Abstract
ErbB3 harbors weak kinase activity, but strongly activates downstream phosphatidylinositol 3-kinase/Akt signaling through heterodimerization with and activation by other ErbB receptor tyrosine kinases. We report here that ErbB3 loss in the luminal mammary epithelium of mice impaired Akt and MAPK signaling and reduced luminal cell proliferation and survival. ERBB3 mRNA expression levels were highest in luminal mammary populations and lowest in basal cell/stem cell populations. ErbB3 loss in mammary epithelial cells shifted gene expression patterns toward a mammary basal cell/stem cell signature. ErbB3 depletion-induced gene expression changes were rescued upon activation of Akt and MAPK signaling. Interestingly, proliferation and expansion of the mammary basal epithelium (BE) occurred upon ErbB3 targeting in the luminal epithelium, but not upon its targeting in the BE. Multiple cytokines, including interleukin 6, were induced upon ErbB3 depletion in luminal epithelium cells, which increased growth of BE cells. Taken together, these results suggest that ErbB3 regulates the balance of differentiated breast epithelial cell types by regulating their growth and survival through autocrine- and paracrine-signaling mechanisms.
Keywords: mammary epithelial differentiation, ErbB3
Aberrant regulation of the ErbB family of receptor tyrosine kinases (RTKs) and their ligands is common in human cancers (1–4). This family consists of four members: HER1/ErbB1/EGFR (epidermal growth factor receptor), HER2/ErbB2/Neu, HER3/ErbB3, and HER4/ErbB4. Except for ErbB3, which has weak kinase activity, the ErbB RTKs exhibit dimerization-induced phosphorylation and catalytic activation. In response to ligand binding, ErbBs form homodimers and heterodimers with other ErbB coreceptors. ErbB3 relies on transphosphorylation by heterodimeric partners to induce signal transduction (5–7).
ErbB RTKs are required for breast development, although each receptor bears a unique spatiotemporal expression pattern. ErbB2 loss in the mammary epithelium delays ductal elongation during puberty and disorganizes cells within terminal end buds (TEBs) (8–10). EGFR and ErbB4 are not required for mammary ductal development. Rather, EGFR is expressed in the basal epithelium (BE) and in the mammary stroma, and ErbB4 is necessary for milk production (11, 12). Although classical knockout of mouse ErbB3 results in embryonic lethality (13), transplant experiments showed that ErbB3 drives growth of the mammary epithelium during puberty (8). Although the mechanism(s) by which ErbB2 and ErbB3 regulate growth of the ductal epithelium are currently unknown, such knowledge will impact our understanding of the earliest events contributing to the formation of ErbB2/HER2-amplified breast cancers, which account for 20–30% of all breast cancers. ErbB3-ErbB2 heterodimers are the most potent oncogenic ErbB-signaling pair due in part to strong ErbB3-induced phosphatidylinositol 3-kinase (PI3K) activation in response to ErbB3 tyrosine phosphorylation at six PI3K interaction motifs (14, 15).
To understand the role of ErbB3 in mammary gland development, we knocked out ERBB3 in mammary epithelial cells (MECs) and tumors using a mouse mammary tumor virus (MMTV)-driven Cre/lox system (ErbB3MMTV-KO) (16), which expresses Cre recombinase primarily in the mammary luminal epithelium (LE). We discovered that ErbB3 is required in the LE, but not in the BE, to support cell proliferation and survival. Loss of ErbB3 decreased MEK/MAPK and PI3K/Akt signaling and impaired differentiation of MECs along the luminal lineage. Definitive LE markers were decreased in the absence of ErbB3, and rescued upon reactivation of Akt and MEK. In contrast, the BE exhibited increased cell proliferation when ErbB3 was lost from the LE, suggesting communication between these two epithelial compartments. ErbB3-depleted LE cells produced mitogenic cytokines, which increased BE growth. These data demonstrate that ErbB3 maintains the LE at the luminal progenitor stage and regulates the balance of differentiated epithelial cell types within the mammary gland through both autocrine and paracrine mechanisms.
Results
ErbB3 Directs Growth, Survival, and Organization of the Developing Mammary Epithelium.
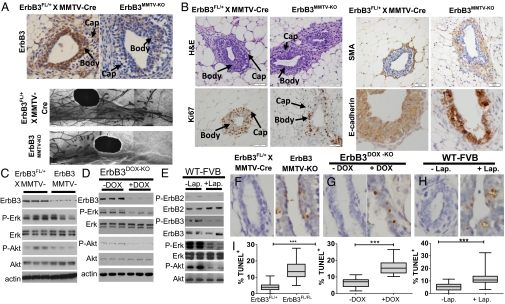
The mammary ductal epithelium begins lengthening distally through the mammary fat pad during puberty. Proliferation of the mammary epithelium and invasion through the fat pad occur primarily within club-shaped multicell-layered TEBs located at the distal-most aspects of the growing ducts. TEBs consist of two main cell layers: the cap layer, which gives rise to the BE, and the body layer, which gives rise to the LE. ErbB3 protein expression was higher in TEB body cells than in cap cells (Fig. 1A). ErbB3 was substantially reduced in TEBs of ErbB3MMTV-KO mice, which use MMTV-Cre transgene expression in the LE to cause genomic recombination at floxed ErbB3 alleles in ErbB3FL/FL mice (16). Ductal lengthening during puberty was delayed in 8-wk-old ErbB3MMTV-KO virgin female mice compared with heterozygous ErbB3FL/+ × MMTV-Cre controls (Fig. 1A), although ducts permeated the full length of the mammary fat pads by 16 wk of age in ErbB3MMTV-KO female mice (Fig. S1); ErbB3flox/+ heterozygotes showed no change in mammary phenotype compared with wild-type (WT) mice (16) and were used as controls. Decreased thickness of the TEB body cell layer was evident in ErbB3MMTV-KO samples (Fig. 1B) due in part to decreased cellular proliferation as measured by Ki67 immunohistochemistry (IHC). In heterozygous TEBs, E-cadherin IHC-defined body cells organized in a multilayered club-shaped pattern. TEBs in ErbB3MMTV-KO mice displayed thinning E-cadherin+ body cell layers with undulating patterns of disorganization. Although smooth muscle actin (SMA) identified a single layer of cap cells in heterozygous controls, ErbB3MMTV-KO TEBs harbored multiple layers of SMA+ cap cells. These results suggest ErbB3 loss in TEBs disrupts structural organization and the body/cap cell ratio, which may contribute to reduced ductal growth (Fig. 1A).
Fig. 1.
ErbB3 phosphorylation induces PI3K and MAPK signaling to promote cell growth, survival, and organization of mammary LE. (A) (Upper) IHC detection of ErbB3 in TEBs from 6-wk-old mice demonstrates expression in body cell layers with the lowest expression in the cap layer. ErbB3 expression is lost in ErbB3MMTV-KO TEBs. B, body layer; C, cap layer. (Lower) Whole-mount hematoxylin-stained mammary glands from 8-wk-old ErbB3MMTV-KO mice display reduced rate of ductal lengthening during puberty upon ErbB3 loss. (B) Histological analysis of ErbB3MMTV-KO and ErbB3Flox/+ control mammary glands from 6-wk-old mice by hematoxylin and eosin (H&E) staining and IHC detection of Ki67, SMA, and E-cadherin shows alterations in TEB body cell proliferation and structural organization. B, body layer; C, cap layer. (C–E) Western analysis of whole mammary lysates from virgin female mice was performed for the indicated proteins and phosphoproteins. (C) Six-week-old ErbB3MMTV-KO and ErbB3Flox/+ × MMTV-Cre mice. (D) Twelve-week-old ErbB3DOX-KO mice treated 7 d with or without DOX. (E) Six-week-old WT mice treated 3 wk with or without lapatinib. (F–H) TUNEL analysis of mammary glands. (F) Six-week-old ErbB3MMTV-KO and ErbB3Flox/+ × MMTV-Cre mice. (G) Twelve-week old ErbB3DOX-KO mice treated 7 d with or without DOX. (H) Six-week-old WT mice treated 3 wk with or without lapatinib. (Scale bars, 50 μm.) Average percentage of total epithelial nuclei that were TUNEL+ (±SD) was calculated from five random 400× fields/sample; n = 7 per condition, compared by t test.
ErbB3 Phosphorylation and Signaling Drive Cell Survival in the Mature and Pubertal Luminal Mammary Epithelium.
Loss of ErbB3 in the mammary epithelium of 6-wk-old virgin female ErbB3MMTV-KO mice resulted in decreased P-Akt in the mammary gland (Fig. 1C). Because Akt is phosphorylated in response to PI3K activation, these results suggest that ErbB3 is required for PI3K activation and Akt signaling in the LE. Similarly, acute doxycycline (DOX)-induced ErbB3 depletion in the adult LE achieved using double-transgenic mice expressing DOX-inducible Cre (MMTV-rtTA × TetOp-Cre) (17, 18) crossed with ErbB3FL/FL mice to produce ErbB3DOX-KO mice also decreased P-Akt levels and decreased P-MAPK levels in the mammary gland (Fig. 1D). ErbB3 tyrosine phosphorylation was blocked by treating WT females with the ErbB1/2 inhibitor lapatinib (100 mg/kg/d). Lapatinib treatment decreased P-ErbB2, P-ErbB3, P-Akt, and P-MAPK compared with controls (Fig. 1E), suggesting that PI3K and MAPK signaling in the mammary gland requires heterodimeric activation of ErbB3.
Because Akt regulates cell survival, we examined apoptosis in mammary glands using TUNEL analysis. Constitutive or inducible loss of ErbB3 increased LE cell death in pubertal or adult mice, respectively (Fig. 1 F and G). Also, inhibition of ErbB3 phosphorylation using lapatinib increased the fraction of TUNEL+ cells (Fig. 1H). Fig. 1 demonstrates that ErbB3 is required within the quiescent mature LE to maintain cell survival and that ErbB3 phosphorylation by ErbB family members drives PI3K and MAPK signaling and cell survival in the untransformed mammary epithelium.
Mammary Gland ErbB3 Expression Is Highest in Luminal Epithelium.
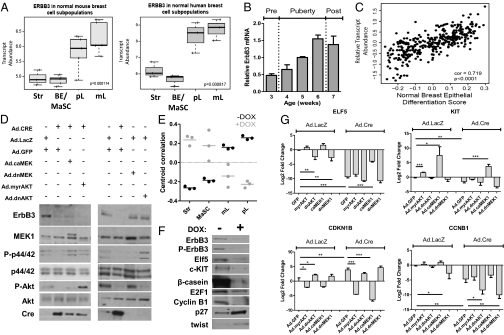
Antibodies against cell-surface markers of distinct mammary epithelial populations are used to sort freshly digested mammary glands into epithelial subpopulations by flow cytometry. Using this approach on mouse and human tissues, gene expression analysis of each mammary epithelial subpopulation was performed (19, 20). We analyzed available datasets and found that low levels of ERBB3 mRNA were present in the BE/mammary stem cell (BE/MaSC) population, whereas robust ERBB3 expression was detected in the mature luminal population (“mL” in Fig. 2A). Committed luminal progenitors also expressed high levels of ERBB3, consistent with the notion that ERBB3 expression is induced in the mammary LE population prior to commitment to the luminal lineage. In addition, mammary ERBB3 expression was higher in mid-to-late puberty (6–7 wk) in mice, when specification and maintenance of the LE is maximized (Fig. 2B). Given that luminal breast cancers are thought to arise from transformed luminal breast epithelial cells and that ErbB3 expression is highest in untransformed LE cells, we examined ERBB3 mRNA expression in a panel of human breast cancers. Interestingly, ERBB3 expression positively correlated with more differentiated breast cancers (r2 = 0.72, P < 0.0001) defined by the previously published luminal differentiation genomic model (21) (Fig. 2C). These expression data suggest that ErbB3 correlates with luminal differentiation of the mammary epithelium.
Fig. 2.
ErbB3 expression in mammary LE is required to maintain its differentiated molecular phenotype. (A) Examination of ERBB3 relative mRNA across normal mouse (Left) and human (Right) mammary cell fractions demonstrated that ErbB3 expression is highest in LE cell populations. Str, stromal; BE/MaSC, basal epithelium/mammary stem cell; pL, luminal progenitor; mL, mature luminal cell. Differences in ErbB3 expression were tested by ANOVA. (B) ERBB3 mRNA expression is maximal during late puberty in mice. Data and plot were extracted and generated from the National Center for Biotechnology Information Gene Expression Omnibus dataset GDS2721 using the probeset 1434606_at. Details of the study were previously published (34). (C) Positive correlation between ERBB3 gene expression in normal breast specimens and a molecular score of the degree of luminal differentiation (20). (D) Western analysis of ErbB3DOX-KO organoids treated with or without DOX × 7 d confirmed results of cDNA microarrays showing altered expression of the genes encoding Elf5, cyclin B1, E2F1, β-casein, p27, and Twist. (E) Rank correlation of expression patterns comparing mammary subpopulation signatures to gene expression data derived from ErbB3-deficient PMECs and control WT cells. The Spearman's correlation is plotted for each treated sample (n = 3) and control (n = 3) across all four mammary subpopulation signatures. Bars represent the mean correlation of the three experiments. (F and G) ErbB3flox/flox PMECs were infected ex vivo with Ad.Cre or Ad.LacZ and with adenovirally encoded myrAkt, dnAkt, caMek1, or dnMek1. Ad.GFP was used as a secondary negative control for dual infections. Seven days later, cell lysates and RNA were harvested. (F) Western analysis of cell lysates for the indicated proteins. (G) qRT-PCR to measure ELF5, KIT, CDKN1B, and CCN1B. *P < 0.05, ***P < 0.01, ***P < 0.001.
ErbB3 Is Required to Maintain Expression of an LE Signature.
We examined gene expression changes occurring in response to ErbB3 depletion in primary mammary epithelial organoids grown in 3D Matrigel, allowing us to assess molecular changes occurring in the mammary epithelium without potentially confounding stromal gene expression changes (22, 23). DOX-induced loss of ErbB3 decreased Akt phosphorylation in ErbB3DOX-KO primary MECs (PMECs) in monolayer culture (Fig. S2A). ErbB3DOX-KO organoids recapitulated the phenotypic consequences of ErbB3 loss seen in vivo, including formation of smaller acinar structures with less complexity, increased cell death, and decreased proliferation (Fig. S2 B–D). Gene expression analysis of organoids treated for 10 d with or without DOX ex vivo identified 403 genes with altered expression (equal to or more than twofold, false discovery rate-adjusted P ≤ 0.05) in response to ErbB3 ablation. Gene Ontology analysis implicated the products of many such genes in cell cycle progression, including up-regulation of cell cycle inhibitors (e.g , DDIT3, GADD45a, CDKN1B) and down-regulation of cell cycle activators (e.g , CCNB1, PLK1, CCNE1) in response to ErbB3 ablation. Many such genes are included in a “proliferation cluster,” a core set of genes identified by cDNA profiling whose expression correlated with rapid cell proliferation in large breast cancer datasets (24). Western analysis of ErbB3DOX-KO MECs confirmed up-regulation of the cell cycle inhibitor p27 (CDKN1B) and down-regulation of cyclin B1 (CCNB1) (Fig. 2D).
Genes associated with luminal differentiation were also down-regulated in ErbB3-deficient mammary glands, including the milk protein β-casein, which was also down-regulated at the protein level (Fig. 2D). Decreased gene expression of E74-like factor 5 (ELF5), a transcription factor required for growth and differentiation of the luminal alveolar population (25–27), and the RTK gene KIT were also observed. Elf5 and c-KIT have emerged as definitive markers of the luminal progenitor population (19). Elf5 and c-KIT down-regulation was confirmed by Western blot (Fig. 2D).
Next, we used the previously described gene expression signatures for distinct cell types within the hierarchical model of mammary epithelial differentiation (Fig. 2 A and B) to query expression data derived from organoid cultures expressing or lacking ErbB3. Untreated organoids retaining ErbB3 displayed expression patterns that correlated positively with expression signatures from mature luminal cells and luminal progenitors (Fig. 2E) and negatively correlated with the BE/MaSC signature. However, DOX-induced loss of ErbB3 in organoids shifted gene expression patterns, resulting in a negative correlation with luminal signatures, but a positive correlation with the BE/MaSC signature.
ErbB3-PI3K and -MAPK Signaling Regulate Expression of Luminal Markers.
We investigated the signaling pathways downstream of ErbB3 that regulate expression of luminal molecular markers and proliferation cluster genes. ErbB3flox/flox PMECs were infected ex vivo with adenoviral Cre (Ad.Cre) or lacZ control (Ad.LacZ) in combination with adenoviral constitutively active Akt (myrAkt), dominant-negative Akt (dnAkt), active MEK1 (caMEK1), or dominant-negative MEK1 (dnMEK1). Western analysis demonstrated that Cre-mediated loss of ErbB3 in Ad.Cre-infected PMECs decreased P-Akt and P-MAPK (Fig. 2F). Expression of myrAkt1 and caMEK1 restored P-Akt and P-MAPK levels, respectively. Conversely, P-Akt and P-MAPK were decreased upon expression of dnAkt1 and dnMek1, respectively, even in the presence of ErbB3.
Expression of ELF5 and KIT were chosen as surrogate markers of luminal differentiation, as these are definitive markers of luminal progenitors and were decreased upon ErbB3 depletion (Fig. 2D). Transcript levels of ELF5 and KIT were reduced upon ErbB3 ablation in PMECs (Fig. 2G), but were partially rescued upon expression of caMEK1 and myrAkt, although caMEK1 produced a greater effect on ELF5 and KIT up-regulation compared with myrAkt. Conversely, ELF5 and KIT expression were inhibited upon expression of dnAkt1 and dnMEK1 despite continued expression of ErbB3.
Down-regulation of CCNB1 and up-regulation of CDKN1B were seen in ErbB3-depleted cells. Expression of caMEK1 or myrAkt prevented down-regulation of CCNB1 and up-regulation CDKN1B in response to ErbB3 ablation. Conversely, dnMEK1 and dnAkt increased expression of CDKN1B and decreased CCNB1, despite continued expression of ErbB3. These results highlight the importance of ErbB3-PI3K signaling within the luminal lineage and establish an important role for ErbB3-MAPK signaling in controlling proliferation in this compartment.
Expansion of the Basal Epithelium in Response to ErbB3 Depletion.
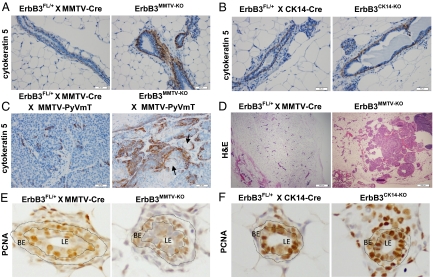
Because ErbB3 loss expanded the SMA+ cap cell layer, which gives rise to the mature BE, we used cytokeratin 5 (CK5) staining to detect BE cells in ErbB3MMTV-KO mammary glands. In 12-wk-old virgin female mice, the CK5+ BE was a single cell layer in heterozygous controls, but was expanded to multiple cell layers in age-matched ErbB3MMTV-KO samples (Fig. 3A). To test if loss of ErbB3 in the BE could also directly expand the BE population, we crossed ErbB3Flox/Flox mice to transgenic mice expressing Cre recombinase via the basal cytokeratin 14 (CK14) promoter (ErbB3CK14-KO mice). ErbB3 loss in CK14-expressing basal cells did not disrupt mammary ductal elongation (Fig. S3A) and did not alter TEB cellular organization in ErbB3CK14-KO mice (Fig. S3B). Ki67 IHC did not reveal changes in cell proliferation due to BE knockout of ErbB3. Importantly, the SMA+ cap cell layer in developing TEBs appeared normal, and the CK5+ basal cell population was unaltered in ErbB3CK14-KO mice compared with heterozygous controls.
Fig. 3.
Expansion of the basal epithelium in untransformed mammary glands and mammary tumors lacking ErbB3. (A–C) Representative images of CK5 IHC in 12- to 20-wk-old virgin females. (A) ErbB3MMTV-KO and ErbB3FL/+ × MMTV-Cre. (B) ErbB3CK14-KO and ErbB3FL/+ × CK14-Cre. (C) ErbB3MMTV-KO × MMTV-PyVmT and ErbB3FL/+ × MMTV-Cre × MMTV-PyVmT. n = 7/group. Arrows in C indicate keratinizing squamous metaplasia. (D) Representative image of squamous metaplasia observed in ErbB3MMTV-KO mammary glands, but not in ErbB3FL/+ × MMTV-Cre glands. (E and F) Representative images of PCNA IHC in ErbB3MMTV-KO, ErbB3FL/+ × MMTV-Cre, ErbB3CK14-KO, and ErbB3FL/+ × CK14-Cre mammary glands. Basal epithelium (BE) is outlined. Quantification is provided in Fig. S3D.
We next examined CK5 expression in ErbB3-deficient MMTV-PyVmT tumors (28). The CK5+ population exhibited profound expansion in ErbB3MMTV-KO × PyVmT tumors compared with heterozygous controls (Fig. 3C). Keratinizing squamous metaplasia was evident in 7/20 ErbB3-deficient MMTV-PyVmT tumors, but was not identified in ErbB3flox/+ × MMTV-PyVmT tumors (Fig. 3C). Similarly, keratinizing squamous transdifferentiation of the mammary epithelium was seen in 3/12 ErbB3MMTV-KO mice (Fig. 3D), but was not observed in ErbB3CK14-KO samples (0/12). Therefore, loss of ErbB3 in the LE alters the balance of luminal and basal cells in both normal and transformed mammary epithelium.
It is possible that ErbB3 loss in LE cells indirectly promotes BE growth. In support of this idea, increased BE proliferation was observed in ErbB3MMTV-KO mammary glands compared with heterozygous controls [assessed using proliferating cell nuclear antigen (PCNA) IHC] (Fig. 3E), but not in ErbB3CK14-KO glands (Fig. 3F). Fewer PCNA+ LE cells were seen in ErbB3MMTV-KO mammary glands compared with heterozygous controls and with ErbB3CK14-KO samples, consistent with the decreased body cell proliferation seen in ErbB3-deficient TEBs (Fig. 1B).
IL-6 Secreted by ErbB3-Deficient Luminal Cells Increases Myoepithelial Cell Growth.
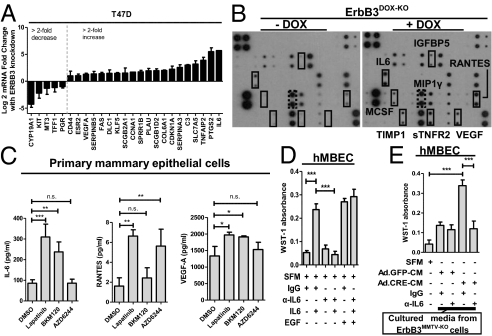
We next tested the hypothesis that secreted factors from luminal cells in response to ErbB3 loss could drive expansion of the BE. Using a quantitative PCR array platform, we identified profound up-regulation of genes encoding secreted factors in ErbB3-siRNA–transfected T47D human luminal breast cancer cells (Fig. 4A), such as IL-6 and vascular endothelial growth factor-A (VEGF-A). Conditioned media from T47D luminal breast cancer cells transfected with siRNA targeting ERBB3 and from DOX-treated ErbB3DOX-KO PMECs was analyzed by cytokine array (Fig.4B and Fig. S4). ErbB3 loss induced secretion of IL-6, VEGF, and other highlighted cytokines. Inhibition of ErbB3 phosphorylation in ErbB3DOX-KO PMECs using the ErbB1/2 inhibitor lapatinib increased secretion of IL-6, VEGF-A, and RANTES/CCL5 (Fig. 4C). Inhibition of PI3K using the pan-p110 inhibitor BKM120 similarly increased secretion of IL-6 and VEGF-A, but not of RANTES. The MEK1 inhibitor AZD6244 increased RANTES secretion from ErbB3DOX-KO MECs, but did not significantly alter expression of IL-6 or VEGF-A. Therefore, interruption of ErbB3-PI3K and ErbB3-MAPK signaling in LE cells increases the secretion of cytokines in distinct ways.
Fig. 4.
IL-6 produced by ErbB3-deficient luminal epithelium increases growth of basal epithelium. (A) RNA from T47D cells transfected with control or ErbB3-specific siRNA was used for reverse transcription and quantitative PCR for 84 individual transcripts. Values shown are the mean fold-change (log2) relative to control-siRNA–transfected cells, normalized to GAPDH. Only transcripts demonstrating greater than twofold changes are shown. Samples were analyzed in triplicate, and experiments were repeated three times. Bars represent mean of three experiments ± SD. (B) Primary ErbB3DOX-KO MECs were cultured in serum-free medium (SFM) for 7 d with or without DOX. Cultured media was assessed by cytokine array to detect secreted factors. Cytokines outlined in solid black indicate DOX-induced cytokines, and dashed lines indicate cytokines down-regulated by DOX. (C) Primary ErbB3DOX-KO PMECs were cultured in SFM ± lapatinib (1 μM), BKM120 (0.5 μM), or AZD6244 (1 μM). Cultured media were collected and assayed by ELISA to measure IL-6 (Left), RANTES (Center), and VEGF-A (Right). Results indicate mean ± SD (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001. (D) Human mammary BE cells were cultured in SFM with neutralizing antibody (Ab) against IL-6 (10 μg/mL) or control IgG and then treated +/− IL-6 (10 ng/mL) or EGF (10 ng/mL). Cell viability was measured after 96 h using WST-1 assay. Bars represent mean ± SD. ***P < 0.001. (E) ErbB3FL/FL × MMTV-PyVmT mammary tumor cells were infected with Ad.Cre or Ad.GFP and then cultured for 7 d in SFM. Conditioned media were collected, filtered, and used to culture human mammary BE cells for 96 h and BE cell viability was measured by WST-1 assay. Where indicated, BE cells were pretreated for 1 h with neutralizing IL-6 Ab (10 μg/mL) or control IgG. Bars represent mean ± SD. ***P < 0.001.
Recombinant human IL-6 (10 ng/mL) in serum-free medium increased growth of primary human mammary basal epithelial cells (hMBEC) 5.2-fold compared with untreated cells (Fig. 4D), an effect that was impaired by a neutralizing IL-6 antibody (10 μg/mL). EGF was used as a positive control and similarly enhanced growth of BE cells, but was not inhibited by IL-6 antibody. Conditioned media from ErbB3-deficient LE cells infected with Ad.Cre increased growth of hMBECs to a greater extent than did media from ErbB3-expressing controls infected with Ad.GFP (Fig. 4E). Addition of IL-6 antibody to cultured media from ErbB3-deficient tumor cells abrogated its ability to increase BE cell growth, demonstrating that IL-6 is secreted by ErbB3-deficient breast cells increasing growth of neighboring BE cells.
Discussion
The data presented here suggest (i) that ErbB3 signaling specifies and/or maintains the luminal phenotype of breast epithelium and (ii) that loss of ErbB3 from the LE drives expansion of the BE subpopulation. These conclusions are supported by the decreased presence of body cells within ErbB3-deficient TEBs (Fig. 1B), increased LE cell death upon ErbB3 loss or impaired ErbB3 phosphorylation (Fig. 1F), and an ErbB3 loss-induced shift in genome-wide expression patterns away from previously defined luminal signatures (Fig. 2E). These observations are consistent with the fact that ErbB3 expression is highest in mature luminal and luminal progenitor cells and lowest in the basal cell subpopulations of the breast (Fig. 2A). In addition, the luminal progenitor population markers ELF5 and KIT (19, 20) are decreased in response to loss of ErbB3, suggesting that hierarchical differentiation of the mammary epithelium along the luminal lineage requires ErbB3 for luminal specification and/or maintenance before expansion of committed luminal progenitors.
ErbB3 ablation from the mammary LE increased cell death while decreasing cell growth. This is in contrast to a report suggesting that mammary epithelial ErbB3 loss decreased cell survival but did not alter cell growth (8). The reasons underlying this discrepancy are currently unclear, although numerous differences in the models used may contribute (8). For example, differing genetic backgrounds used in the two studies may be a factor. Also, results presented here describe development of intact mammary glands in the context of a competent immune system, compared with the previous report, which used orthotopically transplanted embryonic mammary buds in immunocompromised mice (8). Finally, ErbB3 loss in this study was directed to specific mammary epithelial compartments: the LE (via MMTV-Cre) or the BE (via CK14-Cre). In contrast, the previously published model (8) lacked ErbB3 in all mammary epithelial populations. We have shown here that ErbB3 impacts distinct mammary epithelial populations in profoundly different ways, potentially contributing to this phenotypic discrepancy.
MMTV-PyVmT tumors express high levels of the luminal cytokeratins 8 and 18 and low levels of CK5, a cytokeratin associated with basal-like breast cancers, consistent with expression analyses clustering the MMTV-PyVmT tumor model with the luminal subtype of human breast cancers (29). Although loss of ErbB3 in the mammary epithelium decreases the rate of tumor formation in MMTV-PyVmT mice (28), ErbB3-deficient tumors eventually formed, exhibiting an increased CK5+ tumor cell population (Fig. 3C). Because BE and LE cells arise from common stem cells, it is possible that loss of ErbB3 prevents differentiation along the luminal lineage, forcing cells to differentiate into the basal lineage as a default. Our results do not disprove this possibility, but strongly support an alternative scenario in which ErbB3 loss from the LE causes cytokine secretion, which causes growth of neighboring basal cells (Fig. 4).
Interestingly, ErbB3 loss increased CK5+ cells in both untransformed mammary epithelia and mammary tumors (Fig. 3A), suggesting that ErbB3 directs cell fate decisions in cancers. This could have important implications regarding molecular classifications of breast cancers. Advances in molecular analysis of primary tumors make clear that different subtypes of human breast cancer exist (30). Increasing evidence suggests that the molecular subtype of a given breast cancer may be a reflection of the cell type from which that cancer originates (20, 31–33). Therefore, it is critical to understand the signaling pathways that define the epithelial ontogeny of the mammary gland and how these pathways may be used within cancers that arise from each cell type. Our results demonstrate that ErbB3 is required within the luminal lineages of the breast. Further study of how ErbB3 and other differentiation signals may influence cell fate decisions within preneoplastic mammary glands will support our understanding of how tumors adopt specific molecular and clinical phenotypes, information that may be used to treat or prevent breast cancer.
Materials and Methods
All mouse experiments were approved by the Vanderbilt Institutional Animal Care and Use Committee. All models used, including genetically engineered mouse models and cell lines, are described in SI Materials and Methods. Detailed methods for Western analysis, RT-PCR, quantification of cell growth, and histological analyses can be found in SI Materials and Methods. Detailed materials and methods can be found in SI Materials and Methods. Additionally, Figs. S1–S4, associated legends, and references can also be found in Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01CA143126 (to R.S.C.), R01CA80195 (to C.L.A.), and F32CA121900 and K99CA142899 (to T.W.M.); Breast Cancer Specialized Programs of Research Excellence Grants P50CA98131 (to Vanderbilt University) and P50CA058223 (to University of North Carolina-Lineberger Cancer Center); Vanderbilt-Ingram Cancer Center Support Grant P30CA68485; Susan G. Komen for the Cure Grant KG100677 (to R.S.C.); American Cancer Society Clinical Research Professorship CRP-07-234 (to C.L.A.); the Lee Jeans Translational Breast Cancer Research Program (C.L.A.); and Stand Up to Cancer/American Association for Cancer Research Dream Team Translational Cancer Research Grant SU2C-AACR-DT0209 (to C.L.A.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Gene expression microarray data have been uploaded to the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession ID GSE32129).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115802109/-/DCSupplemental.
References
- 1.Abd El-Rehim DM, et al. Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br J Cancer. 2004;91:1532–1542. doi: 10.1038/sj.bjc.6602184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Citri A, Yarden Y. EGF-ERBB signalling: Towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 3.Bazley LA, Gullick WJ. The epidermal growth factor receptor family. Endocr Relat Cancer. 2005;12(Suppl 1):S17–S27. doi: 10.1677/erc.1.01032. [DOI] [PubMed] [Google Scholar]
- 4.Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284:99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- 5.Carraway KL, III, et al. The erbB3 gene product is a receptor for heregulin. J Biol Chem. 1994;269:14303–14306. [PubMed] [Google Scholar]
- 6.Pinkas-Kramarski R, et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HH, Vijapurkar U, Hellyer NJ, Bravo D, Koland JG. Signal transduction by epidermal growth factor and heregulin via the kinase-deficient ErbB3 protein. Biochem J. 1998;334:189–195. doi: 10.1042/bj3340189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson-Fisher AJ, et al. ErbB3 is required for ductal morphogenesis in the mouse mammary gland. Breast Cancer Res. 2008;10:R96. doi: 10.1186/bcr2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrechek ER, White D, Muller WJ. Targeted disruption of ErbB2/Neu in the mammary epithelium results in impaired ductal outgrowth. Oncogene. 2005;24:932–937. doi: 10.1038/sj.onc.1208230. [DOI] [PubMed] [Google Scholar]
- 10.Jackson-Fisher AJ, et al. ErbB2 is required for ductal morphogenesis of the mammary gland. Proc Natl Acad Sci USA. 2004;101:17138–17143. doi: 10.1073/pnas.0407057101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long W, et al. Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development. 2003;130:5257–5268. doi: 10.1242/dev.00715. [DOI] [PubMed] [Google Scholar]
- 12.Wiesen JF, Young P, Werb Z, Cunha GR. Signaling through the stromal epidermal growth factor receptor is necessary for mammary ductal development. Development. 1999;126:335–344. doi: 10.1242/dev.126.2.335. [DOI] [PubMed] [Google Scholar]
- 13.Erickson SL, et al. ErbB3 is required for normal cerebellar and cardiac development: A comparison with ErbB2-and heregulin-deficient mice. Development. 1997;124:4999–5011. doi: 10.1242/dev.124.24.4999. [DOI] [PubMed] [Google Scholar]
- 14.Holbro T, et al. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu S, et al. Gene targeting of ErbB3 using a Cre-mediated unidirectional DNA inversion strategy. Genesis. 2006;44:477–486. doi: 10.1002/dvg.20243. [DOI] [PubMed] [Google Scholar]
- 17.Gunther EJ, et al. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 2002;16:283–292. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- 18.Wagner KU, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 20.Lim E, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fata JE, et al. The MAPK(ERK-1,2) pathway integrates distinct and antagonistic signals from TGFalpha and FGF7 in morphogenesis of mouse mammary epithelium. Dev Biol. 2007;306:193–207. doi: 10.1016/j.ydbio.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 25.Harris J, et al. Socs2 and elf5 mediate prolactin-induced mammary gland development. Mol Endocrinol. 2006;20:1177–1187. doi: 10.1210/me.2005-0473. [DOI] [PubMed] [Google Scholar]
- 26.Oakes SR, et al. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev. 2008;22:581–586. doi: 10.1101/gad.1614608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, et al. Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO J. 2005;24:635–644. doi: 10.1038/sj.emboj.7600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muraoka-Cook RS, et al. ErbB3 ablation impairs PI3K/Akt-dependent mammary tumorigenesis. Cancer Res. 2011;71:3941–3951. doi: 10.1158/0008-5472.CAN-10-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 32.Asselin-Labat ML, et al. Delineating the epithelial hierarchy in the mouse mammary gland. Cold Spring Harb Symp Quant Biol. 2008;73:469–478. doi: 10.1101/sqb.2008.73.020. [DOI] [PubMed] [Google Scholar]
- 33.Lindeman GJ, Visvader JE. Insights into the cell of origin in breast cancer and breast cancer stem cells. Asia Pac J Clin Oncol. 2010;6:89–97. doi: 10.1111/j.1743-7563.2010.01279.x. [DOI] [PubMed] [Google Scholar]
- 34.McBryan J, Howlin J, Kenny PA, Shioda T, Martin F. ERalpha-CITED1 co-regulated genes expressed during pubertal mammary gland development: implications for breast cancer prognosis. Oncogene. 2007;26:6406–6419. doi: 10.1038/sj.onc.1210468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.