Abstract
The transcription factor zinc-finger protein Miz1 represses TNF-α–induced JNK activation and the repression is relieved upon TNF-α stimulation. However, the underlying mechanism is incompletely understood. Here we report that Miz1 interferes with the ubiquitin conjugating enzyme (E2) Ubc13 for binding to the RING domain of TNF-receptor associated factor 2 (TRAF2), thereby inhibiting the ubiquitin ligase (E3) activity of TRAF2 and suppressing TNF-α–induced JNK activation. Upon TNF-α stimulation, Miz1 rapidly undergoes K48-linked polyubiquitination at Lys388 and Lys472 residues and subsequent proteasomal degradation in a TRAF2-dependent manner. Replacement of Lysine 388 and Lysine 472 by arginines generates a nondegradable Miz1 mutant, which significantly suppresses TNF-α–induced JNK1 activation and inflammation. Thus, our results reveal a molecular mechanism by which the repression of TNF-α–induced JNK activation by Miz1 is de-repressed by its own site-specific ubiquitination and degradation, which may account for the temporal control of TNF-α–JNK signaling.
Keywords: TNF receptor complex, protein kinase
The proinflammatory cytokine TNF-α regulates a wide range of biological activities, including inflammation, immune responses, apoptosis, and tumorigenesis (1, 2). TNF-α signaling is mainly mediated by its cytoplasm membrane receptor 1 (TNF-R1) (3). Upon TNF-α stimulation, TNF-R1-associated death domain protein, TNF-receptor associated factor 2 (TRAF2) and TRAF5, receptor interacting protein 1, and two inhibitors of apoptosis, cIAP1/cIAP2, are recruited to TNF-R1 to form TNF-R1 Complex 1, which in turn activates multiple downstream effectors, such as JNK1 (also known as stress-activated protein kinase), p38, and the inhibitor of NF-κB kinase (IKK) complex (4–8). The temporal and spatial regulation of TNF-α signaling determines its biological functions. For example, although prolonged activation of JNK1 is essential for TNF-α–induced cell death when NF-κB activation is impaired (9–17), transient activation of JNK1 is involved in TNF-α–induced inflammation through c-Jun–induced expression of several proinflammatory genes (18) and contributes to inflammation-driven certain types of cancer through cell death-induced “compensatory proliferation” (19–21).
TRAF2 is a key regulator of TNF-α–induced JNK1 activation (8, 22). Although TRAF2 protein is required for TNF-α to activate JNK1, p38, and IKK (4, 5, 7, 8), TRAF2 K63-linked polyubiquitination is required for TNF-α–induced activation of JNK1, but not p38 and IKK (23, 24). The K63-linked polyubiquitination of TRAF2 is positively regulated by the E3 ubiquitin ligases, including TRAF2 itself (25), and is negatively regulated by deubiquitination enzymes, such as CYLD and A20 (26–28). Recently, it has been reported that the transcription factor Miz1 (Myc-interacting zinc-finger protein 1; also known as Z13), which is involved in cell-cycle control and is also a transcription repressor that mediates Myc gene-repression effect (29, 30), represses TRAF2 K63-linked polyubiquitination independently of its transcription activity, resulting in suppression of TNF-α–induced JNK1 activation and apoptosis (31). The repression by Miz1 is both TNF-α and JNK-specific, as it does not inhibit JNK1 activation by other extracellular stimuli examined, such as UV or IL-1β, or TNF-α–induced activation of IKK, p38, and ERK (31). Thus, Miz1 functions as a signal- and pathway-specific modulator or regulator (SMOR) for TNF-α–induced JNK1 activation (31). Interestingly, upon TNF-α stimulation, Miz1 is degraded in a proteasome-dependent manner (31). However, it is unknown how Miz1 suppresses TNF-α–induced TRAF2 K63-linked polyubiquitination and JNK1 activation, and whether the proteasomal degradation of Miz1 is required for relieving the repression by Miz1. Here we report that Miz1 binds to the RING domain of TRAF2 to interfere with the interaction between TRAF2 and the ubiquitin conjudating enzyme (E2) Ubc13, thereby preventing the E3 ligase TRAF2 from regulating its own K63-linked polyubiquitination and JNK1 activation. Upon TNF-α stimulation, Miz1 is polyubiquitinated at Lys388 and Lys472 and subsequently degraded by the proteasome. The Miz1(K388R/K472R) mutant resists proteasomal degradation and significantly suppresses TNF-α–induced JNK1 activation.
Results
Miz1 Inhibits the Ubiquitin Ligase (E3) Activity of TRAF2.
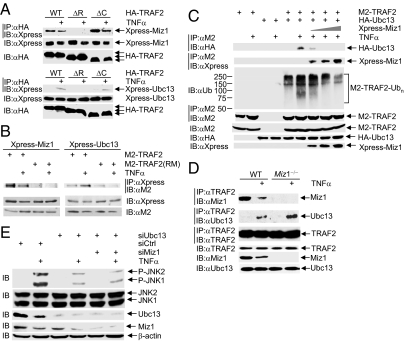
Miz1 selectively inhibits TNF-α–induced JNK activation through suppression of TRAF2 K63-linked polyubiquitination with no known mechanisms (31). We hypothesized that Miz1 may inhibit TRAF2 E3 ligase activity. We found that TNF-α–induced activation of JNK or p38 was significantly reduced in TRAF2-null mouse embryonic fibroblasts (MEFs) (Fig. 1A, compare lane 6 with lane 2), consistent with previous reports (23, 24). Reintroduction of HA-TRAF2, but not the E3 ligase-deficient HA-TRAF2(C49A/C51A) mutant (32), restored TNF-α–induced JNK1 activation (Fig. 1A, compare lane 7 with lane 8). In contrast, both HA-TRAF2 and HA-TRAF2(C49A/C51A) mutant restored TNF-α–induced p38 activation (Fig. 1A, compare lane 7 with lane 8). Thus, TRAF2 E3 ligase activity is essential for TNF-α–induced JNK1 activation.
Fig. 1.
Miz1 inhibits TRAF2 E3 ligase activity. (A and C) TRAF2-null MEFs were transfected with expression vectors encoding HA-TRAF2, the E3 ligase-deficient HA-TRAF2 (C49A/51A) mutant, or empty vector (2 μg each in A and 5 μg each in C), and treated with TNF-α (5 ng/mL) for 15 min (A) or various periods of time (C). Phosphorylation of JNK and p38 and expression levels of JNK, p38, and HA-TRAF2 were analyzed (A). Synthesis of polyubiquitin chains from purified recombinant ubiquitin proteins by HA-TRAF2 or its mutant was determined (B), as described in Methods, except the whole reaction mixture was analyzed (C). (B) The loss of Miz1 augmented TRAF2 E3 ligase activity in synthesizing polyubiquitin chains from purified recombinant ubiquitin proteins (see Methods for details). Cells were stimulate by TNF-α (5 ng/mL) for 15 min. (D) The interaction between purified GST-Miz1 and [35S]TRAF2 or [35S]TRAF6 was analyzed by an in vitro binding assay. The amount of GST was 2 μg and GST-Miz1 was 6 μg, because GST-Miz1 was partially degraded when purified from bacteria. CBB, Coomassie brilliant blue. (E) HEK293 cells were transfected with expression vectors encoding M2-TRAF2, along with WT and various truncation mutants of Xpress-Miz1, as indicated. The interaction between M2-TRAF2 and Xpress-Miz1 (WT and mutants) was analyzed. (F) Miz1-null MEFs were transfected with expression vectors encoding WT or various truncation mutants of Xpress-Miz1 that fail to interact with TRAF2 as shown (E). TNF-α–induced JNK activation was analyzed by immunoblotting using antiphospho-JNK antibody.
Next, we tested whether Miz1 regulates TRAF2 E3 ligase activity. Although immunoprecipitated TRAF2 could no longer catalyze its own K63-linked polyubiquitination for reasons unknown (33), it was still able to act as an E3 to synthesize polyubiquitin chains from purified recombinant ubiquitin proteins in vitro (ex vivo ubiquitin assembly assay) (33). Using this assay, we found that the E3 ligase activity of TRAF2 isolated from Miz1-null MEFs was significantly augmented compared with that isolated from WT fibroblasts (Fig. 1B, Upper). Because protein A-sepharose bead-bound immunoprecipitated TRAF2 had been removed from the reaction mixture by centrifugation before the reaction products in the supernatant were analyzed (Fig. 1A, Lower), the polyubiquitinated products were polyubiquitin chains of K63O ubiquitins, rather than ubiquitinated TRAF2 or TRAF2-associated proteins. It is possible that the activity of other K63-E3 ligases that might be coimmunoprecipitated with TRAF2, rather than TRAF2 itself, were augmented by the loss of Miz1; however, this is unlikely because the immune complex isolated from TRAF2-null MEFs did not possess any detectable E3 ligase activity in the ex vivo ubiquitin assembly assay (Fig. 1C). Under the same conditions, ectopically expressed HA-TRAF2 but not HA-TRAF2(C49A/C51A) was able to synthesize polyubiquitin chains (Fig. 1C). Thus, the E3 ligase activity of TRAF2 immune complex (Fig. 1B, Upper) was not from other E3s.
To determine whether the inhibition of TRAF2 E3 ligase activity requires the interaction between Miz1 and TRAF2, we used an in vitro binding assay. We found that purified GST-Miz1 directly interacted with in vitro translated 35S-labeled TRAF2, but not 35S-labeled TRAF6 (Fig. 1D), suggesting that Miz1 directly binds to TRAF2. Furthermore, truncation experiments revealed that the zinc fingers 7–8 (amino acids 466–514) [Miz1(Δ466–514)], 9–10 (amino acids 522–570) [Miz1(Δ522–570)], and 11–12 (amino acids 578–635) [Miz1(Δ578–635)] were required for the binding of Miz1 to TRAF2 (Fig. 1E). Consistently, TNF-α–induced JNK activation in Miz1-null MEFs was inhibited by ectopically expressed WT Xpress-Miz1, but not the three Miz1 zinc-finger truncation mutants (Fig. 1F). Taken together, Miz1 binds to TRAF2 via its zinc fingers 7–12, thereby inhibiting TRAF2 to activate JNK upon TNF-α stimulation.
Miz1 Interferes with the Binding of Ubc13 to TRAF2.
We were curious about how the binding of Miz1 to TRAF2 inhibits its E3 ligase activity. One possibility is that the binding of Miz1 to TRAF2 may interfere with the interaction between TRAF2 and its E2, such as Ubc13 (23, 25, 34). Like WT TRAF2, TRAF2 ΔC mutant, in which the TRAFC domain has been deleted, interacted with either Miz1 or Ubc13 (Fig. 2A). In contrast, TRAF2 ΔR mutant, in which the RING domain has been deleted, was unable to do so (Fig. 2A). Consistently, both Miz1 and Ubc13 were unable to bind to the TRAF2 (RM) mutant, which is no longer able to interact with TRAF2 RING domain-interacting proteins (Fig. 2B) (23). These data suggest that Miz1 interferes with Ubc13 binding to the RING domain of TRAF2. Consistently, ectopic expression of Xpress-Miz1 inhibited the association between HA-Ubc13 and M2-TRAF2 in a dose-dependent manner, accompanying with the increase in its own association with M2-TRAF2 and decrease in M2-TRAF2 ubiquitination (Fig. 2C). Similar results were obtained when the effect of endogenous Miz1 on the interaction between TRAF2 and Ubc13 was examined. In resting WT fibroblasts, Miz1 already associated with TRAF2 and the association was significantly reduced after cells were stimulated with TNF-α for only 15 min (Fig. 2D). Under the same conditions, TNF-α–induced association of Ubc13 with TRAF2 was significantly increased (Fig. 2D). The association between TRAF2 and Ubc13 was further augmented in TNF-α–stimulated Miz1-null MEFs (Fig. 2D). Interestingly, the interaction between Ubc13 and TRAF2 was not augmented in resting Miz1-null MEFs (Fig. 2D). This finding suggests that the removal of Miz1 is necessary but not sufficient for the stable interaction between Ubc13 and TRAF2. Silencing of Ubc13 with specific siRNA significantly reduced TNF-α–induced JNK activation in WT fibroblasts (Fig. 2E) and also abrogated augmented TNF-α–induced JNK activation in Miz1-null MEFs (Fig. S1), consistent with the notion that Ubc13 is involved in TNF-α–induced JNK1 activation (23). In Ubc13 knockdown cells, further knockdown of Miz1 was no longer able to augment TNF-α–induced JNK1 activation (Fig. 2E). Taken together, these data show that Miz1 inhibits TRAF2 E3 ligase activity through interfering with the binding of Ubc13 to the RING domain of TRAF2, either directly or allosterically.
Fig. 2.
Miz1 interferes with the interaction between TRAF2 and Ubc13. (A) WT fibroblasts were transfected with expression vectors encoding HA-TRAF2, HA-ΔRING-TRAF2 mutant (ΔR), or HA-ΔTRAF-C-TRAF2 mutant (ΔC) (5 μg each), along with Xpress-Miz1 or Xpress-Ubc13 (5 μg each). The interaction between various HA-TRAF2 and Miz1 or Ubc13 was analyzed. (B) HEK293 cells were transfected with expression vectors encoding Xpress-Miz1 or Xpress-Ubc13, along with WT M2-TRAF2 or M2-TRAF2 RING domain mutant (23). The interaction between Xpress-Miz1 or Xpress-Ubc13 with WT or the RING domain mutant of M2-TRAF2 was analyzed. (C) WT fibroblasts were transfected with expression vectors encoding M2-TRAF2, along with HA-Ubc13 (5 μg each), with or without various amounts of Xpress-Miz1 (3, 4, and 6 μg), as indicated. The interaction between M2-TRAF2 and HA-Ubc13 or Xpress-Miz1, as well as ubiquitination of M2-TRAF2, in control or TNFα-stimulated cells was analyzed. (D) The association between TRAF2 and Miz1 or Ubc13 in control or TNFα-treated WT and Miz1-null MEFs were analyzed. (E) The effect of Ubc13 knockdown or double knockdown of Ubc13 and Miz1 on TNF-α–induced JNK phosphorylation was analyzed.
TNF-α Induces K48-Linked Polyubiquitination of TRAF2-Associated Miz1 for Its Proteasomal Degradation.
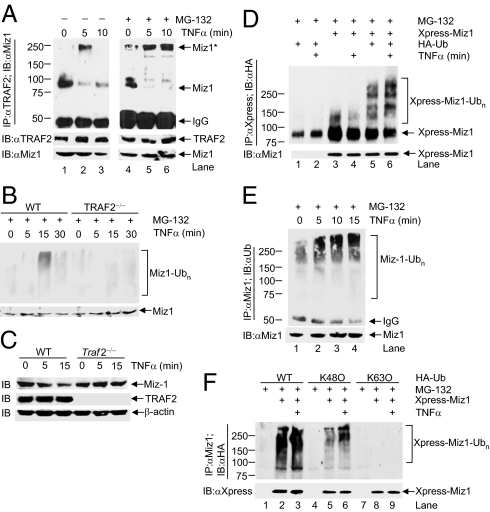
TNF-α typically induces TRAF2 K63-linked polyubiquitination and JNK1 activation in its target cells within minutes (13, 23, 24, 35). If Miz1 is a physiologically relevant repressor, the repression by Miz1 on TRAF2 K63-linked polyubiquitination and JNK1 activation has to be rapidly relieved upon TNF-α stimulation. Indeed, we found that in resting cells Miz1 already associated with TRAF2 (Fig. 3A), migrating as an 87-kDa species in SDS-gel (Fig. 3A, lane 1). Five minutes after TNF-α stimulation, the 87-kDa species of Miz1 was significantly reduced, accompanying with the appearance of a major high molecular-weight species of Miz1 that migrated around 250 kDa in SDS-gel (Fig. 3A, lane 2). Ten minutes after TNF-α stimulation, the 250-kDa species of Miz1 already disappeared, whereas the reduced 87-kDa specie of Miz1 remained unchanged (Fig. 3A, lane 3). The disappearance of the 250-kDa species of Miz1 10 min after TNF-α stimulation was not the result of the decrease in the amount of TRAF2, as there were no detectable changes in the amount of TRAF2 during this period (Fig. 3A). Pretreatment with the proteasome inhibitor MG-132 blocked the disappearance of 250-kDa species of Miz1 but had little effect on the reduction of the 87-kDa species of Miz1 (Fig. 3A, lanes 5 and 6). In addition, TNFα–induced polyubiquitination and degradation of Miz1 was blocked in TRAF2-null MEFs (Fig. 3 B and C), suggesting that Miz1 is degraded in a TRAF2-dependent manner. Thus, TRAF2-associated Miz1 was rapidly modified upon TNF-α stimulation, triggering its proteasomal degradation and thereby relieving its inhibition on TRAF2.
Fig. 3.
TNF-α induces Miz1 K48-linked polyubiquitination of TRAF2-associated Miz1. (A) TRAF2 was immunoprecipitated from WT fibroblasts that have been pretreated without or with MG-132 (50 μM, 2 h) and then treated with TNF-α (5 ng/mL) for various periods of time, as indicated. TRAF2-coimmunoprecipitated Miz1 was detected by immunoblotting using anti-Miz1 antibody. Miz1*, upshifted Miz1. (B and C) TNF-α–induced degradation of Miz1 was dependent on TRAF2. (D and F) TNF-α–induced polyubiquitination of Xpress-Miz1 in the presence of cotransfected HA-Ub (D), or HA-Ub, HA-K63O-Ub, or HA-K48O-Ub (F) (5 μg each) was analyzed. (E) TNF-α–induced ubiquitination of Miz1 in the presence of MG-132 (see Methods for details).
Next, we tested whether K48-linked polyubiquitination is involved in this process. We found that in MG-132 pretreated WT fibroblasts, Xpress-Miz1 was already modestly polyubiquitinated when cotransfected with HA-Ub (Fig. 3D, lane 5), which is a quite common phenomenon seen with overexpressed proteins cotransfected with ubiquitin. The polyubiquitination of Xpress-Miz1 was further augmented by TNF-α (Fig. 3D, lane 6). The polyubiquitination of endogenous Miz1 was also significantly increased as early as 5 min after TNF-α stimulation and peaked at 15 min in the presence of MG-132 (Fig. 3E, lanes 2, 3, and 4). In HeLa cells pretreated with MG-132, treatment with TNF-α for 15 min induced polyubiquitination of ectopically expressed Xpress-Miz1 with cotransfected HA-ubiquitin or HA-Ub(K48O), in which all lysine residues except lysine 48 in ubiquitin have been replaced by arginines, but not with Ub(K63O), in which all lysine residues except lysine 63 in ubiquitin have been replaced by arginines (Fig. 3F). This finding indicates that the polyubiquitin chains on Miz1 were primarily linked through K48 of ubiquitin. Taken together, the data show that TNF-α induces rapid Miz1 K48-linked polyubiquitination, triggering its proteasomal degradation.
TNF-α Induces K48-Linked Polyubiquitination of Miz1 at Lysine388 and Lysine472.
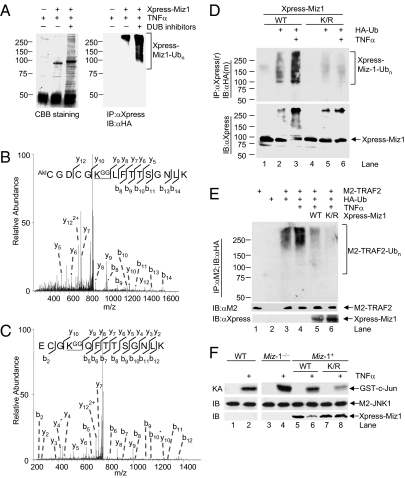
To identify TNF-α–induced K48-linked polyubiquitination sites in Miz1, we used the approach of mass spectrometry. Xpress-Miz1 immunoprecipitated from transfected HeLa cells was separated by SDS-gel electrophoresis and visualized by Coomassie brilliant blue staining (Fig. 4A), and its polyubiquitination was confirmed by immunoblotting analysis using anti-HA antibody (Fig. 4A). The gel region corresponding to ubiquitinated Xpress-Miz1 was excised, digested with trypsin, and analyzed by MS. Protein sequence database searching of the resulting MS/MS data using the MASCOT algorithm identified K388 and K472 as two ubiquitin-adduct sites (Fig. 4 B and C), which were evidenced by an increase of 114 Da at the ubiquitinated lysine residues caused by the addition of two glycine residues from ubiquitin chain (36, 37). The ubiquitination sites were exclusively mapped from the adjacent y types of fragment ions of the peptides (Fig. 4 B and C).
Fig. 4.
Miz1 is polyubiquitinated at Lys388 and Lys472 upon TNF-α stimulation. (A) HeLa cells transfected with expression vectors encoding Xpress-Miz1, along with HA-Ub or empty vector (5 μg each) were treated with TNF-α (5 ng/mL, 15 min) and harvested in the presence or absence of deubiquitination enzyme (DUB) inhibitors (Ub aldehyde 5 μM and N-ethylmaleimde 8 mM). CBB, coomassie brilliant blue. (B and C) MS/MS spectra of two ubiquitinated Miz1 peptides. (D) WT fibroblasts were transfected with expression vector encoding Xpress-Miz1 (WT or K388/473R mutant) (5 μg each), along with HA-Ub (5 μg) and then stimulated without or with TNF-α (5 ng/mL, 15 min). Ubiquitinated Xpress-Miz1 was analyzed as described in Fig. 3. (E) The effect of Xpress-Miz1 and Miz1(K388/472R) mutant on ubiquitination of cotransfected M2-TRAF2 was analyzed, as described previously (31). (F) WT and Miz1-null MEFs transfected with expression vector encoding M2-JNK1 (1 μg), along with or without Xpress-Miz1 (WT or K388/472R mutant) or empty vector (2 μg each) were treated without or with TNF-α (5 ng/mL, 15 min). M2-JNK1 activity and expression of M2-JNK1 and Xpress-Miz1 were determined.
Next we determined the role of K388 and K472 in TNF-α–induced K48-linked polyubiquitination of Miz1. TNF-α significantly inuduced polyubiquitination of Xpress-Miz1 (Fig. 4D). In contrast, TNF-α–induced polyubiquitination of Xpress-Miz1(K388R/K472R), in which K388 and K472 had been replaced by arginines, was negligible (Fig. 4D, Upper). Consistently, Xpress-Miz1 was partially degraded upon TNF-α stimulation but Xpress-Miz1(K388R/K472R) was resistant (Fig. 4D, Lower). Thus, K388 and K472 are primary TNF-α–induced K48-linked polyubiquitination sites of Miz1.
To test whether Xpress-Miz1(K388R/K472R) mutant is a more potent suppressor for TNF-α–induced TRAF2 K63-linked polyubiquitination and JNK1 activation, HeLa cells were stimulated with TNF-α for 15 min, at which time TRAF2 was mainly K63-linked polyubiquitinated (31). Although TNF-α–induced polyubiquitination of M2-TRAF2 was suppressed by Xpress-Miz1 (Fig. 4E, compare lane 5 with lane 4), M2-TRAF2 polyubiquitination was further suppressed by Xpress-Miz1(K388R/K472R) (Fig. 4E, compare lane 6 with lane 5). Because both WT and mutant Miz1 bound to TRAF2 with similar affinity (Fig. S2), the stronger suppression by Xpress-Miz1(K388R/K472R) is likely a result of its resistance to TNF-α–induced degradation (Fig. 4E, lowest panel, compare lanes 6 and 5; also see Fig. 4F). Miz1(K388R/K472R) mutant was also a more potent inhibitor of TNF-α–induced JNK1 activation. Immune complex kinase assays showed that M2-JNK1 activity was augmented in Miz1 null MEFs compared with that in WT fibroblasts (Fig. 4F, compare lane 4 with lane 2). Ectopic expression of Xpress-Miz1 reduced the augmented M2-JNK1 activity back to the level similar to TNF-α–induced M2-JNK1 activity in WT fibroblasts (Fig. 4F, compare lane 6 with lane 2), and Xpress-Miz1(K388R/K472R) mutant further reduced M2-JNK1 activity (Fig. 4F, compare lane 8 with lane 6). This reduction was not the result of the difference in expression of Xpress-Miz1 and its K388R/K472R mutant before TNF-α stimulation, as analyzed by immunoblotting using anti-Xpress antibody (Fig. 4F, compare lane 5 with lane 7); rather, it is because WT Xpress-Miz1 underwent degradation upon TNF-α stimulation (Fig. 4F, compare lane 6 with lane 5), but Xpress-Miz1(K388R/K472R) was not (Fig. 4F, compare lane 8 with lane 7). Thus, K48-linked polyubiquitination of Miz1 at K388 and K472 is required for its proteasomal degradation and subsequent TRAF2 K63-linked polyubiquitination and JNK1 activation in response to TNF-α stimulation.
Loss of Miz1 Accelerates TNF-α–Induced Proinflammatory Gene Expression.
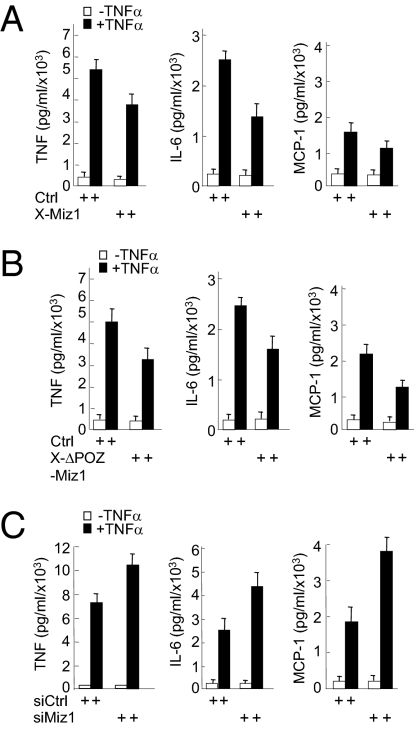
JNK plays a critical role in TNF-α–induced inflammation (12, 13). Because Miz1 negatively regulates TNF-α–induced JNK1 activation, we were curious about whether Miz1 modulates TNF-α–induced inflammation. To test this idea, primary mouse bone marrow-derived macrophages were isolated and subsequently infected with retroviral vectors encoding WT Xpress-Miz1 or ΔPOZ-Miz1, which is deficient in transcription and repression (38), siMiz1, or empty vector, followed by treatment without or with TNF-α. Analysis by the inflammation cytometric bead array revealed that TNF-α–induced production of proinflammatory cytokines, including TNF-α, IL-1β, and monocyte chemoattractant protein-1 (MCP-1), were significantly reduced in the macrophages overexpressing Xpress-Miz1 or Xpress-ΔPOZ-Miz1 (Fig. 5 A and B). Conversely, silencing of Miz1 by siRNA potentiated TNF-α–induced production of the proinflammatory cytokines (Fig. 5C). Taken together, these data suggest that Miz1 is a physiological regulator for TNF-α signaling.
Fig. 5.
The loss of Miz1 accelerates TNF-α–induced inflammation. (A–C) Bone marrow-derived macrophages infected with retroviral vector encoding Xpress-Miz1, Xpress-Miz1(ΔPOZ), or empty vector as indicated (A and B), or the control siRNA or siMiz1 (C) were treated without or with TNF-α (10 ng/mL, 8 h). Production of TNF-α, IL-6, and MCP-1 were analyzed using inflammation cytometric bead array (mouse inflammation kit; BD Biosciences). The results of A–C are presented as means SEs and represent three independent experiments.
Discussion
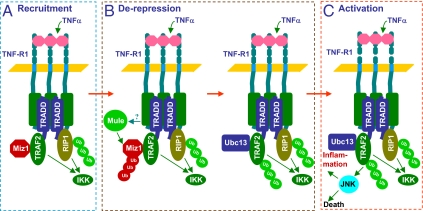
TNF-α is a pleiotropic proinflammatory cytokine that regulates immune responses, inflammation, cell death, and tumorigenesis through activation of multiple downstream signaling pathways, including NF-κB, JNK, and caspases (2, 3). It is incompletely understood whether the JNK pathway can be selectively regulated without affecting the activation of other signaling pathways by TNF-α or affecting JNK activation by other stimuli, such as UV and IL-1β. Recently, it has been reported that Miz1 can function as signal- and pathway-SMORs to selectively regulate TNF-α–induced JNK activation and cell death (14, 31). However, the underlying mechanism is unknown. In this report, we demonstrate that Miz1 inhibited TRAF2 E3 ligase activity by interfering with the binding of Ubc13 to TRAF2, thereby inhibiting TRAF2 K63-linked polyubiquitination and subsequent JNK1 activation (Figs. 1 and 2). We found that TNF-α induced K48-linked polyubiquitination of Miz1 primarily at K388 and K472 residues, and the ubiquitination was responsible for Miz1 proteasomal degradation (Figs. 3 and 4). The loss of Miz1 accelerated TNF-α–induced production of proinflammatory cytokines by primary bone marrow-derived macrophages (Fig. 5). These results prompted us to put forward a “de-repression” model for TNF-α–JNK1 signaling (Fig. 6). According to this model, in resting cells the binding of Miz1 to the RING domain of TRAF2 interferes with the interaction between TRAF2 and Ubc13, thereby repressing TRAF2 E3 ligase activity and TRAF2 K63-linked polyubiquitination. Upon TNF-α stimulation, the recruitment of TRAF2 to TNF-R1 is not sufficient for JNK1 activation, because its E3 ligase activity is repressed by Miz1. Miz1 needs to undergo K48-linked polyubiquitination and subsequently proteasomal degradation to provide the physical space for Ubc13 to bind to TRAF2, so that TRAF2 can undergo K63-linked polyubiquitination for selective activation of JNK and inflammation (Fig. 6). This de-repression mechanism may be critical for the specificity and temporal/strength control of the signaling by TNF-α and perhaps other members of TNF family.
Fig. 6.
Schematic illustration of the de-repression model by which Miz1 regulates TNF-α–JNK signaling. (A) Upon TNF-α stimulation, Miz1 is recruited to TNF-R1 Complex 1 in a TRAF2-dependent manner and blocks immediate K63-linked polyubiquitination of TRAF2, so that TRAF2 and ubiquitinated receptor interacting protein 1 (RIP1) activate IKK but not JNK. (B) TNF-α induces Miz1 K48-linked polyubiquitination, which is catalyzed by Mule, and proteasomal degradation, so that Ubc13 can bind to TRAF2 and promote TRAF2 K63-linked polyubiquitination. (C) Polyubiquitinated TRAF2 activates JNK, which contributes to inflammation or cell death, when NF-κB activation is impaired.
Our finding that Miz1 interfered with the binding of Ubc13 to the RING domain of TRAF2, thereby inhibiting the E3 ligase activity of TRAF2 (Fig. 1), contradicts a previous report showing that, based on the structural analysis of the RING domain and the first ring finger of TRAF2 and in vitro binding assays, the TRAF2 RING domain was unable to bind to Ubc13 (39). There are several possible explanations about this apparent controversy. First, it is possible that other parts of TRAF2 may be required for the binding of Ubc13. Second, purified GST-TRAF2 is quite different from the one isolated from TNF-α–stimulated cells. For example, GST-TRAF2 failed to catalyze the free ubiquitin chain synthesis (39). However, TNF-α–activated TRAF2 significantly catalyzed the free ubiquitin-chain synthesis (Fig. 1 B and C), consistent with a previous report (33). Third, the interaction between TRAF2 and Ubc13 appeared to be stimulation-dependent (Fig. 2D), consistent with the previous report (34). Finally, the interaction between TRAF2 and Ubc13 may depend on cofactors that lead to the structural changes of TRAF2, thereby facilitating the interaction between TRAF2 and Ubc13 (25). It is likely that Miz1 inhibits TRAF2 E3 ligase activity by interfering with the binding of Ubc13 to the TRAF2 RING domain, either directly or allosterically. Future studies are needed to explore these possibilities.
Is Ubc13 involved in TNF-α–induced activation of JNK1? Our results show that silencing of Ubc13 impaired JNK1 activation by TNF-α and also abolished the augmentation by Miz1 (Fig. 2E and Fig. S1), consistent with the previous report that Ubc13 was involved in TRAF2 K63-linked polyubiquitination (23). However, genetic disruption of Ubc13 alleles did not block TNF-α–induced JNK activation in MEFs (40). It is possible that other Ubc13-like E2s may compensate for the loss of Ubc13 in Ubc13-null MEFs, although WT fibroblasts transiently expressing siRNA of Ubc13 might have yet to mount the compensatory response. Another possibility is that the involvement of Ubc13 in TNF-α–induced JNK activation may be cell type- and stress-dependent, as TNF-α–induced JNK activation was impaired in Ubc13+/− splenocytes (41).
How is the repression by Miz1 on TNF-α–induced JNK1 activation released? Upon TNF-α stimulation, Miz1 was rapidly up-shifted and then degraded in a proteasome-dependent manner (Fig. 3A). Although we cannot formally exclude that other posttranslational modifications might be involved, the up-shift of Miz1 proteins is most likely a result of its K48-linked polyubiquitination. TNF-α–induced up-shift of Miz1 proteins was dramatically reduced with the Miz1(K388R/K472R) mutant, which was resistant to TNF-α–induced polyubiquitination and degradation (Fig. 4D). During the course of preparing the current article, it was reported that the E3 ligase Mule catalyzes Miz1 K48-linked polyubiquitination to trigger Miz1 degradation (42), further supporting our finding that K48-linked polyubiquitination at K388 and K472 is critical for relieving the repression by Miz1. Interestingly, Mule is not a component of the TNF-R1 complex. Future work is needed to determine how Mule is regulated by TNF-α.
Methods
Immunoprecipitation.
For immunoprecipitation of endogenous or transfected proteins from WT and Miz1-null MEFs or HeLa cells, cells were pretreated without or with MG-132 (25 μM) for 2 h and then treated with TNF-α (5 ng/mL) for various periods of time. Cells were harvested in the lysis buffer (50 mM Hepes, pH 7.6, 250 mM NaCl, 0.1% Nonidet P-40, 1 mM DTT, 20 mM β-glycerol phosphate, 1 mM sodium vanadate, 10 mM p-nitrophenylphosphate, 1 mM PMSP, 1 μg/mL leupeptin, 1 μg/mL aproptinin, and 1 μg/mL pepstatin). After clarification by centrifugation, cell lysates were incubated with specific antibodies and 30 μL (50% vol/vol) of Protein A-Sepharose beads for 4 h at 4 °C. The immune complexes were analyzed by immunoblotting. For analysis of ubiquitination of endogenous Miz1 or transfected Xpress-Miz1 or M2-TRAF2 (cotransfected with HA-Ub), cells were harvested in the lysis buffer as described above. After clarification, the cell extracts were heated at 95 °C for 3 min in the presence of 1% SDS to disrupt protein complexes. The extracts were diluted 10-fold with the lysis buffer to reduce the SDS concentration to 0.1% before immunoprecipitation in combination with immunoblotting analysis.
Supplementary Material
Acknowledgments
We thank Xin Lin and Tong-Chuan He for valuable reagents and protocols that made this work possible; and Mrs. Hanh Chi Do and Mr. Yue Chen for their excellent technical support. This work is supported by in part by National Institutes of Health Grants CA100460 and ES015868 (to A.L.) and GM081603 (to J.L.); National Basic Research Program of China (2012CB910801) and National Natural Science Foundation of China Grant (31130035) (to A.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105176108/-/DCSupplemental.
References
- 1.Tartaglia LA, Goeddel DV. Two TNF receptors. Immunol Today. 1992;13:151–153. doi: 10.1016/0167-5699(92)90116-O. [DOI] [PubMed] [Google Scholar]
- 2.Tracey KJ, Cerami A. Tumor necrosis factor: A pleiotropic cytokine and therapeutic target. Annu Rev Med. 1994;45:491–503. doi: 10.1146/annurev.med.45.1.491. [DOI] [PubMed] [Google Scholar]
- 3.Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 4.Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 5.Liu ZG, Hsu H, Goeddel DV, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-kappaB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 6.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 7.Tada K, et al. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-κ B activation and protection from cell death. J Biol Chem. 2001;276:36530–36534. doi: 10.1074/jbc.M104837200. [DOI] [PubMed] [Google Scholar]
- 8.Yeh WC, et al. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 9.De Smaele E, et al. Induction of gadd45β by NF-kappaB downregulates pro-apoptotic JNK signalling. Nature. 2001;414:308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- 10.Kamata H, et al. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 11.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 12.Lin A. Activation of the JNK signaling pathway: Breaking the brake on apoptosis. Bioessays. 2003;25:1–8. doi: 10.1002/bies.10204. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Minemoto Y, Lin A. c-Jun N-terminal protein kinase 1 (JNK1), but not JNK2, is essential for TNFα-induced c-Jun kinase activation and apoptosis. Mol Cell Biol. 2004;24:10844–10856. doi: 10.1128/MCB.24.24.10844-10856.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Lin A. Wiring the cell signaling circuitry by the NF-κ B and JNK1 crosstalk and its applications in human diseases. Oncogene. 2007;26:3267–3278. doi: 10.1038/sj.onc.1210417. [DOI] [PubMed] [Google Scholar]
- 15.Maeda S, et al. IKKbeta is required for prevention of apoptosis mediated by cell-bound but not by circulating TNFalpha. Immunity. 2003;19:725–737. doi: 10.1016/s1074-7613(03)00301-7. [DOI] [PubMed] [Google Scholar]
- 16.Tang G, et al. Inhibition of JNK activation through NF-kappaB target genes. Nature. 2001;414:313–317. doi: 10.1038/35104568. [DOI] [PubMed] [Google Scholar]
- 17.Tang F, et al. The absence of NF-kappaB-mediated inhibition of c-Jun N-terminal kinase activation contributes to tumor necrosis factor alpha-induced apoptosis. Mol Cell Biol. 2002;22:8571–8579. doi: 10.1128/MCB.22.24.8571-8579.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karin M, Gallagher E. From JNK to pay dirt: Jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life. 2005;57:283–295. doi: 10.1080/15216540500097111. [DOI] [PubMed] [Google Scholar]
- 19.Greten FR, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Sakurai T, Maeda S, Chang L, Karin M. Loss of hepatic NF-κ B activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc Natl Acad Sci USA. 2006;103:10544–10551. doi: 10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baud V, et al. Signaling by proinflammatory cytokines: Oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habelhah H, et al. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. EMBO J. 2004;23:322–332. doi: 10.1038/sj.emboj.7600044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi CS, Kehrl JH. TNF-induced GCKR and SAPK activation depends upon the E2/E3 complex Ubc13-Uev1A/TRAF2. J Biol Chem. 2003;278:15429–15434. doi: 10.1074/jbc.M211796200. [DOI] [PubMed] [Google Scholar]
- 25.Alvarez SE, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia ZP, Chen ZJ. TRAF2: A double-edged sword? Sci STKE. 2005;2005:pe7. doi: 10.1126/stke.2722005pe7. [DOI] [PubMed] [Google Scholar]
- 27.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 28.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 29.Peukert K, et al. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wanzel M, Herold S, Eilers M. Transcriptional repression by Myc. Trends Cell Biol. 2003;13:146–150. doi: 10.1016/s0962-8924(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Zhao Y, Eilers M, Lin A. Miz1 is a signal- and pathway-specific modulator or regulator (SMOR) that suppresses TNF-α-induced JNK1 activation. Proc Natl Acad Sci USA. 2009;106:18279–18284. doi: 10.1073/pnas.0906328106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison MD, Reiley W, Zhang M, Sun SC. An atypical tumor necrosis factor (TNF) receptor-associated factor-binding motif of B cell-activating factor belonging to the TNF family (BAFF) receptor mediates induction of the noncanonical NF-kappaB signaling pathway. J Biol Chem. 2005;280:10018–10024. doi: 10.1074/jbc.M413634200. [DOI] [PubMed] [Google Scholar]
- 33.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 34.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin A, et al. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 36.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 37.Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: The expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herold S, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 39.Yin Q, Lamothe B, Darnay BG, Wu H. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry. 2009;48:10558–10567. doi: 10.1021/bi901462e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto M, et al. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol. 2006;7:962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- 41.Fukushima T, et al. Ubiquitin-conjugating enzyme Ubc13 is a critical component of TNF receptor-associated factor (TRAF)-mediated inflammatory responses. Proc Natl Acad Sci USA. 2007;104:6371–6376. doi: 10.1073/pnas.0700548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, et al. E3 ubiquitin ligase Mule ubiquitinates Miz1 and is required for TNFalpha-induced JNK activation. Proc Natl Acad Sci USA. 2010;107:13444–13449. doi: 10.1073/pnas.0913690107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.