression of Mps1 fused with a kinetochore protein Mis12 could cause persistent activation of the spindle checkpoint (35). The outer plate of the kinetochore would likely be a site suitable for the function of Mph1/Mps1. Indeed, it was reported that Mps1 binds to Ndc80 in budding yeast (36). Interestingly, although Mph1-KD expressed from pREP41 did not cause a tight mitotic arrest (Fig. 1D), it was able to cause a mitotic arrest when overexpressed from pREP81 as a fusion protein with Ndc80, although the efficiency was low (Fig. 3 A–C). This effect was dependent on Mph1 expressed from the native locus (Fig. 3A) and the activity of the endogenous kinase (Fig. S2C). The result would suggest that Mph1-KD-Ndc80-GFP, which is presumably kinetochore bound, functions without its kinase activity in the presence of the wild-type Mph1 in a soluble form. Mph1 may normally play two roles; for example, one to activate Mph1 at unattached kinetochores without the kinase activity and the other to phosphorylate substrates for signaling. It is however equally possible at present that Mph1-KD tethered at kinetochores forms a functional heterodimer with the wild-type Mph1.
In the above experiments Mph1-Ndc80-GFP was expressed from the nmt1 promoter of pREP81, which might allow expression of Mph1-Ndc80-GFP at a level higher than that of the native Mph1. To examine the effect of Mph1-Ndc80-GFP under a physiological condition, we next attempted to express Mph1-Ndc80-GFP from the native promoter of the mph1+ locus. A construct for tagging Ndc80-GFP at the C-terminal of Mph1 by integration was transformed into a strain in which expression of Mad2 could be turned on or off by removing/adding thiamine to the media (the mad2+ gene was replaced with nmt1-mad2+). The spindle checkpoint in this strain was not functional when expression of Mad2 was turned off. We could thereby obtain transformants even when Mph1-Ndc80-GFP expressed from the native locus persistently activated the checkpoint. As expected, we obtained the transformants only when expression of Mad2 was turned off and confirmed that the construct was integrated at the mph1+ locus. Mph1-Ndc80-GFP expressed from the native mph1+ locus caused a growth arrest when expression of Mad2 was turned on in a manner dependent on Bub1, Mad1, the kinase activity of Mph1, and interaction between Slp1 and Mad2 (Fig. 4 A and B). Analysis by immunoblot indicated that the level of Mph1-Ndc80-GFP did not exceed that of Mph1-GFP (Fig. S3A). It also revealed that the mobility of Mph1-Ndc80-GFP was slower when expression of Mad2 was on (Fig. S3A). This mobility shift likely suggests autophosphorylation of Mph1. As shown in Fig. S3 B and C, Mph1-Ndc80-GFP was localized at centromeres with Cnp3 regardless of the expression of Mad2. When Mad2 was not expressed, the cells grew normally, exhibiting the interphase nuclear morphology (Fig. S3B). Upon induction of Mad2, the cells exhibited overcondensed chromosomes (Fig. S3C) and accumulated Slp1 (Fig. S3A), both of which were hallmarks of mitotic arrest. These results indicated that Mph1-Ndc80-GFP expressed at a level comparable to (or lower than) that of Mph1-GFP alone could efficiently maintain the spindle checkpoint active.
Fig. 4.
Effect of kinetochore-tethered Mph1 expressed from the native locus. (A) Mph1 (or Mph1-KD) was expressed from the native mph1+ locus as a fusion protein with GFP or Ndc80-GFP in cells expressing Mad2 conditionally. The plates were incubated at 32 °C for 3 d. (B) Mph1-Ndc80-GFP was expressed as in A in the indicated genetic background. (C and D) Localization of Bub1-tdTomato (red) was determined with Mph1-Ndc80-GFP expressed as in A (green) in cells expressing Mad2 conditionally. DNA was visualized by staining with Hoechst 33342. (E and F) Localization of Mad1-mCherry (red) was determined as in C and D. (Scale bars, 5 μm.)
Recruitment of Bub1 and Mad1 to Centromeres/Kinetochores.
The strain examined above could offer an opportunity to investigate the role of Mph1 to recruit other components of the checkpoint to unattached kinetochores. We first expressed Bub1 labeled with a red fluorescent protein tdTomato and found that it formed foci with Mph1-Ndc80-GFP throughout the cell cycle in the absence of Mad2 (Fig. 4 C and D). Because expression of Mph1-KD-Ndc80-GFP or Mph1-GFP alone did not recruit Bub1 to centromeres/kinetochores (Fig. S3 D and E), the results suggested that localization of Bub1 required the kinase activity of Mph1 localized at centromeres/kinetochores. It is likely that Bub1 itself does not sense whether or not a kinetochore is attached to the spindle, but is recruited to unattached kinetochore marked by Mph1. Because a previous study indicated that Mad3 and Bub3 were recruited to Bub1 forced to localize at telomeres (37), we speculate that these two components would not recognize unattached kinetochores by themselves, but would rather passively be recruited by Mph1 via Bub1.
By a similar strategy, we determined the dependency of Mad1 for its localization at centromeres/kinetochores. As shown in Fig. 4E, when Mad2 was not expressed, Mad1 was not colocalized with Mph1-Ndc80-GFP, suggesting that localization of Mph1 at centromeres/kinetochores was not sufficient for recruitment of Mad1. Upon induction of Mad2, Mad1 accumulated at kinetochores in cells arrested at mitosis (Fig. 4F).
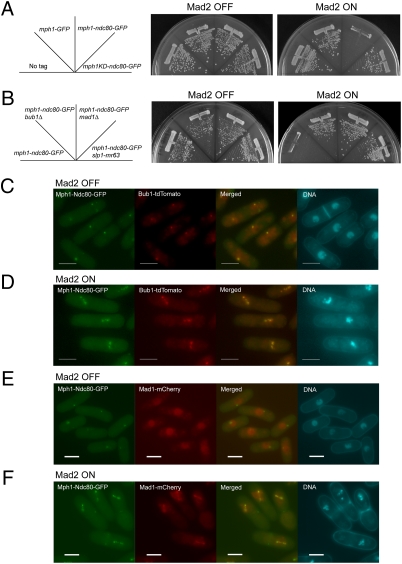
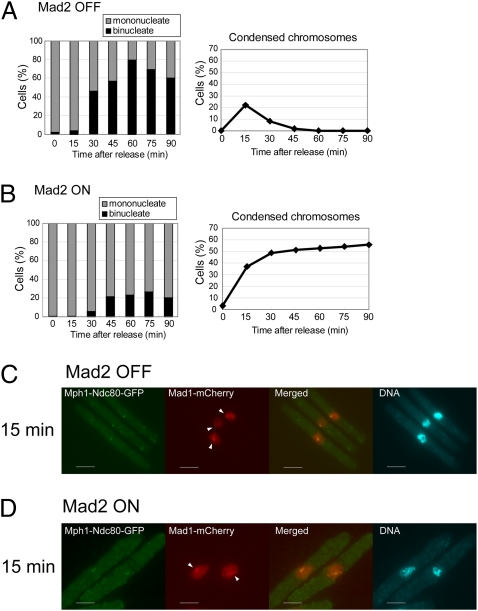
We further examined localization of Mad1 by synchronizing cell growth. The cdc25-22 mutation was introduced into the strain in which three proteins were expressed as follows: Mph1-Ndc80-GFP form the native mph1+ locus, Mad2 from the nmt1 promoter at the mad2+ locus, and Mad1 tagged with a red fluorescent protein mCherry from the native mad1 locus. Block-and-release experiment with the resulting strain was performed as illustrated in Fig. S4A. First, the cells were incubated with thiamine (Mad2 OFF, Fig. S4B) or without thiamine (Mad2 ON, Fig. S4C) at the permissive temperature of 26 °C for 22 h (at this time point, the level of Mad2 was not sufficient to cause a mitotic arrest even when expression of Mad2 was turned on). After subsequent incubation at 36 °C, the cells were arrested at the G2/M boundary with Mad2 at a level sufficient to cause a mitotic arrest (Mad2 ON, Fig. S4B). As shown in Fig. 5 A and B, most of the cells, which were mononucleate, at release from the block (time 0) did not have condensed chromosomes. When Mad2 was turned off, the index of the chromosome condensation reached a peak at 15 min after the release. At 60 min after the release, the percentage of the binucleate cells reached a peak (∼80%), indicating that the cells progressed through mitosis synchronously (Fig. 5A). When Mad2 was turned on, the index of the chromosome condensation gradually increased from 0 to more than 50%. Binucleate cells, which passed through anaphase, however, did not increase. These results indicated that when Mad2 was turned on, the cells, which were initially at the boundary of G2/M, were arrested before anaphase (Fig. 5B).
Fig. 5.
Localization of Mad1 in mitosis. (A and B) The percentages of the binucleate cells (Left) and cells with condensed chromosomes (Right) were determined after release from the arrest at the G2/M boundary. The cells were expressing Mad2 conditionally and Mph1-Ndc80-GFP from the native mph1+ locus. (C and D) Localization of Mad1-mCherry (red) was determined with Mph1-Ndc80-GFP expressed as in A and B 15 min after the release from the arrest at the G2/M boundary. Arrowheads indicate Mad1 foci colocalized with Mph1-Ndc80-GFP. DNA was visualized by staining with DAPI (4′-6-diamino-2-phenylindole). (Scale bars, 5 μm.) Fig. S4 shows images at other time points.
We first observed Mad1 in cells in which expression of Mad2 was turned off. Mad1 was not colocalized with Mph1-Ndc80-GFP before release of the arrest (0 min, Fig. S4B). It was then found as foci in some cells 15 min after the shift to a permissive temperature (Fig. 5C and Fig. S4B). As these foci were colocalized with Mph1-Ndc80-GFP, they were on kinetochores. At a later time point (30 min), Mad1 was found on the spindle. By the completion of anaphase (45 min), Mad1 disappeared from the spindle. The observation would suggest that Mad1, at an initial stage of mitosis, is recruited to kinetochores, which are presumably unattached to the spindle. It was then stripped off from the kinetochores and translocated to the spindle. Mad1 thereby determines its localization autonomously even when Mph1 was fixed to kinetochores. Although Mad1 recruitment is autonomous, we do not exclude a possibility that kinetochore-localized Mph1 might still be necessary but is certainly not sufficient to direct Mad1 recruitment. In a number of studies, Mps1 was shown to be required for recruitment of Mad1 to kinetochores (6, 21, 22). Our result would indicate that an additional component/condition is required for constitutive association of Mad1 with kinetochores. Because expression of Mad2 was turned off, it is possible that Mad2 might be required for stable association of Mad1 to kinetochores.
We next observed Mad1 in cells in which expression of Mad2 was turned on (Fig. 5D and Fig. S4B). By 30 min after the shift to a permissive temperature for the cdc25 mutation, the behavior of Mad1 in cells expressing Mad2 was similar to that in cells lacking Mad2. It however remained as foci or on the spindle 45 and 60 min after the shift to a permissive temperature. On the basis of the results, we speculated that once the checkpoint was activated, Mph1-Ndc80 prevented silencing of the checkpoint. As a result, Mad1 remained as foci at kinetochores or on the spindle.
Whereas kinetochores are considered to be equal in their constituents and overall structure, the components of the spindle checkpoint are selectively recruited to unattached kinetochores. Which of the components of the spindle checkpoint recognizes unattached kinetochores has been an important and long-standing question. Our study suggests that Mph1/Mps1 would be one of the primary components for recognition of the unattached kinetochore. We speculate that a putative receptor may change its affinity for Mph1/Mps1, depending on microtubule occupancy and/or tension. Once Mph1 is captured by the receptor, certain centromere components could be modified by Mph1 for recruitment of Bub1. In our experimental system, in which expression of Mad2 was turned off, Mad1 did not constitutively associate with centromere/kinetochores marked by Mph1. However, it was transiently found at kinetochores (Fig. 5C). The result thereby suggests that even in the absence of Mad2, Mad1 recognizes unattached kinetochore autonomously. It has been shown that recruitment of Mad1 and Mps1 requires mitotic kinases, Plk1 and Aurora B, respectively (38, 39). These kinases may be involved in recognition of unattached kinetochores by modifying Mad1, Mps1, or their receptors.
Materials and Methods
The S. pombe strains used in this study are listed in Table S1. The strains were grown in yeast extract with supplement (YES) media or synthetic Edinburgh minimal media (EMM) with appropriate nutrient supplements as previously described (40). The block-and-release experiment of nda3-KM311 cold-sensitive mutant cells was performed as described previously (33). See SI Materials and Methods for other procedures.
Supplementary Material
Acknowledgments
The authors thank Kevin Hardwick (University of Edinburgh), Shelly Sazer (Baylor College of Medicine), Silke Hauf (Friedrich Miescher Laboratory of the Max Planck Society), Yoshinori Watanabe (University of Tokyo), Chikashi Shimoda (Osaka City University), Mitsuhiro Yanagida (Okinawa Institute of Science and Technology), and the Yeast Genetic Resource Center for strains and plasmids. This work was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114647109/-/DCSupplemental.
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 3.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 5.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 6.Abrieu A, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 7.Stucke VM, Silljé HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters JM. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 10.Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang LH, et al. Budding yeast Cdc20: A target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 12.Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: An effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 13.Sczaniecka M, et al. The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-anaphase-promoting complex (APC/C) J Biol Chem. 2008;283:23039–23047. doi: 10.1074/jbc.M803594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 15.Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howell BJ, et al. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 17.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millband DN, Hardwick KG. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol Cell Biol. 2002;22:2728–2742. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 20.He X, Jones MH, Winey M, Sazer S. Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe. J Cell Sci. 1998;111:1635–1647. doi: 10.1242/jcs.111.12.1635. [DOI] [PubMed] [Google Scholar]
- 21.Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 22.Liu ST, et al. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maciejowski J, et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hewitt L, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tange Y, Niwa O. Schizosaccharomyces pombe Bub3 is dispensable for mitotic arrest following perturbed spindle formation. Genetics. 2008;179:785–792. doi: 10.1534/genetics.107.081695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanoosthuyse V, Meadows JC, van der Sar SJ, Millar JB, Hardwick KG. Bub3p facilitates spindle checkpoint silencing in fission yeast. Mol Biol Cell. 2009;20:5096–5105. doi: 10.1091/mbc.E09-09-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Windecker H, Langegger M, Heinrich S, Hauf S. Bub1 and Bub3 promote the conversion from monopolar to bipolar chromosome attachment independently of shugoshin. EMBO Rep. 2009;10:1022–1028. doi: 10.1038/embor.2009.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauzé E, et al. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisk HA, Winey M. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto A, Hiraoka Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 2003;22:2284–2296. doi: 10.1093/emboj/cdg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol. 2009;19:1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: A cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 34.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 35.Jelluma N, Dansen TB, Sliedrecht T, Kwiatkowski NP, Kops GJ. Release of Mps1 from kinetochores is crucial for timely anaphase onset. J Cell Biol. 2010;191:281–290. doi: 10.1083/jcb.201003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kemmler S, et al. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rischitor PE, May KM, Hardwick KG. Bub1 is a fission yeast kinetochore scaffold protein, and is sufficient to recruit other spindle checkpoint proteins to ectopic sites on chromosomes. PLoS ONE. 2007;2:e1342. doi: 10.1371/journal.pone.0001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi YH, et al. Requirements for protein phosphorylation and the kinase activity of polo-like kinase 1 (Plk1) for the kinetochore function of mitotic arrest deficiency protein 1 (Mad1) J Biol Chem. 2008;283:35834–35844. doi: 10.1074/jbc.M804967200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saurin AT, van der Waal MS, Medema RH, Lens SM, Kops GJ. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat Commun. 2011;2:316. doi: 10.1038/ncomms1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.