Abstract
In recent years, several paramyxoviruses have emerged to infect humans, including previously unidentified zoonoses. Hendra and Nipah virus (henipavirus (HNV)) zoonoses were first identified in 1994 or 1998, causing deaths in animals and humans in Australia or Malaysia, respectively. Other paramyxoviruses, such as menangle virus, tioman virus, human metapneumovirus, and avian paramyxovirus-1, with less morbidity in humans, have also been recently identified. Although the Paramyxoviridae family of viruses has been previously recognized as biomedically and veterinarily important, the recent emergence of these paramyxoviruses has increased our attention to this family. Antiviral drugs can be designed to target specific important determinants of the viral/cell life cycle. Therefore, identifying and understanding the mechanistic underpinnings of viral entry, replication, assembly, and budding will be critical in the development of antiviral therapeutic agents. This review focuses on the molecular mechanisms discovered and the antiviral strategies pursued in recent years for emerging paramyxoviruses, with a concentration on viral entry and exit mechanisms.
Keywords: emerging, henipavirus, paramyxovirus, Nipah, Hendra, inhibitor, antiviral, fusion, entry, assembly
INTRODUCTION
Globalization and human encroachment into native wild-life habitats will likely continue to cause an increase in emerging zoonotic viral diseases. In recent years, members of the Paramyxoviridae viral family have caused some of the deadliest emerging zoonoses. The Paramyxoviridae family comprises important old and new human and animal viral pathogens, and Nipah (NiV) and Hendra (HeV) viruses make up the new henipavirus genus within this family (1–3). Henipaviruses first appeared in the 1990s in Australia, Malaysia and Singapore, causing epidemics that concerned national and international authorities due to the high mortality and morbidity rates in affected animals and humans (4, 5). For most paramyxoviruses, the host range is narrow and cross species transmission events are rare; hence, the recent emergence of the henipaviruses with high virulence and broad host-range is alarming.
Other paramyxoviruses with lower mortality rates and/or fewer incidents in humans have also emerged in recent years, including menangle virus, tioman virus, avian paramyxovirus-1, and human metapneumovirus (HMPV). Nonetheless, the incidence of HMPV in human populations approaches 100%, causing 5–20% of young children to be hospitalized with respiratory tract infections (reviewed in (6)). In addition, although other emerging paramyxoviruses such as the Beilong or J viruses have not been reported to cross species from their putative rodent reservoirs, the ability of Beilong virus to cross contaminate human cell cultures from rodent cell cultures in the same lab raises the spectre of zoonotic spread to humans (7–9). Therefore, understanding the mechanistic underpinnings of viral entry, replication, and assembly of these emerging paramyxoviruses is of critical importance. This review will focus primarily on henipaviruses as most recent molecular and mechanistic studies that inform potential antiviral strategies have been directed against this most lethal group of paramyxoviruses. We will not focus on vaccine approaches, as they have been recently reviewed elsewhere (10–12).
The Paramyxoviridae family
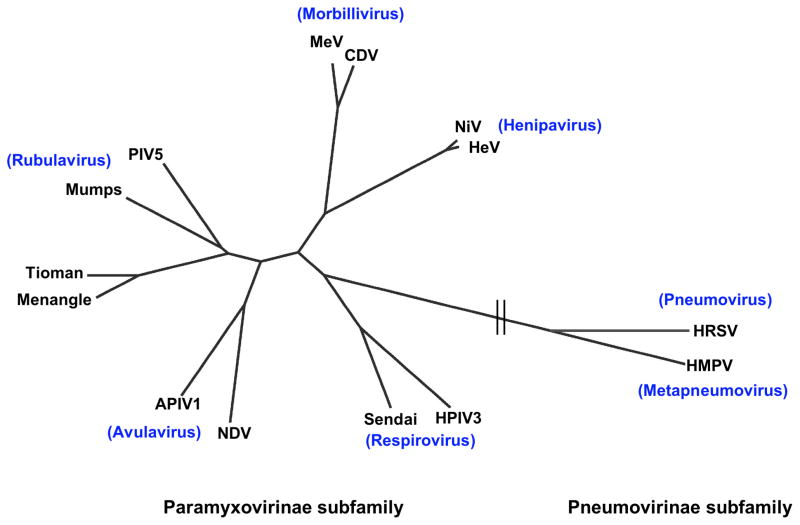
The Paramyxoviridae family has been divided into two subfamilies: Paramyxovirinae and Pneumovirinae (Fig. 1). The Paramyxovirinae subfamily comprises five genera: Respirovirus, Rubulavirus, Avulavirus, Morbillivirus, and Henipavirus. This subfamily includes the important Measles, Mumps, Newcastle disease, Parainfluenza, Hendra and Nipah viruses, among others, although some of the emerging Paramyxovirinae members (e.g. Menangle, Tioman, Beilong, and J) do not formally cluster into these 5 main genera. Some viruses within this subfamily have caused important human diseases for millennia. For example, reports of measles virus like symptoms date back to the 7th century. Although measles virus has now been eradicated from most developed countries through vaccination, it still produces a significant number of deaths globally, with 197,000 deaths reported in 2007 (13). The henipaviruses are the most virulent paramyxoviruses and will be a central focus of this review. The second subfamily, the Pneumovirinae, consists of two genera: Pneumovirus and Metapneumovirus (Fig. 1). This subfamily also includes important old and new human and animal pathogens, such as the human and bovine respiratory syncytial viruses that specifically affect bovine, caprine, and ovine species, and the human and avian metapneumoviruses, among others. An example of an important human pathogen within this subfamily is respiratory syncytial virus, with 64 million infections and 160,000 deaths, primarily infant, per year (14).
Fig. 1. Phylogenetic tree of the Paramyxoviridae family, built using a fusion protein sequence comparison.
The tree was generated from a Cobalt (NCBI) multiple fusion protein sequence alignment, by the fast minimum evolution method, and visualized using the Fig Tree program. Representative members of each genus of the Paramyxovirinae and Pneumovirinae subfamilies are shown. APIV1, avian parainfluenza virus 1; CDV, canine distemper virus; HeV, Hendra virus; HMPV, human metapneumovirus; HPIV3, human parainfluenza virus 3; HRSV, human respiratory syncytial virus; MeV, measles virus; NDV, Newcastle disease virus; NiV, Nipah virus; PIV-5, parainfluenza virus 5.
The emerging henipaviridae genus
HeV and NiV have been classified in a new genus because their genomic lengths and protein homology is sufficiently different from extant genera of paramyxoviruses (4). Their particularly broad tropism and extreme virulence compared to other paramyxoviruses also sets them apart. The henipaviruses naturally infect flying foxes, genus Pteropus, and transmit to humans either via an intermediate host, usually horses for HeV and swine for NiV, or directly from bat-to-humans or from human-to-human, as reported for post-2004 epidemics for NiV in Bangladesh (1, 15–17). HeV has reportedly caused the death of dozens of horses and 3 humans in Australia, through several outbreaks since 1994 (5, 18–21). In contrast, NiV has caused the death of almost 200 humans and high numbers of animals, with 1.1 million pigs culled in the first 1998 Malaysian epidemic alone (4). Since then, flying foxes seropositive for NiV have been detected in Cambodia, Thailand, India, and as far west as Madagascar and Ghana in West Africa (22, 23). NiV causes respiratory and neurological symptoms that often lead to encephalitis and mortality rates from 40% to 92% in humans (2, 24, 25). Additionally, NiV can spread efficiently and cause morbidity in economically important livestock (24). Due to their high virulence and absence of therapeutics or vaccines, henipaviruses are classified as biosafety level 4 (BSL4) pathogens, and NiV is classified a Category C priority pathogen by the NIAID Biodefense Research Agenda for its bio- and agro-terrorism potential (24). These characteristics of the henipaviruses underscore the need for research and treatment development against these perilous pathogens.
Molecular advancements in emerging paramyxovirus biology
Numerous studies have uncovered determinants important for various steps in the paramyxovirus life cycle, and each step represents a potential target for the development of antiviral drugs (Fig. 2). Among the emerging paramyxoviruses, the henipaviruses have been studied most extensively because of their relatively high morbidity rates. In general, after virus binding to the host cell receptor, paramyxoviruses require the cooperation of their two separate attachment and fusion transmembrane glycoproteins (reviewed in (26–29)). However, how the attachment glycoprotein activates the fusion protein, or how the fusion protein senses that it is the right time and place for carrying out its host/viral membrane fusion function, is still a matter of intense investigation. The regulation of the molecular choreography that leads to productive membrane fusion provides a fertile area for the development of therapeutics that can thwart this process.
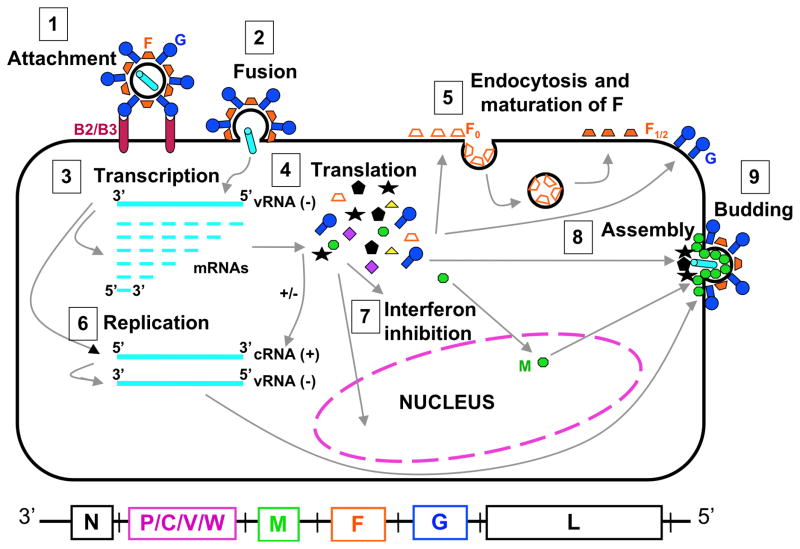
Fig. 2. Henipavirus replication cycle.
Depiction of henipavirus replication: After attachment to the B2/B3 receptor (1) and fusion (2), the virus enters the cell. The negative RNA genome (vRNA−) is a template for transcription of viral mRNAs following a 3–5′ attenuation gradient from N to L (3). mRNAs are translated into proteins (4) while the vRNA− is also a template for cRNA(+), which in turn is a template for vRNA(−) genomes during replication (6). New vRNA(−) genomes will be incorporated into new virions during viral assembly (8). Following translation (4), various viral proteins will function in interferon signaling pathways (7), and F0 will be endocytosed and matured (5). Assembly (8) and budding (9) are orchestrated primarily by the M protein, and N, P, C, M, F, and G, are incorporated into virions.
The development of antiviral therapeutic agents has been facilitated by the elucidation of the molecular mechanisms underlying various steps of the viral life cycle. As an example, insights into the life cycle of HIV-1 have led to approved antiretroviral drugs that target distinct steps: coreceptor antagonists and fusion inhibitors target viral entry, nucleoside and non-nucleoside inhibitors target the viral reverse transcriptase, integrase inhibitors target integration, and protease inhibitors target viral maturation (reviewed in (30, 31)). For the emerging paramyxoviruses, recent discoveries on the molecular mechanisms underpinning several steps of their life cycle, including host receptor usage, membrane fusion and viral entry, viral replication, interferon responses, assembly, and budding, promise to shed light on the development of antiviral therapeutic drugs. These research advances and antiviral therapeutic strategies are discussed here, with a heavier concentration on the viral entry and assembly steps carried out by the F, G, and M viral proteins. The molecular mechanisms and antiviral approaches that target the functions of other non-structural paramyxovirus proteins, particularly the gene products P, V, C, and W, have been previously reviewed in greater detail (11, 32–34).
Molecular mechanisms and antiviral strategies targeting the attachment glycoprotein
The paramyxovirus attachment proteins are type II transmembrane proteins on the surface of virions that mediate attachment of the virus to the cell surface receptor. This attachment protein receptor interaction plays an important role in determining cell tropism. There are several conserved features among all known paramyxovirus attachment proteins (G, H, or HN). They contain a head domain linked to the viral membrane by a stalk domain, and a cytoplasmic tail that is intraviral, or intracellular when the proteins are expressed at the cell surface (Fig. 3). The globular head of HeV-G and NiV-G (HNV-G) has a six-bladed-β-propeller structure common to the head domains of multiple paramyxovirus attachment proteins (35, 36). The oligomeric structure of HNV-G (dimers of dimers) (37) is also thought to resemble the attachment glycoproteins of other paramyxovirinae (28, 38), and it is likely that a finely balanced stoichiometry is required for optimal fusion as endogenous lectins like galectin-1 (see below) that cause inappropriate oligomerization of henipavirus envelope proteins can be detrimental to the fusion process (39).
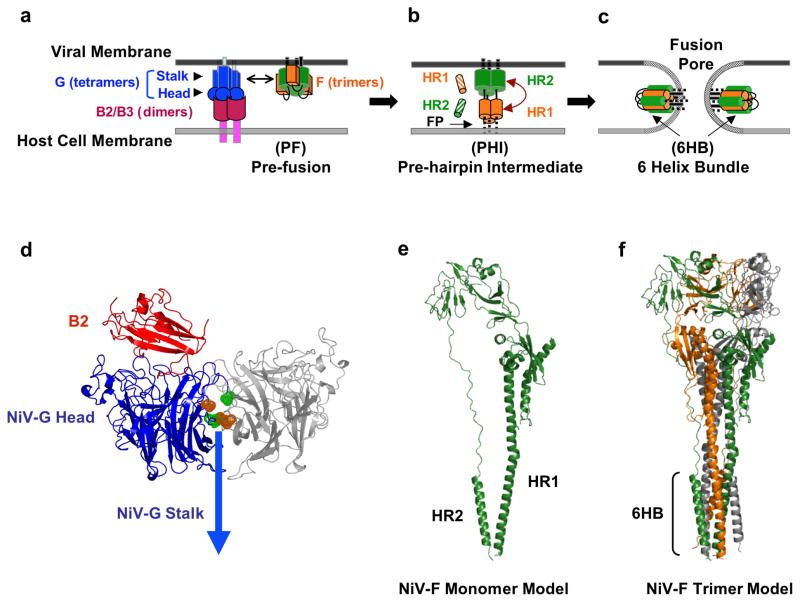
Fig. 3. Membrane Fusion and Viral Entry.
The attachment and membrane fusion steps necessary for viral entry (steps 1 & 2 from Fig. 2) are depicted here in greater detail in three major stages. (a) F is depicted in its pre-fusion, pre-hairpin intermediate, and post-fusion forms. EphrinB2 or ephrinB3 binding to NiV-G initiates a conformational cascade in F. (b) After F is triggered, it forms a pre-hairpin intermediate (PHI), in which a fusion peptide is harpooned into the host cell membrane. The PHI can be captured by peptides that mimic the NiV HR1 (orange striped cylinders) or HR2 regions (green striped cylinders) and bind the F HR2 or HR1 regions, respectively. (c) The HR1 and HR2 region in the PHI coalesce to form the six-helix bundle (6HB) conformation, bringing the viral and cell membranes together and facilitating viral-host membrane fusion and viral entry. At the figure bottom, the henipavirus genomic RNA is represented in its 3–5′ orientation. (d) Ribbon structure of the monomer of NiV-G (blue) head domain (pdb code 2VSM) and its interaction with its ephrinB2 receptor (red), drawn using PYMOL (www.pymol.org) and modeled by aligning the G/B2 monomer with each monomer of the hPIV3 Hemaglutinin-Neuraminidase dimer (pdb code 1V2I) similarly to (46). The second monomer is shown in gray. According to this model, the flexible region in the NiV-G ectodomain (green and orange) may interact with the same region in another monomer and may be involved in receptor-induced G mediated NiV-F triggering (46). (e) Representation of the structure of the NiV-F protein modeled using the HPIV3-F crystal structure (pdb code 1ztm) by the Phyre threding program, as performed in (78). (f) Representation of the trimer of NiV-F monomers from (e), also modeled using the HPIV3-F crystal structure as in (78).
Emerging paramyxovirus receptors
The host receptors for menangle virus, Tioman virus, human metapneumovirus, Beilong or J viruses, which are considered emerging paramyxoviruses with lower morbidities in humans, are unknown (reviewed in (6)). In contrast, the receptors for the henipaviruses were discovered in 2005 and 2006 to be ephrinB2 and ephrinB3, respectively (40–42). These transmembrane proteins are receptor-tyrosine kinases that interact with their endogenous receptors on opposing cells and play critical roles in cell-cell signaling, particularly during angiogenic and neuronal development (43). The distribution of ephrinB2 and ephrinB3 is consistent with the respiratory and neurological symptoms of henipavirus infections, as ephrinB2 and ephrinB3 are highly expressed in endothelial cells that line the microvasculature and in neurons. (40–42). In the CNS, ephrinB3 but not ephrinB2 is expressed in the brain stem, and ephrinB3-mediated entry may account for the brain stem dysfunction that is the ultimate cause of death from NiV encephalitis (42, 44). The identification of NiV and HeV receptors greatly facilitates the rational development of strategies and therapeutics that block virus/receptor binding.
Mechanisms of fusion triggering by the attachment protein
With very few exceptions, the attachment protein of paramyxoviruses is essential for viral entry (Fig. 3). Even for the respiratory syncytial virus, whose attachment protein is not required for membrane fusion, fusion is enhanced in the presence of the attachment glycoprotein. Interestingly, HMPV membrane fusion, and sometimes replication, is not enhanced by the presence of the attachment protein (reviewed in (26, 28)). Thus, the specific role(s) of the attachment protein in promoting viral entry is a subject of intense study (reviewed in (27, 28, 45)).
Several studies in various paramyxoviruses implicate a role of the attachment glycoprotein stalk domain in interacting with and triggering the fusion glycoprotein, which is the ultimate protein that mediates membrane fusion (46–52). Biochemical and biophysical studies suggest that a receptor-induced conformational change in NiV-G, which involves critical residues at the base of the NiV-G head domain and the presence of an intact stalk domain, is important for allosteric triggering of the fusion protein (46). Although no dramatic differences were found between the apo- and receptor-bound structures of NiV-G (36, 53), the stalk domain was not apparent in any of these structures. Perhaps the presence of the stalk allows for proper disassembly of higher ordered oligomers upon receptor binding, which may lead to the exposure of neo-epitopes that functionally trigger the fusion protein. Although the specifics of how HNV-G triggers its own fusion protein is beyond the scope of this review, it is likely that this triggering process is finely tuned (46) and therefore vulnerable to disruption. A better understanding of this triggering process may lead to therapeutics that target conserved features that may limit the development of resistance. For example, anti-HNV-G antibodies that recognize conserved neo-epitopes exposed after receptor binding may be a good candidates for passive immunization strategies (46).
Antiviral strategies that target the attachment protein
There have been a number of Mabs produced against NiV-G and HeV-G with a range of in vitro neutralization activities (IC50 ~40 – 600 ng/ml) (46, 54–56). One of these human Mabs (m102.4), which engages the receptor binding site in NiV or HeV G, appears to be protective in a lethal challenge ferret model when administered intravenously 10 hours post-infection but not 24 hours pre-infection (54). This difference could be due to the relatively low serum stability of m102.4 when administered intravenously, but nevertheless bodes well for the development of m102.4 as a post-exposure therapeutic in resource-sufficient settings. In comparison, Palivizumab, (Synagis®, MedImmune Inc.), an FDA approved MAb therapeutic targeted against the fusion protein of human respiratory syncytial virus (hRSV) has an in vitro IC50 of 363.7 ng/ml (57) and can be administered monthly (for hRSV prophylaxis) via intramuscular injections and still maintain serum concentrations of 100-fold (>40 μg/ml) above its in vitro IC50 in most patients (58). It would be interesting to see if IM injection will increase the effective half-life of m102.4 in vivo.
Soluble ephrinB2 or ephrinB3, or soluble henipavirus G (HNV-G) proteins have also been shown to block virus entry and cell-cell fusion (40–42, 59), although the likely interference with ephrinB function and the antigenicity of HNV-G itself limits the practical utility of these molecules as antivirals. However, the structure of the ephrinB2 or B3 bound HNV-G complex shows a large protein-protein interface but also reveals a lock-and-key binding pocket that may be targeted by small molecule therapeutics (35, 36). For example, Trp125 and Phe120 in the G-H loop of ephrinB2 interact differently with EphB4 than with HNV-G, suggesting a ‘druggable’ pocket to disrupt B2/B3-G interactions specifically (60). A likely caveat to this approach is that a small molecule designed to fit the B2/B3-G binding pocket specifically might still not be able to overcome the strong avidity of oligomeric B2/B3-G interactions. For example, ephrinB2 binds to NiV-G with a sub-nanomolar affinity (Kd ~0.06 nM) (42), suggesting that a drug would have to bind at picomolar concentrations or have a very slow off-rate to compete with B2-G interactions.
Molecular mechanisms and antiviral strategies targeting the fusion (F) glycoprotein
The fusion glycoproteins are synthesized as trimeric precursors that are activated by protease cleavage into a metastable pre-fusion conformation, poised for enabling membrane fusion (Fig. 3). Cleavage generates a new hydrophobic N-terminus, the fusion peptide, which is buried in the metastable pre-fusion F conformation. Upon attachment protein-receptor binding, the fusion cascade is triggered and the fusion peptide in F is harpooned into the target cell membrane in the pre-hairpin intermediate conformation (Fig. 3b). Two helical regions present in the pre-hairpin intermediate, HR1 and HR2, have high affinities for each other and coalesce to form the six-helix bundle (6HB), which brings the viral and target cell membranes together in close apposition allowing viral/target-cell membrane fusion and viral entry.
Maturation of the fusion protein
However, important differences in viral entry and membrane fusion mechanisms, carried out by the F protein, have been highlighted for the emerging paramyxoviruses (26, 29, 32). First, while many paramyxoviral F proteins are cleaved (once or twice) by furin-like cellular proteases during transport through the trans Golgi network (61–65), HMPV and Sendai virus Fs are cleaved by tissue-specific extracellular proteases such as mini-plasmin or tryptase Clara (66, 67), and cell surface henipavirus F is cleaved by cathepsin-L upon their endocytosis (68–71). Specific inhibition of these proteases by antiviral compounds could be envisioned. For example the lack of an acutely lethal phenotype in cathepsin-L knockout mice suggests that short-term inhibition of cathepsin-L in the context of a highly pathogenic virus infection may be a clinically viable option. Recently, a small-molecule oxocarbazate specific inhibitor of cathepsin L was reported effective against Ebola and SARS viruses at subnanomolar concentrations in vitro (72). Although Ebola and SARS viruses directly require cathepsin L cleavage during viral entry, this compound could also prove useful in treating henipavirus infections by preventing the generation of mature F. However, past in vitro vs. in vivo discrepancies between drugs that indirectly inhibit cathepsin-L cleavage have been observed. Chloroquine, normally used to treat malaria, has been shown to inhibit pseudotyped NiV entry, presumably by inhibiting endosomal acidification and indirectly cathepsin-L activity (73). However, chloroquine treatment was found not to prevent NiV infection or disease in ferrets (74), and combined chloroquine and ribavirin treatments did not prevent death in a hamster model of NiV and HeV infection (75). These in vitro vs. in vivo discrepancies suggest that we need to improve our understanding of the role of endocytosis and cathepsin-L cleavage in henipavirus infection.
N-glycans in henipavirus F and galectin-1
Another characteristic of emerging paramyxoviral F proteins is their atypical use of N-glycans. For most paramyxovirus F proteins, specific N-glycans are either necessary for proper protein folding and/or N-glycan removal is deleterious to the fusion process (76, 77). Surprisingly, removal of specific individual or multiple N-glycans from NiV- and HeV-F resulted in marked hyperfusogenicity manifested in fusion and viral entry (78, 79) assays. However, N-glycan removal also increased the sensitivity of NiV-F to antibody neutralization; thus appearing that N-glycans in henipavirus F are kept (at least partially) to serve as a shield against antibody neutralization (78).
NiV-F N-glycans were also found able to mediate binding to galectin-1, an innate immune lectin with many functions that binds to specific galactose-containing carbohydrates on the surface of mammalian cells or pathogens, (reviewed in (80)). Galectin-1 inhibits NiV-mediated cell-cell fusion and syncytia formation, a hallmark of NiV pathogenicity (39). Interestingly, the individual N-glycan in NiV-F (F3), whose removal resulted in the highest level of hyperfusogenicity, also gave rise to the most optimal N-glycan moiety that mediates galectin-1 binding to NiV-F. Endogenous levels of galectin-1 in endothelial cells were sufficient to inhibit NiV envelope mediated syncytia, and galectin-1 binding to the F3 N-glycan in NiV-F inhibited maturation, mobility, and triggering of the F protein (81). While it is unlikely that galectin-1 can be developed as an anti-viral therapeutic because of its pleotropic effects, these reports shed light on the innate immune defenses based on recognition of pathogen associated molecular patterns (PAMPs). Furthermore, 14 single nucleotide polymorphisms have been identified in the genomic locus of galectin-1 (82), which raises the intriguing possibility that genetic variability at this locus may contribute to the range in pathophysiology seen in henipavirus infections.
Blocking the membrane fusion cascade
Blocking viral entry by trapping one of the fusion protein intermediates during the membrane fusion cascade has been a therapeutic approach pursued and utilized for class I fusion protein enveloped viruses. For example, enfuvirtide, sifuvirtide, and their analogs, are peptides that mimic the C-terminal heptad-repeat region (HR2) of class I fusion proteins, and are approved for HIV-1 treatment (reviewed in (83–85)). Since paramyxoviral F proteins undergo equivalent class I fusion protein conformational changes, including pre-hairpin intermediate formation (26, 28, 29, 32, 86), the paramyxovirus HR2 (a.k.a. HRC) peptide has been used to trap the pre-hairpin intermediate (46, 78, 87–93) (Fig. 3b). Although the N-terminal HR1 region-mimicking peptide also inhibits fusion, it is generally a less efficient inhibitor (89), even when artificially trimerized to mimic the trimeric HR1 core (46).
HR2 peptides
For the henipaviruses, the HR2 peptide has been shown to inhibit cell-cell membrane fusion and viral entry in a pseudotyped viral system at nanomolar concentrations (78, 88, 89, 91). Surprisingly, higher levels of inhibition of HeV fusion were observed when using a human paramyxovirus-3 F vs. a HeV-F -derived HR2 peptide, although the mechanism for this phenomenon is unclear (92). Additionally, a second generation of capped and PEGylated HR2 peptides resulted in increased solubility in water, stability, synthesis yields, and possibilities for their use as antiviral agents in vivo (89). Another strategy for increasing HR2 peptide inhibition efficacy has been the addition of cholesterol to the peptide C-terminus. This approach likely brings the peptide into close proximity to the membrane site of action where fusion occurs, reducing the IC50 of HPIV-3 derived peptides on pseudotyped HeV and NiV infections from 10–100 nM to near 1 nM (94). However, the IC50’s for inhibition of live HeV and NiV viruses in vitro were close to 100 nM, and relatively large amounts of HR2-cholesterol peptides (2 mg/kg) were needed to achieve 60% or less survival of hamsters infected with NiV, when used simultaneously or previous to NiV infection. It is likely that large HR2 peptide amounts are needed in order to efficiently “coat” the surfaces of target cells in the host (95).
Anti-F Mabs
Another approach to inhibiting membrane fusion is the blocking of the fusion protein conformational changes required for the fusion cascade by the use of Mabs. Two anti-NiV-F antibodies have been reported to neutralize NiV and HeV in vitro (1.6 – 20 ng) and in a hamster model (180 – 520 μg/animal) (96). Although the binding epitopes of these antibodies have not been characterized, their cross-reactivity with HeV is desirable, suggesting that they may target a conserved region in HNV-F that may limit the generation of escape variants. Moreover, antibodies that bind conformational epitopes critical for membrane fusion are highly desirable, since mutations that annul both Mab binding and the need of conformational changes would be relatively rare. Conformational Mabs against the henipaviruses that preferably bind hyper- or hypo-fusogenic mutants have been reported, but their neutralization activities or their binding epitopes have not been shown (88).
Small-molecule inhibitors
Quinolone derivatives designed based on structure similarities among paramyxovirus F proteins in their HR1/HR2 binding motifs were tested for inhibition of NiV- and measles virus-induced cell fusion. Two of 18 compounds tested were moderately active as inhibitors of NiV-induced cell-cell fusion and NiV infection-induced syncytia at an EC50 of 1–3 μM. These compounds also showed some cytotoxicity in Vero cells (CC50 of 10 to >20 μM using the MTT test), resulting in a selectivity index (CC50/IC50) of ~13 for the compound with the lowest toxicity (97). This SI is relatively poor for a lead compound but may be improved by further structure activity relations (SAR) analysis. Mutants that cause resistance to HR2 peptide binding have been detected for HIV (83–85), and similar mutants may occur after the use of these small-molecule inhibitors that target HR1/HR2 interactions.
Molecular mechanisms and antiviral strategies targeting the matrix (M) protein
Paramyxoviral matrix (M) proteins are structural proteins that directly underlie the viral envelope, and are important for assembly and budding of viral particles (98, 99). Infectious paramyxoviral particles form after all the structural viral components have assembled at selected sites on the cell membrane, and M proteins are known to organize the assembly process. The position of M proteins underneath the cellular plasma membrane allows them to interact with both ribonucleoproteins (RNA genomes bound to nucleocapsid (N or NP) proteins) and viral glycoproteins via their cytoplasmic tails (98, 99). Recently, the atomic structure of the paramyxovirus human respiratory syncytial virus M protein was solved and shown to contain two beta-sheet-rich domains, joined by a short unstructured linker (100). This structure is similar to that of the filovirus Ebola M (101). The joined domains share an extensive positively charged surface, which likely binds to the negatively-charged membrane phospholipid head groups (100). For many paramyxoviruses, transient expression of M proteins alone, without the expression of other viral proteins, is sufficient to form and release viral-like particles (VLPs). Such is the case of hPIV-1(102), Sendai virus (103), NDV (104), measles virus (105, 106), and NiV (107, 108). However, in some cases, M-dependent VLP production is enhanced in the presence of other viral proteins, such as the glycoproteins, the nucleocapsid protein, or the C protein (reviewed in (98)).
Antivirals against M
Since the M protein is critical in paramyxoviral assembly and budding, antiviral agents that target important aspects of M-directed assembly and budding can be envisioned. For example, inhibition of NDV replication by targeting two distinct sites of the M gene using RNAi has been recently reported (109). In addition, for simian virus 5, proteasome inhibitors and expression of dominant-negative VPS4(E228Q) ATPase blocked budding, likely because of the involvement of the ubiquitin-proteasome pathway in budding (110). For NiV, a recent study showed that ubiquitin-regulated nuclear-cytoplasmic trafficking of NiV-M is important for viral budding (111). Therefore, compounds that block M ubiquitinating enzymes, depleting free ubiquitin in the cell (proteasome inhibitors), or that preferentially block nuclear import or export of NiV-M, could be potential anti-henipavirus candidates (Fig. 2). Indeed, bortezomib, an FDA-approved proteasome inhibitor used for treating multiple myeloma, reduced viral titers significantly at an IC50 of 2.7 nM, 100-fold less than the achievable plasma concentration in humans (111). Thus this FDA-approved agent has the potential for being evaluated as an off-label use for henipavirus treatment. Understanding of the cellular components that play important roles in viral assembly and release should also aid the discovery of novel drugs to target these steps of the life cycle of emerging paramyxoviruses.
Molecular mechanisms and antiviral strategies targeting the P/V/C proteins
Interferons (IFN) are part of the innate immune system and constitute one of the first lines of defense against viral pathogens in mammals (112) in the early virus/host battle that determines the establishment of an infection (113). The P gene encodes for the P, C, V, and W proteins, and in the subfamily Paramyxovirinae the P gene products generally have anti-IFN activities (see (32)). In part, P gene antiviral activities are due to their effects in limiting the extent of viral genome replication, since aberrant transcripts activate the retinoic acid inducible gene I (RIG-I) RNA helicase pathway, which activates interferon production (114). For example, the simian virus 5 P protein (115), Sendai C protein (116), measles C protein (117), J-virus and Beilong virus C proteins (114), hPIV-3 C protein (114), and henipavirus C, V, and W proteins (118), have all been shown to inhibit viral genome replication. In addition, all the henipavirus P gene proteins have been shown to inhibit the IFN signaling pathways (reviewed in (119, 120)).
Since restoring interferon responses has been successful in the treatment of cancer, autoimmune, and infectious diseases (121–123), this type of approach may also be suitable against emerging paramyxovirus infections. One study showed that the interferon inducer poly(I)-poly(C12U) (Ampligen®, a mismatched double-stranded RNA) prevented death from NiV infection in a hamster model (124). Ampligen was also observed to be effective against SARS-coronavirus infection in a mouse model (125), and has shown positive effects in HIV patients (126). Congruent with these studies is the finding that NiV and HeV replicate more efficiently in Vero cells, which are defective in IFN responses, compared to other cell lines (127). Therefore, stimulation of interferon production seems to be a promising treatment for henipavirus infections.
Broad-spectrum and other antiviral strategies
Most current antiviral drugs target differences between viral agents and hosts, such as specific viral protein moieties important for viral entry, replication, assembly, budding, etc., conferring specificity for the infected cells. However, targeting specific viral protein moieties is not always the best solution, as viral resistance by mutagenesis is very common when targeting single, or even multiple viral proteins (128, 129). Thus strategies that target non-protein determinants of important steps in the viral life cycle, particularly for a broad assortment of viruses, are highly desirable. For example, broad-spectrum compounds that target the viral membrane fluidity required for viral entry or exit, or RNA replication have recently been explored.
LJ001, a viral membrane inhibitor
Recently, a high-throughput screening assay based on NiV/VSV-pseudotype viral entry inhibition identified a small molecule that intercalates into and irreversibly damages viral membranes, but not cellular membranes, at low micromolar concentrations (130). Studies with lipid biosynthesis inhibitors indicated that LJ001 exploits the differences between static viral membranes and biogenic cellular membranes with reparative capacity. LJ001, a rhodanine derivative, was effective against numerous enveloped viruses, but not nonenveloped viruses, and showed no overt toxicity in vitro or in vivo with an SI of >100. LJ001 inactivated virions while leaving envelope proteins functionally intact, inhibiting a post-binding but pre-fusion step (130). Thus, LJ001 may represent a new class of broad-spectrum antivirals that target physiological rather than physical differences between viral and cellular lipid membranes. A potential mechanism of action would be disruption of the proper balance between saturated and unsaturated phospholipids that is required for the positive to negative membrane curvature transitions during the fusion process (reviewed in (131)). Elucidating the exact mechanism by which LJ001 effectuates its membrane damaging activities will shed light on whether differences between viral and cellular membranes can be exploited by other chemotypes, and help refine medicinal chemistry efforts to improve bioavailability and in vivo efficacy.
Cationic compounds
In another study, a high-throughput screening based on live-virus infection identified three compounds unsuitable for internal administration, but possibly suitable to topical applications (132). These three compounds, gliotoxin, gentian violet, and brilliant green, have been previously used as anti-bacterial and anti-fungal agents, and they showed antiviral activity against NiV, HeV, VSV, and HPIV-3. Additionally, gliotoxin inhibited Influenza A, suggesting a broad-spectrum activity for this compound. Although the mode of action of these cationic compounds is not known, it has been proposed that they directly bind to and inhibit viral membranes (132).
Calcium influx inhibitors
In a recent study that tested licensed pharmaceuticals against henipavirus replication in vitro, compounds that released intracellular calcium stores, calcium chelators, as well as calcium channel and calmodulin antagonists, inhibited henipavirus replication at the micromolar range (133). However, the mechanism that links calcium influx to henipavirus replication is unknown, and in vivo assays have not been reported.
Ribavirin
Ribavirin is a broad-spectrum antiviral used particularly for RSV and hepatitis C, and it is also used for RNA viruses for which there are no other available treatment (134, 135). It is a purine nucleoside analog, and although its exact mechanism(s) of inhibition of viral replication is not completely understood, it is known that ribavirin interferes with RNA metabolism, which is required for virus replication (136). For the emerging paramyxoviruses, various results with ribavirin have been reported. In the first NiV outbreak in Malaysia in 1998–1999, a 36% reduction in mortality in humans was reported (137). In addition, several studies have reported inhibition of henipavirus replication by ribavirin in vitro (73, 75, 124, 138, 139). However, in vivo studies carried out in animal models have not yielded promising results with ribavirin (75, 124). Ribavirin’s inability to cross the blood-brain barrier (BBB) may account for its inadequacy in in vivo studies. It has been previously shown that ribavirin is effective in the brain only when administered intracranially, but not intraperitonially in a hamster model (140). In the Malaysian epidemic, the effect of ribavirin in late-onset NiV encephalitis was not reported (137, 140). In addition, the complex molecular mechanism(s) of inhibition of viral replication by ribavirin, such as induction of error catastrophe and depletion of intracellular GTP pools, may not allow rapid design of more potent analogues (reviewed in (141)).
Chloroquine
Chloroquine (9-aminoquinoline) is used for the treatment of pathogens that require endosome acidification, such as malaria and pH-dependent viruses. Since the henipaviruses require endosomal cleavage of their F protein, it was not surprising that chloroquine was found to be a potential inhibitor of NiV infection in vitro (73–75). However, oral administration of chloroquine did not protect ferrets from lethal NiV infection (74) even though effective serum chloroquine concentrations was achieved, and peritoneal administration of chloroquine alone or in combination with ribavirin did not protect hamsters from lethal NiV or HeV challenges (74, 75). As with ribavirin, the lack of in vivo success with chloroquine may be due to its inability to cross the BBB or inadequate tissue distribution (142), as well as to its effects on the immune system which may not favor the host (143). In vitro vs. in vivo discrepancies in choroquine treatment results have also been reported for influenza, SARS, HIV, and Chikungunya viruses (143).
siRNA
An alternative way to inhibit viral gene expression is the use of small interfering RNA (siRNA) (144). In one recent study, siRNA molecules directed against the L and N genes were tested against minigenome and live henipavirus replication in vitro (145). While some siRNA had effects in both minigenome and live virus replication, some had effects only on minigenome replication, and others on neither. In addition, siRNA targeting more conserved genome sequences, for instance in P, V, or W, have been proposed (145). Although somewhat promising, one disadvantage of this approach is the need of gene therapy-based siRNA delivery methods, which might not be readily available.
Inhibitors of macropinocytosis
A recent report indicates that NiV can enter cells via macropinocytosis (146). This type of entry pathway for NiV necessitates phosphorylation of cytoplasmic domain of ephrinB2, after NiV-G attachment. Although it is not known whether this is a major pathway utilized for NiV entry, drugs that affect macropinocytosis, with the exception of chloroquine, affected NiV entry, but not cell-cell fusion (146). Two of the strongest inhibitors of NiV entry were latrunculin A and the amiloride analogue EIPA (5-(N-ethyl-N-isopropyl)amiloride). While the first one is likely hazardous in vivo, EIPA is a commonly used antihypertensive agent, and can be evaluated for its in vivo efficacy in animal models of henipavirus infections.
Favipiravir (T-705)
Favipiravir is a compound with promising broad-spectrum antiviral activities. Host enzymes metabolize its precursor into a ribofuranosyltriphosphate derivative that selectively inhibits viral RNA-dependent RNA polymerases, by reasons not fully understood (reviewed in (147)). Importantly, it does not inhibit host DNA or RNA synthesis, and is not cytotoxic to mammalian cells. In vivo experiments with T-705 against influenza virus, arenavirus, bunyaviruses, West Nile virus, yellow fever virus, and foot-and-mouth-disease virus, have shown one or more of the following results: protection from death, reduction of viral loads, and limitation of symptoms. In addition, protective effects of T-705 were observed when it was administered 1–7 days post-virus inoculation (see (147)). Although these pathogens were not paramyxoviruses, in vitro succeptibility of respiratory syncytial virus to T-705 has been observed (148), suggesting that favipiravir may serve as an antiviral against emerging paramyxoviruses.
Future of antiviral strategies
The various antiviral strategies discussed in this review are summarized in Table 1. In general, a better understanding of the structures and functions of viral and host proteins involved in the viral life cycle (Fig. 2) will aid in the development of new antiviral therapeutics. In addition, animal model experiments that examine the potential antivirals born from the in vitro studies described above are important, for example, as not all compounds can successfully cross the BBB. Because the emerging virus entry mechanisms have been explored in greater detail than the assembly and budding mechanisms, further progress in the elucidation of these late (and other) steps of the viral life cycle is imperative. Prompt antiviral discovery and characterization against emerging paramyxoviruses should be facilitated by the use of pseudotyped and reverse genetics viral systems.
Table 1.
Effect of antiviral agents on emerging paramyxovirus infections
| Life cycle step | Antiviral | Efficacy In vitro (on live virus) | Refs. |
|---|---|---|---|
| Soluble proteins: | IC50: | (59, 149) | |
| ephrinB2, B3 | ephrinB2: <10 μg/ml | ||
| EPhB3, B4 | ephrinB3: <25 μg/ml | ||
| NiV-G | EPhB3, EPhB4: >100 μg/ml | ||
| HeV-G | NiV-G: 13.2 μg/ml | ||
| HeV-G: 3.3 μg/ml | |||
| Attachment | mMabsa | IC90: 0.27–2.34 ng | (96, 150) |
| α-NiV-G | |||
| IC90, m101: <12.5 μg/ml | (54) | ||
| hMabsb | IC50, m102.4: | ||
| α-HeV-G | 0.04 mg/ml (NiV) | ||
| 0.6 mg/ml (HeV) | |||
| 2nd generation N-PEG NiV HR2 | IC50: 0.46–2.05 nM | (89) | |
| HPIV-3-F HR2 | IC50: 208 nM (NiV) | (91, 92) | |
| 179 nM (HeV) | |||
| Fusion | mMab | IC90: 1.6–425 ng | (96, 150) |
| α-NiV-F | |||
| Matrix | Quinolone derivatives bortezomib | IC50: 0.5–4 μM | (97) |
| IC50: 2.7 nM | (111) | ||
| IFN responses | Poly(I):poly(C12U)c | IC90: <6.25 μg/ml | (124) |
| LJ001 | IC50: ~ 1 μM | (130) | |
| Ribavirind | IC50: ~ 4 μM (~ 1 μg/ml) | (75, 124) | |
| IC90: ~ 100 μM (~25 μg/ml) | |||
| Broad-spectrum and other antivirals | Chloroquinee | IC50: 1 μM | (73–75) |
| IC90: 20–100 μM | |||
| siRNA | >60% Inhibition at 50 nM | (145) | |
| IC50: | (146) | ||
| Macropinocytic inhibitors | Latrunculin A: < 2 μM | ||
| EIPA: ~15 μM | |||
| Favipiravir | EC50, RSV: 260 μM | (148) |
100% Protection in vivo at 100–112 μg.
hMab:m102.4: Protection of 1/3 pre-infused, 3/3 post-infused (ferrets) at dose of 50 mg.
Protection 5/6 animals, at dose of 3 mg/kg q/d.
Survival increased + 1–3 days, at dose of 25–100 mg/kg.
No protection at 50–150 mg/kg.
Abreviations: NiV, Nipah virus; HeV, Hendra virus; Mab, monoclonal antibody; PEG, polyethylene glycol; HR2, heptad repeat 2; EIPA, 5-(N-Ethyl-N-isopropyl)-amiloride; RSV, Respiratory syncytial virus; siRNA, small interfering RNA.
Acknowledgments
Acknowledgements and funding
Supported by grants AI060694, AI069317, AI065359 (Pacific Southwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases), AI082100 and AI07495 from the US National Institutes of Health. We apologize to investigators whose work was not discussed because of space limitations. We sincerely thank our peer reviewers for their valuable comments and suggestions.
References
- 1.Human to-Human Transmission may be Implicated. Wildlife Trust; 2004. NIPAH Virus Breaks out in Bangladesh: Mortality Rates of 60% to 74% www.ewire.com/display.cfm/Wire_ID/2117. [Google Scholar]
- 2.Luby SP, et al. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg Infect Dis. 2009;15(8):1229–35. doi: 10.3201/eid1508.081237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luby SP, et al. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis. 2006;12(12):1888–94. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chua KB, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288(5470):1432–5. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 5.Halpin K, et al. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J Gen Virol. 2000;81(Pt 8):1927–32. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- 6.Virtue ER, Marsh GA, Wang LF. Paramyxoviruses infecting humans: the old, the new and the unknown. Future Microbiol. 2009;4:537–54. doi: 10.2217/fmb.09.26. [DOI] [PubMed] [Google Scholar]
- 7.Jun MH, Karabatsos N, Johnson RH. A new mouse paramyxovirus (J virus) Aust J Exp Biol Med Sci. 1977;55(6):645–7. doi: 10.1038/icb.1977.63. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, et al. Beilong virus, a novel paramyxovirus with the largest genome of non-segmented negative-stranded RNA viruses. Virology. 2006;346(1):219–28. doi: 10.1016/j.virol.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 9.Mesina JE, et al. The pathology of feral rodents in North Queensland. Tropenmed Parasitol. 1974;25(1):116–27. [PubMed] [Google Scholar]
- 10.Bossart KN, Broder CC. Developments towards effective treatments for Nipah and Hendra virus infection. Expert Rev Anti Infect Ther. 2006;4(1):43–55. doi: 10.1586/14787210.4.1.43. [DOI] [PubMed] [Google Scholar]
- 11.Vigant F, Lee B. Hendra and Nipah virus infection: pathology, models, and potential therapies. Infect Disord Drug Targets. doi: 10.2174/187152611795768097. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williamson MM, Torres-Velez FJ. Henipavirus: a review of laboratory animal pathology. Vet Pathol. 2010;47(5):871–80. doi: 10.1177/0300985810378648. [DOI] [PubMed] [Google Scholar]
- 13.WHO. Measles. 2009 www.who.int/mediacentre/factsheets/fs286/en/index.html.
- 14.WHO. Respiratory syncytial virus and parainfluenza viruses. 2009 http://www.who.int/vaccine_research/diseases/ari/en/index2.html.
- 15.Nipah virus outbreak(s) in Bangladesh, January-April 2004. Wkly Epidemiol Rec. 2004;79(17):168–71. [PubMed] [Google Scholar]
- 16.Person-to-person transmission of Nipah virus during outbreak in Fradipur District. Health Science Bull. 2004;2:5–9. [Google Scholar]
- 17.Gurley ES, et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis. 2007;13(7):1031–7. doi: 10.3201/eid1307.061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendra virus, human, equine - Australia (20080821.2606). ProMED-mail. International Society for Infectious Diseases. 2008 www.promedmail.org/pls/otn/f?p=2400:1202:2636675875700709.
- 19.Field HE, et al. A fatal case of Hendra virus infection in a horse in north Queensland: clinical and epidemiological features. Aust Vet J. 2000;78(4):279–80. doi: 10.1111/j.1751-0813.2000.tb11758.x. [DOI] [PubMed] [Google Scholar]
- 20.Field HE, et al. Epidemiological perspectives on Hendra virus infection in horses and flying foxes. Aust Vet J. 2007;85(7):268–70. doi: 10.1111/j.1751-0813.2007.00170.x. [DOI] [PubMed] [Google Scholar]
- 21.Young PL, et al. Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus. Emerg Infect Dis. 1996;2(3):239–40. doi: 10.3201/eid0203.960315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drexler JF, et al. Henipavirus RNA in African bats. PLoS One. 2009;4(7):e6367. doi: 10.1371/journal.pone.0006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iehle C, et al. Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg Infect Dis. 2007;13(1):159–61. doi: 10.3201/eid1301.060791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam SK. Nipah virus-a potential agent of bioterrorism? Antiviral Res. 2003;57(1–2):113–9. doi: 10.1016/s0166-3542(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 25.Tan CT, Wong KT. Nipah encephalitis outbreak in Malaysia. Ann Acad Med Singapore. 2003;32(1):112–7. [PubMed] [Google Scholar]
- 26.Dutch RE. Entry and fusion of emerging paramyxoviruses. PLoS Pathog. 2010;6(6):e1000881. doi: 10.1371/journal.ppat.1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamb RA, Jardetzky TS. Structural basis of viral invasion: lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17(4):427–36. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb RA, Paterson RG, Jardetzky TS. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology. 2006;344(1):30–7. doi: 10.1016/j.virol.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith EC, et al. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 2009;276(24):7217–27. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menendez-Arias L. Molecular basis of human immunodeficiency virus drug resistance: an update. Antiviral Res. 2010;85(1):210–31. doi: 10.1016/j.antiviral.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Stellbrink HJ. Novel compounds for the treatment of HIV type-1 infection. Antivir Chem Chemother. 2009;19(5):189–200. doi: 10.1177/095632020901900502. [DOI] [PubMed] [Google Scholar]
- 32.Eaton BT, et al. Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol. 2006;4(1):23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuentes SM, et al. Phosphorylation of paramyxovirus phosphoprotein and its role in viral gene expression. Future Microbiol. 2010;5:9–13. doi: 10.2217/fmb.09.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez JJ, Horvath CM. Host evasion by emerging paramyxoviruses: Hendra virus and Nipah virus v proteins inhibit interferon signaling. Viral Immunol. 2004;17(2):210–9. doi: 10.1089/0882824041310568. [DOI] [PubMed] [Google Scholar]
- 35.Bowden TA, et al. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J Virol. 2008;82(23):11628–36. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu K, et al. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci U S A. 2008;105(29):9953–8. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bishop KA, et al. Residues in the stalk domain of the Hendra virus G glycoprotein modulate conformational changes associated with receptor binding. J Virol. 2008;82(22):11398–11409. doi: 10.1128/JVI.02654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowden TA, et al. Dimeric architecture of the Hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J Virol. 2010;84(12):6208–17. doi: 10.1128/JVI.00317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levroney EL, et al. Novel innate immune functions for galectin-1: galectin-1 inhibits cell fusion by Nipah virus envelope glycoproteins and augments dendritic cell secretion of proinflammatory cytokines. J Immunol. 2005;175(1):413–20. doi: 10.4049/jimmunol.175.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonaparte MI, et al. From The Cover: Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A. 2005;102(30):10652–7. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Negrete OA, et al. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436(7049):401–5. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 42.Negrete OA, et al. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2006;2(2):e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133(1):38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Goh KJ, et al. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med. 2000;342(17):1229–35. doi: 10.1056/NEJM200004273421701. [DOI] [PubMed] [Google Scholar]
- 45.White JM, et al. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aguilar HC, et al. A Novel Receptor-induced Activation Site in the Nipah Virus Attachment Glycoprotein (G) Involved in Triggering the Fusion Glycoprotein (F) J Biol Chem. 2009;284(3):1628–35. doi: 10.1074/jbc.M807469200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng R, et al. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology. 1995;209(2):457–69. doi: 10.1006/viro.1995.1278. [DOI] [PubMed] [Google Scholar]
- 48.Lee JK, et al. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem. 2008;283(24):16561–72. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melanson VR, Iorio RM. Amino acid substitutions in the F-specific domain in the stalk of the newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J Virol. 2004;78(23):13053–61. doi: 10.1128/JVI.78.23.13053-13061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paal T, et al. Probing the spatial organization of measles virus fusion complexes. J Virol. 2009;83(20):10480–93. doi: 10.1128/JVI.01195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanabayashi K, Compans RW. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J Virol. 1996;70(9):6112–8. doi: 10.1128/jvi.70.9.6112-6118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsurudome M, et al. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology. 1995;213(1):190–203. doi: 10.1006/viro.1995.1559. [DOI] [PubMed] [Google Scholar]
- 53.Bowden TA, et al. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol. 2008;15(6):567–72. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- 54.Bossart KN, et al. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog. 2009;5(10):e1000642. doi: 10.1371/journal.ppat.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Z, et al. Exceptionally potent cross-reactive neutralization of Nipah and Hendra viruses by a human monoclonal antibody. J Infect Dis. 2008;197(6):846–53. doi: 10.1086/528801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Z, et al. Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J Virol. 2006;80(2):891–9. doi: 10.1128/JVI.80.2.891-899.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu H, et al. Development of motavizumab, an ultra-potent antibody for the prevention of respiratory syncytial virus infection in the upper and lower respiratory tract. J Mol Biol. 2007;368(3):652–65. doi: 10.1016/j.jmb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 58.Fenton C, Scott LJ, Plosker GL. Palivizumab: a review of its use as prophylaxis for serious respiratory syncytial virus infection. Paediatr Drugs. 2004;6(3):177–97. doi: 10.2165/00148581-200406030-00004. [DOI] [PubMed] [Google Scholar]
- 59.Bossart KN, et al. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J Virol. 2005;79(11):6690–702. doi: 10.1128/JVI.79.11.6690-6702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee B, Ataman ZA, Jin L. Evil versus ‘eph-ective’ use of ephrin-B2. Nat Struct Mol Biol. 2008;15(6):540–2. doi: 10.1038/nsmb0608-540. [DOI] [PubMed] [Google Scholar]
- 61.Begona Ruiz-Arguello M, et al. Effect of proteolytic processing at two distinct sites on shape and aggregation of an anchorless fusion protein of human respiratory syncytial virus and fate of the intervening segment. Virology. 2002;298(2):317–26. doi: 10.1006/viro.2002.1497. [DOI] [PubMed] [Google Scholar]
- 62.Garten W, et al. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie. 1994;76(3–4):217–25. doi: 10.1016/0300-9084(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 63.Gonzalez-Reyes L, et al. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci U S A. 2001;98(17):9859–64. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ortmann D, et al. Proteolytic cleavage of wild type and mutants of the F protein of human parainfluenza virus type 3 by two subtilisin-like endoproteases, furin and Kex2. J Virol. 1994;68(4):2772–6. doi: 10.1128/jvi.68.4.2772-2776.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe M, et al. Engineered serine protease inhibitor prevents furin-catalyzed activation of the fusion glycoprotein and production of infectious measles virus. J Virol. 1995;69(5):3206–10. doi: 10.1128/jvi.69.5.3206-3210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murakami M, et al. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur J Biochem. 2001;268(10):2847–55. doi: 10.1046/j.1432-1327.2001.02166.x. [DOI] [PubMed] [Google Scholar]
- 67.van den Hoogen BG, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7(6):719–24. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diederich S, et al. The Nipah Virus Fusion Protein Is Cleaved within the Endosomal Compartment. J Biol Chem. 2005;280(33):29899–903. doi: 10.1074/jbc.M504598200. [DOI] [PubMed] [Google Scholar]
- 69.Diederich S, Thiel L, Maisner A. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology. 2008;375(2):391–400. doi: 10.1016/j.virol.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pager CT, et al. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology. 2006;346(2):251–7. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pager CT, Dutch RE. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J Virol. 2005;79(20):12714–20. doi: 10.1128/JVI.79.20.12714-12720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shah PP, et al. A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol Pharmacol. 2010;78(2):319–24. doi: 10.1124/mol.110.064261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Porotto M, et al. Simulating henipavirus multicycle replication in a screening assay leads to identification of a promising candidate for therapy. J Virol. 2009;83(10):5148–55. doi: 10.1128/JVI.00164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pallister J, et al. Chloroquine administration does not prevent Nipah virus infection and disease in ferrets. J Virol. 2009;83(22):11979–82. doi: 10.1128/JVI.01847-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Freiberg AN, et al. Combined chloroquine and ribavirin treatment does not prevent death in a hamster model of Nipah and Hendra virus infection. J Gen Virol. 2010;91(Pt 3):765–72. doi: 10.1099/vir.0.017269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bagai S, Lamb RA. Individual roles of N-linked oligosaccharide chains in intracellular transport of the paramyxovirus SV5 fusion protein. Virology. 1995;209(1):250–6. doi: 10.1006/viro.1995.1251. [DOI] [PubMed] [Google Scholar]
- 77.von Messling V, Cattaneo R. N-linked glycans with similar location in the fusion protein head modulate paramyxovirus fusion. J Virol. 2003;77(19):10202–12. doi: 10.1128/JVI.77.19.10202-10212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aguilar HC, et al. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol. 2006;80(10):4878–89. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carter JR, et al. Role of N-linked glycosylation of the Hendra virus fusion protein. J Virol. 2005;79(12):7922–5. doi: 10.1128/JVI.79.12.7922-7925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Camby I, et al. Galectin-1: a small protein with major functions. Glycobiology. 2006;16(11):137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- 81.Garner OB, et al. Endothelial galectin-1 binds to specific glycans on nipah virus fusion protein and inhibits maturation, mobility, and function to block syncytia formation. PLoS Pathog. 2010;6(7):e1000993. doi: 10.1371/journal.ppat.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iida A, et al. Fine-scale SNP map of an 11-kb genomic region at 22q13.1 containing the galectin-1 gene. J Hum Genet. 2005;50(1):42–5. doi: 10.1007/s10038-004-0218-4. [DOI] [PubMed] [Google Scholar]
- 83.He Y, et al. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J Biol Chem. 2008;283(17):11126–34. doi: 10.1074/jbc.M800200200. [DOI] [PubMed] [Google Scholar]
- 84.Makinson A, Reynes J. The fusion inhibitor enfuvirtide in recent antiretroviral strategies. Curr Opin HIV AIDS. 2009;4(2):150–8. doi: 10.1097/COH.0b013e32832498d8. [DOI] [PubMed] [Google Scholar]
- 85.Poveda E, Briz V, Soriano V. Enfuvirtide, the first fusion inhibitor to treat HIV infection. AIDS Rev. 2005;7(3):139–47. [PubMed] [Google Scholar]
- 86.Xu Y, et al. Crystallization and preliminary crystallographic analysis of the fusion core from two new zoonotic paramyxoviruses, Nipah virus and Hendra virus. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 6):1161–4. doi: 10.1107/S0907444904009515. [DOI] [PubMed] [Google Scholar]
- 87.Aguilar HC, et al. A quantitative and kinetic fusion protein-triggering assay can discern distinct steps in the nipah virus membrane fusion cascade. J Virol. 2010;84(16):8033–41. doi: 10.1128/JVI.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aguilar HC, et al. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J Virol. 2007;81(9):4520–32. doi: 10.1128/JVI.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bossart KN, et al. Inhibition of Henipavirus fusion and infection by heptad-derived peptides of the Nipah virus fusion glycoprotein. Virol J. 2005;2(1):57. doi: 10.1186/1743-422X-2-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lambert DM, et al. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci U S A. 1996;93(5):2186–91. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Porotto M, et al. Molecular determinants of antiviral potency of paramyxovirus entry inhibitors. J Virol. 2007;81(19):10567–74. doi: 10.1128/JVI.01181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Porotto M, et al. Inhibition of hendra virus fusion. J Virol. 2006;80(19):9837–49. doi: 10.1128/JVI.00736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. Embo J. 2001;20(15):4024–34. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Porotto M, et al. Viral entry inhibitors targeted to the membrane site of action. J Virol. 2010;84(13):6760–8. doi: 10.1128/JVI.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Porotto M, et al. Inhibition of Nipah Virus Infection In Vivo: Targeting an Early Stage of Paramyxovirus Fusion Activation during Viral Entry. PLoS Pathog. 2010;6(10):e1001168. doi: 10.1371/journal.ppat.1001168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guillaume V, et al. Antibody prophylaxis and therapy against Nipah virus infection in hamsters. J Virol. 2006;80(4):1972–8. doi: 10.1128/JVI.80.4.1972-1978.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Niedermeier S, et al. A small-molecule inhibitor of Nipah virus envelope protein-mediated membrane fusion. J Med Chem. 2009;52(14):4257–65. doi: 10.1021/jm900411s. [DOI] [PubMed] [Google Scholar]
- 98.Harrison MS, Sakaguchi T, Schmitt AP. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol. 2010;42(9):1416–29. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takimoto T, Portner A. Molecular mechanism of paramyxovirus budding. Virus Res. 2004;106(2):133–45. doi: 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 100.Money VA, et al. Surface features of a Mononegavirales matrix protein indicate sites of membrane interaction. Proc Natl Acad Sci U S A. 2009;106(11):4441–6. doi: 10.1073/pnas.0805740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dessen A, et al. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 2000;19(16):4228–36. doi: 10.1093/emboj/19.16.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Coronel EC, et al. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J Virol. 1999;73(8):7035–8. doi: 10.1128/jvi.73.8.7035-7038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sugahara F, et al. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology. 2004;325(1):1–10. doi: 10.1016/j.virol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 104.Pantua HD, et al. Requirements for the assembly and release of Newcastle disease virus-like particles. J Virol. 2006;80(22):11062–73. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pohl C, et al. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J Gen Virol. 2007;88(Pt 4):1243–50. doi: 10.1099/vir.0.82578-0. [DOI] [PubMed] [Google Scholar]
- 106.Runkler N, et al. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell Microbiol. 2007;9(5):1203–14. doi: 10.1111/j.1462-5822.2006.00860.x. [DOI] [PubMed] [Google Scholar]
- 107.Ciancanelli MJ, Basler CF. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol. 2006;80(24):12070–8. doi: 10.1128/JVI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Patch JR, et al. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol J. 2007;4:1. doi: 10.1186/1743-422X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yin R, et al. Inhibition of Newcastle disease virus replication by RNA interference targeting the matrix protein gene in chicken embryo fibroblasts. J Virol Methods. 2010;167(1):107–11. doi: 10.1016/j.jviromet.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 110.Schmitt AP, et al. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol. 2005;79(5):2988–97. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang YE, et al. Ubiquitin-regulated nuclear-cytoplasmic trafficking of the Nipah virus matrix protein is important for viral budding. PLoS Pathog. 2010;6(11):e1001186. doi: 10.1371/journal.ppat.1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weber F, Kochs G, Haller O. Inverse interference: how viruses fight the interferon system. Viral Immunol. 2004;17(4):498–515. doi: 10.1089/vim.2004.17.498. [DOI] [PubMed] [Google Scholar]
- 113.Yokota S, Okabayashi T, Fujii N. The battle between virus and host: modulation of Toll-like receptor signaling pathways by virus infection. Mediators Inflamm. 2010;2010:184328. doi: 10.1155/2010/184328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Magoffin DE, Mackenzie JS, Wang LF. Genetic analysis of J-virus and Beilong virus using minireplicons. Virology. 2007;364(1):103–11. doi: 10.1016/j.virol.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 115.Dillon PJ, Parks GD. Role for the phosphoprotein P subunit of the paramyxovirus polymerase in limiting induction of host cell antiviral responses. J Virol. 2007;81(20):11116–27. doi: 10.1128/JVI.01360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cadd T, et al. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70(8):5067–74. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reutter GL, et al. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology. 2001;285(1):100–9. doi: 10.1006/viro.2001.0962. [DOI] [PubMed] [Google Scholar]
- 118.Sleeman K, et al. The C, V and W proteins of Nipah virus inhibit minigenome replication. J Gen Virol. 2008;89(Pt 5):1300–8. doi: 10.1099/vir.0.83582-0. [DOI] [PubMed] [Google Scholar]
- 119.Fontana JM, Bankamp B, Rota PA. Inhibition of interferon induction and signaling by paramyxoviruses. Immunol Rev. 2008;225:46–67. doi: 10.1111/j.1600-065X.2008.00669.x. [DOI] [PubMed] [Google Scholar]
- 120.Ramachandran A, Horvath CM. Paramyxovirus disruption of interferon signal transduction: STATus report. J Interferon Cytokine Res. 2009;29(9):531–7. doi: 10.1089/jir.2009.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Foster G, Mathurin P. Hepatitis C virus therapy to date. Antivir Ther. 2008;13(Suppl 1):3–8. [PubMed] [Google Scholar]
- 122.Maher SG, et al. Interferon: cellular executioner or white knight? Curr Med Chem. 2007;14(12):1279–89. doi: 10.2174/092986707780597907. [DOI] [PubMed] [Google Scholar]
- 123.Pfeffer LM, et al. Biological properties of recombinant alpha-interferons: 40th anniversary of the discovery of interferons. Cancer Res. 1998;58(12):2489–99. [PubMed] [Google Scholar]
- 124.Georges-Courbot MC, et al. Poly(I)-poly(C12U) but not ribavirin prevents death in a hamster model of Nipah virus infection. Antimicrob Agents Chemother. 2006;50(5):1768–72. doi: 10.1128/AAC.50.5.1768-1772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barnard DL, et al. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir Chem Chemother. 2006;17(5):275–84. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- 126.Thompson KA, et al. Results of a double-blind placebo-controlled study of the double-stranded RNA drug polyI:polyC12U in the treatment of HIV infection. Eur J Clin Microbiol Infect Dis. 1996;15(7):580–7. doi: 10.1007/BF01709367. [DOI] [PubMed] [Google Scholar]
- 127.Aljofan M, et al. Characteristics of Nipah virus and Hendra virus replication in different cell lines and their suitability for antiviral screening. Virus Res. 2009;142(1–2):92–9. doi: 10.1016/j.virusres.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Phillips AN, et al. Risk of extensive virological failure to the three original antiretroviral drug classes over long-term follow-up from the start of therapy in patients with HIV infection: an observational cohort study. Lancet. 2007;370(9603):1923–8. doi: 10.1016/S0140-6736(07)61815-7. [DOI] [PubMed] [Google Scholar]
- 129.Pillay D. The priorities for antiviral drug resistance surveillance and research. J Antimicrob Chemother. 2007;60(Suppl 1):i57–8. doi: 10.1093/jac/dkm159. [DOI] [PubMed] [Google Scholar]
- 130.Wolf MC, et al. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A. 2010;107(7):3157–62. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438(7068):590–6. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 132.Aljofan M, et al. Antiviral activity of gliotoxin, gentian violet and brilliant green against Nipah and Hendra virus in vitro. Virol J. 2009;6:187. doi: 10.1186/1743-422X-6-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Aljofan M, et al. Off Label Antiviral Therapeutics for Henipaviruses: New Light Through Old Windows. J Antivir Antiretrovir. 2010;2(1):1–10. doi: 10.4172/jaa.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Olszewska W, Openshaw P. Emerging drugs for respiratory syncytial virus infection. Expert Opin Emerg Drugs. 2009;14(2):207–17. doi: 10.1517/14728210902946399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Snell NJ. Ribavirin--current status of a broad spectrum antiviral agent. Expert Opin Pharmacother. 2001;2(8):1317–24. doi: 10.1517/14656566.2.8.1317. [DOI] [PubMed] [Google Scholar]
- 136.Parker WB. Metabolism and antiviral activity of ribavirin. Virus Res. 2005;107(2):165–71. doi: 10.1016/j.virusres.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 137.Chong HT, et al. Treatment of acute Nipah encephalitis with ribavirin. Ann Neurol. 2001;49(6):810–3. doi: 10.1002/ana.1062. [DOI] [PubMed] [Google Scholar]
- 138.Aljofan M, et al. Development and validation of a chemiluminescent immunodetection assay amenable to high throughput screening of antiviral drugs for Nipah and Hendra virus. J Virol Methods. 2008;149(1):12–9. doi: 10.1016/j.jviromet.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wright PJ, Crameri G, Eaton BT. RNA synthesis during infection by Hendra virus: an examination by quantitative real-time PCR of RNA accumulation, the effect of ribavirin and the attenuation of transcription. Arch Virol. 2005;150(3):521–32. doi: 10.1007/s00705-004-0417-5. [DOI] [PubMed] [Google Scholar]
- 140.Honda Y, et al. Effect of ribavirin on subacute sclerosing panencephalitis virus infections in hamsters. Antimicrob Agents Chemother. 1994;38(4):653–5. doi: 10.1128/aac.38.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Leyssen P, De Clercq E, Neyts J. Molecular strategies to inhibit the replication of RNA viruses. Antiviral Res. 2008;78(1):9–25. doi: 10.1016/j.antiviral.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Koreeda A, et al. Immunohistochemical demonstration of the distribution of chloroquine (CQ) and its metabolites in CQ-poisoned mice. Arch Toxicol. 2007;81(7):471–8. doi: 10.1007/s00204-007-0185-6. [DOI] [PubMed] [Google Scholar]
- 143.Savarino A, et al. Effects of chloroquine on viral infections: an old drug against today’s diseases? Lancet Infect Dis. 2003;3(11):722–7. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–33. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mungall BA, et al. Inhibition of Henipavirus infection by RNA interference. Antiviral Res. 2008;80(3):324–31. doi: 10.1016/j.antiviral.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pernet O, et al. Nipah virus entry can occur by macropinocytosis. Virology. 2009;395(2):298–311. doi: 10.1016/j.virol.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 147.Furuta Y, et al. T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 2009;82(3):95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Furuta Y, et al. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob Agents Chemother. 2002;46(4):977–81. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bossart KN, et al. Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology. 2008;372(2):357–71. doi: 10.1016/j.virol.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 150.Guillaume V, et al. Acute Hendra virus infection: Analysis of the pathogenesis and passive antibody protection in the hamster model. Virology. 2009;387(2):459–65. doi: 10.1016/j.virol.2009.03.001. [DOI] [PubMed] [Google Scholar]
Further reading
- Dutch RE. Entry and fusion of emerging paramyxoviruses. PLoS Pathogens. 2010;6(6):e1000881. doi: 10.1371/journal.ppat.1000881. Brief review on entry of emerging paramyxoviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Ataman ZA, Jin L. Evil versus ‘eph-ective’ use of ephrin-B2. Nat Struct Mol Biol. 2008;15(6):540–2. doi: 10.1038/nsmb0608-540. Review on Henipavirus receptor usage during entry. [DOI] [PubMed] [Google Scholar]
- Smith EC, et al. Viral entry mechanisms: the increasing diversity of Paramyxovirus entry. FEBS J. 2009;276(24):7217–7227. doi: 10.1111/j.1742-4658.2009.07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Review on diversity of Paramyxovirus entry Williamson MM, Torres-Velez FJ. Henipavirus: a review of laboratory animal pathology. Review. Vet Pathol Online. 2010;47:871. doi: 10.1177/0300985810378648. Review on animal Henipavirus studies.
- Vigant F, Lee B. Hendra and Nipah virus infection: pathology, models, and potential therapies. Infect Disord Drug Targets. doi: 10.2174/187152611795768097. (In Press) Review on Henipavirus antivirals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtue ER, Marsh GA, Wang L-F. Paramyxoviruses infecting humans: the old, the new and the unknown. Future Medicine. 2009;4(5):537–554. doi: 10.2217/fmb.09.26. Review on human-infecting Paramyxoviruses. [DOI] [PubMed] [Google Scholar]