Abstract
Fifteen years of genetic research in Parkinson’s disease (PD) have led to the identification of several monogenic forms of the disorder and of numerous genetic risk factors increasing the risk to develop PD. Monogenic forms, caused by a single mutation in a dominantly or recessively inherited gene, are well-established, albeit relatively rare types of PD. They collectively account for about 30% of the familial and 3%–5% of the sporadic cases. In this article, we will summarize the current knowledge and understanding of the molecular genetics of PD. In brief, we will review familial forms of PD, basic genetic principles of inheritance (and their exceptions in PD), followed by current methods for the identification of PD genes and risk factors, and implications for genetic testing.
Currently, 28 chromosomal regions are thought to be related to Parkinson’s disease (PD). Six contain genes with mutations that conclusively, albeit rarely, cause monogenic PD.
In 1996, the mapping and subsequent identification of the first mutations responsible for Parkinson’s disease (PD) indisputably showed that PD may be hereditary (Polymeropoulos et al. 1996, 1997). In the two years to follow, genetic links of PD to two new chromosomal regions were reported, and linkage to the first gene was excluded in a large number of families (Munoz et al. 1997; Scott et al. 1997, 1999; Farrer et al. 1998). Thus, it became clear that PD is a genetically heterogeneous and most likely, complex disorder.
Just how complex it is, is underlined by the notion that today, nearly 15 years later, we know of 28 distinct chromosomal regions more or less convincingly related to PD. Only six of these specific regions contain genes with mutations that conclusively cause monogenic PD; that is, a form of the disease for which a mutation in a single gene is sufficient to cause the phenotype. Even collectively, mutations in these six genes explain only a limited number (3%–5%) of sporadic disease occurrences. Rather, the etiology of PD is multifactorial, which probably results from an elaborate interplay of mostly unknown factors: several genes, modifying effects by susceptibility alleles, environmental exposures and gene-environment interactions (e.g., influence of environmental agents on gene expression), and their direct impact on the developing and aging brain.
In this article, we will summarize the current knowledge and understanding of the molecular genetics of PD and outline its basic principles. First, we will describe the present genetic “classification” of PD and address some of the existing inconsistences thereof. Second, we will explain basic genetic principles of inheritance of monogenic disorders and will define the exceptions that a researcher trying to define the inheritance pattern of PD in a particular family needs to be aware of. Third, we will describe state-of-the-art methods for the identification of new PD genes and risk factors. Finally, we will cover the most important genes contributing to the pathogenesis of PD and discuss which patients should be considered for diagnostic genetic testing.
GENETIC CLASSIFICATION OF PD
In the current PD genetics nomenclature, 18 specific chromosomal regions, also called chromosomal locus, are termed PARK (to denote their putative link to PD), and numbered in chronological order of their identification (PARK1, PARK2, PARK3, etc.) (Table 1). In addition to being an incomplete list of known PD-related genes, this classification system, unfortunately, has a number of inconsistencies. It comprises confirmed loci, as well as those for which linkage or association could not be replicated (nonconfirmed). The causative gene has not yet been identified for all of the loci, nor do all of the identified genes contain causative or disease-determining mutations (i.e., variations in some of these genes are considered genetic risk factors increasing the risk to develop PD rather than being a sufficient cause). Finally, one locus, PARK4, was designated as a novel chromosomal region associated with PD (Farrer et al. 1999; Singleton et al. 2003) but was later found to be identical with PARK1 (SNCA-associated PD) (Singleton et al. 2003). It is noteworthy that some of the loci have been identified by genetic linkage analysis in large families, some based on the known function of the protein product of the gene they contain, yet others have been established by genome-wide association studies performed on a population level. A list of the PARK PD-related genes and loci is given in Table 1, along with their clinical classification, inheritance pattern (where applicable), gene (when known), status (confirmed/nonconfirmed), and mode of identification.
Table 1.
PARK-designated PD-related loci
| Symbol | Gene locus | Disorder | Inheritance | Gene | Status and remarks | Mode of identification |
|---|---|---|---|---|---|---|
| PARK1 | 4q21-22 | EOPD | AD | SNCA | Confirmed | Linkage analysis |
| PARK2 | 6q25.2–q27 | EOPD | AR | Parkin | Confirmed | Linkage analysis |
| PARK3 | 2p13 | Classical PD | AD | Unknown | Unconfirmed; may represent a risk factor; gene not found since first described in 1998 | Linkage analysis |
| PARK4 | 4q21–q23 | EOPD | AD | SNCA | Erroneous locus (identical to PARK1) | Linkage analysis |
| PARK5 | 4p13 | Classical PD | AD | UCHL1 | Unconfirmed (not replicated since described in 1998) | Functional candidate gene approach |
| PARK6 | 1p35–p36 | EOPD | AR | PINK1 | Confirmed | Linkage analysis |
| PARK7 | 1p36 | EOPD | AR | DJ-1 | Confirmed | Linkage analysis |
| PARK8 | 12q12 | Classical PD | AD | LRRK2 | Confirmed; variations in LRRK2 gene include risk-conferring variants and disease-causing mutations | Linkage analysis |
| PARK9 | 1p36 | Kufor-Rakeb syndrome; atypical PD with dementia, spasticity, and supranuclear gaze palsy | AR | ATP13A2 | Confirmed; but complex phenotype that would not be mistaken for early-onset or classical parkinsonism | Linkage analysis |
| PARK10 | 1p32 | Classical PD | Risk factor | Unknown | Confirmed susceptibility locus; gene unknown since first described in 2002 | Linkage analysis |
| PARK11 | 2q36-27 | Late-onset PD | AD | Unknown; not GIGYF2 | Not independently confirmed; possibly represents a risk factor; gene not found since first described in 2002 | Linkage analysis |
| PARK12 | Xq21–q25 | Classical PD | Risk factor | Unknown | Confirmed susceptibility locus; possibly represents a risk factor; gene not found since first described in 2003 | Linkage analysis |
| PARK13 | 2p12 | Classical PD | AD or risk factor | HTRA2 | Unconfirmed | Candidate gene approach |
| PARK14 | 22q13.1 | Early-onset dystonia-parkinsonism | AR | PLA2G6 | Confirmed | Linkage analysis (homozygosity mapping) |
| PARK15 | 22q12–q13 | Early-onset parkinsonian-pyramidal syndrome | AR | FBX07 | Confirmed | Linkage analysis |
| PARK16 | 1q32 | Classical PD | Risk factor | Unknown | Confirmed susceptibility locus | Genome-wide association studies |
| PARK17 | 16q11.2 | Classical PD | AD | VPS35 | Confirmed | Exome sequencing |
| PARK18 | 3q27.1 | Classical PD | AD | EIF4G1 | Unconfirmed; recently published (Chartier-Harlin et al. 2011) | Linkage analysis |
AD, autosomal dominant; AR, autosomal recessive.
GENETIC PRINCIPLES AND EXCEPTIONS THEREOF IN FAMILIAL PD
The majority of PD cases are sporadic, i.e., only about 10% of patients report a positive family history (Thomas and Beal 2007). Out of the six genes unequivocally linked to heritable, monogenic PD, mutations in SNCA (PARK1 = 4), and LRRK2 (PARK8) are responsible for autosomal-dominant PD forms, and mutations in Parkin (PARK2), PINK1 (PARK6), DJ-1 (PARK7), and ATP13A2 (PARK9) are accountable for PD that displays an autosomal recessive (AR) mode of inheritance.
In general, the inheritance patterns of human disorders are identified by examining the way the disorders are transmitted in the family of the index patient. Such a pedigree analysis requires a careful assembly of the disease records of the family members over several generations, and if possible, examination and sample collection from affected and unaffected individuals from the pedigree. All of the currently known monogenic PD forms are autosomal (dominant or recessive), which means that they are linked with regions on autosomes (chromosomes other than sex chromosomes).
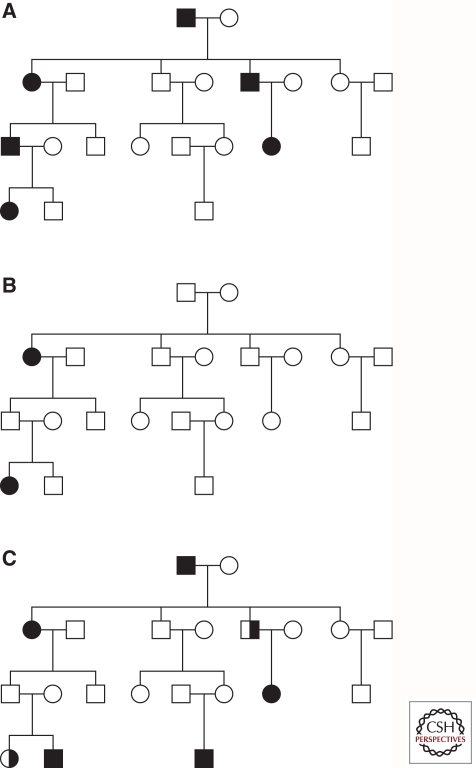
In autosomal-dominant disorders, one mutated allele of the gene is enough to cause the disease. Thus, defining features seen in a family tree with an autosomal-dominant inheritance pattern are (1) every affected person in the pedigree must have at least one affected parent, (2) at least one affected individual is present in every generation, and (3) on average, an affected individual will transmit the mutant gene to half of his or her children (Fig. 1A). In AR disorders two mutations (the same—homozygous, or different—compound-heterozygous), one on each gene copy (allele), are necessary to cause the phenotype. Heterozygous mutation carriers (individuals carrying a pathogenic mutation only on one allele) are phenotypically normal, i.e., unaffected. A clear AR mode of inheritance has three determining characteristics: (1) affected family members have two unaffected parents, both of whom are heterozygous, (2) children of the affected individual are also unaffected and heterozygous, and (3) as both of the affected patients’ parents are expected to be heterozygous carriers of the mutated gene, only one in four children (25%) is affected (Fig. 1B). Thus, the most striking difference between these two types of pedigrees is that in AR, a “generation skipping” is present. Of note, in autosomal-linked pedigrees, the number of affected males and females is nearly equal, whereas families with an unequal representation of affected men and women can imply a sex chromosome-linked inheritance pattern.
Figure 1.
Pedigrees showing different inheritance patterns. (A) Autosomal dominant disease inheritance pattern. Every affected person in the pedigree has one affected parent, one affected individual is present in every generation, and on average, an affected individual transmits the mutant gene (and thus the disease) to half of his or her children. (B) Autosomal recessive disease inheritance pattern. The affected individuals have two unaffected parents (both heterozygous mutation carriers), children of the affected individual are also unaffected and heterozygous, and only one of the four siblings (25%) is affected. (C) Autosomal dominant pedigree complicated by the presence of reduced penetrance (not all individuals with a mutation are affected), variable expressivity (not all individuals with a mutation show the expected disorder phenotype), and phenocopies (some individuals show clinical symptoms of the disorder without carrying the disease-causing mutation). Black symbols - affected individuals; white symbols - unaffected individuals; half-filled symbols - individuals with only resting tremor and/or bradykinesia.
Why is only one mutation sufficient to cause an autosomal-dominant disorder, whereas in recessive forms both alleles have to be altered? When only one allele is mutated and the other one is normal (wild-type), the mutated allele, depending on the type of mutations it carries, can provoke an autosomal-dominant phenotype through one of three scenarios: (1) haploinsufficiency, i.e., a single copy of a normal allele is incapable of providing sufficient protein production to assure normal function; (2) a dominant negative effect, i.e., a nonfunctional mutant polypeptide is produced that interferes with the function of the normal allele; (3) gain-of-function mutations change the gene product in such a way that it gains a new and abnormal function, causing the phenotype.
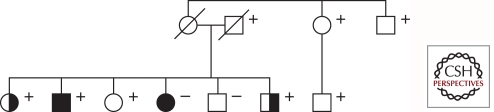
In clinical practice, however, pedigrees rarely follow the aforementioned well-defined Mendelian inheritance patterns, i.e., they are frequently complicated by reduced penetrance, variable expressivity, and phenocopy phenomena, as illustrated in Figures 1C and 2. Penetrance of the disease-causing mutation is defined through the proportion of individuals carrying this mutation who manifest the disorder. Penetrance depends on both the genotype (e.g., the presence and effect of modifier genes on the disease-causing gene) and the environment. Expressivity describes the degree (severity) to which a penetrant mutation is phenotypically expressed in an individual. Traditionally, a phenocopy was defined as an environmentally induced, nonhereditary phenotype that mimics one produced by a gene. In recent years, usage of the term “phenocopy” has shifted to now also include (1) patients with mutations different from the main genetic cause in a given family and (2) patients with an unknown environmental or genetic cause of the disorder, in both cases resulting in the same clinical syndrome or phenotype as in the carriers of the familial mutation (Klein et al. 2011). In addition, a large number of PD patients carrying only a single heterozygous mutation in the recessive genes Parkin or PINK1, raises the intriguing question of whether the much more common heterozygous mutations contribute to the development of parkinsonism in a subset of cases. One possible explanation is that different mutations, based on their type and position in the mutant protein, lead to biochemical consequences of varying severity (loss of function, gain of toxic function). Several lines of evidence suggest that heterozygous Parkin and PINK1 mutations are indeed a susceptibility factor, however, this topic is highly debated. Important insights into the role of heterozygous mutations come from the detailed neurological assessment of heterozygous relatives of index patients that are known to carry two mutations (Khan et al. 2005; Criscuolo et al. 2006; Hedrich et al. 2006; Hiller et al. 2007; Eggers et al. 2010). Notably, the identification of affected individuals in successive generations makes it unlikely that a second mutation in the respective gene had been missed, as may be the case in case-only and case-control studies.
Figure 2.
Pedigree of a PD family that comprises affected members with and without the LRRK2 p.G2019S mutation. Five mutation carriers are unaffected, showing reduced penetrance, two mutation carriers are affected with dystonia, showing variable expressivity, and one affected family member does not have the p.G2019S mutation in LRRK2. Black symbols - affected individuals; white symbols - unaffected individuals; half-filled symbols - individuals with dystonia; + - mutation carriers.
In summary, reduced penetrance, variable expressivity, affected single heterozygous mutation carriers, and phenocopies pose problems when trying to decide if a patient has a positive family history of PD, or when trying to determine which gene-identification or genetic-testing method to use. Owing to incomplete penetrance, autosomal-dominant disorders may seem to “skip generations,” and thus they might be falsely categorized as AR. In addition, because of a very mild or even clinically distinct phenotype, resulting from the variable expressivity, some mutation carriers might be misdiagnosed and not considered affected by the same disease. Also, some patients classified as affected by the same disease as patients in the rest of the family, may actually not carry the (same) disease-causing genotype. Finally, some patients affected by an AR form of disease and carrying only a single heterozygous mutation may cause the pedigree to erroneously look autosomal dominant.
IDENTIFICATION OF NEW GENES AND RISK FACTORS FOR PD
New PD-linked genes or PD risk factors can be identified by gene mapping or candidate gene approaches. Gene mapping in human diseases is the localization of genes underlying the clinical phenotypes of the disease on the basis of correlation with DNA variants (polymorphic markers), without the need for prior hypotheses about biological function. Genetic mapping methods include linkage analysis and genome-wide association studies. Alternatively, based on their known function, levels of expression, or mode of interaction (candidate gene approach), some genes can be considered plausible candidates, and as such, tested for in cohorts of patients.
The gene underlying any heritable form of human disease can be mapped and identified by linkage analysis if the DNA samples from a sufficient number of affected and unaffected family members are available. The first step in classical linkage analysis is to hypothesize the mode of inheritance of the disease on the basis of a constructed pedigree. However, this is frequently complicated by reduced penetrance and other phenomena discussed in the previous section. Linkage analysis is based on the tendency of a disease-causing sequence change and markers at specified loci to be inherited together (“linked”) as a consequence of their physical proximity on a single chromosome. The measure of the likelihood that the disease-associated gene and any single marker are genetically linked rather than unlinked is called the lod (the logarithm of the odds) score. A table of lod scores represents the result of a linkage analysis for a Mendelian trait, with a lod score > +3 considered evidence of linkage, whereas one that is <−2 considered evidence against linkage. After the linked region is narrowed to the smallest possible interval, sequencing of the genes in the region follows. The advent of recent sequencing technology, most notably next-generation sequencing (whole genome or whole exome, i.e., the coding part of the genome) facilitates gene identification. It is now possible to obtain the entire sequence of a patient’s genome in a fast and cost-efficient manner and to compare it to the reference genome. However, analysis of such large amounts of data is not an easy task and it is often difficult to clearly identify the disease-causing change and prove its pathogenicity among thousands of detected variants. Importantly, each person differs from the current reference genome by putative loss-of-function mutations in 250–300 genes (Durbin et al. 2010).
Genome-wide association studies (GWAS) represent another example of how a long-known idea used in a novel, technologically considerably upgraded setting can facilitate the discovery of genes associated with human disorders. In GWAS, the identification of genetic risk factors for the development of PD is achieved by analyzing as many as 500,000 different single-nucleotide polymorphisms (SNPs) in large groups of sporadic PD patients (a few thousands) and healthy individuals, and comparing SNP frequencies in the two groups. If certain variants are more frequent in PD patients, they are considered to be “associated” with the disease. These genetic variants are used to indicate the region of the human genome where the PD-causing change is likely to be situated. Unlike GWAS, candidate gene-association studies are based on the hypothesis that a particular gene may be associated with PD owing to its function and attempts to find this association. Although GWAS are mainly hypothesis-free, in earlier days many PD risk factors (some also classified as PARKs) have been identified in this way but only a few of them could subsequently be replicated in independent studies.
MONOGENIC FORMS OF PD
SNCA (PARK1–4)
SNCA was the first gene with mutations reported to cause autosomal-dominant PD. Patients with SNCA mutations usually have early-onset PD (EOPD, age of onset ≤50 yr) with an initially good response to levodopa treatment. However, the disease has a rapid progression and often presents with dementia and cognitive decline, and sometimes with atypical features such as central hypoventilation and myoclonus. Lewy bodies are present and spread through the substantia nigra, locus ceruleus, hypothalamus, and cerebral cortex (Polymeropoulos et al. 1996).
Nevertheless, mutations in SNCA are overall rare and thus far, only three different missense mutations as well as duplications and triplications of the entire gene have been reported (Klein and Schlossmacher 2006). Out of the three missense mutations, the first identified, p.A53T, seems to be by far the most frequent one and was found in one Italian, eight Greek, two Korean, and one Swedish family (Polymeropoulos et al. 1997; Athanassiadou et al. 1999; Spira et al. 2001; Ki et al. 2007; Choi et al. 2008; Puschmann et al. 2009). The p.A30P and p.E46K mutations were identified in one family each (Kruger et al. 1998; Zarranz et al. 2004). Seventeen duplications of the entire coding region of SNCA have been reported to date, 13 in PD families and four in sporadic cases, in one of which it was shown that the mutation arose de novo (Chartier-Harlin et al. 2004; Ibanez et al. 2004; Nishioka et al. 2006; Fuchs et al. 2007; Ahn et al. 2008; Brueggemann et al. 2008; Ikeuchi et al. 2008; Troiano et al. 2008; Uchiyama et al. 2008; Ibanez et al. 2009). Triplications of the SNCA gene were found in three independent families (Singleton et al. 2003; Farrer et al. 2004; Ibanez et al. 2009). Interestingly, one of the families with a triplication was a branch of one family with a duplication.
Penetrance of the missense mutations appears to be high and is suggested to be as high as 85% for the p.A53T mutation (Polymeropoulos et al. 1996). However, penetrance of gene amplifications is reduced and estimated at 33% in one family with a duplication (Nishioka et al. 2006). Intriguingly, dependence of the clinical symptoms on gene dosage has been suggested with an increased number of SNCA copies (SNCA triplications vs. duplications) being associated with an earlier onset, more severe phenotype, and faster disease progression (Fuchs et al. 2007; Ross et al. 2008).
The SNCA gene has six exons encoding an abundant 140-amino acid cytosolic protein, α-synuclein. α-synuclein consists of three domains: (i) the amino-terminal region (amino acids 7–87) contains seven imperfect repeats, each 11 amino acids in length, and is partially overlapping with (ii) a central hydrophobic domain (amino acids 61–95), and (iii) an acidic, negatively charged carboxy-terminal domain (amino acids 96–140). Thus, all three missense mutations impair the amino-terminal domain. Although natively unfolded, with almost no secondary structure, once it binds to the phospholipid membranes, through its amino-terminal repeats (amino acids 71–82), α-synuclein adopts structures rich in α-helical character (Giasson et al. 2001). Interestingly, the three-point mutants tend to form stable β sheets and thus exacerbate the formation of toxic oligomers, protofibrils, and fibrils (Bertoncini et al. 2005). Therefore, it is believed that the missense SNCA mutations cause PD through a toxic gain of function (Bertoncini et al. 2005), and Lewy bodies may represent the attempt to purge the cell of toxic damaged α-synuclein (Chen and Feany 2005). Wild-type α-synuclein is selectively translocated into lysosomes for degradation (Cuervo et al. 2004), and inhibitors of the lysosomal enzyme β-glucocerebrosidase, mutations in which represent a well-validated risk factor for PD, modulate α-synuclein levels (Manning-Bog et al. 2009). Very recently, it has been shown that the bidirectional effect of α-synuclein and β-glucocerebrosidase forms a positive feedback loop that leads to accumulation of α-synuclein (Mazzulli et al. 2011). Functional loss of β-glucocerebrosidase causes the accumulation of glucocerebroside, which directly influences aggregation of α-synuclein by stabilizing oligomeric intermediates (Mazzulli et al. 2011). In turn, α-synuclein inhibits the lysososmal activity of β-glucocerebrosidase, as shown in neurons and idiopathic PD brain (Mazzulli et al. 2011).
LRRK2 (PARK8)
Mutations in the LRRK2 gene are the most frequent known cause of late-onset autosomal-dominant and sporadic PD, with a mutation frequency ranging from 2% to 40% in different populations (Brice 2005; Lesage et al. 2006; Ozelius et al. 2006). Clinically, LRRK2-linked PD usually shows mid-to-late onset and progresses slowly. Patients respond favorably to levodopa therapy, and dementia is not common. Neuropathological findings are mostly inconsistent, showing both Lewy body (and sometimes tau- and ubiquitin-containing inclusions) pathology and pure nigral degeneration without Lewy bodies, with or without neurofibrillary tangles (Giasson et al. 2006).
LRRK2 is a large gene that consists of 51 exons. It encodes the 2527-amino acid cytoplasmic protein leucine-rich repeat kinase 2 (LRRK2) that consists of a leucine-rich repeat toward the amino terminus of the protein and a kinase domain toward the carboxyl terminus with various conserved domains in between. There are more than 50 different missense and nonsense mutations reported in LRRK2 to date (Nuytemans et al. 2010) and at least 16 of them (including the six recurrent mutations—p.R114C, p.R1441G, p.R1441H, p.Y1699C, p.G2019S, and p.I2020T) seem to be pathogenic. These pathogenic changes are clustered in 10 exons, mostly encoding the carboxy-terminal region of the protein. By far the most frequent and best-studied mutation is c.6055G >A (p.G2019S) that accounts for as many as 40% of cases of Arab descent (Lesage et al. 2006), about 20% of Ashkenazi Jewish patients (Ozelius et al. 2006), and 1%–7% of PD patients of European origin (Clark et al. 2006; Zabetian et al. 2006). Interestingly, three different founders have been described for the p.G2019S mutation and at least 29 patients have been reported to carry the mutation in the homozygous state (Klein and Lohmann-Hedrich 2007). Owing to a founder effect, the p.R1441G is very frequent in Basques (Simon-Sanchez et al. 2006; Gorostidi et al. 2009) and p.I2020T in Japanese patients (Tomiyama et al. 2006). Whereas p.G2019S shows reduced penetrance, sometimes estimated to be as low as 24%, the p.R1441 mutation is highly penetrant (95% at the age of 75 yr) (Haugarvoll et al. 2008).
The pathogenic mechanism leading to PD caused by LRRK2 mutations is still uncertain. LRRK2 is a large protein with many domains capable of protein–protein interactions, and thus it is plausible that changes in these domains would influence the LRRK2’s relationship with other proteins, i.e., currently unknown interactors with which it forms complexes or which it phosphorylates. In addition, various mutations affect its kinase activity as shown for the p.G2019S and p.I2020T mutants (MacLeod et al. 2006).
Parkin (PARK2)
Parkin was the second identified PD gene and the first gene irrefutably causing an AR form of the disorder. The disease usually starts in the third or fourth decade of the patients’ life, and is usually slowly progressive with an excellent response to dopaminergic treatment. However, some of the Parkin-mutation carriers have an onset even in childhood, and homozygous mutations in Parkin are the most frequent cause of juvenile PD (age of onset ≤21 yr). The clinical phenotype of Parkin-, PINK1-, and DJ-1-linked PD is indistinguishable. Reported postmortem examinations indicate that the substantia nigra shows neuronal loss and gliosis, however, it is frequently lacking Lewy bodies.
A large number and wide spectrum of Parkin mutations have been detected, including alterations of all 12 exons, across various ethnic groups. Parkin mutations are the most common known cause of EOPD, accounting for up to 77% of the familial cases with an age of onset <30 yr (Lucking et al. 2000), and for 10%–20% of EOPD patients in general (Klein and Lohmann-Hedrich 2007). To our knowledge, 887 exonic mutations have been described to date, comprising 147 different changes. About a third (293/887) of all exonic mutations are single-nucleotide changes, 13% (119/887) are small deletions, and 54% (475/887) are deletions or duplications of one or several exons (A Grünewald and C Klein, in prep.). However, because methods for the detection of exon rearrangements used to be labor intensive and expensive in the first years after the identification of Parkin, they were frequently omitted. Thus, the number of exon rearrangements is likely even underestimated.
Parkin is the second largest gene in human genome and codes for a 465-amino acid protein with a modular architecture. The Parkin protein functions as an E3 ubiquitin ligase in the process of ubiquitination, a form of posttranslational modification that conjugates ubiquitin protein(s) to lysine residue(s) of target proteins, which in turn determines their cellular fate. The amino-terminal ubiquitin-like domain (UBL) of Parkin shares 62% homology with ubiquitin and plays an important role in stabilizing the structure and controlling the expression levels of Parkin. The carboxy-terminal domain consists of three RING (really interesting new-gene) domains (amino acids 145–215 [RING0], 237–292 [RING1], and 417–448 [RING3]) and one IBR (in-between-ring) domain (amino acids 327–378) and is responsible for the interaction with the ubiquitination machinery.
About half of the published changes affect the region spanning exons 2–4 (52%, 459/887) that codes for the UBL domain, the linker region, and the very beginning of the RING0 domain. Although the largest total number of mutations was identified in exon 3 (257/887), exon 1 is remarkable for the highest mutation density with 2.4 mutations per bp (followed by exon 4 with 1.3 mutations per bp). In exon 2, the largest diversity of mutations was found, comprising 27 different variations. A deletion of exon 3 is the most frequent mutation in the Parkin gene (88/887, reported in 17 different studies). The second most common change is the c.924C>T single-nucleotide mutation in exon 7 (RING1), which was detected 75 times in 13 different screens (A Grünewald and C Klein, in prep.).
From the beginning of 2004 until July 2009, screening results of 4841 patients, mostly EOPD cases, were published. Parkin mutations were detected in about 8% of these. More than half of all mutation-positive cases (55%, 207/378) carried a single heterozygous change. Twenty-five percent (94/378) of them were compound-heterozygous and the remaining 20% (77/378) harbored a homozygous mutation (A Grünewald and C Klein, in prep.).
PINK1 (PARK6)
Mutations in the phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1) gene are the second most common cause of AR EOPD. The frequency of PINK1 mutations is in the range of 1%–9%, with considerable variation across different ethnic groups (Healy et al. 2004; Rogaeva et al. 2004; Valente et al. 2004; Bonifati et al. 2005; Klein et al. 2005; Li et al. 2005).
Interestingly, in contrast to Parkin, the majority of PINK1 mutations reported are either missense or nonsense mutations, and, to date only three families with whole-exon deletions (exons 4–8 [Cazeneuve et al. 2009], 6–8 [Li et al. 2005], and 7 [Camargos et al. 2009]) and one with a heterozygous whole-gene deletion have been reported (Marongiu et al. 2007). More than 60 different missense and nonsense mutations were found in >170 patients, affecting all 8 PINK1 exons at nearly equal frequencies (in each of the exons 5–10 different mutations were reported). The largest total number of mutations was found in exon 7 (in >50 patients), and the most frequent mutation is p.Q456X. Although only a quarter of the PINK1 mutations are truncating (vs. 3/4 missense), >40% of the total number of patients carry this type of mutation.
PINK1 is a 581 amino acid ubiquitously expressed protein kinase. It consists of an amino-terminal 34 amino acid mitochondrial targeting motif, a conserved serine–threonine kinase domain (amino acids 156–509; exons 2–8), and a carboxy-terminal autoregulatory domain. Two-thirds of the reported mutations in PINK1 are loss-of-function mutations affecting the kinase domain, demonstrating the importance of PINK1’s enzymatic activity in the pathogenesis of PD. Interestingly, recent studies provided evidence that PINK1 and Parkin function in a common pathway for sensing and selectively eliminating damaged mitochondria from the mitochondrial network. PINK1 is stabilized on mitochondria with lower membrane potential, and as such, it recruits Parkin from the cytosol. Once recruited to mitochondria, Parkin becomes enzymatically active and initiates the autophagic clearance of mitochondria by lysosomes, i.e., mitophagy (Youle and Narendra 2011).
DJ-1 (PARK7)
DJ-1 is the third gene associated with AR PD, and it is mutated in about 1%–2% of EOPD cases (Pankratz et al. 2006). Given that DJ-1-linked PD seems to be rare, very few patients have been reported in the literature. However, about 10 different point mutations and exonic deletions have been described, mostly in the homozygous or compound-heterozygous state. Given that the mutations are so uncommon, there is not enough data to draw any conclusion about the possible role of heterozygous mutations in this gene.
The seven coding exons of the DJ-1 gene code for a 189-amino acid-long protein that is ubiquitously expressed and functions as a cellular sensor of oxidative stress (Canet-Aviles et al. 2004; Junn et al. 2005). The DJ-1 protein forms a dimeric structure under physiologic conditions (Macedo et al. 2003), and it seems that most of the disease-causing mutants (p.L166P, p.E64D, p.M26I, and p.D149A) heterodimerize with wild-type DJ-1 (Takahashi-Niki et al. 2004). In addition, the mutated proteins are frequently not properly folded, unstable, and promptly degraded by the proteasome. Thus, their neuroprotective function and antioxidant activity are reduced (Anderson and Daggett 2008; Malgieri and Eliezer 2008).
ATP13A2 (PARK9)
Homozygous and compound-heterozygous mutations in ATP13A2 have been found to cause an AR atypical form of PD named Kufor-Rakeb syndrome (Ramirez et al. 2006). This syndrome has juvenile onset with rapid disease progression, accompanied by dementia, supranuclear gaze palsy, and pyramidal signs.
ATP13A2 is a large gene comprised of 29 exons coding for an 1180-amino acid protein. The ATP13A2 protein is normally located in the lysososmal membrane and it has 10 transmembrane domains and an ATPase domain (Ramirez et al. 2006). About 10 different pathogenic mutations have been found in the homozygous or compound-heterozygous state, directly or indirectly affecting transmembrane domains. Most of the mutations produce truncated proteins that are unstable and are retained in the endoplasmic reticulum and subsequently degraded by the proteasome. No exonic deletions or deletions or multiplications of the entire gene have been found, to date. Several single heterozygous missense mutations are known, but their role in PD pathogenicity is currently not clear.
GENES WITH A POSSIBLE ROLE IN PD
Apart from the genes causing the six monogenic forms of PD, changes in a large number of additional genes were considered PD-causative and identified by linkage analysis or a candidate gene approach. Some of these genes even attained a “PARKI” designation (UCHL1 [PARK5], GYGYF2 [PARK11], OMI/HTRA2 [PARK13], PLA2G6 [PARK14], and FBXO7 [PARK15]). However, as discussed in the Genetic Classification of PD section, the link of some of these genes to PD is uncertain and not confirmed. Furthermore, mutations in some PARKs (i) cause PD that is an inconsistent or only a minor feature of a more complex phenotype or (ii) are a very rare cause of PD (responsible for only a few PD occurrences). In addition, mutations in synphilin-1, NR4A2/Nurr1, POLG, mortalin, and recently presenilin-associated rhomboid-like protein (PARL) were considered pathogenic based on the known function or expression/protein interaction pattern of the proteins they encode. Nevertheless, they too, are now recognized as only a minor contributor to the pool of genetic PD if at all.
ASSOCIATED GENES (RISK FACTORS)
Variants in several PARK-designated (SNCA, UCHL1, LRRK2, PARK 16, GAK) and a few other genes (MAPT, GBA, NAT2, INOS2A, GAK, HLA-DRA, and APOE) have been associated with an increased risk of developing PD. Such risk factors polymorphisms/mutations were mostly identified based on GWAS and functional candidate approaches. Interestingly, polymorphic length and SNP variations in SNCA have repeatedly been shown to be among the most significant PD risk factors, closely followed by the occurrence of the p.G2385R and p.R1628P missense SNPs in the LRRK2 gene. In addition to SNCA and LRRK2 that can be involved both in monogenic disease and act as a risk factor, β-glucocerebrosidase (GBA) deserves special attention as a well-validated PD-associated risk factor.
The GBA gene encodes a lysosomal enzyme β-glucocerebrosidase with an important role in glycolipid metabolism. Loss-of-function mutations in β-glucocerebrosidase cause an accumulation of glucocerebroside that results in a wide spectrum of symptoms involving the liver, blood, bone marrow, spleen, lungs, and the nervous system, known as Gaucher disease. Gaucher disease is inherited autosomal recessively and, to date, about 300 missense, nonsense, and frame-shift disease-causing mutations have been identified (Hruska et al. 2008). Interestingly, GBA mutations have been found to increase the risk of developing PD and are found in 8%–14% of autopsy-proven diagnoses of PD (Goker-Alpan et al. 2004; Lwin et al. 2004; Eblan et al. 2005; Sidransky 2006) and both homozygous and heterozygous GBA mutations appear to predispose to classical parkinsonism (Sidransky 2006). In addition, relatives of patients with Gaucher disease carrying heterozygous GBA mutations have an increased incidence of PD (Goker-Alpan et al. 2004; Halperin et al. 2006).
In 2009, in a large collaborative effort, 16 centers from Europe, Americas, and Asia analyzed selected GBA mutations in >5000 (14% Ashkenazi Jews) PD patients and nearly 5000 (8% Ashkenazi Jews) healthy controls without family history of PD and sequenced the entire coding region in a subset of subjects (Sidransky et al. 2009). Mutant GBA alleles were found in 19.6% of Ashkenazi Jews and in 6.9% of non-Ashkenazi Jewish patients. In addition, the percentage of mutations was five times higher in patients in comparison to controls. Age at onset was found to be lower among subjects with GBA mutations than in those without GBA mutations. The recent findings summarized in the Monogenic Forms of PD-SNCA (PARK1 = 4) section of this article shed light on how GBA mutations increase the risk of developing PD.
GENETIC TESTING: WHOM TO TEST AND HOW?
The answer to the question of who should be tested is not trivial. To date, no formal testing guidelines have been developed by the Movement Disorder Society or any other PD alliance group. Out of all monogenic forms, mutations in LRRK2, Parkin, and PINK1 are the most likely to be encountered in clinical practice. In the following scenarios, genetic testing might prove useful to minimize further patient workup, to clarify treatment approaches, and/or to assist with future family planning: juvenile-onset PD irrespective of family history; early-onset PD with atypical features and/or a positive family history of this disease; or late-onset PD with a strong family history of PD (Klein and Schlossmacher 2006). Guidelines published by the European Federation of the Neurological Sciences recommend screening LRRK2 for mutations in Europeans showing dominant inheritance of PD, testing for the LRRK2 p.G2019S mutation in familial and sporadic cases of PD in specific populations, and analysis of Parkin, PINK1, and DJ-1 in patients aged <35 yr with recessively inherited PD (Harbo et al. 2009).
Most patients with early-onset PD are strongly in favor of genetic testing to further understand their disease and to make informed life decisions (Jacobs et al. 2001). A frequent argument offered against genetic testing for PD is that the outcome of such testing does not affect patient management. We believe this notion should be carefully reconsidered, as the identification of a PD gene mutation—especially in a patient with early-onset PD—frequently has an important impact that goes beyond a reduction in diagnostic uncertainty. Indeed, the identification of specific mutations can provide information on prognosis and will affect treatment choices in cases of PD that were initially suspected to be psychogenic. One other pertinent consideration relating to the debate surrounding genetic testing is that various other tests are performed for patients with parkinsonism (and not questioned) that do not alter how patients are treated, such as repeated MRI and single-photon-emission computed tomography to differentiate multiple system atrophy from idiopathic PD.
Despite the arguments presented above, genetic testing for PD genes should not be recommended lightly or out of academic curiosity. Testing should always be offered in the framework of genetic counseling and based on an informed decision made by the patient. Of note, direct-to-consumer testing involving, for example, testing for the p.G2019S mutation in LRRK2 is becoming increasingly available and sought after by patients or even healthy individuals. Therefore, neurologists will have to be prepared for patients seeking post-test counseling for a “genetic diagnosis of PD” or for healthy individuals having an “elevated risk” to develop PD based on the presence of risk SNPs. Such susceptibility testing should be strongly discouraged, and only the advent of neuroprotective or gene-specific therapies is likely to profoundly change our current views on the utility of genetic testing for PD.
CONCLUDING REMARKS
Genetic forms of PD are overall rare but of major importance for a better understanding of the pathophysiology of idiopathic PD. Owing to the clinical, and in many cases, also pathological similarity, genetic PD serves as an excellent human model for the much more common idiopathic condition and enables the identification of at-risk individuals in the earliest, even premotor phases of the disease. All known forms can be tested for in the diagnostic setting, however, no causative treatment approaches have yet become available after 15 years of genetic PD research. Future perspectives for the next decade include the development of improved human cellular models for drug screening and regenerative medicine, generation of animal models that replicate the clinical and pathological findings in humans more faithfully, and finally, the development of cause-directed therapies.
ACKNOWLEDGMENTS
C.K. is supported by a career development award from the Volkswagen and from the Hermann and Lilly Schilling Foundation. A.W. is supported by the Fritz Thyssen Foundation and a career development grant from the University of Lübeck.
Footnotes
Editor: Serge Przedborski
Additional Perspectives on Parkinson’s Disease available at www.perspectivesinmedicine.org
REFERENCES
- Ahn TB, Kim SY, Kim JY, Park SS, Lee DS, Min HJ, Kim YK, Kim SE, Kim JM, Kim HJ, et al. 2008. α-Synuclein gene duplication is present in sporadic Parkinson disease. Neurology 70: 43–49 [DOI] [PubMed] [Google Scholar]
- Anderson PC, Daggett V 2008. Molecular basis for the structural instability of human DJ-1 induced by the L166P mutation associated with Parkinson’s disease. Biochemistry 47: 9380–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanassiadou A, Voutsinas G, Psiouri L, Leroy E, Polymeropoulos MH, Ilias A, Maniatis GM, Papapetropoulos T 1999. Genetic analysis of families with Parkinson disease that carry the Ala53Thr mutation in the gene encoding α-synuclein. Am J Hum Genet 65: 555–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoncini CW, Fernandez CO, Griesinger C, Jovin TM, Zweckstetter M 2005. Familial mutants of α-synuclein with increased neurotoxicity have a destabilized conformation. J Biol Chem 280: 30649–30652 [DOI] [PubMed] [Google Scholar]
- Bonifati V, Rohe CF, Breedveld GJ, Fabrizio E, De Mari M, Tassorelli C, Tavella A, Marconi R, Nicholl DJ, Chien HF, et al. 2005. Early-onset parkinsonism associated with PINK1 mutations: Frequency, genotypes, and phenotypes. Neurology 65: 87–95 [DOI] [PubMed] [Google Scholar]
- Brice A 2005. Genetics of Parkinson’s disease: LRRK2 on the rise. Brain 128: 2760–2762 [DOI] [PubMed] [Google Scholar]
- Brueggemann N, Odin P, Gruenewald A, Tadic V, Hagenah J, Seidel G, Lohmann K, Klein C, Djarmati A 2008. Re: α-synuclein gene duplication is present in sporadic Parkinson disease. Neurology 71: 1294. [DOI] [PubMed] [Google Scholar]
- Camargos ST, Dornas LO, Momeni P, Lees A, Hardy J, Singleton A, Cardoso F 2009. Familial Parkinsonism and early onset Parkinson’s disease in a Brazilian movement disorders clinic: Phenotypic characterization and frequency of SNCA, PRKN, PINK1, and LRRK2 mutations. Mov Disord 24: 662–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA, Cookson MR 2004. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci 101: 9103–9108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazeneuve C, San C, Ibrahim SA, Mukhtar MM, Kheir MM, Leguern E, Brice A, Salih MA 2009. A new complex homozygous large rearrangement of the PINK1 gene in a Sudanese family with early onset Parkinson’s disease. Neurogenetics 10: 265–270 [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, et al. 2004. α-Synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 364: 1167–1169 [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin M-C, Dachsel JC, Vilarino-Guell C, Lincoln SJ, Lepretre F, Hulihan MM, Kachergus J, Milnerwood AJ, Tapia L, Song MS, et al. 2011. Translation initiator EIF4G1 mutations in familial Parkinson disease. Am J Hum Genet 89: 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Feany MB 2005. α-Synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat Neurosci 8: 657–663 [DOI] [PubMed] [Google Scholar]
- Choi JM, Woo MS, Ma HI, Kang SY, Sung YH, Yong SW, Chung SJ, Kim JS, Shin HW, Lyoo CH, et al. 2008. Analysis of PARK genes in a Korean cohort of early-onset Parkinson disease. Neurogenetics 9: 263–269 [DOI] [PubMed] [Google Scholar]
- Clark LN, Wang Y, Karlins E, Saito L, Mejia-Santana H, Harris J, Louis ED, Cote LJ, Andrews H, Fahn S, et al. 2006. Frequency of LRRK2 mutations in early- and late-onset Parkinson disease. Neurology 67: 1786–1791 [DOI] [PubMed] [Google Scholar]
- Criscuolo C, Volpe G, De Rosa A, Varrone A, Marongiu R, Mancini P, Salvatore E, Dallapiccola B, Filla A, Valente EM, et al. 2006. PINK1 homozygous W437X mutation in a patient with apparent dominant transmission of parkinsonism. Mov Disord 21: 1265–1267 [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D 2004. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science 305: 1292–1295 [DOI] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eblan MJ, Walker JM, Sidransky E 2005. The glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med 352: 728–731 [DOI] [PubMed] [Google Scholar]
- Eggers C, Schmidt A, Hagenah J, Bruggemann N, Klein JC, Tadic V, Kertelge L, Kasten M, Binkofski F, Siebner H, et al. 2010. Progression of subtle motor signs in PINK1 mutation carriers with mild dopaminergic deficit. Neurology 74: 1798–1805 [DOI] [PubMed] [Google Scholar]
- Farrer M, Wavrant-De Vrieze F, Crook R, Boles L, Perez-Tur J, Hardy J, Johnson WG, Steele J, Maraganore D, Gwinn K, et al. 1998. Low frequency of α-synuclein mutations in familial Parkinson’s disease. Ann Neurol 43: 394–397 [DOI] [PubMed] [Google Scholar]
- Farrer M, Gwinn-Hardy K, Muenter M, DeVrieze FW, Crook R, Perez-Tur J, Lincoln S, Maraganore D, Adler C, Newman S, et al. 1999. A chromosome 4p haplotype segregating with Parkinson’s disease and postural tremor. Hum Mol Genet 8: 81–85 [DOI] [PubMed] [Google Scholar]
- Farrer M, Kachergus J, Forno L, Lincoln S, Wang DS, Hulihan M, Maraganore D, Gwinn-Hardy K, Wszolek Z, Dickson D, et al. 2004. Comparison of kindreds with parkinsonism and α-synuclein genomic multiplications. Ann Neurol 55: 174–179 [DOI] [PubMed] [Google Scholar]
- Fuchs J, Nilsson C, Kachergus J, Munz M, Larsson EM, Schule B, Langston JW, Middleton FA, Ross OA, Hulihan M, et al. 2007. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology 68: 916–922 [DOI] [PubMed] [Google Scholar]
- Giasson BI, Murray IV, Trojanowski JQ, Lee VM 2001. A hydrophobic stretch of 12 amino acid residues in the middle of α-synuclein is essential for filament assembly. J Biol Chem 276: 2380–2386 [DOI] [PubMed] [Google Scholar]
- Giasson BI, Covy JP, Bonini NM, Hurtig HI, Farrer MJ, Trojanowski JQ, Van Deerlin VM 2006. Biochemical and pathological characterization of Lrrk2. Ann Neurol 59: 315–322 [DOI] [PubMed] [Google Scholar]
- Goker-Alpan O, Schiffmann R, LaMarca ME, Nussbaum RL, McInerney-Leo A, Sidransky E 2004. Parkinsonism among Gaucher disease carriers. J Med Genet 41: 937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorostidi A, Ruiz-Martinez J, Lopez de Munain A, Alzualde A, Masso JF 2009. LRRK2 G2019S and R1441G mutations associated with Parkinson’s disease are common in the Basque Country, but relative prevalence is determined by ethnicity. Neurogenetics 10: 157–159 [DOI] [PubMed] [Google Scholar]
- Halperin A, Elstein D, Zimran A 2006. Increased incidence of Parkinson disease among relatives of patients with Gaucher disease. Blood Cells Mol Dis 36: 426–428 [DOI] [PubMed] [Google Scholar]
- Harbo HF, Finsterer J, Baets J, Van Broeckhoven C, Di Donato S, Fontaine B, De Jonghe P, Lossos A, Lynch T, Mariotti C, et al. 2009. EFNS guidelines on the molecular diagnosis of neurogenetic disorders: General issues, Huntington’s disease, Parkinson’s disease and dystonias. Eur J Neurol 16: 777–785 [DOI] [PubMed] [Google Scholar]
- Haugarvoll K, Rademakers R, Kachergus JM, Nuytemans K, Ross OA, Gibson JM, Tan EK, Gaig C, Tolosa E, Goldwurm S, et al. 2008. Lrrk2 R1441C parkinsonism is clinically similar to sporadic Parkinson disease. Neurology 70: 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy DG, Abou-Sleiman PM, Gibson JM, Ross OA, Jain S, Gandhi S, Gosal D, Muqit MM, Wood NW, Lynch T 2004. PINK1 (PARK6) associated Parkinson disease in Ireland. Neurology 63: 1486–1488 [DOI] [PubMed] [Google Scholar]
- Hedrich K, Hagenah J, Djarmati A, Hiller A, Lohnau T, Lasek K, Grunewald A, Hilker R, Steinlechner S, Boston H, et al. 2006. Clinical spectrum of homozygous and heterozygous PINK1 mutations in a large German family with Parkinson disease: Role of a single hit? Arch Neurol 63: 833–838 [DOI] [PubMed] [Google Scholar]
- Hiller A, Hagenah JM, Djarmati A, Hedrich K, Reetz K, Schneider-Gold C, Kress W, Munchau A, Klein C 2007. Phenotypic spectrum of PINK1-associated parkinsonism in 15 mutation carriers from 1 family. Mov Disord 22: 145–147 [DOI] [PubMed] [Google Scholar]
- Hruska KS, LaMarca ME, Scott CR, Sidransky E 2008. Gaucher disease: Mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA). Hum Mutat 29: 567–583 [DOI] [PubMed] [Google Scholar]
- Ibanez P, Bonnet AM, Debarges B, Lohmann E, Tison F, Pollak P, Agid Y, Durr A, Brice A 2004. Causal relation between α-synuclein gene duplication and familial Parkinson’s disease. Lancet 364: 1169–1171 [DOI] [PubMed] [Google Scholar]
- Ibanez P, Lesage S, Janin S, Lohmann E, Durif F, Destee A, Bonnet AM, Brefel-Courbon C, Heath S, Zelenika D, et al. 2009. α-Synuclein gene rearrangements in dominantly inherited parkinsonism: Frequency, phenotype, and mechanisms. Arch Neurol 66: 102–108 [DOI] [PubMed] [Google Scholar]
- Ikeuchi T, Kakita A, Shiga A, Kasuga K, Kaneko H, Tan CF, Idezuka J, Wakabayashi K, Onodera O, Iwatsubo T, et al. 2008. Patients homozygous and heterozygous for SNCA duplication in a family with parkinsonism and dementia. Arch Neurol 65: 514–519 [DOI] [PubMed] [Google Scholar]
- Jacobs H, Latza U, Vieregge A, Vieregge P 2001. Attitudes of young patients with Parkinson’s disease towards possible presymptomatic and prenatal genetic testing. Genet Couns 12: 55–67 [PubMed] [Google Scholar]
- Junn E, Taniguchi H, Jeong BS, Zhao X, Ichijo H, Mouradian MM 2005. Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc Natl Acad Sci 102: 9691–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NL, Horta W, Eunson L, Graham E, Johnson JO, Chang S, Davis M, Singleton A, Wood NW, Lees AJ 2005. Parkin disease in a Brazilian kindred: Manifesting heterozygotes and clinical follow-up over 10 years. Mov Disord 20: 479–484 [DOI] [PubMed] [Google Scholar]
- Ki CS, Stavrou EF, Davanos N, Lee WY, Chung EJ, Kim JY, Athanassiadou A 2007. The Ala53Thr mutation in the α-synuclein gene in a Korean family with Parkinson disease. Clin Genet 71: 471–473 [DOI] [PubMed] [Google Scholar]
- Klein C, Lohmann-Hedrich K 2007. Impact of recent genetic findings in Parkinson’s disease. Curr Opin Neurol 20: 453–464 [DOI] [PubMed] [Google Scholar]
- Klein C, Schlossmacher MG 2006. The genetics of Parkinson disease: Implications for neurological care. Nat Clin Pract Neurol 2: 136–146 [DOI] [PubMed] [Google Scholar]
- Klein C, Djarmati A, Hedrich K, Schafer N, Scaglione C, Marchese R, Kock N, Schule B, Hiller A, Lohnau T, et al. 2005. PINK1, Parkin, and DJ-1 mutations in Italian patients with early-onset parkinsonism. Eur J Hum Genet 13: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Klein C, Chuang R, Marras C, Lang AE 2011. The curious case of phenocopies in families with genetic Parkinson’s disease. Mov Disord 10.1002/mds.23853 [DOI] [PubMed] [Google Scholar]
- Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O 1998. Ala30Pro mutation in the gene encoding α-synuclein in Parkinson’s disease. Nat Genet 18: 106–108 [DOI] [PubMed] [Google Scholar]
- Lesage S, Durr A, Tazir M, Lohmann E, Leutenegger AL, Janin S, Pollak P, Brice A 2006. LRRK2 G2019S as a cause of Parkinson’s disease in North African Arabs. N Engl J Med 354: 422–423 [DOI] [PubMed] [Google Scholar]
- Li Y, Tomiyama H, Sato K, Hatano Y, Yoshino H, Atsumi M, Kitaguchi M, Sasaki S, Kawaguchi S, Miyajima H, et al. 2005. Clinicogenetic study of PINK1 mutations in autosomal recessive early-onset parkinsonism. Neurology 64: 1955–1957 [DOI] [PubMed] [Google Scholar]
- Lucking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denefle P, Wood NW, et al. 2000. Association between early-onset Parkinson’s disease and mutations in the parkin gene. French Parkinson’s Disease Genetics Study Group. N Engl J Med 342: 1560–1567 [DOI] [PubMed] [Google Scholar]
- Lwin A, Orvisky E, Goker-Alpan O, LaMarca ME, Sidransky E 2004. Glucocerebrosidase mutations in subjects with parkinsonism. Mol Genet Metab 81: 70–73 [DOI] [PubMed] [Google Scholar]
- Macedo MG, Anar B, Bronner IF, Cannella M, Squitieri F, Bonifati V, Hoogeveen A, Heutink P, Rizzu P 2003. The DJ-1L166P mutant protein associated with early onset Parkinson’s disease is unstable and forms higher-order protein complexes. Hum Mol Genet 12: 2807–2816 [DOI] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A 2006. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron 52: 587–593 [DOI] [PubMed] [Google Scholar]
- Malgieri G, Eliezer D 2008. Structural effects of Parkinson’s disease linked DJ-1 mutations. Protein Sci 17: 855–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning-Bog AB, Schule B, Langston JW 2009. α-Synuclein-glucocerebrosidase interactions in pharmacological Gaucher models: A biological link between Gaucher disease and parkinsonism. Neurotoxicology 30: 1127–1132 [DOI] [PubMed] [Google Scholar]
- Marongiu R, Brancati F, Antonini A, Ialongo T, Ceccarini C, Scarciolla O, Capalbo A, Benti R, Pezzoli G, Dallapiccola B, et al. 2007. Whole gene deletion and splicing mutations expand the PINK1 genotypic spectrum. Hum Mutat 28: 98. [DOI] [PubMed] [Google Scholar]
- Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, Sidransky E, Grabowski GA, Krainc D 2011. Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell 146: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz E, Oliva R, Obach V, Marti MJ, Pastor P, Ballesta F, Tolosa E 1997. Identification of Spanish familial Parkinson’s disease and screening for the Ala53Thr mutation of the α-synuclein gene in early onset patients. Neurosci Lett 235: 57–60 [DOI] [PubMed] [Google Scholar]
- Nishioka K, Hayashi S, Farrer MJ, Singleton AB, Yoshino H, Imai H, Kitami T, Sato K, Kuroda R, Tomiyama H, et al. 2006. Clinical heterogeneity of α-synuclein gene duplication in Parkinson’s disease. Ann Neurol 59: 298–309 [DOI] [PubMed] [Google Scholar]
- Nuytemans K, Theuns J, Cruts M, Van Broeckhoven C 2010. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: A mutation update. Hum Mutat 31: 763–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozelius LJ, Senthil G, Saunders-Pullman R, Ohmann E, Deligtisch A, Tagliati M, Hunt AL, Klein C, Henick B, Hailpern SM, et al. 2006. LRRK2 G2019S as a cause of Parkinson’s disease in Ashkenazi Jews. N Engl J Med 354: 424–425 [DOI] [PubMed] [Google Scholar]
- Pankratz N, Pauciulo MW, Elsaesser VE, Marek DK, Halter CA, Wojcieszek J, Rudolph A, Shults CW, Foroud T, Nichols WC 2006. Mutations in DJ-1 are rare in familial Parkinson disease. Neurosci Lett 408: 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Higgins JJ, Golbe LI, Johnson WG, Ide SE, Di Iorio G, Sanges G, Stenroos ES, Pho LT, Schaffer AA, et al. 1996. Mapping of a gene for Parkinson’s disease to chromosome 4q21-q23. Science 274: 1197–1199 [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. 1997. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276: 2045–2047 [DOI] [PubMed] [Google Scholar]
- Puschmann A, Ross OA, Vilarino-Guell C, Lincoln SJ, Kachergus JM, Cobb SA, Lindquist SG, Nielsen JE, Wszolek ZK, Farrer M, et al. 2009. A Swedish family with de novo α-synuclein A53T mutation: Evidence for early cortical dysfunction. Parkinsonism Relat Disord 15: 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez A, Heimbach A, Grundemann J, Stiller B, Hampshire D, Cid LP, Goebel I, Mubaidin AF, Wriekat AL, Roeper J, et al. 2006. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet 38: 1184–1191 [DOI] [PubMed] [Google Scholar]
- Rogaeva E, Johnson J, Lang AE, Gulick C, Gwinn-Hardy K, Kawarai T, Sato C, Morgan A, Werner J, Nussbaum R, et al. 2004. Analysis of the PINK1 gene in a large cohort of cases with Parkinson disease. Arch Neurol 61: 1898–1904 [DOI] [PubMed] [Google Scholar]
- Ross OA, Braithwaite AT, Skipper LM, Kachergus J, Hulihan MM, Middleton FA, Nishioka K, Fuchs J, Gasser T, Maraganore DM, et al. 2008. Genomic investigation of α-synuclein multiplication and parkinsonism. Ann Neurol 63: 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WK, Staijich JM, Yamaoka LH, Speer MC, Vance JM, Roses AD, Pericak-Vance MA 1997. Genetic complexity and Parkinson’s disease. Deane Laboratory Parkinson Disease Research Group. Science 277: 387–388 [DOI] [PubMed] [Google Scholar]
- Scott WK, Yamaoka LH, Stajich JM, Scott BL, Vance JM, Roses AD, Pericak-Vance MA, Watts RL, Nance M, Hubble J, et al. 1999. The α-synuclein gene is not a major risk factor in familial Parkinson disease. Neurogenetics 2: 191–192 [DOI] [PubMed] [Google Scholar]
- Sidransky E 2006. Heterozygosity for a Mendelian disorder as a risk factor for complex disease. Clin Genet 70: 275–282 [DOI] [PubMed] [Google Scholar]
- Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, et al. 2009. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med 361: 1651–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Marti-Masso JF, Sanchez-Mut JV, Paisan-Ruiz C, Martinez-Gil A, Ruiz-Martinez J, Saenz A, Singleton AB, Lopez de Munain A, Perez-Tur J 2006. Parkinson’s disease due to the R1441G mutation in Dardarin: A founder effect in the Basques. Mov Disord 21: 1954–1959 [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. 2003. α-Synuclein locus triplication causes Parkinson’s disease. Science 302: 841. [DOI] [PubMed] [Google Scholar]
- Spira PJ, Sharpe DM, Halliday G, Cavanagh J, Nicholson GA 2001. Clinical and pathological features of a Parkinsonian syndrome in a family with an Ala53Thr α-synuclein mutation. Ann Neurol 49: 313–319 [PubMed] [Google Scholar]
- Takahashi-Niki K, Niki T, Taira T, Iguchi-Ariga SM, Ariga H 2004. Reduced anti-oxidative stress activities of DJ-1 mutants found in Parkinson’s disease patients. Biochem Biophys Res Commun 320: 389–397 [DOI] [PubMed] [Google Scholar]
- Thomas B, Beal MF 2007. Parkinson’s disease. Hum Mol Genet 16 (Spec No 2): R183–R194 [DOI] [PubMed] [Google Scholar]
- Tomiyama H, Li Y, Funayama M, Hasegawa K, Yoshino H, Kubo SI, Sato K, Hattori T, Lu CS, Inzelberg R, et al. 2006. Clinicogenetic study of mutations in LRRK2 exon 41 in Parkinson’s disease patients from 18 countries. Mov Disord 21: 1102–1108 [DOI] [PubMed] [Google Scholar]
- Troiano AR, Cazeneuve C, Le Ber I, Bonnet AM, Lesage S, Brice A 2008. Re: α-Synuclein gene duplication is present in sporadic Parkinson disease. Neurology 71: 1295. [DOI] [PubMed] [Google Scholar]
- Uchiyama T, Ikeuchi T, Ouchi Y, Sakamoto M, Kasuga K, Shiga A, Suzuki M, Ito M, Atsumi T, Shimizu T, et al. 2008. Prominent psychiatric symptoms and glucose hypometabolism in a family with a SNCA duplication. Neurology 71: 1289–1291 [DOI] [PubMed] [Google Scholar]
- Valente EM, Salvi S, Ialongo T, Marongiu R, Elia AE, Caputo V, Romito L, Albanese A, Dallapiccola B, Bentivoglio AR 2004. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann Neurol 56: 336–341 [DOI] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP 2011. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabetian CP, Hutter CM, Yearout D, Lopez AN, Factor SA, Griffith A, Leis BC, Bird TD, Nutt JG, Higgins DS, et al. 2006. LRRK2 G2019S in families with Parkinson disease who originated from Europe and the Middle East: Evidence of two distinct founding events beginning two millennia ago. Am J Hum Genet 79: 752–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, et al. 2004. The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann Neurol 55: 164–173 [DOI] [PubMed] [Google Scholar]