Abstract
Metazoans and plants use pattern recognition receptors (PRRs) to sense conserved microbial-associated molecular patterns (MAMPs) in the extracellular environment. In plants, the bacterial MAMPs flagellin and elongation factor Tu (EF-Tu) activate distinct, phylogenetically related cell surface pattern recognition receptors of the leucine-rich repeat receptor kinase (LRR-RK) family called FLS2 and EF-Tu receptor, respectively. BAK1 is an LRR-RK coreceptor for both FLS2 and EF-Tu receptor. BAK1 is also a coreceptor for the plant brassinosteroid (BR) receptor, the LRR-RK BRI1. Binding of BR to BRI1 primarily promotes cell elongation. Here, we tune the BR pathway response to establish how plant cells can generate functionally different cellular outputs in response to MAMPs and pathogens. We demonstrate that BR can act antagonistically or synergistically with responses to MAMPs. We further show that the synergistic activities of BRs on MAMP responses require BAK1. Our results highlight the importance of plant steroid homeostasis as a critical step in the establishment of plant immunity. We propose that tradeoffs associated with plasticity in the face of infection are layered atop plant steroid developmental programs.
Keywords: brassinosteroid signaling, plant immune system signaling, signaling crosstalk
Innate immune systems of plants and animals rely on pattern recognition receptors (PRRs) to produce an appropriate physiological response upon detection of nonself molecules (1, 2). PRRs respond to conserved microbe-associated molecular patterns (MAMPs) (1, 2). In Arabidopsis, the majority of PRRs are leucine-rich repeat (LRR)-receptor kinases (RKs). MAMPs often play no role in pathogenesis, but rather are indispensable for core microbial functions (1). MAMPs are typically conserved among diverse sets of pathogens. Well studied MAMPs in plant immune system studies include a 22-aa peptide derived from flagellin (flg22) and a bioactive 18-aa peptide derived from the translational elongation factor Tu (elf18), peptidoglycans (PGNs), and chitin, a component of fungal cell walls. MAMPs elicit a suite of defense responses including the accumulation of reactive oxygen intermediates, deposition of callose, large-scale transcriptional reprogramming, and biosynthesis of microstatic and/or microcidal secondary metabolites. This response constitutes MAMP-triggered immunity (MTI), which is sufficient to slow or halt microbial proliferation (1–3).
Recent studies have unveiled a web of interactions between the plant immune system and growth regulating hormones like auxin, gibberellins, and ethylene (2). However, there is little evidence for direct physical convergence points linking hormone and defense response signaling systems (4). One convergence point that could link growth-promoting hormone responses to MAMP signaling is the LRR-RK BRI1-Associated Kinase 1 (BAK1) (2, 5). BAK1 was identified as an interactor of the LRR-RK Brassinosteroid (BR)-Insensitive 1 (BRI1), which binds brassinolide (BL), the most potent steroid hormone in plants. Upon BL binding, BRI1 autophosphorylates and activates BAK1 by transphosphorylation, thereby enhancing signaling competency through reciprocal BRI1 transphosphorylation (6). Similarly, Flagellin-Sensitive 2 (FLS2), the receptor for the bacterial flagellum peptide flg22, associates with BAK1 immediately upon ligand binding (7). BAK1 recruitment to specific cell surface signaling systems involve its extracellular LRRs and differential phosphorylation-dependent events for proper signal transduction (7–9). Hence, BAK1 is an adapter recruited downstream of ligand perception for several cell surface signaling pathways. To date, BAK1 function in innate immunity appears to be genetically independent from its function in BR signaling (10). Here, we revisit the interplay between the plant immune system and plant steroids and demonstrate that BR biosynthesis and signaling can be rate-limiting modulators of BAK1-mediated MAMP responses.
Results
Maintenance of BR Homeostasis Is Critical for flg22-Induced MTI Signaling.
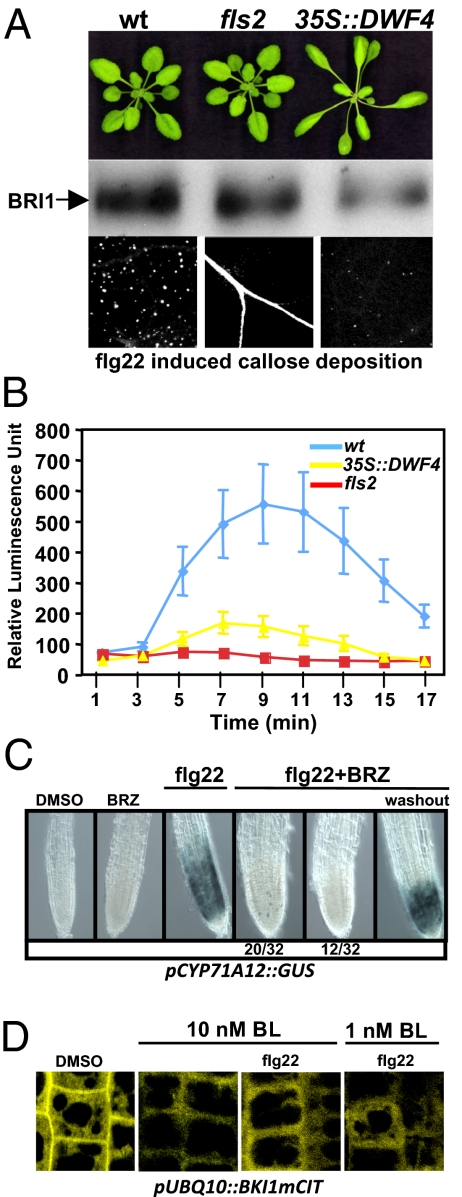
Exogenous application of extremely high concentrations of BL unmasked a role for BRs in the control of a range of weak immune responses (11). This function of BR as a modulator of plant immunity is uncharacterized to our knowledge. We used transgenic Arabidopsis plants ectopically overexpressing DWARF4 (35S::DWF4) to investigate whether PRR signaling is altered under conditions of excess BR biosynthesis (12). DWF4 encodes a C-22 hydroxylase, which is a rate-limiting step of BL biosynthesis. 35S::DWF4 plants display elongated organ phenotypes, and dramatically reduced responses to flg22 (Fig. 1 A and B and Fig. S1). Thus, increasing the endogenous pool of bioactive BR antagonizes flg22-induced responses. Loss-of-function BR biosynthetic or signaling mutants display dramatic changes in cell elongation, resulting in severe dwarfism (13), complicating assays for immune system function. We therefore transiently reduced BR biosynthesis by using brassinazole (BRZ), a triazole compound that reversibly and specifically blocks DWF4 activity (14). To monitor flg22 output in the root, we used a CYP71A12 gene–GUS reporter transgenic line (pCYP71A12::GUS) (15) (Fig. 1C). We subjected pCYP71A12::GUS seedlings to BRZ treatment and monitored GUS reporter gene activation upon flg22 treatment. In the presence of BRZ, pCYP71A12 response to flg22 was greatly diminished (Fig. 1C). Removal of BRZ before flg22 treatment (i.e., washout) restored full induction of pCYP71A12-GUS (Fig. 1C). Thus, transient depletion of endogenous BR pools strongly suppresses flg22 signaling in root cells. We tested whether flg22 signaling can also modulate BR responses. We used displacement of the BRI1 inhibitory protein BKI1 from the plasma membrane (PM) as a readout of early BR activation (16). Our results show that sustained activation of the FLS2 pathway could not block BKI1 displacement from the PM, even at low doses of BL (Fig. 1D). Thus, sustained FLS2 signaling does not modulate early BR signaling outputs. Collectively, our results suggest the existence of one-way cross-talk between the balanced activities of BR biosynthesis and FLS2 signaling.
Fig. 1.
BR biosynthesis modulates MAMP signaling. (A) Top: Images representative of Arabidopsis WT Col-0, fls2, and 35S::DWF4. Middle: Microsomal protein extracts prepared from genotypes listed at the top were subjected to anti-BRI1 immunoblot analysis. Bottom: Aniline blue-stained callose deposits in the leaves of the genotypes listed at the top treated with 1 μM flg22. (B) Oxidative burst triggered by 1 μM flg22 in WT Col-0 (blue), fls2 (red), and 35S::DWF4 (yellow) leaf discs measured in relative luminescence units. (C) GUS stains of CYP71A12::GUS line. Seedlings grown in the presence or absence of 5 μM BRZ were left untreated or treated with 1 μM flg22 for 12 h before GUS staining. Washout indicates removal of BRZ during flg22 treatment. Numbers at the bottom indicate the number of roots tested that fall into each category among the 32 roots assayed when BRZ was used in conjunction with flg22. In this assay, 20 roots of 32 displayed a highly attenuated response in the form of small blue spots. (D) Subcellular dynamics of BKI1 upon BL treatment is not affected by flg22 treatments. Subcellular localization of BKI1mCIT is shown in root meristem epidermal cells. BKI1mCIT is localized to the PM and cytosol in the absence of BL treatment (i.e., DMSO) and relocates rapidly from the PM to the cytosol following BL application. Note that BKI1 relocalization to the cytosol after BL treatment is not affected by flg22 treatment even when low concentrations of BL are used (10 nM and 1 nM). This experiment was repeated two times with similar results.
Overexpression of BRI1 Antagonizes BAK1-Mediated PRR Signaling and Cell Death.
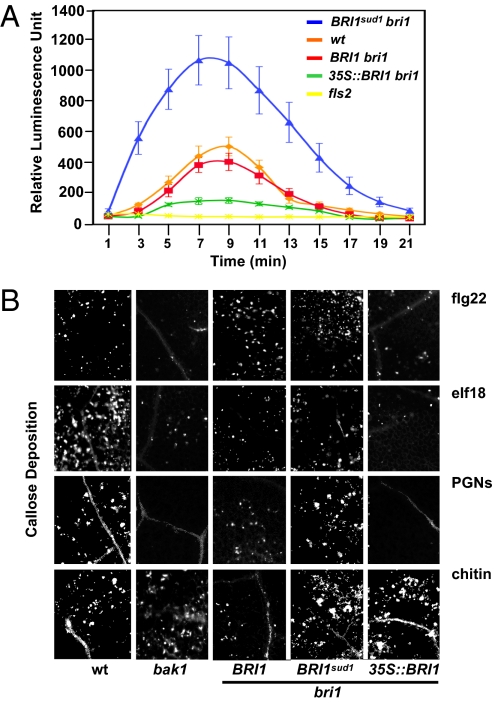
Next, we tested whether BR signaling could be limiting for MTI responses when signaling is activated via BRI1. Plants expressing BRI1 under the control of the strong CaMV 35S promoter (35S::BRI1) failed to respond to flg22 treatment, similar to bak1 and fls2 plants (Fig. 2 A and B and Figs. S2 and S3). We extended these findings to signaling pathways activated by two additional MAMPs (elf18 and PGN), which also require BAK1 for signaling (Fig. 2B). Conversely, 35S::BRI1 plants displayed WT responses to chitin, a MAMP that triggers BAK1-independent responses. Hence, only BAK1-mediated MTI is impaired in 35S::BRI1 plants. The lack of BAK1-mediated MTI response in 35S::BRI1 plants could be the consequence of a limiting pool of BAK1 that is unavailable for MAMP signaling. To test this, we moderately increased BAK1 dosage by introducing a BAK1-HA transgene into 35S::BRI1 plants and tested flg22 responsiveness (Fig. 3A). The presence of extra copies of BAK1 restored flg22 sensitivity in 35S::BRI1 plants. Thus, there is a direct link between restoration of FLS2 function and BAK1 dosage in the specific context of heightened BRI1 levels. Collectively, our results demonstrate that increased BR signaling triggered by BRI1 overexpression can antagonize the activities of at least three BAK1-dependent MAMP signaling pathways.
Fig. 2.
BR signaling modulates MTI. (A) Oxidative burst in relative luminescence units, triggered by 1 μM flg22 in leaf discs of the genotypes listed on the top right corner. (B) Aniline blue-stained callose deposits in leaves of the genotypes listed at the bottom treated with 1 μM flg22 or elf18 and 100 μg/mL of PGNs or chitin.
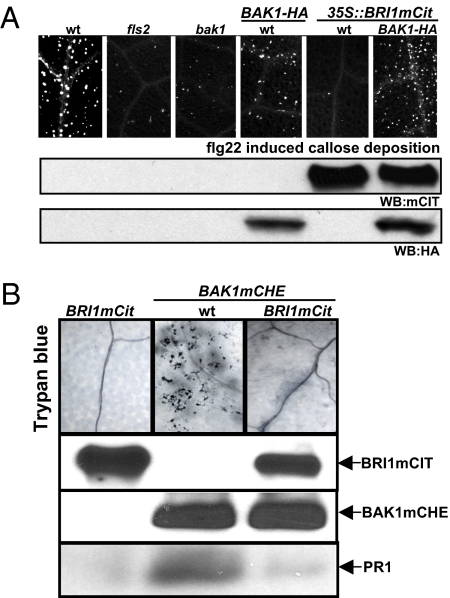
Fig. 3.
Overexpression of BRI1 antagonizes BAK1-mediated MAMP and cell death responses. (A) Top: Aniline blue-stained callose deposits in leaves of the genotypes listed treated with 1 μM flg22. Middle, Bottom: Microsomal protein extracts prepared from the genotypes listed at the top were subjected to an anti-GFP (WB:mCIT) or to an anti-HA (WB:HA) immunoblot analysis to detect the accumulation of mCit or HA tagged proteins. (B) Top: Trypan blue stain of leaves from the genotypes listed at the top. Genotypes of parental and resulting F1 lines are indicated at the top. The control lane (Left) represents F1 plants obtained through a cross between BRI1mCIT and Col-0. Trypan blue stains of F1 plants from the following crosses: BRI1mCit × Col-0 (control; Left), BAK1mCHE × Col-0 (Middle), and BAK1mCHE × BRI1mCit (Right). Two middle panels: Microsomal protein extracts were prepared from the genotypes listed at the top and subjected to anti-GFP and anti-DsRED immunoblot analysis to detect mCitrine and mCherry tagged proteins, respectively. Bottom: Total protein extracts were prepared from the genotypes listed at the top and were subjected to anti-PR1 immunoblot analysis.
In some instances, our BAK1-HA transgenic plants displayed morphological phenotypes reminiscent of those associated with plants that develop spontaneously plant cell death (17). To confirm our observations, we used the native promoter to express a mCitrine epitope-tagged BAK1 in WT (BAK1mCIT) and bak1 (BAK1mCIT bak1) plants. BAK1mCIT accumulates to equivalent levels in bak1 and WT plants and complements the BR- and flg22-dependent phenotypes of bak1 plants (9) (Fig. S4). The increased dose of BAK1 in WT, but not in bak1, resulted again in plants displaying morphological phenotypes reminiscent of spontaneously discrete necrotic lesions (17), which we confirmed with trypan blue staining in BAK1mCIT plants (Fig. S4). Plants displaying ectopic cell death often accumulate constitutively Pathogenesis-Related (PR) proteins and are called cpr mutants (17). PR1 protein accumulated to readily detectable levels in BAK1mCIT plants but not in BAK1mCIT bak1 plants (Fig. S4). Thus, increased BAK1 dosage drives inappropriate cell death responses. By contrast, we never observed plants displaying a cpr morphology when the levels of a dominant allele of BAK1, bak1elg, were increased (Fig. S4). Because the elg mutation generates a BAK1 variant that more strongly associates with BRI1 (9), we reasoned that the phenotypes associated with heightened BAK1 levels could be suppressed by altering BRI1 gene dosage. We used transgenic plants expressing mCherry epitope-tagged BAK1 under its native promoter in Columbia-0(Col-0) WT (BAK1mCHE) (16). BAK1mCHE plants displayed phenotypes similar to those observed in BAK1mCIT plants. Thus, the inappropriate cell death phenotypes observed in our transgenic lines are specific to BAK1 levels, rather than being related merely to the BAK1 epitope tag. We crossed BAK1mCHE plants to WT BRI1mCIT plants and monitored ectopic cell death and PR protein accumulation. Importantly, increasing the dose of BRI1 in the context of heightened BAK1 levels suppressed almost entirely the BAK1 overexpression phenotypes (Fig. 3B). Thus, increased BR signaling mediated by BRI1 overexpression can antagonize the inappropriate cell death signaling triggered by increased BAK1 dosage.
BRI1sud1 Plants Display Enhanced flg22-Induced Signaling.
Our data suggest that the BR-independent functions of BAK1 in plant immunity can collide with its BR-dependent role in growth and development when BRI1 dosage is increased. To test whether the loss of MAMP signaling in 35S::BRI1 plants was a result of increased BRI1 levels, or of increased BRI1 activity, we used the hypermorphic BRI1sud1 allele (Fig. S2). BRI1sud1 plants phenocopied every BR-dependent morphological aspect of 35S::BRI1 plants, but displayed dramatically increased responses to flg22 (Fig. 2A and Figs. S2 and S3A). BRI1sud1 protein accumulates to approximately WT levels (Fig. S2). Thus, the antagonistic effects on MAMP signaling we observed in 35S::BRI1 plants are likely the consequence of increased levels of BRI1, and not of increased intrinsic BRI1 signaling activity.
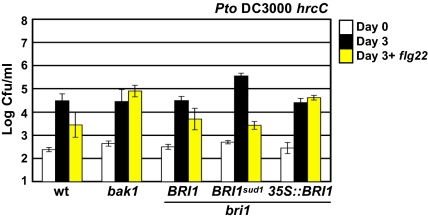
The unexpected result that BRI1sud1 plants displayed enhanced responses to flg22 treatment prompted us to investigate this response during a plant–microbe interaction. We used the hemibiotrophic bacterial pathogen, Pseudomonas syringae pv. tomato (Pto) DC3000, and the isogenic Pto DC3000hrcC mutant, which is deficient in its type III secretion system (T3SS). The T3SS allows bacterial effectors delivery to host cells for maximal virulence (18). Flg22 infiltrated into Arabidopsis leaves induces MTI that can inhibit P. syringae growth (19). Flg22 treatments weakly inhibited Pto DC3000 growth on all genotypes tested, with the exception of BRI1sud1, which exhibited enhanced response, and fls2, which, as expected, exhibited none (Fig. S3). When Pto DC3000hrcC was infiltrated together with flg22, bacterial growth was inhibited approximately 10 fold on BRI1 plants and approximately 100 fold on BRI1sud1 plants (Fig. 4). Thus, the FLS2 signaling pathway in BRI1sud1 plants can be further stimulated and acts effectively to limit bacterial growth. In contrast, we did not observe flg22-dependent inhibition of Pto DC3000hrcC growth on 35S::BRI1 or bak1 plants. We propose that the loss of FLS2 signaling in 35S::BRI1 plants is likely the consequence of the inability of BAK1 to contribute to flg22 signaling. Our results with DC3000hrcC imply that increased BR signaling via BRI1sud1 primes BR and flg22 signaling through BAK1. Accordingly, when BRI1 levels are increased (i.e., 35S::BRI1) the stimulatory effects of BR on flg22 signaling are negated, presumably by increased recruitment of BAK1 to BRI1.
Fig. 4.
BR signaling modulates flg22-dependent disease resistance. Growth of PtoDC3000 hrcC was measured in the genetic backgrounds indicated at the bottom of the chart. Four-week-old plants were infiltrated with 105 cfu/mL PtoDC3000 hrcC in the absence (black bars) or presence (yellow bars) of 1 μM flg22. The number of bacteria per area of leaf was determined at 0 and 3 d postinoculation (Experimental Procedures) and plotted on a log10 scale. Values are mean cfu/mL ± 2 SE.
We used three molecular readouts to monitor the basal activity of FLS2 in BRI1sud1 plants: (i) its phosphorylation status, (ii) its internalization and subsequent disappearance, and (iii) the induction of downstream target genes (20, 21). By using an anti-FLS2 antibody, we immunoprecipitated FLS2 from unchallenged WT and BRI1sud1 plants (Fig. S5). We noted that FLS2 in BRI1sud1 plants is already phosphorylated in the absence of flg22, and accumulates to lower levels (Fig. S5). Thus, the basal activity of FLS2 is increased in BRI1sud1 plants in the absence of flg22, and FLS2 accumulation is not rate-limiting for the enhanced flg22 responses. We could not observe reliably increased internalization of FLS2 in endosomal compartments in BRI1sud1 plants (Fig. S5) (21). This suggests that the stimulatory effect of BRI1sud1 on FLS2 is mild.
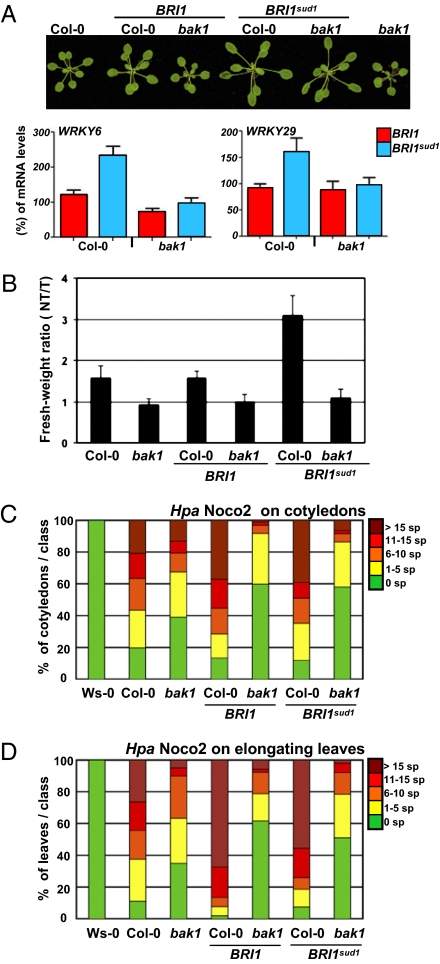
MAMP signals perceived at the cell surface often terminate on the promoters of WRKY transcription factor genes (20). To further confirm that FLS2 signaling is primed in BRI1sud1 plants, we quantified the expression levels of several WRKY genes. All the WRKY genes tested showed a relative increase in their expression levels in BRI1sud1 plants (Fig. S5). This change in WRKY gene expression might not specifically reflect FLS2 signaling, as MAMP-driven transcriptional reprogramming is generic and results in the activation of a largely overlapping set of defense genes (1). Rather, we envision that the induction of basal defense genes in unchallenged BRI1sud1 plants is a general consequence of inappropriate BAK1-mediated signaling. To test this hypothesis, we created isogenic lines expressing BRI1sud1 in the WT and bak1 genetic backgrounds and looked at the steady-state expression of WRKY genes in 30 independent T1 plants. Our results demonstrated that the effects of BRI1sud1 on defense gene expression are BAK1-dependent, whereas the effects on cell elongation are not (Fig. 5A). We attribute this to the fact that BAK1 function in BR signaling can be partially compensated by the LRR-RK Somatic Embryogenesis Receptor Kinase 1 (SERK1) (22). To further confirm that the enhanced flg22 responses observed in BRI1sud1 plants are bak1-dependent, we assayed flg22-mediated seedling growth inhibition (SGI) in BRI1sud1 bak1 double-mutant plants (Fig. 5B). BRI1sud1 bak1 plants showed no SGI, similar to bak1 plants. Thus, the increased flg22-dependent SGI response in BRI1sud1 is BAK1-dependent. Together, our results indicate that the increased activity of the BR signaling pathway, triggered by the BRI1sud1 variant, requires BAK1 to enhance MAMP-dependent responses.
Fig. 5.
Enhanced BR signaling promotes growth of an obligate biotrophic pathogen in a BAK1-dependent manner. (A) Images of representative rosette stage Arabidopsis plants with genotypes listed at the top. Bottom: Quantitative RT-PCR analyses of WRYK6 and WRKY29 transcripts from plants expressing BRI1 (red) or BRI1sud1 (blue) in WT or bak1 genetic backgrounds. (B) Average fresh-weight ratio of seedlings grown in water (NT) or 1 μM flg22 (T). Genotypes are listed at the bottom. (C and D) Twelve-day-old seedlings of the genetic backgrounds indicated at the bottom of the chart were inoculated with conidiospores of the virulent Hpa isolate Noco2 at 32,000 spores/mL. Sporangiophores were counted 4 d after inoculation on cotyledons (C) and on first true leaves (D) for each of the indicated genetic backgrounds. Means, sample sizes, and 2 × SE are presented in Table S1. The experiment was repeated twice. sp, sporangiophores per cotyledon (C) or sporangiophores per leaf (D).
BR Signaling Mediates BAK1-Dependent Changes in Susceptibility to Obligate Biotrophic Pathogen.
To test if imbalance in BR signaling could be exploited by obligate biotrophic pathogens to further their fitness and growth, we challenged BRI1sud1 plants with the virulent oomycete Hyaloperonospora arabidopsidis (Hpa) isolate Noco2. bak1 displayed a significant increase in resistance to the virulent Hpa isolate Noco2 in comparison with WT Col-0 (Fig. 5 C and D and Table S1) (23). Sporulation on BRI1sud1 was not different from that of BRI1 plants (Fig. 5 C and D and Table S1). Morphologically, BRI1 plants display very mild BR gain-of-function phenotypes, possibly because of accumulation greater than WT endogenous levels. BRI1 and BRI1sud1 allowed reproducible, strong increases in sporulation compared with Col-0(Fig. 5 C and D and Table S1).
Thus, an increase in BR signaling driven by the extra gene dosage of BRI1 or the hypermorphic activity of BRI1sud1 enhances susceptibility to Noco2. Importantly, this effect was more pronounced on the first true leaves, organs in which BR-driven cell elongation programs are hyperactive (Fig. 5D). Furthermore, the increased sporulation of Noco2 driven by heightened BR signaling was BAK1-dependent, as the BRI1 bak1 and BRI1sud1 bak1 double mutants supported dramatically less sporulation. These plants supported even less sporulation than bak1 (Fig. 5 C and D and Table S1). Thus, increased BR signaling in the absence of bak1 is able to suppress Noco2 sporulation. Importantly, BRI1 and BRI1sud1 accumulated to approximately equivalent levels in Col-0 and bak1 plants (Fig. S6). RPP4-mediated resistance to the avirulent Hpa isolate Emwa1 was compromised in a BAK1-dependent manner in BRI1 and BRI1sud1 plants as well (Fig. S6 and Table S1) (24). These findings suggest that a slight increase in BR signaling in the presence of BAK1 enhances susceptibility to Hpa, whereas the same slight increase in BR signaling reduces susceptibility in bak1. This is consistent with our proposal that BAK1 can act as mediator for the activities of BRs on PRR responses but further indicates that BR can also act on plant defenses by using a BAK1-independent mechanism.
Discussion
Despite the involvement of various hormone-regulated responses in plant defense, relatively little is known about the molecular mechanisms that connect pathogen perception to ongoing developmental programs in the infected cell or organ (2, 4). We studied the interaction of growth promotion by BRs with plant immune system activities by taking advantage of the fact that BRI1 activity and BR concentrations are highly regulated, rate-limiting factors for BR-dependent responses. We therefore could manipulate BR signaling to dissect its overlap with the plant immune system. Our data suggest a model in which BRs can reset or prime MTI responses. Our model refines current models of independent signaling functions for BR and MTI outputs, and suggests that there is a potential physiological intersection between the signaling pathways that control body size and innate immunity in plants (10). As such, pathway cross-communication simply cannot be tackled by studying signaling systems in isolation (25, 26).
In WT Arabidopsis, BRs and BRI1 exist at concentrations that allow cells to easily modulate their elongation state in response to challenging environmental conditions via BR synthesis (26). However, to set endogenous BR levels and BRI1 sensitivity to a particular activity range, BR biosynthesis is tightly modulated by a feedback regulatory loop controlled by BR signaling (27). A deviation from this range could have detrimental consequences on the adaptive plasticity of plants. In light of our model, we speculate that this homeostatic loop also coordinates BR biosynthesis and signaling to keep the plant immune system properly sensitized. Atop this BR feedback regulatory loop, BR homeostasis is also achieved through circadian control and other environmental factors (e.g., light, temperature, and the circadian clock) (28). Our results showing that flg22-mediated responses can be modified by the availability of BRs suggest that the temporal integration of the BR pathway could directly influence the efficiency of MTI in a circadian manner. Thus, our results build on recent findings revealing a fundamental link between the circadian clock and plant immunity (29). We posit the existence of a narrow range of BR concentrations that sets the immune response for rapid deployment. It will be important to determine precisely how the inducible responses of the plant immune system interact with BR biosynthesis and signaling to integrate defenses responses over normal growth and development programs.
Because MTI and BR responses are coupled, we predict that BR and MAMP signaling pathways do not respond to ligand concentrations directly but rather indirectly, via the ratios of respective ligand concentrations. BAK1 has the attributes required to couple the relative phenotypic outputs in this model: BAK1 interacts physically with several PRRs and BRI1 (5, 20). BAK1 does not appear to bind ligand, and thus cannot directly measure ligand concentrations (13). BAK1 pools are dynamic and, most importantly, our data show that BAK1 dosage is important for two different signaling events to occur in a sustained way. Our data showing that BRI1 overexpression antagonizes BAK1-mediated MAMP signaling and inappropriate BAK1-mediated cell death is in agreement with our proposed model. We suggest that the capacity of BAK1 to evaluate the ratios of ligands can be skewed by ligand-independent activation of BR signaling: overexpressed BRI1 (35S::BRI1) could possibly titrate BAK1 from PRRs, activating BR response by mass action. Accordingly, when intracellular BR levels are increased (e.g., in 35S::DWF4), the inhibitory effects of BRs on FLS2 could take place through increased recruitment of BAK1 to BRI1. However, Albrecht and colleagues (30) have demonstrated that exogenous application of high concentrations of BR inhibits flg22-dependent signaling via a BAK1-independent mechanism. Thus, the reduced flg22 responses associated with an increase in endogenous BR levels through DWF4 overexpression could also operate through a BAK1-independent mechanism. However, comparing and understanding the effects of endogenously increased BR pools and exogenously applied BRs on flg22 response may be complex. For instance, exogenously applied BRs taken up by Arabidopsis roots can rescue shoot dwarfism, yet, it is known that BRs synthesized in the roots of WT plants cannot rescue the shoot dwarfing phenotype of a BR biosynthetic mutant (31). Nonetheless, our data complement those of Albrecht et al., who also clearly demonstrate that BR and MTI signaling are coupled in a unidirectional antagonistic manner (30).
Collectively, our results demonstrate that BR signaling can dictate the output of a plant–pathogen interaction. Hence, pathogens may perturb PRRs to exploit their crosstalk with BR signaling. To test this, we challenged our BR response gain-of-function plants with oomycetes and bacterial and fungal plant pathogens. The poised activation state of BRI1sud1 led to enhanced susceptibility to hemibiotrophic pathogens, despite enhanced MTI output responses. This enhanced susceptibility to hemibiotrophic pathogens was not apparent in plants in which BR signaling was achieved through increased BRI1 dosage. In contrast, very slight activation of the BR pathway triggered both BAK1-dependent and BAK1-independent changes during interactions with an obligate biotrophic pathogen. This is consistent with our proposal that BAK1 can act as mediator for the synergistic activities of BRs. These data are also consistent with the work of Albrecht et al. (30), who propose that BR signaling modulates plant immunity in a BAK1-independent manner. We propose that biotrophic pathogens have evolved virulence mechanisms to detect or create physiological states in which BR concentrations are optimal for pathogen success, perhaps centered on the modification of BR biosynthesis or signaling (32). Further studies aimed at understanding how BRs impact the concerted array of plant hormone signaling relationships with innate immune function will be instrumental to determine how the plant cell integrates normal growth signals with immune system function.
Experimental Procedures
Confocal Microscopy.
flg22 and BL treatments were as described previously (9). Confocal microscopy was performed with a Leica SP/2 inverted microscope, and image analysis was performed as previously described (16).
Protein Analysis.
Equal loading was ensured by Bradford protein quantification before loading. Monoclonal anti-GFP (Roche), anti-HA (Roche), and polyclonal anti-CHERRY (DsRed polyclonal; Clontech) were used at a dilution of 1:2,000. Polyclonal anti-PR1 antibodies were provided by Dan Kliebenstein (University of California, Davis, CA) and used at a dilution of 1:5,000.
MAMP Responses Assays.
These assays were as described (9). pCYP71A12::GUS staining assays following treatment with 1 μM flg22 were as described previously (15). Seedlings grown in the presence or absence of 5 μM BRZ (Chemiclones) were untreated or treated with 1 μM flg22 for 12 h before GUS staining. Washout indicates removal of BRZ during a short period before flg22 treatment. Representative examples from approximately 30 roots per condition are shown.
Pathogens and Cell Death Assays.
P. syringae (Pto DC3000 and hrcC) assays are described (18, 19). Hpa isolates Emwa1 and Noco2 were propagated as described (33) on the susceptible Arabidopsis ecotypes Ws and Col-0, respectively. Trypan blue staining to visualize cell death was performed as described previously (33).
Supplementary Material
Acknowledgments
We thank Gregory Vert and Marc Nishimura for discussions. Anti-PR1 and anti-FLS2 antibodies were gifts from Dan Kliebenstein and Cyril Zipfel, respectively. Yves Millet in the Ausubel laboratory provided the pCYP71A12::GUS line. We thank Cyril Zipfel and Sacco de Vries for sharing data and ideas before publication. This work was supported by National Science Foundation Grants IOS-0649389 (to J.C.) and IOS-0929410 (to J.L.D.) and National Institutes of Health Grant GM066025 (to J.L.D.), and by the Howard Hughes Medical Institute (J.C.). Y.B. was a Howard Hughes Medical Institute fellow of the Life Sciences Research Foundation and the recipient of the Phillipe Foundation award. Y.J. was supported by fellowships from the European Molecular Biology Organization, the F.M. Kirby Foundation, and the Stern Foundation. E.B-P. was supported by PhD and SWE fellowships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior and Conselho Nacional de Desenvolvimento Científico e Tecnológico, respectively, and by the Salk Institute. J.L.D. is a Howard Hughes Medical Institute–Gordon and Betty Moore Foundation Plant Science Investigator.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 7.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112840108/-/DCSupplemental.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nat Chem Biol. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 3.Ronald PC, Beutler B. Plant and animal sensors of conserved microbial signatures. Science. 2010;330:1061–1064. doi: 10.1126/science.1189468. [DOI] [PubMed] [Google Scholar]
- 4.Spoel SH, Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Vert G. Plant signaling: Brassinosteroids, immunity and effectors are BAK. Curr Biol. 2008;18:R963–R965. doi: 10.1016/j.cub.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Schulze B, et al. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem. 2010;285:9444–9451. doi: 10.1074/jbc.M109.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwessinger B, et al. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011;7:e1002046. doi: 10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaillais Y, Belkhadir Y, Balsemão-Pires E, Dangl JL, Chory J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA. 2011;108:8503–8507. doi: 10.1073/pnas.1103556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinchilla D, Shan L, He P, de Vries S, Kemmerling B. One for all: The receptor-associated kinase BAK1. Trends Plant Sci. 2009;14:535–541. doi: 10.1016/j.tplants.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakashita H, et al. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003;33:887–898. doi: 10.1046/j.1365-313x.2003.01675.x. [DOI] [PubMed] [Google Scholar]
- 12.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belkhadir Y, Chory J. Brassinosteroid signaling: A paradigm for steroid hormone signaling from the cell surface. Science. 2006;314:1410–1411. doi: 10.1126/science.1134040. [DOI] [PubMed] [Google Scholar]
- 14.Sekimata K, et al. Brz220 interacts with DWF4, a cytochrome P450 monooxygenase in brassinosteroid biosynthesis, and exerts biological activity. Biosci Biotechnol Biochem. 2008;72:7–12. doi: 10.1271/bbb.70141. [DOI] [PubMed] [Google Scholar]
- 15.Millet YA, et al. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell. 2010;22:973–990. doi: 10.1105/tpc.109.069658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaillais Y, et al. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25:232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorrain S, Vailleau F, Balagué C, Roby D. Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8:263–271. doi: 10.1016/S1360-1385(03)00108-0. [DOI] [PubMed] [Google Scholar]
- 18.Kim MG, et al. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 19.Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 20.Zipfel C. Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol. 2009;12:414–420. doi: 10.1016/j.pbi.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Geldner N, Robatzek S. Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 2008;147:1565–1574. doi: 10.1104/pp.108.120287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albrecht C, Russinova E, Kemmerling B, Kwaaitaal M, de Vries SC. Arabidopsis somatic embryogenesis receptor kinase proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol. 2008;148:611–619. doi: 10.1104/pp.108.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemmerling B, et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol. 2007;17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 24.Knoth C, Ringler J, Dangl JL, Eulgem T. Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol Plant Microbe Interact. 2007;20:120–128. doi: 10.1094/MPMI-20-2-0120. [DOI] [PubMed] [Google Scholar]
- 25.Sato M, et al. Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog. 2010;6:e1001011. doi: 10.1371/journal.ppat.1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaillais Y, Chory J. Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Biol. 2010;17:642–645. doi: 10.1038/nsmb0610-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael TP, et al. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 2008;6:e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, et al. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albrecht C, et al. Brassinosteroids inhibit pathogen-associated molecular pattern–triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA. 2012;109:303–308. doi: 10.1073/pnas.1109921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Symons GM, Reid JB. Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol. 2004;135:2196–2206. doi: 10.1104/pp.104.043034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kemen E, et al. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holt BF, III, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.