Abstract
Interactions of cytochrome c (cyt c) with cardiolipin (CL) are important for both electron transfer and apoptotic functions of this protein. A sluggish peroxidase in its native state, when bound to CL, cyt c catalyzes CL peroxidation, which contributes to the protein apoptotic release. The heterogeneous CL-bound cyt c ensemble is difficult to characterize with traditional structural methods and ensemble-averaged probes. We have employed time-resolved FRET measurements to evaluate structural properties of the CL-bound protein in four dansyl (Dns)-labeled variants of horse heart cyt c. The Dns decay curves and extracted Dns-to-heme distance distributions P(r) reveal a conformational diversity of the CL-bound cyt c ensemble with distinct populations of the polypeptide structures that vary in their degree of protein unfolding. A fraction of the ensemble is substantially unfolded, with Dns-to-heme distances resembling those in the guanidine hydrochloride-denatured state. These largely open cyt c structures likely dominate the peroxidase activity of the CL-bound cyt c ensemble. Site variations in P(r) distributions uncover structural features of the CL-bound cyt c, rationalize previous findings, and implicate the prime role of electrostatic interactions, particularly with the protein C terminus, in the CL-induced unfolding.
Keywords: membrane, redox protein, fluorescence
Release of the mitochondrial heme protein cytochrome c (cyt c) into the cytosol is a critical step in activation of caspases in apoptosis (1). Interactions of cyt c with the mitochondrial phospholipid cardiolipin (CL) play an important role in mediating this release (2). Although a sluggish peroxidase in its native state, when bound to CL, cyt c catalyzes CL peroxidation, which in turn contributes to mitochondrial membrane permeabilization (2). Redistribution of CL during apoptosis and increase in the concentration of reactive oxygen species (ROS) are thought to activate the protein peroxidase function (3).
Several studies have examined the interaction of cyt c with CL and phospholipid membrane mimics revealing partial unfolding of the protein and dissociation of the native ligand Met80 (Fig. 1) from the heme (4, 5). Upon lipid binding, the protein retains some of its secondary structure and adopts a loosely packed tertiary structure (6–8). However, the disordered nature of this ensemble has been a challenge to traditional structural methods and details of CL-bound cyt c conformations have remained elusive.
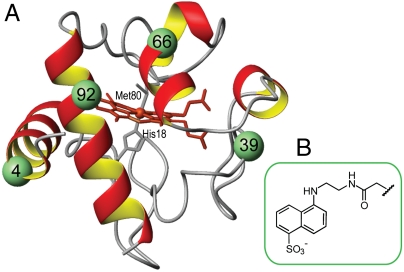
Fig. 1.
(A) Structure of horse heart cyt c showing the labeled positions. (B) Structure of the Dns label.
Different heme ligation states and multiple possible modes of the protein-lipid interactions further complicate structural analyses of this heterogeneous protein ensemble. In addition to the main low-spin bis-His heme species (heme is coordinated by His18 and His26 or His33), resonance Raman studies have found subpopulations of high-spin five- and six-coordinate hemes (9). Several cyt c-CL interaction models have been proposed. Cationic residues Lys72, Lys73, Lys86, and Lys87 are thought to participate in electrostatic binding to anionic phospholipids (10, 11), with hydrogen bonding between Asn52 and protonated acidic phosholipids further stabilizing CL binding at this site (10, 12). Another electrostatic binding site involving residues Lys22, Lys25, His26, Lys27, and His33 has been proposed to play a role at low pH (13). EPR studies have found a partial penetration of cyt c into the lipid bilayer (14), while another study suggested that a hydrophobic acyl chain could also bind into the protein interior (15).
Resolution of conformational heterogeneity is critical for the accurate structural and functional description of the CL-bound cyt c conformations as ensemble-averaging may mask the true features of the species involved. Measurements of time-resolved fluorescence resonance energy transfer (TR-FRET) yield distributions of the distance P(r) between a fluorescent donor (D) and an energy acceptor (A) providing structural information about heterogeneous states (16). The technique has become a valuable tool in characterization of unfolded protein ensembles and folding intermediates, including those in cyt c (17, 18).
Herein we have used four dye-labeled variants of horse heart cyt c to investigate the conformational properties of the protein in the CL-bound state and the forces that govern its interactions with CL. A small dansyl (Dns) dye has been attached to mutant Cys residues at positions 4, 39, 66, and 92 (Fig. 1) that correspond to the protein regions of different stability (19). Dns fluorescence (donor, D) is quenched by FRET to the cyt c covalently bound heme (acceptor, A). Analyses of TR-FRET have revealed multiple types of CL-bound cyt c conformations, with distinct differences in the degree of protein unfolding. Among these conformations, a subpopulation of highly unfolded structures may be responsible for the bulk of the protein peroxidase activity.
Results
Cyt c Fluorescent Variants.
The Dns dye was coupled to a Cys residue in each of the four mutants of horse heart cyt c E4C, K39C, E66C, and E92C. The labeling locations were designed to sample different regions of the cyt c structure and minimize structural perturbations upon mutation and labeling. A Zn analog, without Dns labels but with a fluorescent porphyrin group (ZnP) served as a valuable control in our experiments. Zn substitution is noninvasive and produces a variant with stability similar to that of ferric cyt c (20).
The combined effects of Cys mutation and Dns labeling did not appreciably alter the secondary structure of the protein (Fig. S1). Moreover, stabilities of the variants were minimally affected by these modifications (Table S1). Absorption, CD, and fluorescence spectroscopy measurements delivered identical values, within the error bounds, of the free energy of folding ΔGf. Importantly, results of GuHCl unfolding experiments suggest that Dns modification in each of the four variants have similar effects on the protein global stability.
Cyt c-CL Binding.
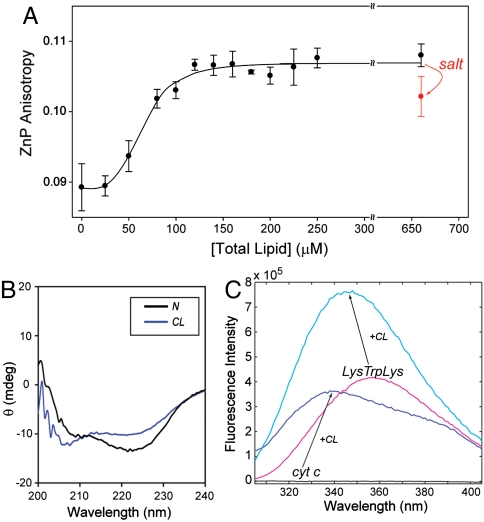
Fluorescence anisotropy of Zncyt increases upon addition of CL liposomes, consistent with the binding of the protein to a larger vesicle (Fig. 2A). The measurements in a 25 mM Hepes at pH 7.4 yielded an apparent binding constant Ka of 1.5 × 104 M-1. In agreement with previous reports (4, 21), binding to CL liposomes also resulted in changes in the cyt c heme absorption spectrum (Fig. S2), and increase in both the Trp59 fluorescence intensity and protein peroxidase activity.
Fig. 2.
(A) Fluorescence anisotropy of ZnP (λex = 423 nm, λem = 588 nm, [Zncyt] = 2 μM, and [cyt c] = 3 μM) with added TOCL/DOPC liposomes as a function of total lipid concentration. Results of multiple independent experiments have indicated that at 660 μM lipid, addition of 150 mM NaCl consistently leads to a decrease in the ZnP anisotropy value. The line is a fit to the Hill binding equation with Ka = 1.5 × 104 M-1. (B) CD spectra of cyt c ([cyt c] = 5 μM, l = 0.2 cm) in a 25 mM TES buffer at pH 7.4 with (CL-bound, blue) and without (native, black) TOCL/DOPC liposomes at 660 μM total lipid. (C) Fluorescence spectra (λex = 295 nm) of cyt c and LysTrpLys in a 25 mM Hepes buffer at pH 7.4 with (cyt c—blue; LysTrpLys—cyan) and without (cyt c—black; LysTrpLys—magenta) TOCL/DOPC liposomes at 660 μM total lipid.
All fluorescence experiments in this study were performed with 5 μM cyt c and liposomes at 660 μM total lipid, conditions at which changes in cyt c spectra and peroxidase activity level off (4, 21). Under these conditions, Met80 is no longer coordinated to the heme (21); the disappearance of the 695 nm absorption band at high lipid-protein ratios confirmed this finding (Fig. S2A). A minor blue shift of the Soret band to 407 nm and an increase in the band intensity support change in the heme ligation (Fig. S2B), likely to a nonnative His26 or 33. Far-UV CD spectra indicate that there are some perturbations in the protein secondary structure (Fig. 2B), which are larger than those previously seen with lower concentrations of CL (5). Increase in the Trp59 fluorescence signal reflects increase in the average Trp59-to-heme distance upon protein unfolding. The λmax of this signal at 337 nm is characteristic of the hydrophobic environment of Trp59. The fact that a model peptide LysTrpLys shows a comparable blue shift (λmax = 343 nm) upon binding to CL (Fig. 2C) suggests that this hydrophobic environment may not necessarily be provided by the protein alone but also by the lipid.
At chosen positions away from the known CL interaction sites (10, 11), a small Dns label is unlikely to interfere with CL binding. Ultracentrifugation pelleting experiments have confirmed that all the labeled cyt c variants behave similar to the wild-type (Fig. S2C).
Site Variations in Cyt c upon CL Binding.
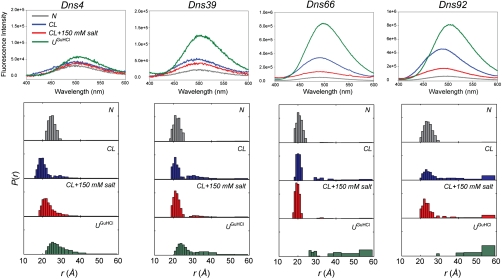
Measurements of integrated fluorescence intensities have revealed changes in protein conformation upon lipid binding. Dns fluorescence is quenched in the native folded protein. Upon addition of CL liposomes, Dns fluorescence intensities increase for all the variants (Fig. 3). The spectra are consistent with the protein state that is on average more loosely packed than the native state but less extensively unfolded than the GuHCl-denatured state. All variants exhibited small blue shifts of λmax for Dns fluorescence compared to λmax in the model N-acetyl Cys-Dns compound. The largest shifts of ∼8 and 14 nm from λmax in the unfolded proteins were observed for Dns66 and Dns92, respectively, suggesting that these sites are situated in a more hydrophobic environment.
Fig. 3.
Fluorescence spectra (λex = 336 nm) and distributions of D-A distances P(r) for four Dns-labeled variants of cyt c in a 25 mM Hepes buffer at pH 7.4 [native (N), gray]; with TOCL/DOPC liposomes at 660 μM total lipid [CL-bound (CL), blue]; with TOCL/DOPC liposomes at 660 μM total lipid and 150 mM NaCl (CL + salt, red); and in 5.8 ± 0.2 M GuHCl solution at pH 7.4 [GuHCl-unfolded (UGuHCl), green]. The corresponding P(k) distributions are shown in Fig. S3. At distances longer than 1.5 × R0 = 59 Å, energy transfer rate constants and D-A distances cannot be determined reliably; the structures with r≥1.5R0 are represented by a single bar.
TR-FRET measurements yielded distinct decays for the folded, GuHCl-unfolded, and CL-bound proteins. In the absence of specific information about the structure and heterogeneity of the CL-bound state, we have chosen model-independent regularization approaches to extract P(k) and P(r) distributions from the observed kinetics (Fig. 3, Fig. S3) (17). Analyses of the TR-FRET data for folded variants yielded D-A distances consistent with existing structural data (Table S2) (22). Distributions of D-A distances P(r) for the GuHCl-denatured variants are in accord with the previous reports (18, 23, 24) and reveal a heterogeneous nature of the unfolded ensemble. Compact structures arising from misligation and other intramolecular interactions are particularly prominent in Dns39, where the labeling site is close to the implicated ligands His26 and His33.
For all the four Dns variants, the P(k) and P(r) distributions for the CL-bound state indicate distinct populations of disparate structures, some resembling the GuHCl-denatured structures and others appearing more compact. With the exception of Dns4, the fast decay rates and corresponding D-A distances in the compact structures are very close to those for the native state. Based on the association constant Ka, the heterogeneous protein population under our experimental conditions is not due to the bound and unbound cyt c, but rather to different CL-bound cyt c conformers. Furthermore, the lack of Met80-heme ligation, a hallmark of the cyt c native state, argues against a population of unbound cyt c under these conditions.
For Dns4, increased quenching rates suggest more efficient FRET between the Dns label and the heme, owing to a change in D-A distances or an altered orientation of the Dns dipole. Both these scenarios point to the involvement of the N-terminal helix in binding to the lipid. Residue 4 is separated by only nine residues from the heme, and upon loss of contacts between N- and C-terminal helices it may come close to the heme. This new positioning could possibly result from a twisting of the N-terminal helix with respect to the heme as this region interacts with the lipid. The lack of a prominent blue shift for Dns4 argues against deep insertion of this label into the lipid bilayer. Furthermore, the anisotropy data suggest efficient Dns fluorescent depolarization for the majority of the Dns4 ensemble. However, if the probe is immobilized even for the subset of lipid-bound conformations, the orientational freedom of the Dns dipole with respect to the heme would still be restricted, affecting the R0 value. Therefore, we have considered the upper R0 limit (κ2 = 4, R0 = 53 Å); this analysis has yielded D-A distances in compact Dns4 structures similar to those in the native protein (Fig. S3).
Highly extended structures with Dns-heme distances similar to those in the GuHCl-denatured protein are prominent members of the polypeptide ensemble (Fig. 3). Higher populations of extended structures for the Dns92 variant suggest a greater degree of polypeptide unfolding in the vicinity of site 92.
Effects of Ionic Strength on Cyt c Conformational Ensemble.
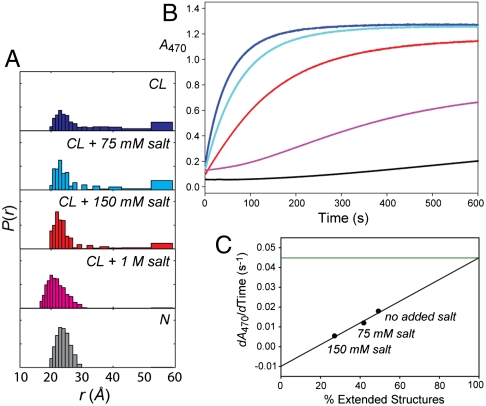
In order to elucidate the nature of CL-cyt c interactions, ionic strength effects were probed. Increase in the concentration of salt diminishes binding between the negatively-charged CL and the positively-charged cyt c (4, 5). As revealed by Zncyt anisotropy measurements (Fig. 2A), at 660 μM total lipid and salt concentration of 150 mM, about 20% of the cyt c molecules are no longer bound to the liposomes. When NaCl is added, the Dns fluorescence shifts back to the red (Fig. 3), indicating more hydrophilic environment of the probe. Salt influences P(r) distributions, decreasing the population of highly unfolded structures (Fig. 4, Fig. S4). The population of compact structures instead grows, and these conformations become more native-like. These changes are consistent with salt-induced dissociation of cyt c from liposomes.
Fig. 4.
(A) Distributions of D-A distances P(r) from TR-FRET for Dns92 in the native state and with TOCL/DOPC liposomes at 660 μM total lipid at indicated concentrations of NaCl. (B) Peroxidase activity of cyt c in the native state and with TOCL/DOPC liposomes at 660 μM total lipid at indicated concentrations of NaCl, monitored with a guaiacol oxidation assay. (C) Initial rates versus population of extended cyt c structures (from Dns92 TR-FRET). A linear fit predicts an initial rate dA470/dTime of 0.045 s-1 at 100% of extended strucures, almost 100-fold slower than the value of 3.9 s-1 determined experimentally for the protein in 6 M GuHCl at pH 7.4.
Addition of salt to the CL-bound cyt c yields identical TR-FRET curves to those obtained with the protein incubated in the salt-containing liposome solutions from the start (Fig. S5). This finding indicates that the CL-induced unfolding is reversible, consistent with previous reports (25).
The cyt c peroxidase activity increases with increase in the relative populations of extended structures at different concentrations of salt (Fig. 4). However, the projected rate of guaiacol oxidation at a 100% population of extended structures is still about 100-fold slower than that for the the GuHCl-unfolded protein. Residual protein structure and the protein-lipid interface likely limit access of H2O2 and the substrate guaiacol to the heme.
Although the cyt c ensemble is comprised of only compact polypeptide structures at very high salt concentrations of 1 M, they are not identical to those of the native protein. The differences in the visible CD spectra (Fig. S5) and residual peroxidase activity (Fig. 4B) further demonstrate that this ensemble is not fully native.
Discussion
Cyt c-membrane interactions are important for both electron transfer and apoptotic functions of this protein. Association of cyt c with mitochondrial membranes affects the protein reactivity with physiological redox partners (26) and its ability to relocate into the cytosol during apoptosis (3). The mitochondrial phospholipid CL binds tightly to cyt c and is particularly effective in unfolding the protein (4). CL-induced conformational changes in cyt c confer peroxidase activity on the protein. The unleashed peroxidase selectively catalyzes CL peroxidation, which is an important transformation in the initial stages of apoptosis (2). Interestingly, CL is also required for full activity of cyt c oxidase, and it has been suggested that cyt c-CL interactions may potentially recruit cyt c to this redox enzyme (27).
The unique structural features of CL, its dianionic charge and the accessibility of phosphate groups at the surface of the lipid membranes (28), promote strong interactions of this lipid with the positively-charged cyt c. The binding, however, is complex. While there are changes in the cyt c heme environment detectable even at very low CL/cyt c ratios (5), higher lipid concentrations are needed to induce changes in intramolecular distances. Increase in the available lipid surface area of the model liposomes at higher CL/cyt c ratios is likely an important factor for protein unfolding.
Our results clearly show that CL liposomes cause a substantial unfolding of cyt c, with a population of the ensemble with Dns-to-heme distances similar to those in the denatured protein. High mobility of the displaced ligand Met80 (29), facile binding and efficient escape of carbon monoxide CO in the Fe(II)-CO photolysis experiments (30), as well as a negative heme redox potential (31), corroborate a wide opening of the heme pocket in the extended structures. The dynamic behavior of lipid- and surfactant-bound cyt c structures previously seen in the NMR exchange experiments (25, 32, 33) also agrees with the presence of largely unfolded polypeptides.
However, such unfolding is not uniform across the species in the ensemble. At low ionic strength, when the entire protein population is presumably bound to CL liposomes, the TR-FRET data has revealed the existence of different cyt c conformations that vary in the degree of the polypeptide compaction. A number of previous observations, including distinct environments of Met65 revealed by NMR (29) and multiphasic kinetics of CO rebinding (30), are in accord with the heterogeneous nature of this ensemble. The revealed Dns-to-heme P(r) distributions argue against the popular model of a uniformly compact molten globular state for the CL-bound cyt c and instead suggest the prime role of highly unfolded structures in the observed peroxidase activity of this ensemble. Importantly, such extended structures persist even at ionic strength of 150 mM (Fig. 3) associated with the intermembrane space (34).
Site variations in population of extended structures, as well as the lower peroxidase activity of CL-bound cyt c compared to that of the GuHCl-denatured protein, illustrate that some structural preferences exist even for highly unfolded conformers. Only a partial decrease of ∼25% in the CD signal at 222 nm (Fig. 2B) suggests presence of the α-helical structure in these species. The greater population of extended structures in Dns92 corraborates the strong affinity for anionic phospholipids of the previously implicated electrostatic binding site A (10, 12) to which the residue 92 is close. However, our data suggests that CL-induced conformational changes involve other regions of cyt c as well. Observed populations of extended structures in Dns39 are consistent with polypeptide unfolding next to Pro44 (35) and supports the recent model of structural changes in the protein 37–61 loop (36). Changes in the N-terminal helix have not been noted prior to this work. The presence of extended structures in both Dns4 and Dns92 variants suggests that critical stabilizing interactions between the N- and C-termini have been broken for some of the protein conformers, and supports the existence of extended “open” structures in the ensemble. The finding further highlights that the CL-bound cyt c is not a molten globular state as the N- and C- terminal contacts are known to be maintained in this cyt c state under classical conditions of low pH and high concentration of salt (37).
Regions of different folding stability, foldons, have been identified in native cyt c (19). With the exception of site 92, the populations of extended structures (Fig. 3) do correlate with stability of the foldons (Table S3). The N- and C-helices belong to the same foldon in the native protein, yet the populations of extended structures differ for Dns4 and Dns92 in the CL-bound state. Comparison of Dns66 and Dns92 results suggests additional unfolding of the polypeptide fragment between residues 66 and 92. This region corresponds to a lower stability Met80-containing loop, whose unfolding provides an explanation for the increased protein peroxidase activity. Unfolding of this region could account for long Dns92-heme distances even in structures where the N- and C-interhelical contacts are preserved. However, the higher population of extended structures in Dns92 may also suggest the importance of new stabilizing interactions with the lipid in defining the conformational properties of CL-bound cyt c c rather than stability of the native fold alone.
As the salt concentration increases, the populations of extended structures and the variances of the P(r) distributions decrease, indicating the importance of electrostatic interactions in mediating strong binding and protein unfolding (Fig. 4). Electrostatic interactions likely guide the formation of hydrophobic contacts with the lipid. With a disrupted cyt c native structure, particularly in the vicinity of sites 66 and 92, additional hydrophobic interactions with CL acyl chains are possible (Fig. S6). Observed unfolding of these polypeptide segments and larger blue shifts of Dns fluorescence in Dns66 and Dns92 compared to other variants are consistent with such an interpretation. The C-terminal sequence of cyt c has a high density of polar and charged residues, and theoretical studies have shown that this region has a tendency to fray when new electrostatic interactions are introduced (38). It is conceivable that electrostatic interactions with the negatively-charged CL membranes unfold this region of the protein, assisting in forming further hydrophobic contacts with the lipid. Interestingly, a recent study of cyt c oligomerization suggests an ease of displacement of the C-terminal helix and associated perturbation of Met80-heme interactions (39). With the CL-induced break-up of the critical interfacial contacts between N- and C-helices, additional unfolding of the protein is possible (Fig. 5). Future work with additional site-specific probes will examine this possibility and uncover the overall sequence of cyt c unfolding steps.
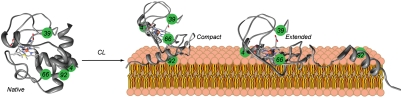
Fig. 5.
A cartoon illustrating populations of different cyt c conformations in the CL-bound ensemble.
Previous studies have revealed some penetration of cyt c into CL and other anionic phospholipid bilayers (14, 40), however, this topic is controversial (11, 29). It appears that the mode of cyt c-lipid binding and the possibility of insertion depend on the lipid composition and surface coverage (9, 41). Estimates of the heme location with dye-labeled CL in the similar TOCL/DOPC lipid vesicles have placed the heme within 9–17 Å from the center of the bilayer (42). It is unlikely, however, that any of the four Dns labels studied here insert deeply into the bilayer. Binding to CL introduced minimal λmax shifts in Dns fluorescence, while fluorescence of a model Cys-Dns compound undergoes a dramatic shift to 460 nm in hydrophobic solvents and upon insertion into liposomes (42). Furthermore, reversibility of the CL-cyt c binding and high mobility of the Dns probes from fluorescence anisotropy experiments support the positioning of the protein backbone predominantly on the membrane surface. A unique structure of CL with four bulky acyl chains provides opportunities for hydrophobic interactions already on the surface of CL liposomes (43).
The nonnative character of the cyt c ensemble in the presence of CL and 1 M salt is intriguing. The protein conformations are compact yet the P(r) distributions are broader than those for the native protein (both at high and low salt concentrations, Fig. S4) and resemble those seen for the cyt c molten globule state (16). Increased light scattering by the larger lipid vesicles at high salt concentrations may partially contribute to the observed changed in the decay curves. However, a residual peroxidase activity for this ensemble suggests also increased accessibility of the heme group within these compact structures compared to the native state. Evidently, salt does not fully eliminate cyt c interactions with CL. High ionic strength favors hydrophobic interactions but in the absence of cyt c unfolding these interactions may be different from those at low ionic strength. One possibility is the insertion of a CL acyl chain, lipid anchorage (15), in the protein interior of these compact structures. Intrinsic conformational strain of CL side chains (28) may be further enhanced in small, curved small unilamellar vesicles membranes, reducing the barrier for their transfer from the liposome into the protein interior. The lower intensity of the negative CD band at 416 nm (Fig. S5) is consistent with this model, although its presence also implies that only a very small fraction of the protein ensemble is perturbed by such interactions with CL. High concentrations of salt, typically above 1.5 M for the TOCL mixtures, could also induce a nonlamellar phase formation in CL membranes (28), exposing acyl groups on the membrane surface and affecting their interactions with cyt c. However, it is not clear if these changes in CL membrane morphology play a role under our experimental conditions.
Conclusions
Abundant literature on cyt c-CL interactions with different views on the mode of protein-lipid binding and degrees of cyt c structural perturbations so far has not been rationalized in terms of one consistent model. Distributions of structures from TR-FRET measurements offer an explanation of how different models can coexist. Observations of the heterogeneous CL-bound cyt c ensemble and its transformation with experimental conditions highlight the multitude of different structures that populate this ensemble and illustrate the role of conformational dynamics for the protein peroxidase function.
Dye-to-heme P(r) distributions and emission energies in four site-specifically labeled variants shed light on the structural features and diversity of cyt c polypeptides. Driven by electrostatic interactions, CL promotes dramatic unfolding of cyt c. Although very open, the formed cyt c conformations likely preserve some intramolecular interactions such as residual α-helicity. The loss of the tertiary contacts, particularly involving the protein C terminus, explains the role of the previously implicated CL binding site and the associated increase in the cyt c peroxidase activity. The vast differences in the polypeptide compactness, and thus the heme accessibility of cyt c species, suggest that peroxidase activity of the CL-bound cyt c ensemble may be dominated by largely open structures. The structural and mechanistic insights revealed in this study will be instrumental for a better understanding of cyt c-CL interactions and the design of unique ways to manipulate the apoptotic peroxidase function of cyt c.
Materials and Methods
Preparation of Dns Derivatives.
Details of protein expression and purification are provided in SI Materials and Methods. Protein labeling with the thiol-reactive reagent 1,5-IAEDANS (Invitrogen) and purification of dye-labeled products was done as described (44). MALDI-TOF mass spectrometry confirmed the labeling.
Zinc-Substituted Cyt c.
Iron-free cyt c was prepared from commercial cyt c (Sigma, C2506) according to Ensign, et al. (24). Zinc reconstitution was done in the presence of 3 M GuHCl at pH 7.5 and 50 °C, and zinc-substituted cyt c (Zncyt) was refolded by buffer exchange to 10 mM Tris HCl pH 7.5 (buffer A). Purification was done on a HiTrap SP HP column (GE Healthcare) equilibrated with buffer A. The protein was eluted with a shallow gradient from 0 to 50% buffer B containing 10 mM Tris-HCl pH 7.5 with 0.75 M NaCl.
Preparation of Lipid Vesicles and Protein-Liposome Binding Experiments.
Details of preparation and characterization of vesicles are provided in SI Materials and Methods. The vesicle stock solution was diluted with a 25 mM Hepes buffer pH 7.4 with or without NaCl and the protein was added last. The samples were equilibrated at room temperature for at least 30 min before data collection. Concentration of cyt c in all experiments was 5 μM. All Dns-labeled and Zn-substituted cyt c samples were handled in the dark.
Assuming a spherical vesicle shape and the cross-sectional areas of TOCL and DOPC to be 120 and 70 Å2, respectively (45, 46), an average vesicle in our experiments is comprised of 16,000–24,000 lipid molecules and thus at 660 μM total lipid and 5 μM protein 120–185 cyt c molecules bind to the same vesicle. Intermolecular fluorescence quenching can be excluded because of the absence of FRET from the Dns to the ZnP for a mixture of Dns-labeled cyt c and Zncyt in the presence of our TOCL/DOPC vesicles. Furthermore, distances longer than ones in the GuHCl-unfolded protein (in particular with variants Dns4 and Dns39) have not been observed, and identical results were obtained with Dns-labeled and a mixture of labeled and unlabeled proteins.
Spectroscopic Measurements.
All experiments were done at 21 ± 2 °C. Absorption and circular dichroism (CD) spectra were recorded with an Agilent 8453 diode-array spectrophotometer and a Jasco J-715 spectropolarimeter, respectively. Steady-state fluorescence spectra were measured with a Horiba Jobin Yvon Fluorolog-3 spectrofluorimeter equipped with automatic polarizers. Absorption spectra were referenced against a corresponding blank and the scatter in the fluorescence spectra was removed by subtracting a blank spectrum.
Fluorescence lifetimes were measured by time-correlated single photon counting at 10,000 counts using a NanoLED-375L diode laser (λex = 375 nm, < 70 ps pulsewidth) as the excitation source and a fast TBX-04 detector. Measurements were done under magic angle conditions and Dns emission was observed at 500 nm. For measurements of time-resolved fluorescence anisotropy r(t), a manual polarizer was placed before the excitation beam and automatic polarizers were employed for emission detection. The vertically (V) and horizontally (H) polarized emission decays were collected in 60 s intervals until the difference in counts IVV-IVH reached 40,000. A dilute solution of LUDOX AS-40 (Aldrich) was used to collect an instrument response function (IRF).
Data analysis of the TR-FRET was done in MATLAB (MathWorks) as previously described (16, 47); for specific details see SI Materials and Methods. Critical distance R0 calculations and analyses of the orientation factor κ2 effects on the D-A distances are provided in SI Materials and Methods.
Equilibrium Unfolding.
The unfolding curves were obtained from absorption, CD, and fluorescence measurements (Dns for the labeled variants or Trp59 for the wild-type protein) as previously described (18). GuHCl concentrations were checked for accuracy with refractive index measurements.
Peroxidase Assays.
Peroxidase activity was assayed with guaiacol (Sigma) by measuring absorption at 470 nm of the formed tetraguaiacol product (48). Cyt c preincubated in a liposome solution for at least 2 h was added to a cuvette containing a freshly prepared mixture of guaiacol and hydrogen peroxide, H2O2. The sample was mixed by pipetting and measurements with an Agilent 8453 spectrophotometer were immediately started. Peroxidase activity in GuHCl solution was measured with a Bio-Logic MOS-200 stopped-flow instrument (44). Final concentrations were 5 μM cyt c, 10 mM guaiacol, and 5 mM H2O2.
Supplementary Material
Acknowledgments.
We thank Jay R. Winkler for sharing his MATLAB codes, Kenneth Tichauer and Frederic Leblond for their help with IRF deconvolution, Kara L. Bren for the horse heart cyt c plasmid, William Wickner for his advice about liposome binding assays, and P. Jack Hoopes for access to his dynamic light scattering instrument. This work was supported by Dartmouth College startup funds, a Burke Research Initiation Award (E.V.P.), the Dartmouth Class of 1962 Junior Faculty Fellowship (E.V.P.), and National Institutes of Health (NIH) (GM098502). We are grateful to the following sponsors for providing undergraduate fellowships: the German Academic Exchange Service Research Internships in Science and Engineering (DAAD RISE) program (J.H.), Dr. John L. Zabriskie Jr. (A.M.M.), and the Dartmouth Dean of the Faculty for an Anna Thompson Burnap Student Research Award (J.R.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112312108/-/DCSupplemental.
References
- 1.Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 2.Kagan VE, et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 3.Iverson SL, Orrenius S. The cardiolipin-cytochrome c interaction and the mitochondrial regulation of apoptosis. Arch Biochem Biophys. 2004;423:37–46. doi: 10.1016/j.abb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Belikova NA, et al. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry. 2006;45:4998–5009. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinibaldi F, et al. Insights into cytochrome c-cardiolipin interaction. Role played by ionic strength. Biochemistry. 2008;47:6928–6935. doi: 10.1021/bi800048v. [DOI] [PubMed] [Google Scholar]
- 6.Patriarca A, et al. ATP acts as a regulatory effector in modulating structural transitions of cytochrome c: implications for apoptotic activity. Biochemistry. 2009;48:3279–3287. doi: 10.1021/bi801837e. [DOI] [PubMed] [Google Scholar]
- 7.Pinheiro TJ, Elöve GA, Watts A, Roder H. Structural and kinetic description of cytochrome c unfolding induced by the interaction with lipid vesicles. Biochemistry. 1997;36:13122–13132. doi: 10.1021/bi971235z. [DOI] [PubMed] [Google Scholar]
- 8.Heimburg T, Marsh D. Investigation of secondary and tertiary structural changes of cytochrome c in complexes with anionic lipids using amide hydrogen exchange measurements: an FTIR study. Biophys J. 1993;65:2408–2417. doi: 10.1016/S0006-3495(93)81299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oellerich S, Lecomte S, Paternostre M, Heimburg T, Hildebrandt P. Peripheral and integral binding of cytochrome c to phospholipids vesicles. J Phys Chem B. 2004;108:3871–3878. [Google Scholar]
- 10.Rytömaa M, Kinnunen PK. Evidence for two distinct acidic phospholipid-binding sites in cytochrome c. J Biol Chem. 1994;269:1770–1774. [PubMed] [Google Scholar]
- 11.Kostrzewa A, Páli T, Froncisz W, Marsh D. Membrane location of spin-labeled cytochrome c determined by paramagnetic relaxation agents. Biochemistry. 2000;39:6066–6074. doi: 10.1021/bi992559l. [DOI] [PubMed] [Google Scholar]
- 12.Rytömaa M, Kinnunen PKJ. Reversibility of the binding of cytochrome c to liposomes: implications for lipid-protein interactions. J Biol Chem. 1995;270:3197–3202. doi: 10.1074/jbc.270.7.3197. [DOI] [PubMed] [Google Scholar]
- 13.Kawai C, et al. pH-dependent interaction of cytochrome c with mitochondrial mimetic membranes: the role of an array of positively charged amino acids. J Biol Chem. 2005;280:34709–34717. doi: 10.1074/jbc.M412532200. [DOI] [PubMed] [Google Scholar]
- 14.Heimburg T, Marsh D. Protein surface-distribution and protein-protein interactions in the binding of peripheral proteins to charged lipid membranes. Biophys J. 1995;68:536–546. doi: 10.1016/S0006-3495(95)80215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalanxhi E, Wallace CJ. Cytochrome c impaled: investigation of the extended lipid anchorage of a soluble protein to mitochondrial membrane models. Biochem J. 2007;407:179–187. doi: 10.1042/BJ20070459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pletneva EV, Gray HB, Winkler JR. Nature of the cytochrome c molten globule. J Am Chem Soc. 2005;127:15370–15371. doi: 10.1021/ja0555318. [DOI] [PubMed] [Google Scholar]
- 17.Pletneva EV, Gray HB, Winkler JR. Snapshots of cytochrome c folding. Proc Natl Acad Sci USA. 2005;102:18397–18402. doi: 10.1073/pnas.0509076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pletneva EV, Gray HB, Winkler JR. Many faces of the unfolded state: conformational heterogeneity in denatured yeast cytochrome c. J Mol Biol. 2005;345:855–867. doi: 10.1016/j.jmb.2004.10.085. [DOI] [PubMed] [Google Scholar]
- 19.Maity H, Maity M, Englander SW. How cytochrome c folds, and why: submolecular foldon units and their stepwise sequential stabilization. J Mol Biol. 2004;343:223–233. doi: 10.1016/j.jmb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Tremain SM, Kostic NM. Molten globule and other conformational forms of zinc cytochrome c. Effect of partial and complete unfolding of the protein on its electron-transfer reactivity. Inorg Chem. 2002;41:3291–3301. doi: 10.1021/ic010893b. [DOI] [PubMed] [Google Scholar]
- 21.Kapralov AA, et al. The hierarchy of structural transitions induced in cytochrome c by anionic phospholipids determines its peroxidase activation and selective peroxidation during apoptosis in cells. Biochemistry. 2007;46:14232–14244. doi: 10.1021/bi701237b. [DOI] [PubMed] [Google Scholar]
- 22.Bushnell GW, Louie GV, Brayer GD. High-resolution 3-dimensional structure of horse heart cytochrome c. J Mol Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 23.Lee AJ, Ensign AA, Krauss TD, Bren KL. Zinc porphyrin as a donor for FRET in zinc cytochrome c. J Am Chem Soc. 2010;132:1752–1753. doi: 10.1021/ja909106p. [DOI] [PubMed] [Google Scholar]
- 24.Ensign AA, Jo I, Yildirim I, Krauss TD, Bren KL. Zinc porphyrin: a fluorescent acceptor in studies of Zn-cytochrome c unfolding by fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2008;105:10779–10784. doi: 10.1073/pnas.0802737105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spooner PJR, Watts A. Reversible unfolding of cytochrome c upon interaction with cardiolipin bilayers. 1. Evidence from deuterium NMR measurements. Biochemistry. 1991;30:3871–3879. doi: 10.1021/bi00230a010. [DOI] [PubMed] [Google Scholar]
- 26.Craig DB, Wallace CJ. Studies of 8-azido-ATP adducts reveal two mechanisms by which ATP binding to cytochrome c could inhibit respiration. Biochemistry. 1995;34:2686–2693. doi: 10.1021/bi00008a036. [DOI] [PubMed] [Google Scholar]
- 27.Vik SB, Georgevich G, Capaldi RA. Diphosphatidylglycerol is required for optimal activity of beef heart cytochrome c oxidase. Proc Natl Acad of Sci USA. 1981;78:1456–1460. doi: 10.1073/pnas.78.3.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis RN, McElhaney RN. The physicochemical properties of cardiolipin bilayers and cardiolipin-containing lipid membranes. Biochim Biophys Acta. 2009;1788:2069–2079. doi: 10.1016/j.bbamem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Spooner PJ, Watts A. Cytochrome c interactions with cardiolipin in bilayers: a multinuclear magic-angle spinning NMR study. Biochemistry. 1992;31:10129–10138. doi: 10.1021/bi00156a037. [DOI] [PubMed] [Google Scholar]
- 30.Kapetanaki SM, et al. Interaction of carbon monoxide with the apoptosis-inducing cytochrome c-cardiolipin complex. Biochemistry. 2009;48:1613–1619. doi: 10.1021/bi801817v. [DOI] [PubMed] [Google Scholar]
- 31.Basova LV, et al. Cardiolipin switch in mitochondria: shutting off the reduction of cytochrome c and turning on the peroxidase activity. Biochemistry. 2007;46:3423–3434. doi: 10.1021/bi061854k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertini I, et al. Cytochrome c and SDS: a molten globule protein with altered axial ligation. J Mol Biol. 2004;336:489–496. doi: 10.1016/j.jmb.2003.12.045. [DOI] [PubMed] [Google Scholar]
- 33.Pinheiro TJ, Cheng H, Seeholzer SH, Roder H. Direct evidence for the cooperative unfolding of cytochrome c in lipid membranes from H-(2)H exchange kinetics. J Mol Biol. 2000;303:617–626. doi: 10.1006/jmbi.2000.4159. [DOI] [PubMed] [Google Scholar]
- 34.Cortese JD, Voglino AL, Hackenbrock CR. Ionic strength of the intermembrane space of intact mitochondria as estimated with fluorescein-BSA delivered by low pH fusion. J Cell Biol. 1991;113:1331–1340. doi: 10.1083/jcb.113.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jemmerson R, et al. A conformational change in cytochrome c of apoptotic and necrotic cells is detected by monoclonal antibody binding and mimicked by association of the native antigen with synthetic phospholipid vesicles. Biochemistry. 1999;38:3599–3609. doi: 10.1021/bi9809268. [DOI] [PubMed] [Google Scholar]
- 36.Balakrishnan G, et al. A conformational switch to b-sheet structure in cytochrome c leads to heme exposure. Implications for cardiolipin peroxidation and apoptosis. J Am Chem Soc. 2007;129:504–505. doi: 10.1021/ja0678727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marmorino JL, Lehti M, Pielak GJ. Native tertiary structure in an A-state. J Mol Biol. 1998;275:379–388. doi: 10.1006/jmbi.1997.1450. [DOI] [PubMed] [Google Scholar]
- 38.Weinkam P, Pletneva EV, Gray HB, Winkler JR, Wolynes PG. Electrostatic effects on funneled landscapes and structural diversity in denatured protein ensembles. Proc Natl Acad of Sci USA. 2009;106:1796–1801. doi: 10.1073/pnas.0813120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirota S, et al. Cytochrome c polymerization by successive domain swapping at the C-terminal helix. Proc Natl Acad of Sci USA. 2010;107:12854–12859. doi: 10.1073/pnas.1001839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi S, Swanson JM. Interaction of cytochrome c with cardiolipin: an infrared spectroscopic study. Biophys Chem. 1995;54:271–278. doi: 10.1016/0301-4622(94)00151-9. [DOI] [PubMed] [Google Scholar]
- 41.Domanov YA, Molotkovsky JG, Gorbenko GP. Coverage-dependent changes of cytochrome c transverse location in phospholipid membranes revealed by FRET. Biochim Biophys Acta. 2005;1716:49–58. doi: 10.1016/j.bbamem.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Merrill AR, Cohen FS, Cramer WA. On the nature of the structural change of the colicin E1 channel peptide necessary for its translocation-competent state. Biochemistry. 1990;29:5829–5836. doi: 10.1021/bi00476a026. [DOI] [PubMed] [Google Scholar]
- 43.Mileykovskaya E, et al. Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett. 2001;507:187–190. doi: 10.1016/s0014-5793(01)02948-9. [DOI] [PubMed] [Google Scholar]
- 44.Pound GJ, Pletnev AA, Fang X, Pletneva EV. A small fluorophore reporter of protein conformation and redox state. Chem Commun. 2011;47:5714–5716. doi: 10.1039/c1cc10896d. [DOI] [PubMed] [Google Scholar]
- 45.Goormaghtigh E, Chatelain P, Caspers J, Ruysschaert J-M. Evidence of a specific complex between adriamycin and negatively-charged phospholipids. Biochim Biophys Acta. 1980;597:1–14. doi: 10.1016/0005-2736(80)90145-5. [DOI] [PubMed] [Google Scholar]
- 46.Pinheiro TJT, Duralski AA, Watts A. Phospholipid headgroup-headgroup electrostatic interactions in mixed bilayers of cardiolipin with phosphatidylcholines studied by 2H NMR. Biochemistry. 1994;33:4896–4902. doi: 10.1021/bi00182a018. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, et al. Direct visualization reveals dynamics of a transient intermediate during protein assembly. Proc Natl Acad of Sci USA. 2011;108:6450–6455. doi: 10.1073/pnas.1019051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diederix RE, Ubbink M, Canters GW. Peroxidase activity as a tool for studying the folding of c-type cytochromes. Biochemistry. 2002;41:13067–13077. doi: 10.1021/bi0260841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.