Abstract
Plants and animals use innate immunity as a first defense against pathogens, a costly yet necessary tradeoff between growth and immunity. In Arabidopsis, the regulatory leucine-rich repeat receptor-like kinase (LRR-RLK) BAK1 combines with the LRR-RLKs FLS2 and EFR in pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and the LRR-RLK BRI1 in brassinosteroid (BR)-mediated growth. Therefore, a potential tradeoff between these pathways mediated by BAK1 is often postulated. Here, we show a unidirectional inhibition of FLS2-mediated immune signaling by BR perception. Unexpectedly, this effect occurred downstream or independently of complex formation with BAK1 and associated downstream phosphorylation. Thus, BAK1 is not rate-limiting in these pathways. BRs also inhibited signaling triggered by the BAK1-independent recognition of the fungal PAMP chitin. Our results suggest a general mechanism operative in plants in which BR-mediated growth directly antagonizes innate immune signaling.
Keywords: flagellin sensing 2, brassinosteroid insensitive 1, BRI1-associated kinase 1, cross-talk
Plants continuously adapt to changing environments using surface-localized transmembrane receptor-like kinases (RLKs), of which different members control aspects of growth, development, and innate immunity (1–3). Intriguingly, RLKs involved in different pathways share common regulators, suggesting a potential cross-talk mechanism. The Arabidopsis regulatory leucine-rich repeat RLK (LRR-RLK) BAK1/SERK3 is a prime candidate for a tradeoff mediator. BAK1 interacts with and is a positive regulator of the growth hormone brassinosteroid (BR) receptor, the LRR-RLK BRI1 (4, 5). BRI1 can also complex with SERK1 and BKK1/SERK4 that play partially redundant roles with BAK1 in BR responses (6–8). BRI1 interacts with the inhibitory protein BKI1 that is displaced following BRI1 activation, followed by recruitment of BAK1 into the BRI1 complex (9). This leads to further BRI1 activation and phosphorylation of cytoplasmic BSKs ultimately culminating at the transcription factors BZR1 and BES1/BZR2 (10).
In innate immunity, BAK1 is a positive regulator forming a rapid ligand-induced complex with the LRR-RLKs FLS2 (11, 12) and EFR (13), the pattern-recognition receptors (PRRs) perceiving the bacterial pathogen-associated molecular patterns (PAMPs) flagellin (flg22) and EF-Tu (elf18), respectively. Additional SERKs can be recruited by FLS2 with BKK1 as major regulator besides BAK1 (13). BAK1 also positively regulates other PRR-dependent pathways (12, 14–16). However, innate immune responses triggered by PAMPs such as fungal chitin do not depend on BAK1 (14, 17). Together with BKK1, BAK1 also controls cell death (7, 18).
Signaling downstream of BAK1 differs between BRI1 and FLS2 pathways. BIK1 is bound to FLS2 and dissociates in a BAK1-dependent manner upon flg22 binding. BIK1 and paralogues positively regulate most PAMP-triggered immunity (PTI) responses downstream of FLS2 (19, 20). FLS2 is ubiquitinated by the BAK1-associated ubiquitin ligases PUB12 and PUB13 and degraded (21). FLS2 activation leads to rapid bursts of calcium and reactive oxygen species (ROS), activation of MAP kinases and calcium-dependent protein kinases (CDPKs), ultimately leading to PTI (22).
Upon BR binding, BRI1 auto- and transphosphorylates BAK1, leading to increased BAK1 autophosphorylation, which in turn transphosphorylates BRI1, resulting in optimal BRI1 activation (23). Activation of FLS2 or EFR by their corresponding ligand also leads to phosphorylation of the ligand-binding RLKs and BAK1. BAK1 can provide signaling specificity in a phosphorylation-dependent manner (24).
Thus, BAK1 may be a rate-limiting positive regulator, acting as a decision node between different pathways. BRI1 signaling output can be enhanced by over-expression or hyperactive alleles of BRI1 or positive regulators (8, 25–28), genetic or chemical inactivation of negative regulators (9, 29), or exogenous application of BR (30). This study addresses the hypotheses that BAK1 may cross-regulate or is rate-limiting in the BRI1 and FLS2/EFR pathways. We used primarily WT Arabidopsis plants to reflect as faithfully as possible the natural situation under which tradeoff between development and immunity may occur.
Results and Discussion
Activation of BAK1 by BRs Does Not Lead to Immune Responses.
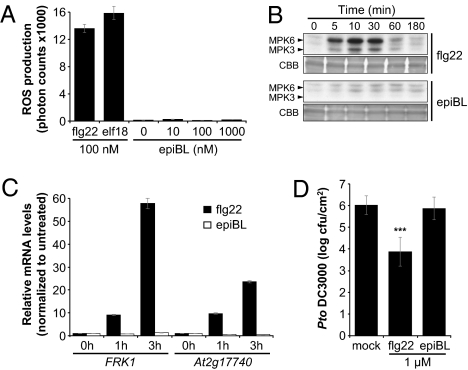
BRs have been implicated in tolerance to pathogens (31–33). Therefore, we tested whether BRs induce responses associated with PTI. Based on the sequential phosphorylation model between BRI1 and BAK1 (23), activation of BAK1 by BRI1 could render the other receptor (i.e., FLS2) more active. An early PAMP response is the rapid and transient production of ROS. To enable comparison between treatments and/or genotypes, the amount of ROS produced is plotted as the total amount of photons detected in the luminol-based assay during 40 min. Whereas treatment with the PAMPs flg22 and elf18 induced a clear ROS burst in WT (Columbia; Col-0) Arabidopsis leaf discs, no ROS was detected after treatment with the biologically active 24-epibrassinolide (epiBL), even at high concentration (Fig. 1A and Fig. S1A). This was not caused by a previous response to endogenous BRs, as treatment with the BR biosynthetic inhibitor brassinazole (34) before epiBL treatment also did not give ROS (Fig. S1B). The leaves used in these experiments were responsive to epiBL as measured by repression of the BR biosynthetic gene CPD (Fig. S2). Flg22 treatment of Arabidopsis seedlings activates MAPKs, which are immunologically detectable within minutes (Fig. 1B). No significant MAPK activation could be observed after treatment with epiBL (Fig. 1B). PAMP perception is associated with rapid transcriptional reprogramming (35), and FRK1 and At2g17740 are commonly used PTI marker genes (14). In contrast to flg22, no changes in FRK1 and At2g17740 transcript levels were observed after epiBL treatment (Fig. 1C). In rice and tobacco, pretreatment with BL induces resistance to several pathogens (31). We therefore tested if pretreatment of Arabidopsis leaves with epiBL could induce resistance to Pseudomonas syringae pv. tomato (Pto) DC3000. Whereas pretreatment for 24 h with 1 μM flg22 reduced the replication of Pto DC3000 by approximately two log units (Fig. 1D) (35), no significant difference in Pto DC3000 numbers recovered from leaves pretreated with 1 μM epiBL was observed (Fig. 1D). Similarly, treatment with BL did not increase resistance to the fungus Alternaria brassicicola (18). Clearly, active BRI1-mediated BR signaling does not induce PTI responses in WT Arabidopsis, despite the participation of BAK1 in both pathways.
Fig. 1.
EpiBL perception does not induce PTI responses. (A) Oxidative burst triggered by flg22, elf18, or epiBL in Col-0 leaf discs. ROS production is presented as total photon counts during 40 min of treatment. Values are mean ± SE (n = 20). (B) Activation profile of MAPKs in response to a time-course treatment with 1 μM flg22 or epiBL in 2-wk-old Col-0 seedlings. Arrowheads indicate phosphorylated MPK3 and MPK6. Blots stained with colloidal brilliant blue (CBB) are presented to show equal loading. (C) Quantitative RT-PCR analysis of FRK1 and At2g17740 expression in 2-wk old Col-0 seedlings treated with 100 nM flg22 or epiBL for 0, 1, or 3 h. Transcript levels are normalized to the U-box gene and are presented as relative to the value at 0 h. Values are mean ± SD (n = 3). (D) Growth of Pto DC3000 in Col-0 leaves pretreated with water, 1 μM flg22, or epiBL for 24 h and then syringe-infiltrated with 105 cfu/mL of bacteria. Bacterial growth was determined 2 d after inoculation. Values are mean ± SE (n = 8; ***P < 0.001). Similar results were observed in at least two independent experiments.
Activation of BAK1 by PAMP Perception Does Not Modulate BR Responses.
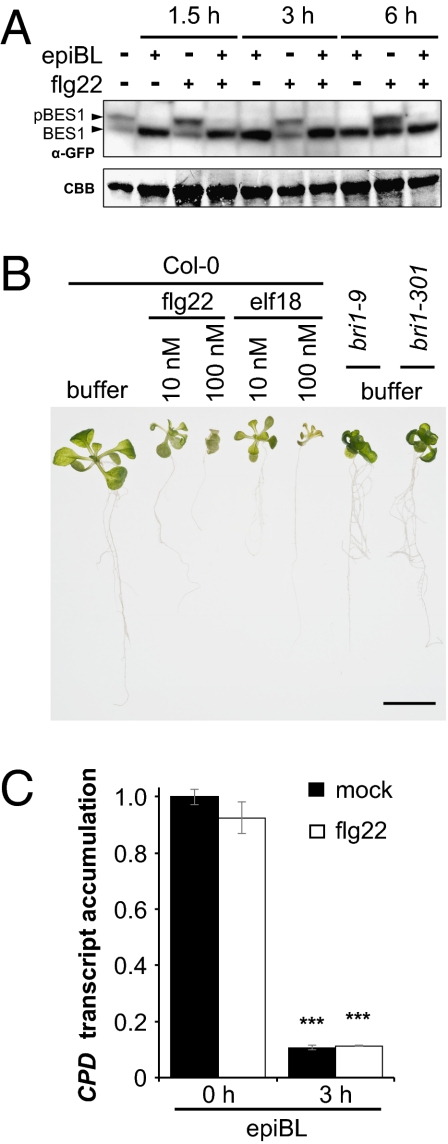
We next tested whether activation of BAK1 following flg22 and/or elf18 perception can modulate BR signaling. Dephosphorylation of the transcription factor BES1 is an early marker of BR perception (25). Treatment of transgenic seedlings expressing BES1-GFP with flg22 did not induce BES1 dephosphorylation, and did not affect epiBL-induced BES1 dephosphorylation (Fig. 2A). Similarly, the activation of multiple BAK1-dependent pathways by cotreatment with flg22 and elf18 had no effect on epiBL-induced BES1 dephosphorylation (Fig. S3). The apparent increase in the abundance of nonphosphorylated BES1 in seedlings treated with flg22 for 6 h (Fig. 2A) was not observed in other independent experiments (e.g., Fig. S3) and is therefore not reproducible.
Fig. 2.
Activation of BAK1 by PAMP perception does not modulate BR signaling. (A) BES1-GFP phosphorylation (detected as band shift) after the indicated times of seedling treatment with 1 μM epiBL and/or 1 μM flg22 by using anti-GFP antibodies. Blot stained with CBB is presented to show equal loading. (B) Phenotype of 2-wk-old Col-0, bri1-9, and bri1-301 Arabidopsis seedlings treated with buffer or different concentrations of flg22 or elf18 for 8 d. (Scale bar: 1 cm.) (C) Quantitative RT-PCR of CPD gene expression in 2-wk-old Col-0 seedlings grown with or without 10 nM flg22 for 1 wk and then treated with 1 μM epiBL for the indicated time. Transcript levels are normalized to the U-box gene and are presented as relative to mock treatment at time 0. Values are mean ± SD (n = 3; ***P < 0.001). Similar results were observed in at least three independent experiments.
Prolonged treatment with flg22 or elf18 leads to seedling growth inhibition, a response that could potentially result from inhibition of BRI1-mediated growth. However, flg22- or elf18-treated seedlings did not show typical impaired BR perception attributes such as curled dark green leaves, reduced petioles, and suppressed hypocotyl elongation (Fig. 2B). Given that bak1 mutants have only a minor rosette phenotype compared with bri1 alleles (4, 5, 23), and that the assay used is not quantitative, we tested if flg22- or elf18-treated seedlings were affected in BR responsiveness by measuring BR-marker gene CPD expression. Arabidopsis seedlings pretreated for 1 wk with flg22 remained fully responsive to endogenous and exogenously applied BRs (Fig. 2C). Therefore, flg22 or elf18 perception does not enhance or inhibit BR signaling.
BRs Inhibit flg22- and elf18-Induced Responses.
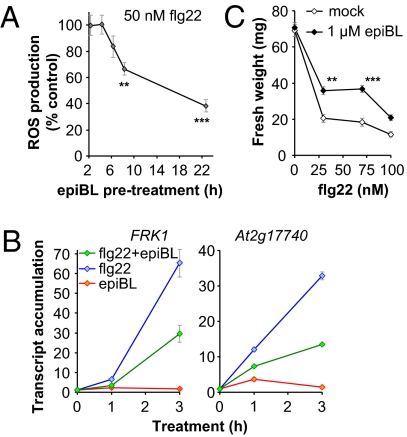
We next asked whether prior or simultaneous treatment with epiBL affects flg22 or elf18 triggered PTI responses in WT leaves. After 5 h of epiBL pretreatment, we observed a marked decrease in ROS production triggered by flg22 (Fig. 3A) or elf18 (Fig. S4). In addition, the expression of PTI marker genes was approximately halved when WT seedlings were simultaneously treated with epiBL and flg22 for 3 h (Fig. 3B). Similarly, seedling growth inhibition triggered by up to 100 nM flg22 was clearly suppressed by cotreatment with epiBL, whereas epiBL by itself did not increase seedling growth in these conditions (Fig. 3C). Together with the inability of FLS2 activation by flg22 to initiate or suppress BR signaling (Fig. 2 A and C), these results point to a unidirectional inhibition of several PTI outputs by epiBL perception.
Fig. 3.
EpiBL perception inhibits PTI signaling. (A) Effect of pretreatment with 1 μM epiBL for the indicated time on ROS triggered by 50 nM flg22 in Col-0 leaf discs. Total ROS production during 40 min of treatment is expressed as percentage of flg22-treated Col-0 without epiBL treatment. Values are mean ± SE (n = 20). (B). Quantitative RT-PCR of FRK1 and At2g17740 gene expression in 2-wk-old Col-0 seedlings treated with 1 μM flg22 and/or 1 μM epiBL for the indicated time. Transcript levels are normalized to the U-box gene and are presented as relative to time 0. Values are mean ± SD (n = 3). (C) Seedling growth inhibition induced by increasing concentrations of flg22 in absence (◇) or presence (◆) of 1 μM epiBL. Values are mean ± SD (n = 12; **P < 0.01 and ***P < 0.001). Similar results were observed in at least two independent experiments.
BAK1 Is Not Rate-Limiting Between BRI1 and FLS2 Pathways.
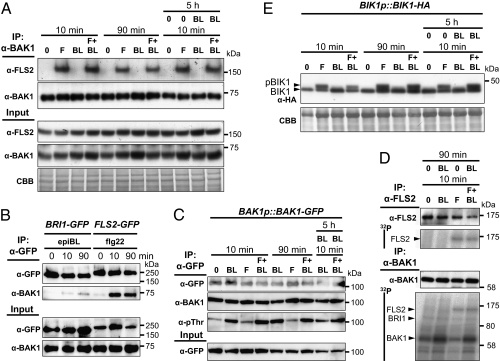
The simplest explanation for BRs to inhibit the FLS2 pathway is that FLS2–BAK1 complexes are not formed as a result of recruitment of BAK1 into BRI1 complexes. This hypothesis assumes that BAK1 is rate-limiting in PTI but not in BR signaling. Therefore, WT Arabidopsis seedlings were treated with flg22, epiBL, or both, and subjected to coimmunoprecipitation experiments using anti-FLS2 and anti-BAK1 antibodies. After 10 min, flg22 induced complex formation between FLS2 and BAK1 (Fig. 4A). Consistent with the ligand dependency of FLS2–BAK1 oligomerization (11, 36), epiBL alone did not induce formation of this complex, nor did cotreatment with flg22 and epiBL lead to a change compared with flg22 alone (Fig. 4A). Oligomerization between BRI1 and BAK1 is usually studied 90 min after BL treatment (23). Flg22-induced FLS2 and BAK1 oligomers could still be observed after 90 min of flg22 treatment, albeit at a lower level (Fig. 4A). Importantly, the presence of epiBL together with flg22 for 90 min did not affect the amount of FLS2 immunoprecipitated with BAK1 (Fig. 4A). Even a 5-h pretreatment with epiBL did not affect the amount of FLS2-BAK1 oligomers after a 10-min flg22 treatment (Fig. 4A). Similar results were obtained in reverse coimmunoprecipitation experiments (Fig. S5) or lines expressing FLS2-GFP (Fig. S6). No alteration of FLS2 amount was observed in seedlings (pre)treated with epiBL (Fig. 4A and Figs. S5 and S6, input). We then compared the amount of native BAK1 that can be pulled down by C-terminally GFP-tagged BRI1 or FLS2 in Arabidopsis transgenic plants (Fig. 4B). These results show that only a very small amount of BAK1 was present in a ligand-dependent complex with BRI1-GFP, whereas a large amount of BAK1 was sequestered by FLS2-GFP. The amount of BAK1 available for recruitment by both FLS2 and EFR was not limiting, as cotreatment with flg22 and elf18 did not impair the amount of BAK1 in complex with FLS2 (Fig. S7). Cotreatment with epiBL and flg22 precluded the use of a single plant line for assessing the relative amounts of BAK1 associating with either receptor. To ensure that the observed difference in BRI1- or FLS2-mediated recruitment of BAK1 is not a result of a difference between BRI1 and FLS2 protein concentration, we used quantitative Western analysis (37). In seedlings grown under the same conditions, the amount of BRI1-eGFP is 17.5 ± 6.1 pmol g−1 fresh weight (FW) and the amount of FLS2-3myc-GFP is 8.0 ± 1.9 pmol g−1 FW (Fig. S8 A and B). The immunoprecipitation (IP) efficiency within the two different backgrounds is highly reproducible at 41 ± 5% (Fig. S8 C and D), whereas, after simultaneous epiBL and flg22 application, less than 5% of the BAK1 pool is recruited by BRI1 (Fig. S8E). Therefore, the observed impairment of flg22-induced responses by BR perception is not caused by a lack of BAK1.
Fig. 4.
EpiBL perception does not affect the dynamic and the activity of the FLS2–BAK1–BIK1 complex. (A) IP of BAK1 in Col-0 seedlings treated with 1 μM flg22 (F) and/or epiBL (BL) for 10 min or 90 min (with or without 5 h epiBL pretreatment). Coimmunoprecipitated proteins were further analyzed by using anti-FLS2 or anti-BAK1 antibodies. (B) IP of GFP-tagged proteins from BRI1-eGFP or FLS2-3myc-GFP seedlings treated with 1 μM epiBL or flg22 (respectively) for 0, 10, and 90 min. Coimmunoprecipitated proteins were further analyzed by using anti-GFP or anti-BAK1 antibodies. (C) IP of BAK1-GFP proteins from transgenic seedlings treated as in A. Immunoblot is analyzed by using anti-BAK1 and anti-pThr antibodies. (D) IP of FLS2 or BAK1 from Col-0 seedlings treated with 1 μM flg22 (F) and/or epiBL (BL) for 10 min (with or without 90 min epiBL pretreatment). Immunoprecipitated proteins were then incubated in presence of radioactive [32P]γ-ATP. Immunoblots are analyzed by using anti-FLS2 or anti-BAK1 antibodies. In vitro phosphorylation is revealed in the autoradiogram (i.e., 32P). (E) BIK1 phosphorylation (detected as a band shift) in BIK1-HA seedlings treated as in A. The blots stained with CBB are presented to show equal loading. Molecular weights (in kDa) are indicated. Similar results were observed in at least two independent experiments.
EpiBL Inhibition of FLS2 Is Independent or Downstream of BAK1-BIK1.
2Although FLS2 and BAK1 kinase activity is not required for heteromerization (24, 36), it is essential for downstream signaling (24, 36, 38). Thus, we asked whether cotreatment with epiBL and flg22 affects FLS2 and/or BAK1 phosphorylation. By using a BAK1p::BAK1-GFP line, BAK1 phosphorylation status was determined by using antiphosphothreonine (anti-pThr) antibodies. Of note, we observed in some experiments that BAK1-GFP could form homo-oligomers with the endogenous BAK1, but this was not always reproducible. Importantly, whereas a large amount of BAK1 is recruited by FLS2 (Fig. 4B), only a very small fraction is phosphorylated (Fig. 4C). In contrast, treatment with epiBL for 10 or 90 min leads to strong BAK1 phosphorylation (Fig. 4C), which prevented us from testing if epiBL co- or pretreatment affects flg22-induced BAK1 phosphorylation. In a complementary strategy, we performed IP followed by in vitro radioactive kinase assays to reveal the phosphorylation status of FLS2 and BAK1 after flg22 and/or epiBL treatment. In this experiment, in vitro FLS2 phosphorylation could be detected in immunoprecipitated FLS2 from flg22-treated but not from mock- or epiBL-treated seedlings (Fig. 4D, Upper). Pretreatment with epiBL for 90 min did not inhibit flg22-induced FLS2 phosphorylation (Fig. 4D, Upper). When BAK1 was immunoprecipitated, enhanced FLS2 phosphorylation was observed within 10 min of flg22 perception (Fig. 4D, Lower), as well as increased BAK1 phosphorylation resulting from BRI1 activation (Fig. 4D, Lower). As seen in Fig. 4C, epiBL-induced BAK1 phosphorylation was higher than that triggered by flg22. Importantly, as also seen in FLS2 immunoprecipitates, the phosphorylation status of BAK1-associated FLS2 was not affected by epiBL pretreatment (Fig. 4D, Lower). Therefore, epiBL-induced inhibition of FLS2 signaling is not associated with reduced phosphorylation of FLS2 or of FLS2-associated BAK1. Next, we tested if BIK1, a FLS2 substrate and positive regulator of PTI signaling (19, 20), could be the target of the epiBL-mediated inhibition of PTI signaling. However, no effect of epiBL on flg22-induced BIK1 phosphorylation, either after simultaneous treatment or after a 5-h pretreatment (Fig. 4E), was observed.
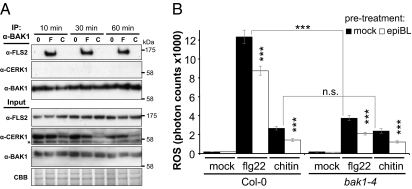
Finally, we tested if epiBL perception also inhibits signaling triggered by the fungal PAMP chitin. This PAMP is perceived via the LysM-RLK CERK1 and induces PTI marker genes in a BAK1-independent manner (14, 17, 39, 40). In agreement with its genetic dispensability for chitin-induced gene expression (14), we show that BAK1 does not form a ligand-dependent complex with CERK1 (Fig. 5A). Importantly, epiBL pretreatment inhibited the chitin-induced ROS burst (Fig. 5B), similarly to that observed for the flg22- and elf18-induced ROS burst (Figs. 3A and 5B and Fig. S4). Notably, the epiBL-mediated inhibition of the flg22- and chitin-induced ROS bursts also occurred in the null bak1-4 mutant.
Fig. 5.
EpiBL perception inhibits BAK1-independent chitin-mediated PTI signaling. (A) IP of BAK1 in Col-0 seedlings treated with water (“0”), 1 μM flg22 (“F”), or 1 mg/mL chitin (“C”) for the indicated time. Coimmunoprecipitated proteins were further analyzed by using anti-FLS2 or anti-CERK1 antibodies. The star indicates a nonspecific band. The blot stained with CBB is presented to show equal loading. (B) Oxidative burst in leaf discs from 5-wk old Col-0 and bak1-4 plants pretreated for 16 h with mock or 1 μM epiBL, then supplemented with 1 μM flg22, 1 mg/mL chitin, or mock. ROS production is presented as total photon counts during 40 min of treatment, and values are mean ± SE (n = 20; ***P < 0.001).
Collectively, our results demonstrate that BR signaling inhibits FLS2-mediated signaling downstream or independently of the FLS2–BAK1–BIK1 complex.
Conclusions
We have shown that activation of the BRI1 pathway leads to inhibition of PTI signaling mediated by several PRRs. This reveals a potential tradeoff between RLK-mediated growth and innate immunity in plants. BR perception in rice was shown to increase resistance to biotic and abiotic stresses (31, 33). Consistently, BRs activate the expression of several immunity-related genes, including PR1 and AtMYB30 (33, 41). Conversely, the BR-activated transcription factor BZR1 represses the promoters of several immune genes, including FLS2 (42). However, our observation that the FLS2 protein level is unaffected by epiBL treatment does not support this finding.
Because the LRR-RLK BAK1 serves in multiple signaling pathways (43), it was an attractive candidate to regulate tradeoff between immunity and growth already at receptor level. It is still unclear how this tradeoff is regulated. Here we have shown that the most likely scenario, in which BAK1 protein is present in rate-limiting amounts, cannot provide the explanation. Also, there is no direct effect of active PAMP signaling on the BR pathway, ruling out a bidirectional cross-talk mechanism as occurs between EGF and insulin signaling (44). Instead, an asymmetric mechanism operating downstream or independently of the common component BAK1 modulates early immune signaling mediated by PAMP recognition. Although BAK1 has been proposed to mediate interplay between BR and PAMP signaling (12), we provided evidence that the growth-inhibiting effect of PAMP perception does not operate through antagonism of BR signaling. Our experiments suggest that different pools of BAK1 exist that are not freely interchangeable. BAK1 recruited by FLS2 seems to be different from that recruited by BRI1, as BL did not displace the amount of BAK1 immunoprecipitated with FLS2. Alternatively, this could be explained by the fact that more BAK1 is recruited into FLS2 complexes than into the BRI1 complex, or that the complexes are more stable. This idea is corroborated by the observed weak impact of BAK1 loss-of-function mutations on BR sensitivity (4, 5, 23), whereas BAK1 plays a more important role in FLS2 signaling (11, 12, 24), and by the finding that the role of BAK1 in BR and PTI signaling can be mechanistically uncoupled (24).
Tradeoffs between endogenous hormonal pathways and responses to exogenous cues have been proposed previously. For example, the nuclear growth-repressing DELLA proteins negatively regulate gibberellic acid (GA) signaling, but are important to mediate the balance between the immune hormones salicylic acid and jasmonic acid in response to biotic and abiotic stresses (45–48). Also, auxin and salicylic acid signaling antagonize each other (49, 50). Notably, flg22 perception inhibits auxin signaling by inducing the expression of the microRNA miR393 that targets auxin receptors to increase disease resistance to biotrophic pathogens (49). Similarly, flg22 perception dampens GA signaling by stabilizing DELLA proteins that are negative regulators of this pathway (47). Given the important role of endogenous hormones in modulating plant defense, several pathogens produce GA or auxin as potential virulence strategies (51). Interestingly, the obligate biotrophic oomycete Albugo laibachii appears to encode a complete BR biosynthesis pathway (52). Together with our results, this leads to the tentative hypothesis that BR production may contribute to the ability of this pathogen to suppress plant immunity. On the plant side, recent work shows that enhanced BR signaling as observed in gain-of-function mutations in BAK1 (27) and in BRI1 (28), both impair responses to flagellin, supporting this idea.
Therefore, BR signaling may play an important role in the modulation of plant immunity during plant growth, by regulating immune signaling downstream of BIK1, and is a potential site of manipulation by pathogens during infection.
Materials and Methods
Plant Materials and Growth.
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the WT control. Mutants, transgenic lines, and growth conditions are described in SI Materials and Methods.
Chemicals.
Flg22 and elf18 peptides were purchased from Peptron, and chitin oligosaccharide from Yaizu Suisankagaku. epiBL was purchased from Xiamen Topusing Chemical and prepared as 20 mM stock solution in ethanol. Brassinazole was purchased from Sigma and prepared as 10 mM stock solution in DMSO.
Measurement of ROS Generation.
Oxidative-burst measurement was performed as previously described (53). ROS was elicited with flg22, elf18, or epiBL, and elicitation in the absence of any PAMP (water treatment) was included in all experiments as negative control. Twenty leaf discs from ten 5-wk-old plants were used for each condition. Luminescence was measured over 40 min by using a high-resolution photon counting system (HRPCS218; Photek) coupled to an aspherical wide lens (Sigma).
Seedling Growth Inhibition Assay.
Seedling growth inhibition was assessed as previously described in (54). In brief, 5-d-old Arabidopsis seedlings were grown in liquid Murashige–Skoog medium containing 1% sucrose supplemented with flg22 or elf18 peptides. Seedlings were weighted 8 d after treatment.
RNA Isolation and Quantitative RT-PCR.
Total RNA was prepared from six 2-wk-old seedlings grown in liquid medium or from four leaf discs (38.5 mm2 each, from 5-wk-old plants) floated overnight in water before treatment. RNA extraction, cDNA synthesis, and quantitative RT-PCR were performed as described in SI Materials and Methods.
Induced Resistance to Bacteria.
Induced resistance assays were realized as described previously (35). Briefly, water, 1 μM flg22, or 1 μM epiBL was infiltrated with a needleless syringe into leaves of 5-wk-old Arabidopsis plants. After 24 h, the same leaves were syringe-infiltrated with 105 cfu/mL of Pto DC3000. Bacterial growth was determined 2 d after inoculation by plating serial dilutions of leaf extracts on L agar with appropriate antibiotics.
MAPK Activation.
Activation profile of the MAPKs MPK3 and MPK6 in response to 1 μM flg22 or 1 μM epiBL was performed as described previously (53). Phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204) rabbit monoclonal antibodies (Cell Signaling) were used according to the manufacturer's protocol.
Protein Extraction and IP Experiments.
Protein extraction and IP using WT Arabidopsis seedlings were performed as described previously (24). Protein extraction and IP by using Ws-0/FLS2p::FLS2-3myc-GFP and Col-0/BAK1p::BAK1-GFP transgenic lines was performed as described in SI Materials and Methods. Proteins were separated by SDS/PAGE 10% and further analyzed by Western blot by using rabbit polyclonal anti-FLS2 antibodies (17), rabbit polyclonal anti-CERK1 antibodies (17), rabbit polyclonal anti-BAK1 antibodies (36), mouse monoclonal anti-GFP antibodies coupled to horseradish peroxidase (Miltenyi Biotec), and rabbit polyclonal anti-pThr antibodies (Zymed-Invitrogen). Phosphorylation statuses of BES1-GFP and BIK1-HA and in vitro phosphorylation of immunoprecipitated proteins were analyzed as described in SI Materials and Methods. Quantitative Western blotting was performed as described previously (37).
Statistical Analysis.
Statistical significances based on one-way ANOVA analyses were determined with Prism 5.01 software (GraphPad).
Supplementary Material
Acknowledgments
We thank J. Chory, N. Geldner, S. Robatzek, and J. M. Zhou for sharing biological materials; D. Weijers and all the members of the laboratory of C.Z. for fruitful discussion and comments on the manuscript; and Joanne Chory and Jeffery Dangl for sharing results and comments before publication. D.C. thanks T. Boller for his professional support. This research was funded by the Gatsby Charitable Foundation (C.Z. and J.P.R), United Kingdom Biotechnology and Biological Sciences Research Council Grants BB/G024936/1 (“ERA-PG PRR CROP,” to C.Z.) and BB/G024944/1 (“ERA-PG Pathonet,” to C.Z.), Swiss National Science Foundation Grant 31003A-120655 (to D.C.), Marie Curie Training ITN network BRAssinosteroid Venture Increasing StudentS’ International MObility Grant 215118 (to S.C.d.V.), and the Department of Agrotechnology and Food Sciences of Wageningen University (S.C.d.V. and C.A.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 7.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109921108/-/DCSupplemental.
References
- 1.Shiu S-H, Bleecker AB. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci STKE. 2001;2001:re22. doi: 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- 2.De Smet I, Voss U, Jürgens G, Beeckman T. Receptor-like kinases shape the plant. Nat Cell Biol. 2009;11:1166–1173. doi: 10.1038/ncb1009-1166. [DOI] [PubMed] [Google Scholar]
- 3.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 4.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 5.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 6.Karlova R, et al. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell. 2006;18:626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He K, et al. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol. 2007;17:1109–1115. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 8.Albrecht C, Russinova E, Kemmerling B, Kwaaitaal M, de Vries SC. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid dependent and independent signaling pathways. Plant Physiol. 2008;148:611–619. doi: 10.1104/pp.108.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaillais Y, et al. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25:232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clouse SD. Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23:1219–1230. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 12.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux M, et al. The arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to Hemibiotrophic and Biotrophic pathogens. Plant Cell. 2011;23:2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan L, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fradin EF, et al. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009;150:320–332. doi: 10.1104/pp.109.136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaparro-Garcia A, et al. The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen phytophthora infestans in Nicotiana benthamiana. PLoS ONE. 2011;6:e16608. doi: 10.1371/journal.pone.0016608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimenez-Ibanez S, et al. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- 18.Kemmerling B, et al. The BRI1-Associated Kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol. 2007;17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 19.Lu D, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA. 2010;107:496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Lu D, et al. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011;332:1439–1442. doi: 10.1126/science.1204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Curr Opin Microbiol. 2011;14:54–61. doi: 10.1016/j.mib.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Schwessinger B, et al. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011;7:e1002046. doi: 10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 26.Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007;21:1598–1602. doi: 10.1101/gad.1561307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaillais Y, Belkhadir Y, Balsemão-Pires E, Dangl JL, Chory J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA. 2011;108:8503–8507. doi: 10.1073/pnas.1103556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belkhadir Y, et al. Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci USA. 2012;109:297–302. doi: 10.1073/pnas.1112840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mora-García S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grove MD, et al. Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature. 1979;281:216–217. [Google Scholar]
- 31.Nakashita H, et al. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003;33:887–898. doi: 10.1046/j.1365-313x.2003.01675.x. [DOI] [PubMed] [Google Scholar]
- 32.Krishna P. Brassinosteroid-mediated stress responses. J Plant Growth Regul. 2003;22:289–297. doi: 10.1007/s00344-003-0058-z. [DOI] [PubMed] [Google Scholar]
- 33.Divi UK, Rahman T, Krishna P. Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 2010;10:151. doi: 10.1186/1471-2229-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asami T, Yoshida S. Brassinosteroid biosynthesis inhibitors. Trends Plant Sci. 1999;4:348–353. doi: 10.1016/s1360-1385(99)01456-9. [DOI] [PubMed] [Google Scholar]
- 35.Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 36.Schulze B, et al. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem. 2010;285:9444–9451. doi: 10.1074/jbc.M109.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilma van Esse G, et al. Quantification of the Brassinosteroid Insensitive1 receptor in planta. Plant Physiol. 2011;156:1691–1700. doi: 10.1104/pp.111.179309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006;20:537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miya A, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wan J, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, et al. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009;58:275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chinchilla D, Shan L, He P, de Vries S, Kemmerling B. One for all: The receptor-associated kinase BAK1. Trends Plant Sci. 2009;14:535–541. doi: 10.1016/j.tplants.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zielinski R, et al. The crosstalk between EGF, IGF, and insulin cell signaling pathways—computational and experimental analysis. BMC Syst Biol. 2009;3:88. doi: 10.1186/1752-0509-3-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 46.Achard P, Renou J-P, Berthomé R, Harberd NP, Genschik P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol. 2008;18:656–660. doi: 10.1016/j.cub.2008.04.034. [DOI] [PubMed] [Google Scholar]
- 47.Navarro L, et al. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol. 2008;18:650–655. doi: 10.1016/j.cub.2008.03.060. [DOI] [PubMed] [Google Scholar]
- 48.Hou X, Lee LYC, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 49.Navarro L, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 50.Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol. 2007;17:1784–1790. doi: 10.1016/j.cub.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 51.Grant MR, Jones JDG. Hormone (dis)harmony moulds plant health and disease. Science. 2009;324:750–752. doi: 10.1126/science.1173771. [DOI] [PubMed] [Google Scholar]
- 52.Kemen E, et al. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 2011;9:e1001094. doi: 10.1371/journal.pbio.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boutrot F, et al. Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA. 2010;107:14502–14507. doi: 10.1073/pnas.1003347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nekrasov V, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28:3428–3438. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.