Abstract
The molecular pathways that are activated and contribute to physiological remodeling of skeletal muscle in response to endurance exercise have not been fully characterized. We previously reported that ∼800 gene transcripts are regulated following 6 wk of supervised endurance training in young sedentary males, referred to as the training-responsive transcriptome (TRT) (Timmons JA et al. J Appl Physiol 108: 1487–1496, 2010). Here we utilized this database together with data on biological variation in muscle adaptation to aerobic endurance training in both humans and a novel out-bred rodent model to study the potential regulatory molecules that coordinate this complex network of genes. We identified three DNA sequences representing RUNX1, SOX9, and PAX3 transcription factor binding sites as overrepresented in the TRT. In turn, miRNA profiling indicated that several miRNAs targeting RUNX1, SOX9, and PAX3 were downregulated by endurance training. The TRT was then examined by contrasting subjects who demonstrated the least vs. the greatest improvement in aerobic capacity (low vs. high responders), and at least 100 of the 800 TRT genes were differentially regulated, thus suggesting regulation of these genes may be important for improving aerobic capacity. In high responders, proangiogenic and tissue developmental networks emerged as key candidates for coordinating tissue aerobic adaptation. Beyond RNA-level validation there were several DNA variants that associated with maximal aerobic capacity (V̇o2max) trainability in the HERITAGE Family Study but these did not pass conservative Bonferroni adjustment. In addition, in a rat model selected across 10 generations for high aerobic training responsiveness, we found that both the TRT and a homologous subset of the human high responder genes were regulated to a greater degree in high responder rodent skeletal muscle. This analysis provides a comprehensive map of the transcriptomic features important for aerobic exercise-induced improvements in maximal oxygen consumption.
Keywords: training responsive transcriptome, aerobic capacity, low responder
complex organisms have evolved to incorporate the benefits of the steep thermodynamic gradient afforded by atmospheric oxygen (44) and to deal with the obligatory reactive nature of oxygen chemistry (50). This suggests that the strong link between low aerobic capacity and risk of disease reflects gene-environment interactions that may have evolved to regulate the safe transport and use of molecular oxygen for the purpose of energy transfer. The ability to increase the oxygen transport and utilization capacity in mammals, as stimulated by exercise training, is extremely variable (8, 37, 57, 62), and this heterogeneity affords an opportunity to dissect out the molecular governors of aerobic capacity adaptation (6). It is a primary assumption of the present study that heterogeneity in aerobic capacity is the consequence of variation distributed across a large number of genes. Single genetic variants can be factors in cardiorespiratory adaptation (3), but each gene is expected to contribute only weakly to normal physiological variation. Determination of gene-product expression (42) on the other hand integrates the influence of multiple genomic sources of variation (21, 55), thus providing a powerful approach that can be complemented by genomewide or targeted sequence analysis.
Recently, the view of conventional genomics has been reappraised to recognize the potential importance of noncoding DNA sequences (29). MicroRNAs (miRNAs) are an example of a class of noncoding RNA molecules, which act as posttranscriptional regulators of protein synthesis from mRNA (12, 15). miRNAs are ∼22-nucleotide RNA molecules that bind to the 3′-untranslated region (UTR) of a target mRNA to repress translation, or alternatively promote mRNA degradation. So far, miRNAs are known to play a role in, for example, organism development (40), cell differentiation (14, 18, 43), and cancer (10). However, a role for miRNAs in differentiated tissue during changes in aerobic capacity in humans has not yet been documented. We hypothesized that modulation of miRNA abundance will play an important role in controlling the phenotypic changes in human skeletal muscle during gains in aerobic capacity.
We previously reported an average 14% improvement in maximal aerobic capacity (V̇o2max) in response to 6 wk of endurance training, while individual responses ranged from a ∼3% decline to ∼28% improvement (62). Gene ontology (GO) analysis using EASE (28) indicated that a large number of the 700 upregulated genes (from a total of 800 regulated) were related to extracellular matrix [>50 genes; false discovery rate (FDR) < 1%] and immune response-related genes (>60 genes; FDR < 1%) (58). The present article utilizes this novel database of transcript changes to discover the potential regulatory molecules (e.g., transcription factors/miRNAs) that coordinate this complex network of genes and examines in closer detail the biology of the training-responsive transcriptome (TRT). In particular we focus on the gene networks formed from those genes for which altered expression appears to be required for physiological adaptation to occur in vivo. In the present analysis, we determined how variations in aerobic training-induced gene expression changes in skeletal muscle are associated with variable adaptability for increased aerobic capacity in response to 6 wk of endurance training in humans. We then profiled a new rodent model, where animals were selected and bred for their ability to increase their aerobic capacity in response to treadmill running. Further, we compared the list of genes that were regulated more substantially when physiological change was greatest (57), with a panel of single-nucleotide polymorphisms (SNPs) from the HERITAGE Family Study (7).
MATERIALS AND METHODS
Experimental factors.
Twenty-four young sedentary healthy men volunteered to take part in the study; full details have previously been reported (58). Each of the volunteers provided written informed consent. The study was approved by the Ethics Committee at the Karolinska Institutet, Stockholm, Sweden, and conformed to the Declaration of Helsinki. Subjects performed a 6-wk fully supervised training program consisting of four 45-min cycling sessions per week at an intensity corresponding to 70% of pretraining V̇o2max (100% compliance). Physiological measurements and vastus lateralis muscle biopsies were taken before the training program and 24 h after the last training session, as previously described (56, 57). This article represents a detailed analysis of the genomics data generated. Genotyping data are derived from the HERITAGE Family Study. The HERITAGE study cohort in this analysis consists of 473 white subjects (230 males and 243 females) from 99 nuclear families who completed at least 58 of the prescribed 60 exercise training sessions. The study design and inclusion criteria have been described previously (7).
Affymetrix microarray process.
Total RNA was extracted from frozen muscle using TRIzol reagent. Frozen pieces were homogenized for 60 s in 1 ml of TRIzol using a 7-mm Polytron aggregate (PT-DA 2107, Kinematica AG, Switzerland) adapted to a Polytron homogenizer (PT-2100) running at maximum speed. Total RNA concentration was determined at least at two different dilutions on a spectrophotometer. RNA concentration and quality were controlled using a Bioanalyser. In vitro transcription (IVT) was made using the Bioarray high-yield RNA transcript labeling kit (P/N 900182, Affymetrix). Unincorporated nucleotides from the IVT reaction were removed using the RNeasy column (Qiagen). Hybridization, washing, staining, and scanning of the arrays were performed according to manufacturer's instructions (Affymetrix). The array design can be found at https://www.affymetrix.com. As a means to control the quality of the individual arrays, all arrays were examined using hierarchical clustering and NUSE to identify outliers before statistical analysis in addition to the standard quality assessments including scaling factors and housekeeper 5′/3′ ratios. The raw data have been submitted to GEO database (GSE18583).
Array analysis methods.
The U133+2 platform microarray data were subjected to global normalization using MAS5.0. Using the MAS5.0-generated present-absent calls can improve the sensitivity of the differential gene expression analysis by improving the power while potentially removing some genuinely expressed genes (16). We chose to retain probe sets with a minimum of 12 from 48 chips declaring a “present” detection, on the basis that there will be subject-to-subject variability and that some genes may be only expressed either before or following training. The normalized log2-file was analyzed with the Significance Analysis of Microarrays (SAM) in R (http://www-stat.stanford.edu/∼tibs/SAM/) (25). SAM provides a list of “significant” genes and an estimate of the FDR, which represents the percentage of genes that could be identified by chance and is comparable to a P value corrected for the number of initial comparisons, a process called multiple testing correction. SAM assigns a score to each gene/probe set as an index of relative difference between groups. For the data presented in the manuscript, genes were considered significantly changed following training when a delta value corresponding to the number of false significant genes of 5% (q value) and an average fold change (FC) of 1.5 were achieved.
We have previously demonstrated that it can be difficult to predict the impact of applying arbitrary filtering criteria before statistical analysis (34). We therefore relied on several statistical models to present, analyze, and interpret our data with and without arbitrary prefiltering of gene lists (22, 49, 60). Using SAM analysis we identified significantly modulated genes (1.5 FC and FDR < 5%). We then analyzed the ∼1,000 differentially expressed probe sets using the EASE gene ontology analysis tool (28) to identify overrepresented gene ontology classifications. Gene ontology analysis demonstrated that our differential expression data set generated using the U133+2 platform produced a data set with similar biological features as our earlier study using the U95A-E platform (59) thus providing a substantial confirmation of the differentially expressed gene list. The topGO package (2) from the Bioconductor suite was used to investigate functional enrichment in gene lists. Significant enrichment was assessed using the classicFishers and the elimFishers function in topGO. The former implements a classical Fishers test while the latter removes genes contributing to significance at a given level of the GO hierarchy before testing the next level.
miRNA microarrays.
Total RNA pooled from eight subjects before and after 6 wk of exercise training was used for the initial detection of microRNA expression using locked nucleic acid (LNA)-based arrays (n = 8 arrays). The quality of the total RNA was examined on an Agilent 2100 Bioanalyser and had an absorbance ratio of A260/A280 > 1.7 and RNA integrity number (RIN) scores above 8.0. The Nanodrop ND-1000 Spectrophotometer was used to measure the concentration of the RNA. The microarrays were custom made using the miRCURY LNA microRNA array ready-to-spot probe set V8.1 from Exiqon (Vedbaek, Denmark). The Exiqon probe set consists of 1,488 custom-made capture probes that are enhanced using LNA technology to normalize the melting temperature (Tm) of the capture probes, as insertion of 1 LNA molecule into the capture probes increases the Tm by 2–8°C. The hybridization temperature can thereby be increased, allowing for high specificity but also sensitivity, which until now has been a problem when detecting miRNAs. Probes were diluted to a concentration of 10 μM in 150 mM sodium phosphate (pH 8.5), 0.01% N-lauroylsarcosine (Sigma), and were printed onto Codelink slides (GE Healthcare) using the Biorobotics Microgrid II with split pins; the diameter of each spot was 170 μm. Arrays contained three replicates of each probe, and two separate arrays were printed per slide. Slides were blocked and washed according to the manufacturer's recommended protocol (GE Healthcare).
The total RNA was labeled with Hy3 dye according to the manufacturer's protocol using the miRCURY LNA array labeling kit from Exiqon with the inclusion of the miRCURY LNA array spike-in miRNAs in the labeling reaction. The spike-in microRNA kit consists of synthetic miRNAs at different concentrations that can be used as controls for the labeling reaction. For the labeling reaction, 1.75 μg of total RNA was incubated with the Hy3 dye, labeling enzyme, and spike-in microRNAs, in a total volume of 20 μl, for 1 h at 0°C. The enzyme was then heat-inactivated at 65°C for 15 min. The sample volume was reduced by centrifugal evaporation, 2× hybridization buffer was added, and the samples were incubated at 95°C for 5 min protected from light. The samples were briefly spun down and filtered through a 0.45-μm Durapore filter (Millipore). Samples were spun down again and 20-μl samples were loaded onto the arrays by capillary force using a coverslip (M-series, Erie Scientific). The arrays were incubated at 60°C for 16 h, then washed briefly in 60°C wash buffer A, rinsed in wash buffer B, followed by a 2-min wash in wash buffer B and a 2-min wash in wash buffer C, according to the manufacturer's instructions. The arrays were spun for 2 min at 1,000 rpm followed by immediate scanning using an Agilent G2565BA microarray scanner. Data were analyzed using QuantArray software using the fixed-circle approach. Background-subtracted signal intensities were normalized in R using quantile normalization, followed by SAM. The FDR was < 15%, and only miRNAs modulated more than 1.5-fold were selected.
Detection of microRNA expression by using real-time PCR.
Using the Taqman MicroRNA assays (Applied Biosystems) that detect mature miRNA, the expression of four miRNAs that showed differential expression with the microarray experiment were validated. The assays use a miRNA-specific looped-primer for the reverse transcription reaction, thus extending the miRNA sequence, thereby enabling detection with Taqman real-time PCR. The following miRNAs were validated in individual RNA derived from 16 subjects (8 high responders and 8 low responders) before and after 6 wk of exercise training: mir-1 (cat. no. 4373161), mir-101 (cat. no. 4373159), mir-133a (cat. no. 4373142), and mir-455 (cat. no. 4378098). For each miRNA to be measured, 10 ng of total RNA was reverse transcribed using the miRNA-specific looped primer using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, PN4366597) according to manufacturer's instructions. In short, a master mix was prepared resulting in a final concentration in a 15-μl volume of 1 mM dNTP, 50 U MuLV Multiscribe reverse transcriptase, 1× reverse transcription buffer, and 3.8 U RNase Inhibitor. RNA was added, and aliquots were made followed by addition of the miRNA-specific reverse transcription primer to a final concentration of 1× RT-primer. The reactions were gently mixed and briefly spun down and run in a PTC-100 (MJ Research) with conditions at 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min.
For the qPCR, the TaqMan 2× Universal PCR Master Mix, No AmpErase UNG was used (Applied Biosystems, PN4324020), and the assay was performed according to manufacturer's instructions. The samples were run on a 7900HT Fast Real-Time PCR System (Applied Biosystems) on the 9,600 emulation mode in triplicates of 20 μl/well. Real-time PCR conditions were 95°C for 10 min followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. The miRNA expression levels were normalized to the small nuclear RNA, RNU48 (cat. no. 4373383), which was previously verified not to vary between subjects or samples (22) and did not vary in the present study either. All reactions were run singleplex and analyzed and quantified using the ΔCt method. Data are expressed as percent change from “pre” levels. miRNA data obtained by qPCR were analyzed using a paired t-test to compare differences between ΔCt values (Ct value of target miRNA minus Ct value of RNU48) before and after 6 wk of aerobic exercise training. For all analyses, P < 0.05 was considered significant. Statistical calculations were performed using Excel.
Rodent aerobic capacity models.
Artificial, two-way, selective breeding is being used to develop rat models of low response to training (LRT) and high response to training (HRT). The founder population was 152 rats from the genetically heterogeneous N:NIH stock (27) obtained from the National Institutes of Health (USA). Low and high response lines were established by selectively breeding females and males with either the least or greatest change in total running capacity as a result of exercise training. Ten families were maintained within each selected line, and within-family rotational breeding was practiced to minimize the rate of inbreeding (∼1.25% per generation).
The protocol for selection on exercise training response included: 1) pretest the rats for intrinsic (untrained) maximal treadmill running capacity, 2) train the rats for 8 wk, and 3) posttest the rats for maximal running capacity. The measure for maximal treadmill exercise capacity was based on a speed-ramped treadmill run to exhaustion, a standard procedure in our laboratory (32), patterned after clinical exercise tests (45). The response to exercise training was calculated as the change in running capacity (meters) before and after 8 wk of training beginning when the rats are ∼3 mo of age. Treadmill training occurred 3 days per week for a total of 24 sessions with the treadmill slope constantly at a 15° incline. Treadmill speed started at 10 m/min and was increased every 2 days of training by 1 m/min to reach a peak training speed of 20 m/min on day 21. Training duration was started at 20 min and incremented daily to a final of 31.5 min on day 24. The entire protocol, from the pretesting period through training to the posttesting period, is completed by the time rats reach 6 mo of age.
Male LRT (n = 6) and HRT (n = 5) rats from the 6th generation of selective breeding were used to investigate first evidence for inherent gene expression differentials in soleus muscle samples using qPCR. Tissue samples from rats were harvested when the rats were ∼15 mo of age (after their use as breeders was completed) and homogenized in TRIzol (Invitrogen, Carlsbad, CA) using a motor-driven homogenizer (Polytron, Kinematica, Newark, NY). Total RNA was isolated according to the manufacturer's protocol, dissolved in 30 μl RNase-free water, and quantified using a Spectrophotometer (Pharmacia Biotech). One microgram total RNA in a reaction volume of 40 μl was reverse-transcribed using reverse transcription reagents (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. Random hexamers were used for first-strand cDNA synthesis. Detection of 26 high responder genes (based on conservation of a homologous transcript) was performed using an ABI-PRISM 7900 Sequence Detection system (Applied Biosystems). Primers were designed using Primer Express software (Applied Biosystems) or obtained using the Universal Probe Library (Roche Applied Science). Primers were synthesized by Invitrogen. A preoptimized primer and probe assay for 18S rRNA was used as an endogenous control (Applied Biosystems). Primers and probes were premixed with SYBR GREEN PCR Master Mix (Applied Biosystems) and applied to 96-well MicroAmp Optical barcode plates (Applied Biosystems). cDNA aliquots of 4 μl were added in triplicates. Thermal cycling conditions were: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 65°C. Twofold-dilutions series were performed for all target genes and endogenous controls to determine the amplification efficiency.
Female LRT (n = 3) and HRT rats (n = 3) from generation 10 were used for microarray analysis. Cardiac and skeletal muscle samples from these rats were harvested at 5 mo of age, 24 h after the last training session. Snap-frozen tissue samples were shipped on dry ice for RNA preparation and analysis. All phenotype and breeding procedures were approved by the Animal Care and Use Committee at University of Michigan. Affymetrix microarray analysis was carried out as described above, using the Rat Genome 230 2.0 Array and RNA isolated from soleus and left ventricle (n = 4).
Genotype investigation of the high responder genes in HERITAGE.
The study cohort in this analysis consists of 473 white subjects (230 males and 243 females) from 99 nuclear families who completed at least 58 of 60 exercise sessions of a 20-wk customized training program. The study design as well as inclusion and exclusion criteria have been described previously (7). The study protocol had been approved by each of the Institutional Review Boards of the HERITAGE Family Study research consortium. Written informed consent was obtained from each participant.
Genomic DNA was prepared from permanent lymphoblastoid cells by commercial DNA extraction kit (Gentra Systems, Minneapolis, MN). The SNPs were selected mainly from the International HapMap Caucasian (CEU) database. The tagSNPs were genotyped using the Illumina (San Diego, CA) GoldenGate chemistry and Sentrix Array Matrix technology on the BeadStation 500GX. Genotype calling was done with the Illumina BeadStudio software, and each call was confirmed manually. All the SNPs had call rates above 98% with the average being 99.9%. For quality control purposes, each 96-sample array matrix included one sample in duplicate and 47 samples were genotyped in duplicate on different arrays. None of the SNPs showed Mendelian errors in the HERITAGE families, and all SNPs were in Hardy-Weinberg equilibrium (tested using the exact HWE test implemented in the PEDSTATS software package). In addition, six CEPH control DNA samples (NA10851, NA10854, NA10857, NA10859, NA10860, NA10861; all samples included in the HapMap Caucasian panel) were genotyped. Concordance between the replicates as well as with the SNP genotypes from the HapMap database was 100%.
A chi-square test was used to verify whether the observed genotype frequencies were in Hardy-Weinberg equilibrium. Associations between the individual tagSNPs and cardiorespiratory fitness phenotypes were analyzed using a variance components and likelihood ratio test-based procedure in the QTDT software package. The total association model of the QTDT software utilizes a variance-components framework to combine phenotypic means model and the estimates of additive genetic, residual genetic, and residual environmental variances from a variance-covariance matrix into a single likelihood model. The evidence of association is evaluated by maximizing the likelihoods under two conditions: the null hypothesis (L0) restricts the additive genetic effect of the marker locus to zero (βa = 0), whereas the alternative hypothesis does not impose any restrictions on βa. The quantity of twice the difference of the log likelihoods between the alternative and the null hypotheses {2[ln(L1) − ln (L0)]} is distributed as χ2 with 1 df (difference in number of parameters estimated). V̇o2max training responses were adjusted for age, sex, baseline body weight, and baseline value of V̇o2max.
GSEA analysis.
An alternative approach to analyzing microarray data is to avoid using artificial significance and fold change threshold but rather look at where genes (or more correctly, gene sets) rank in a rank list of all differentially expressed genes on the array. The method known as Gene Set Enrichment Analysis (GSEA) is one such approach (51). We used this to identify the occurrence of transcription factor binding sites in the differentially expressed genes. Gene sets in this case were a list of genes (typically 200–400 in size) that contained a particular DNA binding sequence. In many cases, the protein transcription factor that binds to a particular sequence is known and thus can be immediately identified, and a literature search can generate information to underpin the validity of the finding. In other cases only the sequence is known and the gene set. In these situations the gene set can be analyzed using gene ontology analysis to provide an indication as to the function of the gene set.
Network analysis.
We used the web-based bioinformatics tool, Ingenuity pathway analysis (IPA, http://www.ingenuity.com), which is based on >1.7 million published PubMed articles, to discover networks regulated by exercise training. IPA is a knowledge database generated from the peer-reviewed scientific publications that enables discovery of biological networks in gene expression data and determines the functions most significant to those networks. The ∼1,000 differentially expressed probe sets (i.e., the TRT), high responder gene set, and the rodent muscle differentially expressed genes were used for network analysis. Gene name identifiers or Affymetrix probe set IDs were uploaded into IPA and queried against all other genes stored in the IPA knowledge database. Each Affymetrix probe set ID was mapped to its corresponding gene identifier in the IPA knowledge database. Probe sets representing genes having direct interactions with genes in the IPA knowledge database are called “focus” genes, which were then used as a starting point for generating functional networks. In reporting our findings, we focus on networks with a high confidence limit and thus represent strong evidence for a given biological pathway being activated by aerobic training. It should be noted, however, that while the database extends the interpretation beyond mRNA transcript levels (i.e., network genes are not required to be differentially expressed), the data are finite and reflect current knowledge.
miRNA target prediction and gene ontology class.
The binding of miRNAs to target mRNAs occurs in the “seed” region of the miRNA, representing nucleotide 2–7 of the 5′-end of the mature miRNA and the 3′-UTR of the mRNA. For miRNA target prediction we used a predicted list of conserved miRNA target genes obtained from PicTar (http://pictar.bio.nyu.edu/). When PicTar had no predictions available, a conserved miRNA target gene list from Targetscan (http://www.targetscan.org/, release 4.0) was used, as these predictions generally were similar on this occasion. For miRNAs differentially expressed by exercise training we used Gene Ontology analysis to obtain an overview of the main classes of biological functions of genes predicted to be targets for a specific miRNA. EASE analysis (using version 2.0, 2,006 annotation files) predicts biological functions of target genes for the miRNA of interest and enlists a FDR and an EASE score, which we calculated using 500 permutations/iterations and a 25 FDR percentile. The topGO package (2) from the Bioconductor suite was used to investigate functional enrichment in gene lists as described above.
RESULTS
Generating a molecular profile of aerobic adaptation.
Using significance analysis of microarrays (SAM), 1,000 probe sets representing ∼800 genes (FDR < 4.5%; >1.5 FC) were differentially regulated during in vivo aerobic adaptation [referred to hereafter as the training-responsive transcriptome, TRT; see Supplementary data page 1 (58)]1 when muscle samples from 20 of the top 24 subjects with the best improvements in aerobic capacity were utilized. This group selection was done to maximize the statistical power of the gene chip analysis, as we have already established that the lowest responders in this exercise paradigm do not modulate their mRNA levels consistently with those that demonstrate a biological response (57). Gene ontology (GO) analysis (28) is presented in Supplementary data page 2 with parameters annotated based on PANTHER (54) and level 5 of the GO structure. This analysis was consistent with our previous analysis using a smaller number of subjects and with less complete coverage of the transcriptome (57). Collagens, muscle fiber, and blood vessel development are examples of significant ontologies associated with the TRT (Supplementary data page 2).
Bibliometric network analysis using Ingenuity found integrin signaling was the top-ranked canonical pathway (Supplementary Fig. S1, BH corrected P = 1.06 × 10−8) followed by immune cell regulatory processes. This suggests that communication with the extracellular matrix was regulated in proportion to the observed physiological change (57). In addition, Pathway analysis identified that a tissue remodeling pathway (68) regulated by transforming growth factor (TGF)-β was a central feature of the TRT (Supplementary Fig. S2a). Further, extracellular signal-regulated kinase (ERK) (Supplementary Fig. S2b) and calcium/calmodulin-dependent protein kinase (CamKII) (Supplementary Fig. S2c) also appeared connected to upregulated genes in the TRT. As we have identified a large number of regulated mRNAs, it should be possible to determine, using informatics, which distinct protein and RNA factors regulate the TRT and thus drive a new generation of research on the biological regulation of muscle adaptation to endurance exercise.
Transcriptional and posttranscriptional regulation of the TRT.
Trans-acting DNA binding proteins can interact at cis-acting transcription factor (TF) binding sites. When found to be present in sets of coregulated genes (e.g., the TRT), such binding sites provide evidence that they coordinate changes in mRNA abundance (25). Using the TRT we aimed to use structural features within this gene list to locate 1) transcription factor binding sites; and 2) miRNA target sites, to develop a model of the in vivo process. Our goal was to ascertain genes that may coordinate the TRT in vivo and so deduce potential signals that coordinate the TRT. GSEA identified runt-related transcription factor-1 (RUNX1) binding sites as being highly enriched in the TRT. This is consistent with the observation that RUNX1 protein expression is regulated during skeletal muscle remodeling (63). Table 1 presents additional TF DNA-binding sequences, including paired box gene-3 (PAX3) and sex determining region Y box-9 (SOX9), which also link to aspects of muscle or tissue remodeling.
Table 1.
GSEA of DNA binding sites on the aerobic training-responsive transcriptome
| DNA Target Motif | FDR | Transcription Factor | Top Ranking Biological Profile (GO) | PubMed Information | Predicted miRNA Targeting |
|---|---|---|---|---|---|
| RACCACAR | 0.097 | RUNX1 | Transcription factor activity (P < 0.00001) | Prevents muscle wasting during inactivity | mir-144, mir-101 |
| RGAANNTTC | 0.096 | HSF1 | Cell communication (P < 0.00001) | Heat/stress response/ageing muscle? | None |
| STTTCRNTTT | 0.022 | IRF1 | Immune response (P < 0.00001) | Vascular proliferation, NO production | None |
| CGTSACG | 0.093 | PAX3 | Regulation of transcription (P < 0.00001) | Stem cell myogenesis | mir-1/206, mir-144, mir-92 |
| GGGNNTTTCC | 0.046 | NFkB | Immune response (P < 0.001) | Cytokine/ myo-differentiation | None |
| CATTGTYY | 0.079 | SOX9 | Calmodulin binding (P < 0.0001) | Cell differentiation | mir-101 |
| RGAGGAARY | 0.051 | PU.1/SPI1 | Signal transducer activity (P < 0.001) | Differentiation of circulating stem cells | None |
| YYCATTCAWW | 0.093 | POU1F1 | Transcription factor activity (P < 0.00001) | Hormone regulation | None |
| GGCKCATGS | 0.072 | Unknown | Cell differentiation (P < 0.0001) | ||
| YNTTTNNNANGCARM | 0.067 | Unknown | Development (P = 0.051) | ||
| GGGNRMNNYCAT | 0.057 | Unknown | Organelle organization and biogenesis (P < 0.05) | ||
| RRCCGTTA | 0.045 | Unknown | Protein binding (P < 0.05) | ||
| GCTNWTTGK | 0.078 | Unknown | Development (P < 0.01); extracellular matrix (P < 0.01) |
DNA sequences identified using Gene Set Enrichment Analysis (GSEA) with a false discovery rate (FDR) < 10% and the transcription factors believed to bind the identified sequence. Bfore callout queryPubMed search identified roles of the transcription factors when focusing on muscle physiology. Of the significantly changed microRNAs (miRNAs) following exercise training, several are predicted to target and repress the identified transcription factors. Only modulated miRNAs targeting conserved sites in the 3′-untranslated region (UTR) of the transcription factors are shown, as predicted by Targetscan (http://www.targetscan.org/).
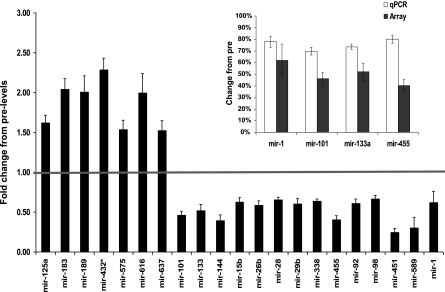
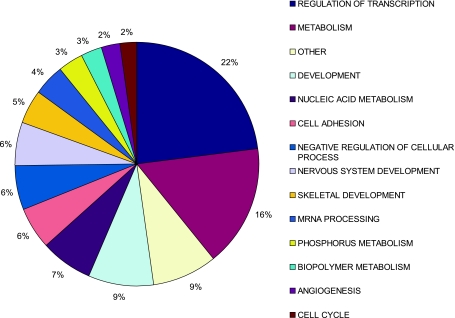
We also screened for all currently known (cloned and verified) miRNAs expressed, both before and after exercise training, using the LNA Exiqon miRNA microarray platform. There was a greater propensity for reduced rather than increased miRNA expression (>1.5 FC, FDR < 15%, 14 miRNAs vs. 7 miRNAs, respectively, Fig. 1) and TaqMan-based qPCR corroborated the changes observed in the array data (Fig. 1, inset). Next, we took all predicted miRNA target genes (33, 35), filtered them for muscle expression using Affymetrix data filtered for absent calls removed (22), and utilized GO analysis (28) to facilitate direct comparison with the Affymetrix GO analysis. Approximately 22% of miRNA target genes belonged to the regulation of transcription ontology (Fig. 2), while ∼16% were incorporated in the metabolism ontology group [consistent with the GSEA analysis of the GO Affymetrix arrays, which demonstrated that coordinated upregulation in OXPHOS genes was the top-ranked gene set (62)]. These findings suggest that loss of miRNA repression of translation increases mitochondrial enzyme expression and in vivo lipid oxidation (62) in the absence of substantial increases in individual metabolic gene mRNA abundance.
Fig. 1.
Differentially expressed microRNAs (miRNAs) in human skeletal muscle following 6 wk of aerobic exercise training. Exiqon locked nucleic acid (LNA) miRNA array data were analyzed using Significance Analysis of Microarrays (SAM) with a false discovery rate (FDR) <15% and fold change (FC) > 1.5. Inset represents miRNA expression validated by qPCR (change from pre: P < 0.01 for all 4 miRNAs). qPCR data are presented as mean percent change from the pretraining expression level ± SE while the array data are plotted as fold change, where <1 equals a reduction in gene expression.
Fig. 2.
Gene Ontology (GO) classes of predicted miRNA gene targets for human miRNAs differentially expressed following 6 wk of aerobic exercise training. The main ontology classes represented were regulation of transcription (∼22% of target genes) and metabolism (∼16% of target genes).
Notably, RUNX1, PAX3, and SOX9 (located using GSEA above) have a total of 33 conserved miRNA target sites in their 3′-UTR (Supplementary data page 4), which include 4 of the 14 miRNAs that were downregulated by aerobic training (Supplementary Fig. S3). Conversely, none of the upregulated miRNAs target RUNX1, PAX3, and/or SOX9. Indeed, loss of expression of multiple miRNAs can potentially increase the protein abundance in vivo (22). This analysis, derived from two independent chip technologies and independent informatic prediction processes, helps confirm RUNX1, PAX3, and SOX9 as potential modulators of muscle aerobic adaptation in vivo. Thus it would appear that in vivo regulated miRNAs bind at conserved (24) target sites in the 3′-UTR of RUNX1-PAX3-SOX9, indicating reduced miRNA directed suppression. This may explain why 167 RUNX1-PAX3-SOX9 target genes were upregulated in our mRNA expression data set (GSEA leading edge genes; Supplementary data pages 5–7 and Supplementary Fig. S3).
“Reverse engineering” of wet-lab experimental findings (mRNA and miRNA) using bibliometric analysis is a potentially powerful approach, relying on large numbers of published gene-gene and protein-protein relationships. We therefore utilized the Ingenuity database (11) to “reverse-engineer” wet-lab experimental findings using bibliometric data and determine 1) whether RUNX1, PAX3, and SOX9 would produce a gene network that recapitulated the mRNA TRT, and 2) whether this would include many genes already known to be regulated by aerobic training. The RUNX1-PAX3-SOX9 bibliometric analysis-derived network contained ∼300 genes that had either one direct or indirect connection. Of this, 15% belonged to the TRT, a figure that rises to 32% when one considers additional genes regulated during aerobic exercise identified from the wider literature (26, 30, 31, 39, 47) (Supplementary Fig. S3 and Supplementary data page 8).
The TRT is modulated in proportion to physiological adaptation.
We utilized the intersubject variability in mRNA expression responses to exercise training to infer cause-effect relationships between physiological and molecular changes. A list of ∼100 genes (Table 2), the “high responder” genes, that were differentially modulated for the lowest (n = 8) and highest responders for aerobic capacity (n = 8) (62) was derived from the TRT. The ontological profile was one of basement membrane proteins and the individual genes that drive this enrichment are also listed in Supplementary data page 9. This ontology analysis was true even when the TRT, rather than the entire muscle transcriptome, was utilized as the GO background file.
Table 2.
Aerobic training-responsive transcriptome gene changes: high vs. low responders (P < 0.05)
| Affymetrix ProbeSet | Gene Symbol | Gene Title | High Responders | Low Responders |
|---|---|---|---|---|
| 211980_at | COL4A1 | Collagen, type IV, alpha 1 | 5.8 ± 0.79† | 3.1 ± 0.29 |
| 228915_at | DACH1 | Dachshund homolog 1 (Drosophila) | 5.3 ± 0.96† | 1.8 ± 0.46 |
| 225681_at | CTHRC1 | Collagen triple helix repeat containing 1 | 4.7 ± 0.78 | 2.8 ± 0.51 |
| 204114_at | NID2 | Nidogen 2 (osteonidogen) | 3.6 ± 0.37* | 2.3 ± 0.26 |
| 211966_at | COL4A2 | Collagen, type IV, alpha 2 | 3.5 ± 0.66* | 1.9 ± 0.08 |
| 226140_s_at | OTUD1 | OTU domain containing 1 | 3.4 ± 1.07* | −1.1 ± 0.23 |
| 200665_s_at | SPARC | Secreted protein, acidic, cysteine-rich (osteonectin) | 3.1 ± 0.40 | 2.2 ± 0.23 |
| 1558214_s_at | CTNNA1 | Catenin (cadherin-associated protein), alpha 1, 102 kDa | 3.0 ± 0.58 | 1.5 ± 0.45 |
| 202202_s_at | LAMA4 | Laminin, alpha 4 | 2.9 ± 0.36* | 1.9 ± 0.12 |
| 205047_s_at | ASNS | Asparagine synthetase | 2.7 ± 0.60* | 1.2 ± 0.18 |
| 230061_at | TM4SF18 | Transmembrane 4 L six family member 18 | 2.7 ± 0.34* | 1.7 ± 0.20 |
| 213139_at | SNAI2 | Snail homolog 2 (Drosophila) | 2.6 ± 0.44* | 1.6 ± 0.14 |
| 202409_at | IGF2 −3′UTR | Insulin-like growth factor 2 (somatomedin A) | 2.5 ± 0.22* | 1.9 ± 0.18 |
| 215277_at | PCDH1 | Protocadherin 1 | 2.5 ± 0.60 | 1.2 ± 0.22 |
| 227415_at | LOC283508 | Hypothetical protein LOC283508 | 2.5 ± 0.37* | 1.4 ± 0.12 |
| 224833_at | ETS1 | v-ets erythroblastosis virus E26 oncogene Homolog 1 (avian) | 2.5 ± 0.38* | 1.4 ± 0.10 |
| 202878_s_at | CD93 | CD93 molecule | 2.4 ± 0.31* | 1.6 ± 0.09 |
| 214660_at | PELO | Pelota homolog (Drosophila) | 2.4 ± 0.24* | 1.5 ± 0.23 |
| 203934_at | KDR | Kinase insert domain receptor (a type III receptor tyrosine kinase) | 2.4 ± 0.32* | 1.5 ± 0.21 |
| 212365_at | MYO1B | Myosin IB | 2.4 ± 0.48* | 1.3 ± 0.12 |
| 233025_at | PDZD2 | PDZ domain containing 2 | 2.4 ± 0.53* | 1.1 ± 0.15 |
| 204821_at | BTN3A3 | Butyrophilin, subfamily 3, member A3 | 2.4 ± 0.55* | 1.1 ± 0.19 |
| 225056_at | SIPA1L2 | Signal-induced proliferation-associated 1 like 2 | 2.4 ± 0.39* | 1.3 ± 0.27 |
| 244840_x_at | DOCK4 | Dedicator of cytokinesis 4 | 2.4 ± 0.49* | −1.1 ± 0.13 |
| 213592_at | APLNR | Apelin receptor | 2.3 ± 0.25 | 1.7 ± 0.22 |
| 202270_at | GBP1 | Guanylate binding protein 1, interferon-inducible, 67 kDa | 2.3 ± 0.29* | 1.4 ± 0.21 |
| 1557286_at | 1557286_at | 2.2 ± 0.44* | 1.1 ± 0.19 | |
| 204115_at | GNG11 | Guanine nucleotide binding protein (G protein), gamma 11 | 2.2 ± 0.17* | 1.7 ± 0.13 |
| 202008_s_at | NID1 | Nidogen 1 | 2.2 ± 0.22* | 1.6 ± 0.16 |
| 1554233_at | C1QTNF9 | C1q and tumor necrosis factor related protein 9 | 2.2 ± 0.32* | 1.2 ± 0.13 |
| 212298_at | NRP1 | Neuropilin 1 | 2.2 ± 0.19* | 1.4 ± 0.19 |
| 203324_s_at | CAV2 | Caveolin 2 | 2.1 ± 0.26* | 1.5 ± 0.13 |
| 236335_at | 236335_at | 2.1 ± 0.20* | 1.5 ± 0.12 | |
| 210105_s_at | FYN | FYN oncogene related to SRC, FGR, YES | 2.1 ± 0.19† | 1.4 ± 0.12 |
| 1553530_a_at | ITGB1 | Integrin, beta 1 | 2.1 ± 0.31* | 1.3 ± 0.11 |
| 217979_at | TSPAN13 | Tetraspanin 13 | 2.1 ± 0.19* | 1.4 ± 0.14 |
| 201655_s_at | HSPG2 | Heparan sulfate proteoglycan 2 | 2.1 ± 0.25 | 1.5 ± 0.07 |
| 210029_at | IDO1 | Indoleamine 2,3-dioxygenase 1 | 2.1 ± 0.25* | 1.4 ± 0.15 |
| 237026_at | 237026_at | 2.1 ± 0.35* | 1.1 ± 0.24 | |
| 1555812_a_at | ARHGDIB | Rho GDP dissociation inhibitor (GDI) beta | 2.1 ± 0.21* | 1.4 ± 0.08 |
| 215034_s_at | TM4SF1 | Transmembrane 4 L six family member 1 | 2.0 ± 0.14* | 1.6 ± 0.15 |
| 229127_at | 229127_at | 2.0 ± 0.22* | 1.3 ± 0.16 | |
| 201215_at | PLS3 | Plastin 3 (T isoform) | 2.0 ± 0.21* | 1.4 ± 0.11 |
| 227443_at | C9orf150 | Chromosome 9 open reading frame 150 | 2.0 ± 0.21† | 1.3 ± 0.09 |
| 213627_at | MAGED2 | Melanoma antigen family D, 2 | 2.0 ± 0.28* | 1.1 ± 0.12 |
| 1566472_s_at | RETSAT | Retinol saturase (all-trans-retinol 13,14-reductase) | 2.0 ± 0.45 | 1.0 ± 0.14 |
| 216438_s_at | TMSB4X/TMSL3 | Thymosin b30eta 4, X-linked /// thymosin-like 3 | 2.0 ± 0.14* | 1.4 ± 0.15 |
| 238025_at | MLKL | Mixed lineage kinase domain-like | 2.0 ± 0.31* | 1.1 ± 0.13 |
| 239849_at | 239849_at | 1.9 ± 0.16† | 1.3 ± 0.11 | |
| 212097_at | CAV1 | Caveolin 1, caveolae protein, 22 kDa | 1.9 ± 0.14† | 1.4 ± 0.10 |
| 202619_s_at | PLOD2 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 1.9 ± 0.41* | −1.1 ± 0.06 |
| 209790_s_at | CASP6 | Caspase 6, apoptosis-related cysteine peptidase | 1.9 ± 0.50 | −1.1 ± 0.13 |
| 201753_s_at | ADD3 | Adducin 3 (gamma) | 1.9 ± 0.19* | 1.3 ± 0.17 |
| 206201_s_at | MEOX2 | Mesenchyme homeobox 2 | 1.9 ± 0.11* | 1.5 ± 0.10 |
| 205619_s_at | MEOX1 | Mesenchyme homeobox 1 | 1.9 ± 0.22* | 1.3 ± 0.14 |
| 204683_at | ICAM2 | Intercellular adhesion molecule 2 | 1.9 ± 0.20* | 1.3 ± 0.09 |
| 201399_s_at | TRAM1 | Translocation associated membrane protein 1 | 1.9 ± 0.16* | 1.3 ± 0.15 |
| 212372_at | MYH10 | Myosin, heavy chain 10, nonmuscle | 1.8 ± 0.17* | 1.3 ± 0.10 |
| 204331_s_at | MRPS12 | Mitochondrial ribosomal protein S12 | 1.8 ± 0.14* | 1.4 ± 0.13 |
| 206332_s_at | IFI16 | Interferon, gamma-inducible protein 16 | 1.8 ± 0.15* | 1.4 ± 0.10 |
| 202345_s_at | FABP5 | Fatty acid binding protein 5 (psoriasis-associated) | 1.8 ± 0.11* | 1.4 ± 0.14 |
| 226022_at | SASH1 | SAM and SH3 domain containing 1 | 1.8 ± 0.22 | 1.3 ± 0.11 |
| 201315_x_at | IFITM2 | Interferon induced transmembrane protein 2 (1-8D) | 1.7 ± 0.11† | 1.3 ± 0.10 |
| 201669_s_at | MARCKS | Myristoylated alanine-rich protein kinase C substrate | 1.7 ± 0.10† | 1.3 ± 0.06 |
| 38671_at | PLXND1 | Plexin D1 | 1.7 ± 0.16 | 1.3 ± 0.10 |
| 217757_at | A2M | Alpha-2-macroglobulin | 1.7 ± 0.09* | 1.4 ± 0.07 |
| 210762_s_at | DLC1 | Deleted in liver cancer 1 | 1.7 ± 0.17* | 1.2 ± 0.06 |
| 1558015_s_at | ACTR2 | ARP2 actin-related protein 2 homolog (yeast) | 1.7 ± 0.23* | 1.1 ± 0.11 |
| 238852_at | 238852_at | 1.7 ± 0.29* | −1.1 ± 0.13 | |
| 201776_s_at | KIAA0494 | KIAA0494 | 1.6 ± 0.12* | 1.3 ± 0.09 |
| 241929_at | CD36 | CD36 | 1.6 ± 0.11† | 1.2 ± 0.06 |
| 213932_x_at | HLA-A | Major histocompatibility complex, class I, A | 1.6 ± 0.11 | 1.3 ± 0.13 |
| 213415_at | CLIC2 | Chloride intracellular channel 2 | 1.6 ± 0.09* | 1.2 ± 0.14 |
| 208836_at | ATP1B3 | ATPase, Na+/K+ transporting, beta 3 polypeptide | 1.6 ± 0.16* | 1.1 ± 0.04 |
| 211911_x_at | HLA-B | Major histocompatibility complex, class I, B | 1.6 ± 0.08† | 1.2 ± 0.05 |
| 209543_s_at | CD34 | CD34 molecule | 1.5 ± 0.12† | 1.1 ± 0.08 |
| 218711_s_at | SDPR | Serum deprivation response (phosphatidylserine binding protein) | 1.5 ± 0.14† | −1.1 ± 0.13 |
| 214091_s_at | GPX3 | Glutathione peroxidase 3 (plasma) | 1.5 ± 0.14 | 1.2 ± 0.06 |
| 200884_at | CKB | Creatine kinase, brain | 1.5 ± 0.07* | 1.3 ± 0.07 |
| 212607_at | AKT3 | v-Akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma) | 1.5 ± 0.14* | 1.1 ± 0.10 |
| 211799_x_at | HLA-C | Major histocompatibility complex, class I, C | 1.5 ± 0.07* | 1.3 ± 0.09 |
| 211006_s_at | KCNB1 | Potassium voltage-gated channel, Shab-related subfamily, member 1 | 1.5 ± 0.18† | −1.2 ± 0.10 |
| 217294_s_at | ENO1 | Enolase 1, (alpha) | 1.5 ± 0.14 | 1.1 ± 0.10 |
| 211529_x_at | HLA-G | Major histocompatibility complex, class I, G | 1.4 ± 0.07* | 1.2 ± 0.06 |
| 235371_at | GLT8D4 | Glycosyltransferase 8 domain containing 4 | 1.3 ± 0.15* | 1.9 ± 0.21 |
| 204036_at | LPAR1 | Lysophosphatidic acid receptor 1 | 1.1 ± 0.08* | 1.8 ± 0.26 |
| 217623_at | MYLK3 | Myosin light chain kinase 3 | −1.5 ± 0.05 | −1.1 ± 0.12 |
| 235309_at | RPS15A | Ribosomal protein S15a | −1.5 ± 0.05* | −1.3 ± 0.03 |
| 231792_at | MYLK2 | Myosin light chain kinase 2 | −1.6 ± 0.05* | −1.2 ± 0.05 |
| 203824_at | TSPAN8 | Tetraspanin 8 | −1.7 ± 0.07* | −1.1 ± 0.12 |
| 227577_at | EXOC8 | Exocyst complex component 8 | −1.8 ± 0.06* | −1.3 ± 0.06 |
| 238625_at | C1orf168 | Chromosome 1 open reading frame 168 | −3.9 ± 0.09* | −1.3 ± 0.17 |
Data are presented as mean fold change ± SE. Differentially expressed genes in the human high and low responders. Approximately 100 genes affected by 6 wk of aerobic exercise training were observed to be differentially expressed in human muscle of high responders when compared with low responders.
P < 0.05;
P < 0.01 for the difference between low and high responders; all remaining genes, P < 0.07.
We utilized topGO (2) to explore the biological processes most overrepresented in the high responder gene list and found that the genes listed contributed mainly to developmental processes, including organ and muscle development (Supplementary Fig. S4). The topGO package allows genes contributing to significance at one level of the GO to be removed before testing higher levels. We used both this method and classical Fisher's tests to examine functional enrichment. The results were similar (Supplementary data page 10). Supplementary Fig. S4 shows the results from the classical Fisher's test. For clarity only “children” categories of the developmental and multiorganismal development ontology “parents” are plotted. Significant GO categories are represented as gray boxes.
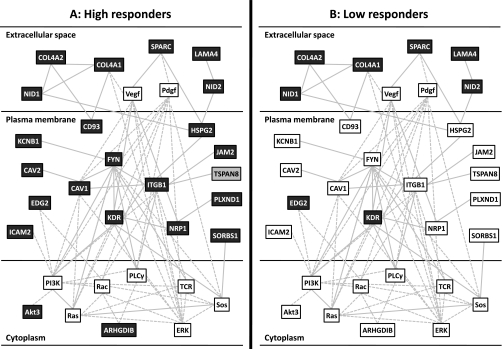
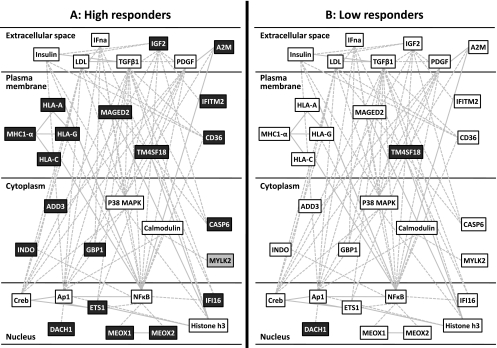
Using Ingenuity, we find the high responder genes are associated with cardiovascular development (network score = 50, Fig. 3A) and embryonic tissue development (network score = 41, Fig. 4A). For these two highest ranking gene networks only few of the genes were regulated >1.5 FC in low responders (Figs. 3B and 4B), and all were regulated to a significantly lesser extent than in the high responders (Table 2). This did not reflect differences in baseline expression (57). While this does not rule out that these networks could be activated in the low responders, it clearly demonstrates that low responders do not activate mRNA changes to a similar extent as, or with a similar time course to, the high responders. Since we know the high responders are clearly more successful at physiological adaptation, this lack of response at 24 h postexercise can be deemed maladaptive or suboptimal. Previously we have shown that adaptational response to aerobic training can be predicted using a set of “predictor genes” (58). Ingenuity IPA identified ∼80 direct or indirect connections (via a single connection gene) to the above top-ranked high responder network genes (Supplementary Fig. S5), implying that while the predictor genes may be largely unresponsive to exercise, they may directly regulate the transcriptional networks activated in proportion to aerobic adaptation.
Fig. 3.
Upregulation of a highly significant Ingenuity pathway analysis (IPA) gene network (IPA network score = 50) in response to endurance training in high responders (A), but not in low responders (B). Aerobic training was found to be associated with differential increases (>1.5 FC, dark gray boxes) and decreases (>1.4 FC, light gray boxes) in gene expression mostly in the high responders to aerobic training. Those genes that were upregulated in the low responders were upregulated to a significantly greater extent in high responders (Table 2). The network involves a number of extracellular matrix genes, VEGF signaling and associates with terms such as “cardiovascular development.” This is consistent with the TopGO analysis presented online in Supplementary Fig. S4.
Fig. 4.
Upregulation of a second highly significant IPA gene network (IPA network score = 41) in response to endurance training in high responders (A), but not in low responders (B). Aerobic training was found to be associated with differential increases (>1.5 FC, dark gray boxes) and decreases (>1.4 FC, light gray boxes) in gene expression almost exclusively in the high responders to aerobic training. This network is regulated by NF-kB, TGF-β, IGF2 and insulin, includes a number of major histocompatibility complex (MHC) genes, and is associated with embryonic tissue development according to IPA.
High responder genes and SNPs in HERITAGE.
We next explored the relationship between genetic variation in the genes that were differentially expressed in the high responder group (HRG) and endurance training-induced changes in V̇o2max in HERITAGE. The 20-wk endurance training program induced an average V̇o2max increase of 396 ml/min with a standard deviation of 216 ml/min and a range from −114 ml/min to +1,097 ml/min in HERITAGE white subjects. The maximal heritability estimate of V̇o2max training response (ΔV̇o2max) reached 47% (6). For the present study, a panel of 3,400 SNPs was genotyped to cover most of the genetic variance in and around (100 kb upstream and downstream) the top 86 high responder genes (Table 2). Twenty-four SNPs associated with responder status (ΔV̇o2max) at P < 0.01, eight of them in the HLA cluster of genes on chromosome 6. The other significant SNPs belonged to ASNS, DLC1, FABP5, NID1, PDZD2, SASH1, SPARC, and TRAM1, which code for proteins largely involved in cell proliferation or tumor suppression. For example ASNS codes for asparagine synthetase, a gene involved with immune cell proliferation (also induced during glucose deprivation) (17). DLC1 binds tensin, a membrane cytoskeletal protein, and is considered a tumor suppressor gene (66). FABP5 promotes cell proliferation (19), and NID1 is a membrane glycoprotein providing a bridge between collagen 4 and laminin with a potential role in tissue recovery (4). PDZD2 is a secreted protein and also a tumor suppressor (52, 53), and it may represent a novel myokine. SASH1 is a candidate tumor suppressor gene (46). SPARC is a matrix-associated protein involved in cell shape and has a tumor suppression function (9), while TRAM1 is secretory protein regulator found in the endoplasmic reticulum (23). The significant HLA cluster is intriguing. HLA-A, HLA-B, HLA-C, and HLA-G belong to the HLA class I heavy chain paralogues. These class I molecules are heterodimers consisting of a light chain (beta-2 microglobulin), and a heavy chain anchored in the membrane. Class I molecules play a central role in the immune system by presenting peptides derived from the endoplasmic reticulum lumen. However, although each of these SNPs significantly associated with responder status at P < 0.01, it should be noted that after taking multiple testing into account using the very conservative Bonferroni correction [using the number of SNPs tested: P < (0.05/86) = 0.00058], none of the SNPs remained statistically significant (see Supplementary data page 11).
Differential activation of TRT genes in rat models of low and high exercise response.
We hypothesized that the high-responder genes identified in humans would be differentially expressed in a novel rat model we selectively bred for either a low response or high response to aerobic exercise training. In 2002, we initiated a large-scale bidirectional selection experiment for the response to exercise training in a laboratory rat population with wide genetic heterogeneity (N:NIH out-crossed stock) (27). A base population (n = 152) was tested for maximal treadmill running capacity before and after receiving the same amount of a moderate standardized treadmill training stimulus over an 8-wk period (3 exercise sessions per week) (61). On average, training produced a 140-m gain in maximal running capacity with interindividual variation ranging in magnitude from −339 to +627 m. Ten male and ten female rats that represented the extreme ends for training response (>1 SD) were bred at each successive generation to develop low response to training (LRT) and high response to training (HRT) selected lines.
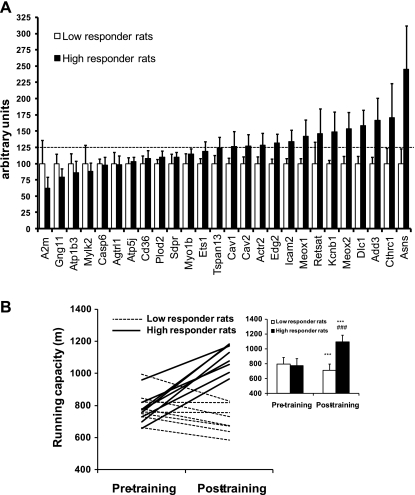
Early during the development of the model (generation 6), we chose 26 TRT genes (based on conservation of a homologous transcript and a significant LOD score for association with metabolic syndrome; see Supplementary data page 12 and Supplementary Table 1) for qPCR analysis to test in soleus muscle from rats characterized by wide differential for meters gained in running capacity after 8 wk of daily aerobic exercise training (215 ± 35 m gain for HRT vs. 219 ± 48 m loss for LRT). Thirteen of these genes demonstrated >125% expression, after physiological phenotyping, in muscle of rats from the high-responder line, and expression of the 26 genes combined was significantly different in the high responder rats compared with the low responder rats (P < 0.0001, Fig. 5A).
Fig. 5.
Rat genetic model for high and low training response to aerobic exercise training. A: differences in basal level of muscle gene expression in 6th-generation rats that demonstrate a high or a low aerobic training response. Of 26 human “high responder genes” analyzed in soleus muscle of rats selectively bred for high and low adaptation to aerobic exercise training, 13 genes already demonstrated >25% higher expression levels in high responder rats (n = 5, black columns) compared with low responder rats (n = 6, white columns) even in this early stage of the breeding model development. As a whole, the 26 genes were expressed at a higher level in the high responder rats (P < 0.0001). Data are presented as mean arbitrary expression units ± SE. B: the change in treadmill running capacity following 8 wk of aerobic exercise training was significantly different (P < 0.001) for rats bred across 10 generations for low (n = 8) and high (n = 8) response to training. Data are presented as meters gained in maximal aerobic running capacity produced by the training mean ± SE. ***P < 0.001 compared with pretraining; ###P < 0.001 compared with low responder rats.
Based on these promising results, gene analysis was subsequently carried out in rats derived from generation 10 of selection. Critically, these high (n = 8) and low (n = 8) responder rats did not significantly differ in body mass (192 ± 17 g vs. 183 ± 15 g) nor intrinsic aerobic running capacity (i.e., maximal running distance before training: 792 ± 99 m vs. 771 ± 91 m). Body mass increased similarly (∼20%) between the pre- and posttraining assessment period in both groups. High responder rats demonstrated a 43 ± 14% increase in aerobic running capacity as a result of training (P < 0.001), while aerobic running capacity declined by 10 ± 6% (P < 0.001) in the low responder cohort (Fig. 5B). We hypothesized that differences in gain in aerobic running capacity between rats selectively bred for exercise responsiveness would be linked to differential activation of the TRT genes. To test this hypothesis, we examined the mRNA expression using Affymetrix gene chips in both skeletal muscle (soleus) and cardiac tissue (left ventricle) from LRT (n = 3) and HRT (n = 3) rats sampled 24 h after the last scheduled treadmill test for running capacity posttraining. We found upregulated TRT genes (n = 457 genes mapped from human to rodent) to be 20% more abundant in the HRT rat soleus, while the downregulated TRT genes (n = 26 genes mapped from human to rodent) were ∼10% less abundant compared with LRT. SAM analysis identified that 176 genes of the TRT were significantly higher (FDR < 1%) expressed in the HRT rodent soleus muscle (none of the downregulated genes reached statistical significance). No differences in cardiac gene expression were found in this study. This pilot gene chip analysis of skeletal muscle yields a striking result, demonstrating common markers between humans and rodents that correlate with gains from exercise training.
DISCUSSION
Aerobic exercise is arguably the most relevant physiological intervention to manipulate to combat cardiovascular disease (64, 65). We present several lines of evidence for the TRT being a miRNA-influenced, RUNX1-PAX3-SOX9 transcription factor-directed gene network. Furthermore, we have identified a subset of 100 genes from the TRT that are modulated in line with greater physiological adaptation. This observation was in part reproduced in a rat model selectively bred for endurance exercise responsiveness. Genomic markers analysis in the HERITAGE study, using the high responder gene list as candidates, identified 16 genes for which genetic variation related to maximal aerobic capacity change. These associations suggest that genetic variants may contribute to the variable gene expression response and ultimately determine the magnitude of aerobic capacity changes (58).
Linking the TRT to oxygen sensing and angiogenesis.
We show that RUNX1, PAX3, and SOX9 are candidate regulators governing in vivo aerobic capacity changes (Supplementary Fig. S3). Given our original aim—to identify the molecular framework for modification of the oxygen transport system in vivo—these findings appear especially intriguing given that RUNX1, PAX3, and SOX9 associate with erythropoiesis (67), oxygen tension (1), and oxidative stress (13), all cellular processes directly responsive to molecular oxygen. Thus our molecular profile connects together for the first time genes known to be individually regulated by oxygen-sensitive mechanisms. We tested the hypothesis that these molecular processes could be regulated by alterations in miRNA expression. The rationale for miRNA-directed posttranscriptional regulation can be argued using RUNX1. RUNX1 expression is regulated by muscle activity (69), plays a critical role regulating skeletal muscle remodeling (63), and interacts with calcineurin to sense alterations in cell calcium levels (36). Although RUNX1 mRNA is unaltered by exercise in our study, there is recent evidence that RUNX1 is regulated at the posttranscriptional level by miRNAs (20). Both mir-101 and mir-144 target conserved sites on RUNX1 and are expressed in human skeletal muscle, and we found these miRNAs to be decreased more than twofold by exercise training. Thus, whereas GSEA identified these key transcriptional regulators (RUNX1-PAX3-SOX9) despite their being individually unaltered at the mRNA level, downregulation of many miRNAs which are predicted to target RUNX1-PAX3-SOX9 (Supplementary data page 4) can explain why using GSEA we observed that hundreds of their target genes are upregulated in skeletal muscle. In addition, a miRNA with a proven role in angiogenesis, mir92a, has been described as a negative regulator of angiogenesis (5), and consistent with this role it was downregulated by endurance training in our study (Fig. 1). Clearly additional studies are required to tease out the relative importance of each miRNA change and which components of the TRT are under direct regulation, and which are not.
Importantly, while we study the variation in transcriptional responses in skeletal muscle, it is plausible that these molecular changes reflect regulatory features present directly in the genomic sequence and hence occur in other organ systems, especially cardiac tissue. Consistent with this idea, a recently established human cardiac hypertrophy transcriptional signature shares many features in common with the TRT (42) including TGF-β activation (e.g., Supplementary Fig. S2a). Manipulation of key control points within the high-responder networks (see below) may allow for the modulation of aerobic capacity in humans for the purpose of prevention or treatment of metabolic and cardiovascular disease. Interestingly, in our rat model we found no evidence for an altered gene expression signature in cardiac tissue in high responders 24 h posttraining. However, although cardiac size did not differ pretraining, at dissection posttraining we found cardiac mass-to-body weight ratio to be 28% higher in the HRT rodents suggesting greater cardiac output could have allowed for improved oxygen delivery and hence greater possible loading of the peripheral muscle.
Linking the TRT to physiological adaptation in vivo.
One major goal was to determine the key molecular changes that underlie gains in aerobic capacity to bring us one step closer to understanding why aerobic capacity protects against metabolic and cardiovascular disease and why there is such a wide range of responsiveness to exercise training. Several novel features of our data set provide numerous new hypothesis testing opportunities. For example, the human high responder network (Fig. 4) features a cluster of human antigen major histocompatibility complex (MHC) class I molecules. The observation that exercise modulates at least 10 MHC genes (and to a larger extent in high responders; Supplementary data pages 1 and 13) adds a very novel dimension through which aerobic training may modulate insulin and growth factor activity at the muscle membrane (38, 48). In the present analysis we reveal that HLA-A, HLA-B, HLA-C, and HLA-G genes may also harbor genetic variation that associated with the degree of gain in V̇o2max. Such genomic analysis using the HERITAGE study offers independent support for the relevance of this cluster of genes on chromosome 6 for gains in maximal aerobic capacity. Indeed, it has been shown that a peptide sequence cleaved from major histocompatibility complex I (MHC I) binds to and prevents the internalization of cell surface receptors (41), thus enhancing the action of hormones such as insulin and insulin-like growth factor 2 (IGF2). Both hormones transpire as potential regulators of the training response networks identified by pathway analysis (Fig. 4), and IGF2 regulation links directly to the 30-gene cluster that we have shown to determine the magnitude of the aerobic training response in humans (58).
Using gain in aerobic capacity as our response parameter, we recently developed a quantitative predictor of response to training by relating ΔV̇o2max to expression of a set of 30 genes at baseline (i.e., pretraining) (58). Bibliometrics (Ingenuity database) demonstrated that the predictor genes made ∼80 known gene-to-gene associations with the high-responder gene networks presented in this article (Supplementary Fig. S5), suggesting that the “predictor” genes participate in the regulation of the complex multigene transcriptional responses described above, which characterize a high potential for aerobic capacity improvement in humans and rodents.
In conclusion, detailed analysis of the in vivo human gene-chip data, SNP analysis, and selective breeding all point toward a powerful gene expression program that characterizes successful adaptation to aerobic training. Development-, tumor biology-, and immunology-related genes appear to provide a bridge between physiological fitness, remodeling capacity, and trainability. It remains to be established whether low responders are at greater potential risk for the development of cardiovascular and metabolic disease.
GRANTS
This study was supported by an Affymetrix Translational Medicine Grant (J. A. Timmons), Swedish Diabetes Society (J. A. Timmons), Chief Scientists Office, Scotland (J. A. Timmons), Lundbeck Foundation (P. Keller), Pfizer Global Research and Development, UK (J. A. Timmons/C. J. Sundberg), Swedish National Centre for Research in Sports (J. A. Timmons/C. J. Sundberg/E. Jansson), Michigan Diabetes Research and Training Center pilot grant (L. G. Koch), funded by NIH5P60-DK-20572 from the National Institute of Diabetes and Digestive and Kidney Diseases and Grant RR-17718 (L. G. Koch/S. L. Britton) from the National Center for Research Resources of the National Institutes of Health (NIH; U.S. Dept. of Health and Human Services). The HERITAGE Family Study has been funded by multiple grants from NIH (HL-45670, HL-47323, HL-47317, HL-47327, HL-47321) to C. Bouchard, T. Rankinen, and Drs. D. C. Rao, Arthur Leon, James Skinner, and Jack Wilmore. C. Bouchard is partially funded by the George A. Bray Chair in Nutrition.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Professor Peter Gahzal at the GTI microarray facility for providing access to array equipment (Dept. Pathway Medicine, Univ. of Edinburgh, United Kingdom). Thanks also for animal care and exercise training provided by Lori Gilligan and Nathan Kanner (Koch/Britton lab, Univ. of Michigan).
Footnotes
Supplementary materials mentioned throughout the text are available with the online version of the present article.
REFERENCES
- 1. Adesida AB, Grady LM, Khan WS, Millward-Sadler SJ, Salter DM, Hardingham TE. Human meniscus cells express hypoxia inducible factor-1alpha and increased SOX9 in response to low oxygen tension in cell aggregate culture. Arthritis Res Ther 9: R69, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607, 2006 [DOI] [PubMed] [Google Scholar]
- 3. An P, Teran-Garcia M, Rice T, Rankinen T, Weisnagel SJ, Bergman RN, Boston RC, Mandel S, Stefanovski D, Leon AS, Skinner JS, Rao DC, Bouchard C. Genome-wide linkage scans for prediabetes phenotypes in response to 20 weeks of endurance exercise training in non-diabetic whites and blacks: the HERITAGE Family Study. Diabetologia 48: 1142–1149, 2005 [DOI] [PubMed] [Google Scholar]
- 4. Baranowsky A, Mokkapati S, Bechtel M, Krugel J, Miosge N, Wickenhauser C, Smyth N, Nischt R. Impaired wound healing in mice lacking the basement membrane protein nidogen 1. Matrix Biol 29: 15–21, 2010 [DOI] [PubMed] [Google Scholar]
- 5. Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324: 1710–1713, 2009 [DOI] [PubMed] [Google Scholar]
- 6. Bouchard C, An P, Rice T, Skinner JS, Wilmore JH, Gagnon J, Perusse L, Leon AS, Rao DC. Familial aggregation of V̇o2max response to exercise training: results from the HERITAGE Family Study. J Appl Physiol 87: 1003–1008, 1999 [DOI] [PubMed] [Google Scholar]
- 7. Bouchard C, Leon AS, Rao DC, Skinner JS, Wilmore JH, Gagnon J. The HERITAGE family study. Aims, design, and measurement protocol. Med Sci Sports Exerc 27: 721–729, 1995 [PubMed] [Google Scholar]
- 8. Bouchard C, Rankinen T. Individual differences in response to regular physical activity. Med Sci Sports Exerc 33: S446–S451; discussion S452–S453, 2001 [DOI] [PubMed] [Google Scholar]
- 9. Brekken RA, Puolakkainen P, Graves DC, Workman G, Lubkin SR, Sage EH. Enhanced growth of tumors in SPARC null mice is associated with changes in the ECM. J Clin Invest 111: 487–495, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99: 15524–15529, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature 437: 1032–1037, 2005 [DOI] [PubMed] [Google Scholar]
- 12. Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, 2nd, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nature Med 13: 613–618, 2007 [DOI] [PubMed] [Google Scholar]
- 13. Chang TI, Horal M, Jain SK, Wang F, Patel R, Loeken MR. Oxidant regulation of gene expression and neural tube development: Insights gained from diabetic pregnancy on molecular causes of neural tube defects. Diabetologia 46: 538–545, 2003 [DOI] [PubMed] [Google Scholar]
- 14. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38: 228–233, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 8: 93–103, 2007 [DOI] [PubMed] [Google Scholar]
- 16. Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol 6: R16, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui H, Darmanin S, Natsuisaka M, Kondo T, Asaka M, Shindoh M, Higashino F, Hamuro J, Okada F, Kobayashi M, Nakagawa K, Koide H, Kobayashi M. Enhanced expression of asparagine synthetase under glucose-deprived conditions protects pancreatic cancer cells from apoptosis induced by glucose deprivation and cisplatin. Cancer Res 67: 3345–3355, 2007 [DOI] [PubMed] [Google Scholar]
- 18. Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 279: 52361–52365, 2004 [DOI] [PubMed] [Google Scholar]
- 19. Fang LY, Wong TY, Chiang WF, Chen YL. Fatty-acid-binding protein 5 promotes cell proliferation and invasion in oral squamous cell carcinoma. J Oral Pathol Med 39: 342–348, 2010 [DOI] [PubMed] [Google Scholar]
- 20. Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, Croce CM, Brunetti E, Grignani F, Peschle C. MicroRNAs 17–5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol 9: 775–787, 2007 [DOI] [PubMed] [Google Scholar]
- 21. Franks PW, Scheele C, Loos RJ, Nielsen AR, Finucane FM, Wahlestedt C, Pedersen BK, Wareham NJ, Timmons JA. Genomic variants at the PINK1 locus are associated with transcript abundance and plasma nonesterified fatty acid concentrations in European whites. FASEB J 22: 3135–3145, 2008 [DOI] [PubMed] [Google Scholar]
- 22. Gallagher IJ, Scheele C, Keller P, Nielsen AR, Remenyi J, Fischer CP, Roder K, Babraj J, Wahlestedt C, Hutvagner G, Pedersen BK, Timmons JA. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in Type 2 Diabetes. Genome Med 2: 9, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gorlich D, Hartmann E, Prehn S, Rapoport TA. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature 357: 47–52, 1992 [DOI] [PubMed] [Google Scholar]
- 24. Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hamilton MT, Booth FW. Skeletal muscle adaptation to exercise: a century of progress. J Appl Physiol 88: 327–331, 2000 [DOI] [PubMed] [Google Scholar]
- 26. Hang J, Kong L, Gu JW, Adair TH. VEGF gene expression is upregulated in electrically stimulated rat skeletal muscle. Am J Physiol Heart Circ Physiol 269: H1827–H1831, 1995 [DOI] [PubMed] [Google Scholar]
- 27. Hansen C, Spuhler K. Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res 8: 477–479, 1984 [DOI] [PubMed] [Google Scholar]
- 28. Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol 4: R70, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566, 2005 [DOI] [PubMed] [Google Scholar]
- 30. Keller P, Keller C, Carey AL, Jauffred S, Fischer CP, Steensberg A, Pedersen BK. Interleukin-6 production by contracting human skeletal muscle: autocrine regulation by IL-6. Biochem Biophys Res Commun 310: 550–554, 2003 [DOI] [PubMed] [Google Scholar]
- 31. Keller P, Penkowa M, Keller C, Steensberg A, Fischer CP, Giralt M, Hidalgo J, Pedersen BK. Interleukin-6 receptor expression in contracting human skeletal muscle: regulating role of IL-6. FASEB J 19: 1181–1183, 2005 [DOI] [PubMed] [Google Scholar]
- 32. Koch LG, Britton SL. Artificial selection for intrinsic aerobic endurance running capacity in rats. Physiol Genomics 5: 45–52, 2001 [DOI] [PubMed] [Google Scholar]
- 33. Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, Kao HL, Gunsalus KC, Pachter L, Piano F, Rajewsky N. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol 16: 460–471, 2006 [DOI] [PubMed] [Google Scholar]
- 34. Larsson O, Wahlestedt C, Timmons JA. Considerations when using the significance analysis of microarrays (SAM) algorithm. BMC Bioinformatics 6: 129, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 115: 787–798, 2003 [DOI] [PubMed] [Google Scholar]
- 36. Liu H, Holm M, Xie XQ, Wolf-Watz M, Grundstrom T. AML1/Runx1 recruits calcineurin to regulate granulocyte macrophage colony-stimulating factor by Ets1 activation. J Biol Chem 279: 29398–29408, 2004 [DOI] [PubMed] [Google Scholar]
- 37. Lortie G, Simoneau JA, Hamel P, Boulay MR, Landry F, Bouchard C. Responses of maximal aerobic power and capacity to aerobic training. Int J Sports Med 5: 232–236, 1984 [DOI] [PubMed] [Google Scholar]
- 38. Malide D, Yewdell JW, Bennink JR, Cushman SW. The export of major histocompatibility complex class I molecules from the endoplasmic reticulum of rat brown adipose cells is acutely stimulated by insulin. Mol Biol Cell 12: 101–114, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McGee SL, Hargreaves M. Exercise and myocyte enhancer factor 2 regulation in human skeletal muscle. Diabetes 53: 1208–1214, 2004 [DOI] [PubMed] [Google Scholar]
- 40. Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol 258: 432–442, 2003 [DOI] [PubMed] [Google Scholar]
- 41. Naranda T, Goldstein A, Olsson L. A peptide derived from an extracellular domain selectively inhibits receptor internalization: target sequences on insulin and insulin-like growth factor 1 receptors. Proc Natl Acad Sci USA 94: 11692–11697, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petretto E, Sarwar R, Grieve I, Lu H, Kumaran MK, Muckett PJ, Mangion J, Schroen B, Benson M, Punjabi PP, Prasad SK, Pennell DJ, Kiesewetter C, Tasheva ES, Corpuz LM, Webb MD, Conrad GW, Kurtz TW, Kren V, Fischer J, Hubner N, Pinto YM, Pravenec M, Aitman TJ, Cook SA. Integrated genomic approaches implicate osteoglycin (Ogn) in the regulation of left ventricular mass. Nat Genet 40: 546–552, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA 103: 8721–8726, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raymond J, Segre D. The effect of oxygen on biochemical networks and the evolution of complex life. Science 311: 1764–1767, 2006 [DOI] [PubMed] [Google Scholar]
- 45. Reinheimer W, Davidson PC, Albrink MJ. Effect of moderate exercise on plasma glucose, insulin, and free fatty acids during oral glucose tolerance tests. J Lab Clin Med 71: 429–437, 1968 [PubMed] [Google Scholar]
- 46. Rimkus C, Martini M, Friederichs J, Rosenberg R, Doll D, Siewert JR, Holzmann B, Janssen KP. Prognostic significance of downregulated expression of the candidate tumour suppressor gene SASH1 in colon cancer. Br J Cancer 95: 1419–1423, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, Meier CA, Bell DR, Kralli A, Giacobino JP, Deriaz O. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes 52: 2874–2881, 2003 [DOI] [PubMed] [Google Scholar]
- 48. Stagsted J, Ziebe S, Satoh S, Holman GD, Cushman SW, Olsson L. Insulinomimetic effect on glucose transport by epidermal growth factor when combined with a major histocompatibility complex class I-derived peptide. J Biol Chem 268: 1770–1774, 1993 [PubMed] [Google Scholar]
- 49. Stephens NA, Gallagher IJ, Rooyackers O, Skipworth RJ, Tan BH, Marstrand T, Ross JA, Guttridge DC, Lundell L, Fearon KC, Timmons JA. Using transcriptomics to identify and validate novel biomarkers of human skeletal muscle cancer cachexia. Genome Med 2: 1, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Strohman R. Thermodynamics—old laws in medicine and complex disease. Nat Biotechnol 21: 477–479, 2003 [DOI] [PubMed] [Google Scholar]
- 51. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tam CW, Cheng AS, Ma RY, Yao KM, Shiu SY. Inhibition of prostate cancer cell growth by human secreted PDZ domain-containing protein 2, a potential autocrine prostate tumor suppressor. Endocrinology 147: 5023–5033, 2006 [DOI] [PubMed] [Google Scholar]
- 53. Tam CW, Liu VW, Leung WY, Yao KM, Shiu SY. The autocrine human secreted PDZ domain-containing protein 2 (sPDZD2) induces senescence or quiescence of prostate, breast and liver cancer cells via transcriptional activation of p53. Cancer Lett 271: 64–80, 2008 [DOI] [PubMed] [Google Scholar]
- 54. Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13: 2129–2141, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Timmons JA. Commentary on viewpoint: Perspective on the future use of genomics in exercise prescription. J Appl Physiol 104: 1250, 2008 [DOI] [PubMed] [Google Scholar]
- 56. Timmons JA, Gustafsson T, Sundberg CJ, Jansson E, Greenhaff PL. Muscle acetyl group availability is a major determinant of oxygen deficit in humans during submaximal exercise. Am J Physiol Endocrinol Metab 274: E377–E380, 1998 [DOI] [PubMed] [Google Scholar]
- 57. Timmons JA, Jansson E, Fischer H, Gustafsson T, Greenhaff PL, Ridden J, Rachman J, Sundberg CJ. Modulation of extracellular matrix genes reflects the magnitude of physiological adaptation to aerobic exercise training in humans. BMC Biol 3: 19, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski MA, Jensen T, Keller P, Scheele C, Vollaard NB, Nielsen S, Akerstrom T, Macdougald OA, Jansson E, Greenhaff PL, Tarnopolsky MA, van Loon LJ, Pedersen BK, Sundberg CJ, Wahlestedt C, Britton SL, Bouchard C. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol 108: 1487–1496, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Timmons JA, Larsson O, Jansson E, Fischer H, Gustafsson T, Greenhaff PL, Ridden J, Rachman J, Peyrard-Janvid M, Wahlestedt C, Sundberg CJ. Human muscle gene expression responses to endurance training provide a novel perspective on Duchenne muscular dystrophy. FASEB J 19: 750–760, 2005 [DOI] [PubMed] [Google Scholar]
- 60. Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, Nedergaard J, Cannon B. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc Natl Acad Sci USA 104: 4401–4406, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Troxell ML, Britton SL, Koch LG. Selected contribution: variation and heritability for the adaptational response to exercise in genetically heterogeneous rats. J Appl Physiol 94: 1674–1681, 2003 [DOI] [PubMed] [Google Scholar]
- 62. Vollaard NB, Constantin-Teodosiu D, Fredriksson K, Rooyackers OE, Jansson E, Greenhaff PL, Timmons JA, Sundberg CJ. Systematic analysis of adaptations in aerobic capacity and submaximal energy metabolism provides a unique insight into determinants of human aerobic performance. J Appl Physiol 106: 1479–1486, 2009 [DOI] [PubMed] [Google Scholar]
- 63. Wang X, Blagden C, Fan J, Nowak SJ, Taniuchi I, Littman DR, Burden SJ. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev 19: 1715–1722, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wei M, Kampert JB, Barlow CE, Nichaman MZ, Gibbons LW, Paffenbarger RS, Jr, Blair SN. Relationship between low cardiorespiratory fitness and mortality in normal-weight, overweight, and obese men. JAMA 282: 1547–1553, 1999 [DOI] [PubMed] [Google Scholar]
- 65. Wisloff U, Najjar SM, Ellingsen O, Haram PM, Swoap S, Al-Share Q, Fernstrom M, Rezaei K, Lee SJ, Koch LG, Britton SL. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science 307: 418–420, 2005 [DOI] [PubMed] [Google Scholar]
- 66. Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C, Singer S, Kuehnel F, Wigler M, Powers S, Zender L, Lowe SW. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev 22: 1439–1444, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yokomizo T, Hasegawa K, Ishitobi H, Osato M, Ema M, Ito Y, Yamamoto M, Takahashi S. Runx1 is involved in primitive erythropoiesis in the mouse. Blood 111: 4075–4080, 2008 [DOI] [PubMed] [Google Scholar]
- 68. Zhu J, Li Y, Shen W, Qiao C, Ambrosio F, Lavasani M, Nozaki M, Branca MF, Huard J. Relationships between transforming growth factor-beta1, myostatin, and decorin: implications for skeletal muscle fibrosis. J Biol Chem 282: 25852–25863, 2007 [DOI] [PubMed] [Google Scholar]
- 69. Zhu X, Yeadon JE, Burden SJ. AML1 is expressed in skeletal muscle and is regulated by innervation. Mol Cell Biol 14: 8051–8057, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.