Abstract
Polycomb group (PcG) proteins exist in multiprotein complexes that modify chromatin to repress transcription. Drosophila PcG proteins Sex combs extra (Sce; dRing) and Posterior sex combs (Psc) are core subunits of PRC1-type complexes. The Sce:Psc module acts as an E3 ligase for monoubiquitylation of histone H2A, an activity thought to be crucial for repression by PRC1-type complexes. Here, we created an Sce knockout allele and show that depletion of Sce results in loss of H2A monoubiquitylation in developing Drosophila. Genome-wide profiling identified a set of target genes co-bound by Sce and all other PRC1 subunits. Analyses in mutants lacking individual PRC1 subunits reveals that these target genes comprise two distinct classes. Class I genes are misexpressed in mutants lacking any of the PRC1 subunits. Class II genes are only misexpressed in animals lacking the Psc-Su(z)2 and Polyhomeotic (Ph) subunits but remain stably repressed in the absence of the Sce and Polycomb (Pc) subunits. Repression of class II target genes therefore does not require Sce and H2A monoubiquitylation but might rely on the ability of Psc-Su(z)2 and Ph to inhibit nucleosome remodeling or to compact chromatin. Similarly, Sce does not provide tumor suppressor activity in larval tissues under conditions in which Psc-Su(z)2, Ph and Pc show such activity. Sce and H2A monoubiquitylation are therefore only crucial for repression of a subset of genes and processes regulated by PRC1-type complexes. Sce synergizes with the Polycomb repressive deubiquitinase (PR-DUB) complex to repress transcription at class I genes, suggesting that H2A monoubiquitylation must be appropriately balanced for their transcriptional repression.
Keywords: Polycomb repression, Sce (dRing), H2A monoubiquitylation, Calypso (Bap1), Drosophila
INTRODUCTION
Polycomb group (PcG) genes encode regulatory proteins that control diverse developmental processes in animals and plants by repressing the transcription of developmental regulator genes. Genetic studies in Drosophila originally identified PcG proteins as repressors that are required for the long-term silencing of HOX genes in cells in which these genes have to remain inactive (Duncan, 1982; Jürgens, 1985; Lewis, 1978; Struhl, 1981). Eighteen different Drosophila proteins have been classified as PcG members on the basis that HOX gene silencing is lost in animals that lack these proteins. PcG proteins compose subunits of four principal protein assemblies: Polycomb repressive complex 1 (PRC1), Polycomb repressive complex 2 (PRC2), Polycomb repressive deubiquitinase (PR-DUB) and Pho repressive complex (PhoRC) (Czermin et al., 2002; Klymenko et al., 2006; Lagarou et al., 2008; Müller et al., 2002; Nekrasov et al., 2007; Scheuermann et al., 2010; Shao et al., 1999). Protein assemblies that are identical or similar to PRC1, PRC2 and PR-DUB have also been identified in mammals (Cao et al., 2002; Kuzmichev et al., 2002; Levine et al., 2002; Misaghi et al., 2009; Sarma et al., 2008; Sowa et al., 2009; Yu et al., 2010).
PcG protein complexes possess specific chromatin-modifying activities that are thought to be crucial for repression of target genes (reviewed by Müller and Verrijzer, 2009; Simon and Kingston, 2009). Specifically, PRC2 tri-methylates lysine 27 in histone H3 (H3-K27me3), and high levels of this modification at target genes correlates with their repression (Cao et al., 2008; Cao et al., 2002; Kahn et al., 2006; Kuzmichev et al., 2002; Nekrasov et al., 2007; Papp and Müller, 2006; Sarma et al., 2008; Schwartz et al., 2006; Schwartz et al., 2010). PRC1 inhibits nucleosome remodeling and transcription on chromatin templates and compacts chromatin in vitro (Francis et al., 2004; Francis et al., 2001; Levine et al., 2002; Shao et al., 1999). PRC1-like complexes that contain only a subset of PRC1 subunits, such as human PRC1L (hPRC1L) and the Drosophila dRing-associated factors (dRAF) complex, function as E3 ligases for the monoubiquitylation (ub) of a specific lysine in histone H2A: H2A-K119 in mammals and H2A-K118 in Drosophila (de Napoles et al., 2004; Lagarou et al., 2008; Wang, H. et al., 2004). The recently identified PR-DUB complex deubiquitylates H2A-K119ub1/H2A-K118ub1 in vitro, and Drosophila mutants lacking PR-DUB show a strong increase of bulk H2A-K118ub levels (Scheuermann et al., 2010). PhoRC does not possess any enzymatic activity but is the only PcG protein complex with sequence-specific DNA-binding activity (Klymenko et al., 2006). PhoRC has been implicated in the targeting of other PcG protein complexes to Polycomb response elements (PREs), possibly in conjunction with other DNA-binding proteins (Mohd-Sarip et al., 2005; Wang, L. et al., 2004; Brown and Kassis, 2010; Klymenko et al., 2006; Kwong et al., 2008; Mohd-Sarip et al., 2006; Oktaba et al., 2008; Schuettengruber and Cavalli, 2009) (reviewed by Müller and Kassis, 2006).
Genome-wide profiling of PcG proteins in Drosophila tissue culture cells and in developing animals revealed that PhoRC, PRC1, PRC2 and PR-DUB all co-occupy PREs of HOX and a large set of other developmental regulator genes (Kwong et al., 2008; Nègre et al., 2006; Oktaba et al., 2008; Scheuermann et al., 2010; Schuettengruber and Cavalli, 2009; Schwartz et al., 2006; Tolhuis et al., 2006). Nevertheless, for most of the non-HOX target genes it is still not known whether and in which cells the PcG machinery is required to repress their transcription and which activities of the different complexes are needed for this repression.
In this study, we investigated how the different chromatin-modifying activities of PRC1 contribute to the transcriptional repression of PRC1 target genes in Drosophila. In particular, we were interested in analyzing the role of H2A monoubiquitylation in repression. The core of Drosophila PRC1 comprises Sex combs extra (Sce; also known as dRing), Polycomb (Pc), the two highly related proteins Polyhomeotic-proximal (Ph-p) and Polyhomeotic-distal (Ph-d), and Posterior sex combs (Psc) or its paralog Suppressor of zeste 2 [Su(z)2] (Francis et al., 2001; Lo et al., 2009; Lo and Francis, 2010; Shao et al., 1999; Strübbe et al., 2011). The ability of PRC1 to inhibit nucleosome remodeling, to repress transcription on chromatin templates and to compact chromatin in vitro is primarily attributed to the Psc-Su(z)2 subunit (Francis et al., 2004; Francis et al., 2001; Lo and Francis, 2010). Psc interacts directly with Sce. In vitro, the Sce:Psc module and the corresponding RING1B (RNF2):BMI1 module of human PRC1 have E3 ligase activity to monoubiquitylate histone H2A in nucleosomes (Buchwald et al., 2006; Lagarou et al., 2008; Li et al., 2006). E3 ligase activity for monoubiquitylation of nucleosomal H2A has also been shown for the PRC1-related dRAF complex that comprises Sce, Psc and Kdm2 but lacks Ph and Pc (Lagarou et al., 2008). However, attempts to show an E3 ligase activity with reconstituted recombinant PRC1 in vitro have so far failed (Lagarou et al., 2008). In addition, a recent study in murine embryonic stem cells proposed that Ring1B compacts HOX gene chromatin and represses their transcription independently of H2A ubiquitylation (Eskeland et al., 2010). The question of whether H2A monoubiquitylation is central to the repression mechanism of PRC1 is therefore still unresolved.
Here, we find that genes that are bound by all four PRC1 core subunits fall into two broad categories with respect to their regulation by PRC1-type complexes. Class I genes require all four PRC1 subunits for repression, whereas repression of class II genes depends on Psc-Su(z)2 and Ph but not on Pc and Sce. Non-covalent chromatin modification by Psc-Su(z)2 and Ph thus appears to play a central role in the repression of all PRC1 target genes, whereas H2A monoubiquitylation by Sce only seems to be crucial for repression of a subset of these targets.
MATERIALS AND METHODS
Fly strains
The following Drosophila strains were used in this study:
w ph504 FRT101/FM7c;
yw; FRT40A FRT42D Asx22P4 y+/SM6b;
yw; FRT40A FRT42D Asx22P4 y+/CyO, twi::EGFP;
w; FRT42D Su(z)21.b8/SM6b;
w; PcXT109 FRT2A/TM6B;
w; FRT82B Sce1/TM6C;
yw; FRT82B Sce33M2/TM6C;
w; FRT82B cu sr SceKO/TM6C;
w; F82B cu sr SceKO/TM3, Sb, Ser, twi::EGFP;
w; FRT40A FRT42D Asx22P4 y+; FRT82B cu sr SceKO/SM5^TM6B;
w hs-nGFP hs-flp FRT101;
yw hs-flp; FRT42D hs-nGFP;
yw hs-flp; hs-nGFP FRT2A;
yw hs-flp; FRT82B hs-nGFP;
yw ey-flp; FRT42D cell lethal ubi-GFP.nls/Cyo;
yw; FRT40 FRT42D y+;
yw; FRT42D Psc-Su(z)2P3C/Cyo-CTG;
yw; ey-GAL4 UAS-flp/Cyo; cell lethal GMR-hid F2A/TM6B; and
yw; ey-GAL4 UAS-flp/Cyo; FRT82B cell lethal GMR-hid/TM6B.
Generation of the SceKO deletion allele
The ends-out recombination strategy was used to disrupt Sce and replace its entire coding region with a miniwhite marker gene following the strategy described previously (Gong and Golic, 2003). In brief, 3.7 kb of Sce 3′ flanking sequences (FlyBase 3R:23502707 to 23506380) and 3.5 kb of 5′ flanking sequences (FlyBase 3R:23507729 to 23511264) were cloned into pw35 (Gong and Golic, 2003). In this construct, the entire coding region of Sce [FlyBase 3R:23506276 to 23507736 (on the minus strand)] is replaced by the miniwhite gene from pw35; only the C-terminal 14 codons of Sce are still present. Following the described strategy (Gong and Golic, 2003), we isolated several independent targeting events that all failed to complement the lethality of Sce1. One of these alleles, SceKO, was selected for in-depth analysis by PCR using primers (5′ to 3′): a-1, CAGTCGCCTGACGCCCATGGAACCACCC; a-2, CCGGAA AGACGACCCTGCTGAATGCC; a-3, ACCTGCTCATCT CGAAGATCTACCCC; b-1, TGATTCTCGGAAGAAAGTGAACTGGG; b-2, ATCCCGGATGGCGATACTTGGATGCC; and b-3, TAGCCATCACCTTCTCCTGGATGGCC (supplementary material Fig. S1).
Knockdown of Sce by RNA interference (RNAi)
Males homozygous for the UAS-anti-Sce hairpin RNA transgene (VDRC transformant line 106328, construct ID 109179) were crossed to P(GawB)T80/CyO-ubiGFP females (Bloomington stock number 1878) to induce degradation of Sce mRNA in the offspring.
Preparation of imaginal disc total protein extracts
Imaginal wing discs from third instar Drosophila larvae were dissected on ice and incubated in SDS gel loading buffer for 5 minutes at 95°C. The suspension was sonicated, debris was pelleted and the supernatant analyzed by SDS-PAGE.
Acid-extraction of histones from Drosophila larval imaginal wing discs
Dissected imaginal wing discs from third instar larvae were homogenized and acid-extraction of histones was performed as described (Scheuermann et al., 2010).
Preparation of embryonic cuticles and immunostaining of embryos and imaginal discs
Preparation of embryonic cuticles, staining of embryos and larval imaginal discs and clonal analysis were performed following standard protocols (Beuchle et al., 2001). Mutant eye-antennal discs were generated using the eyeless-FLP/cell-lethal system as described (Classen et al., 2009).
Antibodies
Antibodies used in this study were Sce (Gorfinkiel et al., 2004; Lagarou et al., 2008; Balicky et al., 2004), Psc (Poux et al., 2001), Ph (Oktaba et al., 2008), Calypso (Scheuermann et al., 2010), Asx (Scheuermann et al., 2010), α-Tubulin (Sigma, T9026), H2A (Millipore, 07-146), H3K27me3 (Upstate, 07-449), Eve (Frasch et al., 1987), Dac (Mardon et al., 1994) (Developmental Studies Hybridoma Bank MR1A), Pros (Srinivasan et al., 1998), ElB (Weihe et al., 2004), Noc (Weihe et al., 2004), Doc2/3 (Reim et al., 2003), En (Developmental Studies Hybridoma Bank 4D9), Wg (Diaz-Benjumea and Cohen, 1995), Cad (Macdonald and Struhl, 1986), Ubx (White and Wilcox, 1984), Abd-B (Celniker et al., 1990) and Lamin (gift from D. Arndt-Jovin, Göttingen, Germany).
Genome-wide profiling of Sce protein in Drosophila using Affymetrix whole-genome tiling arrays
Chromatin preparation from Drosophila larval brain and imaginal discs (wing, haltere and third leg) and chromatin immunoprecipitation (ChIP) were performed essentially as described (Oktaba et al., 2008). To determine Sce-bound regions, six ChIP assays were performed on independent batches of chromatin, three with each of two different antibodies raised against full-length Sce protein (Balicky et al., 2004; Lagarou et al., 2008). The immunoprecipitated material was amplified by ligation-mediated PCR, fragmented, labeled and hybridized to Affymetrix whole-genome microarrays as described (Gambetta et al., 2009). The raw and processed microarray data have been deposited in the ArrayExpress database under accession number E-TABM-988. Data analysis was as described (Oktaba et al., 2008) with the following modifications.
Genome assembly and annotation
We used the third Drosophila melanogaster genome assembly (UCSC dm3 or FlyBase 5.x) and the gff export of FlyBase 5.23 for genome annotation.
Affymetrix probe remapping
We downloaded the Dm35b_MR_v02-3_BDGPv4h.new.bpmap file from the CisGenome website (http://www.biostat.jhsph.edu/~hji/cisgenome/). This file contains the remapped location to genome version 5 (i.e. UCSC dm3) of all the original Affymetrix 25mer sequences, removing those probes that cannot uniquely map to the genome.
Detection of regions that are significantly bound by Sce
A quantile normalization (Bolstad et al., 2003) was applied to normalize together six ChIP hybridizations and three genomic DNA hybridizations. Significantly bound regions were identified using TileMap (Ji and Wong, 2005) with the hidden Markov model. A TileMap score where 60% of the previously identified 237 regions bound by PhoRC and Ph in larva (Gambetta et al., 2009; Oktaba et al., 2008) were recovered was used as threshold for considering significantly bound regions. Additionally, the top 5% regions below cut-off exhibiting overlap with either Pho-, Sfmbt- or Ph-bound regions (Gambetta et al., 2009; Oktaba et al., 2008) were rescued and included in the final high-confidence Sce binding profile. We thus identified 624 regions (405 scoring above cut-off, plus 219 rescued) bound by Sce in brain and larval imaginal discs.
Venn diagram counts
For Venn diagram counts, two or more regions that overlap by at least one base were merged and defined as a ‘common’ region.
Sce-bound regions, target gene assignment and GO slim analysis
The relative distance of the midpoint of each Sce-bound region with respect to the closest transcription start site (TSS) was computed. Target genes were assigned based on TSS-proximal location. Assigned genes to each dataset were tested for enriched GO slim term annotations.
RESULTS
An Sce deletion mutant shows classic homeotic phenotypes
Drosophila Sce encodes a protein of 435 amino acids. Previous studies reported the isolation and characterization of two mutant Sce alleles (Breen and Duncan, 1986; Fritsch et al., 2003; Gaytán de Ayala Alonso et al., 2007; Gorfinkiel et al., 2004). Sce1 encodes a truncated Sce protein that contains the N-terminal 322 amino acids, whereas Sce33M2 encodes a full-length Sce protein with an Arg65Cys mutation (Fritsch et al., 2003; Gorfinkiel et al., 2004). To determine the Sce null mutant phenotype, we used a homologous recombination strategy (Gong and Golic, 2003) to generate SceKO, a knockout allele that deletes the entire Sce coding region (supplementary material Fig. S1).
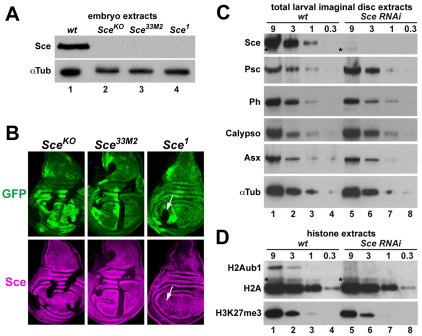
We analyzed Sce expression in protein extracts from SceKO, Sce33M2 or Sce1 homozygous embryos that were derived from germ line clones and therefore lacked maternally deposited wild-type Sce protein. The extracts were probed with an antibody that had been raised against the N-terminal 274 amino acids of the Sce protein (Gorfinkiel et al., 2004). We were unable to detect full-length Sce protein in extracts from any of these Sce mutants (Fig. 1A). The lack of detectable Sce protein in Sce33M2 mutants suggests that the SceR65C protein encoded by this allele is not stable. Staining of imaginal discs that contained clones of Sce mutant cells with anti-Sce antibody corroborated this finding: Sce protein was undetectable not only in SceKO but also in Sce33M2 mutant clones (Fig. 1B). By contrast, the Sce antibody signal was undiminished in Sce1 mutant clones (Fig. 1B), suggesting that the C-terminally truncated Sce protein encoded by this allele is expressed as a stable polypeptide. We were unable to detect the truncated Sce1 protein by western blot analysis because of cross-reactivity of the anti-Sce antibody with an unknown epitope that migrates at the position of the predicted truncated Sce1 protein in embryo extracts (data not shown). Given that SceKO lacks the entire coding region, the phenotype of SceKO mutants represents the null phenotype of Sce. SceKO and Sce33M2 mutant embryos derived from SceKO and Sce33M2 mutant germ cells, respectively, show comparably severe homeotic transformations (supplementary material Fig. S2). This suggests that Sce33M2 is a strong loss-of-function or even a null mutation, consistent with the observation that Sce33M2 mutants lack detectable Sce protein. Intriguingly, Sce1 mutant embryos show more extreme homeotic transformations than SceKO or Sce33M2 mutants, and their phenotype resembles that of Pc mutants (supplementary material Fig. S2). The truncated Sce1 protein lacks the C-terminal Pc-interaction domain (Gorfinkiel et al., 2004; Schoorlemmer et al., 1997; Wang et al., 2010) but contains the N-terminal RING finger domain required for complex formation with Psc-Su(z)2 (Buchwald et al., 2006; Li et al., 2006). It is possible that Sce1 mutants show a stronger PcG phenotype than SceKO mutants because the Sce1 protein sequesters Psc and/or Su(z)2 into non-productive protein assemblies and thereby interferes with their ability to repress target genes.
Fig. 1.
Analysis of Sce protein in Sce mutants and Sce requirement for H2A monoubiquitylation. (A) Total extracts from wild-type (wt, lane 1), SceKO (lane 2), Sce33M2 (lane 3) and Sce1 (lane 4) homozygous Drosophila embryos probed with antibodies against Sce and α-Tubulin 1 (αTub) as loading control. The truncated Sce1 protein is not shown (see text). (B) Wing imaginal discs from SceKO/GFP (left), Sce33M2/GFP (middle) and Sce1/GFP (right) heterozygous third instar larvae with clones of cells that are homozygous for the indicated Sce mutation. Discs were stained with antibody against Sce protein, 96 hours after induction of clones. Clones of Sce homozygous cells are marked by absence of GFP (green). In SceKO and Sce33M2 homozygous cells, Sce protein is undetectable. In Sce1 homozygous cells (arrows), the truncated Sce1 protein is detected at levels that are comparable to those of the full-length wild-type Sce protein. (C) Western blots of serial dilutions (9:3:1:0.3) of total imaginal disc extracts from wild-type and Sce RNAi-treated third instar larvae, probed with antibodies against PRC1 and PR-DUB subunits and α-Tubulin 1 (αTub). Sce signals in lanes 4 and 5 are comparable, suggesting that Sce levels are reduced more than 95% in Sce RNAi-treated larvae. Asterisks indicate a cross-reacting band. (D) Western blots of serial dilutions of histone extracts from imaginal discs of wild-type and Sce RNAi-treated third instar larvae, probed with antibodies against H2A and H3-K27me3. The H2A-K118ub1 band is identified as the monoubiquitylated form of H2A because it migrates at the same position as H2A-K118ub1 generated by monoubiquitylation of recombinant mononucleosomes with Ring1B-Bmi1 complex in vitro and at the same position as H2A-K118ub1 in extracts from PR-DUB mutant embryos in which the levels of H2A-K118ub1 are strongly increased (Scheuermann et al., 2010). Note that the H2A-K118ub1 signal in lane 5 is substantially weaker than in lane 2 and only slightly stronger than that in lane 3, suggesting that H2A-K118ub1 levels are reduced by about 80% in Sce RNAi-treated larvae. Asterisks indicate a cross-reacting band.
Taken together, these analyses show that the complete lack of Sce protein in embryos causes severe homeotic transformations that, nonetheless, are less extreme than the transformations observed in some of the other PcG mutants (see below). The truncated Sce protein expressed from the Sce1 allele also lacks Sce+ function but, in addition, acts in a dominant-negative fashion to exacerbate the loss of HOX gene silencing.
Sce is the major E3 ligase for H2A monoubiquitylation in Drosophila
We next investigated how H2A-K118ub1 levels were affected by removal of the Sce protein. We used an RNAi strategy to knock down Sce protein in imaginal disc tissues. Sce protein levels in imaginal disc cells were reduced by more than 95% after Sce RNAi (Fig. 1C). By contrast, the levels of the PRC1 subunits Psc and Ph and of the PR-DUB subunits Calypso and Additional sex combs (Asx) were unaffected by the Sce RNAi treatment (Fig. 1C). We isolated bulk histones by acid extraction from imaginal disc tissue of wild-type and Sce RNAi-treated larvae and compared the levels of H2A-K118ub1 and H3-K27me3. Both modifications were readily detected in wild-type animals (Fig. 1D). In Sce RNAi-treated animals, H2A-K118ub1 levels were strongly reduced, whereas H3-K27me3 levels were comparable to those in wild-type animals (Fig. 1D). We conclude that Sce is responsible for the bulk of H2A monoubiquitylation in developing Drosophila. This is consistent with studies by Lagarou et al. (Lagarou et al., 2008) who reported that depletion of Sce by RNAi in Drosophila tissue culture cells results in a drastic reduction of H2A-K118ub1 levels.
Mutants lacking PRC1 subunits show distinct morphological defects
Sce or Pc homozygous mutant embryos that are derived from females with a mutant germ line and therefore lack maternally deposited wild-type Sce or Pc protein, respectively, show segmental transformations due to misexpression of HOX genes (supplementary material Fig. S2). By comparison, ph0 embryos, which are mutant for both Ph-p and its paralog Ph-d, or embryos that are homozygous for a small deficiency that deletes both Psc and its paralog Su(z)2 [i.e. Psc-Su(z)2 mutants], show much more severe morphological defects, even if these embryos are derived from females with a heterozygous germ line and therefore contain the maternally deposited wild-type products of these genes (supplementary material Fig. S2) (see Dura et al., 1987; Feng et al., 2011; Martin and Adler, 1993; Smouse et al., 1988). ph0 or Psc-Su(z)2 mutant embryos that also lack maternally deposited Ph or Psc and Su(z)2 proteins are highly abnormal already at the gastrulation stage, arrest development shortly afterwards and form no, or only very rudimentary, embryonic cuticle (supplementary material Fig. S2) (Smouse et al., 1988). Moreover, zygotic expression of wild-type Ph and Psc-Su(z)2 proteins is unable to restore normal development to embryos that lack maternally deposited Ph and Psc-Su(z)2 proteins, respectively (supplementary material Fig. S2) (Smouse et al., 1988). By contrast, zygotic expression of wild-type Sce or Pc protein rescues embryos that lack maternally deposited Sce or Pc proteins, respectively, into viable and fertile adults (not shown) (Breen and Duncan, 1986; Lawrence et al., 1983). Taken together, comparison of the loss-of-function phenotypes of mutants lacking different PRC1 subunits shows that Ph, Psc and Su(z)2 proteins are required for processes that appear to occur normally in the absence of Sce or Pc.
Genome-wide binding maps of Sce, Ph, Psc and Pc identify a set of PRC1-bound genes
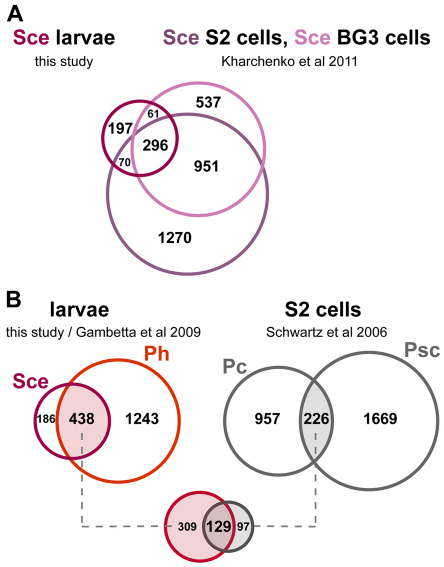
Ph, Psc-Su(z)2, Sce and Pc might regulate different sets of target genes and this could account for the differences in their mutant phenotypes. Here, we focused on target genes that are bound by all four PRC1 subunits and investigated how their expression is affected in the absence of individual subunits. Previous studies reported the genome-wide binding profiles of Psc, Pc and Sce in Drosophila tissue culture cells and that of Ph and Pc in Drosophila embryos and larval imaginal disc cells (Gambetta et al., 2009; Kharchenko et al., 2011; Kwong et al., 2008; Schuettengruber and Cavalli, 2009; Schwartz et al., 2006). To identify genes that are bound by Sce in developing Drosophila, we generated the genome-wide Sce protein-binding profile in imaginal disc and central nervous system (CNS) tissues from third instar larvae. We performed chromatin immunoprecipitation (ChIP) assays with two independent antisera that had been raised against the Sce protein and hybridized the immunoprecipitated material to high-density whole-genome tiling arrays. Only genomic regions significantly enriched by both anti-Sce antibodies were considered. Using a stringent cut-off, we thus obtained a high-confidence set of 624 genomic regions bound by the Sce protein in larval tissues (Fig. 2A,B; supplementary material Table S1). Comparison of this binding profile with the Sce profiles in two different tissue culture cell lines (Kharchenko et al., 2011) showed that 427 of the 624 regions bound by Sce in imaginal disc cells (68%) had also been identified as Sce target sites in tissue culture cells (Fig. 2A).
Fig. 2.
Identification of cis-regulatory regions co-bound by Sce and other PRC1 subunits. (A) Venn diagram showing the overlap of Sce-bound regions identified by genome-wide profiling in larval tissues and in Drosophila S2 and BG3 tissue culture cells. The overlap identifies 427 regions bound by Sce in both larval and tissue culture cells. (B) Venn diagram showing the overlap of Sce- and Ph-bound regions in larval tissues and the overlap of Pc- and Psc-bound regions in Drosophila S2 tissue culture cells. The overlap of 438 regions bound by both Sce and Ph and 226 regions bound by both Pc and Psc identifies 129 regions that are bound by all four PRC1 core subunits.
We then determined the overlap between Sce-bound regions and regions bound by other PRC1 subunits. For this analysis we used the genome-wide binding profiles of Ph in imaginal disc cells (Gambetta et al., 2009) and of Psc and Pc in tissue culture cells (Schwartz et al., 2006). The Ph profile from Gambetta et al. (Gambetta et al., 2009) permitted direct comparison with the Sce profile because both profiles had been generated in imaginal disc tissues dissected from developing third instar larvae. Similarly, the Psc and Pc datasets had both been generated in the same Drosophila S2 tissue culture cell line (Schwartz et al., 2006). Comparison of the Sce and Ph profiles in imaginal disc cells revealed that Ph is co-bound at 438 of the 624 Sce-bound regions (70%; Fig. 2B). Among these 438 regions bound by Sce and Ph, we identified 129 genomic locations that Schwartz et al. (Schwartz et al., 2006) had reported to be bound by both Psc and Pc in S2 cells (29%; Fig. 2B). These 129 regions thus represent genomic sites bound by Sce and Ph in imaginal disc cells and by Psc and Pc in tissue culture cells; they include PREs in HOX and other target genes that have been shown to be misexpressed in mutants lacking PRC1 subunits (see below). Because of the different source of material used for profiling (imaginal disc cells versus tissue culture cells), it is very likely that these 129 regions only represent a fraction of all regions that are bound by PRC1 in Drosophila. However, for simplicity, we shall here refer to the genes associated with these 129 regions as PRC1-bound genes.
Repression of many PRC1-bound genes requires ph and Psc-Su(z)2 but not Sce or Pc
We assessed how the expression of PRC1-bound genes is affected in animals that lack individual PRC1 subunits. For this analysis, we focused on target genes for which it was not known whether they are regulated by PRC1: dachshund (dac), prospero (pros), elbow (elB) and no ocelli (noc). We also included a few non-HOX target genes known to be regulated by the PcG system: engrailed (en), caudal (cad), even-skipped (eve) and Dorsocross 2/3 (Doc2/3) (Beuchle et al., 2001; Busturia and Morata, 1988; Oktaba et al., 2008). Each of the analyzed genes is bound by all four PRC1 subunits (Figs 3, 4; supplementary material Fig. S3, Table S1).
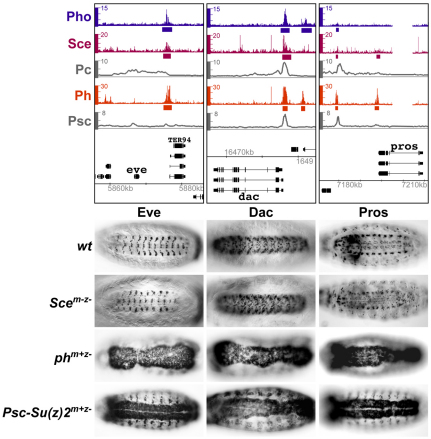
Fig. 3.
Repression of eve, dac and pros in embryos requires ph and Psc-Su(z)2 but not Sce. (Top) ChIP profiles of Sce, Pc, Ph and Psc and, as reference, of Pho, the DNA-binding subunit of PhoRC, at the eve, dac and pros genes. PRC1 subunit profiles are from experiments shown in Fig. 2B and the Pho profile in imaginal disc tissues is the PhoL dataset from Oktaba et al. (Oktaba et al., 2008). Hybridization intensities for oligonucleotide probes are plotted as colored bars (Pho, Sce, Ph) or smoothed profiles (Pc, Psc) above the genomic map (release 5/UCSC dm3, kb coordinates) of Drosophila. In the Pho, Sce and Ph datasets, bars below plots mark regions that the analyses shown in Fig. 2 and in Oktaba et al. (Oktaba et al., 2008) identified as Sce-, Ph- or Pho-bound, respectively. (Below) Ventral views of 14- to 16-hour-old Drosophila embryos stained with antibodies against Eve, Dac or Pros protein.
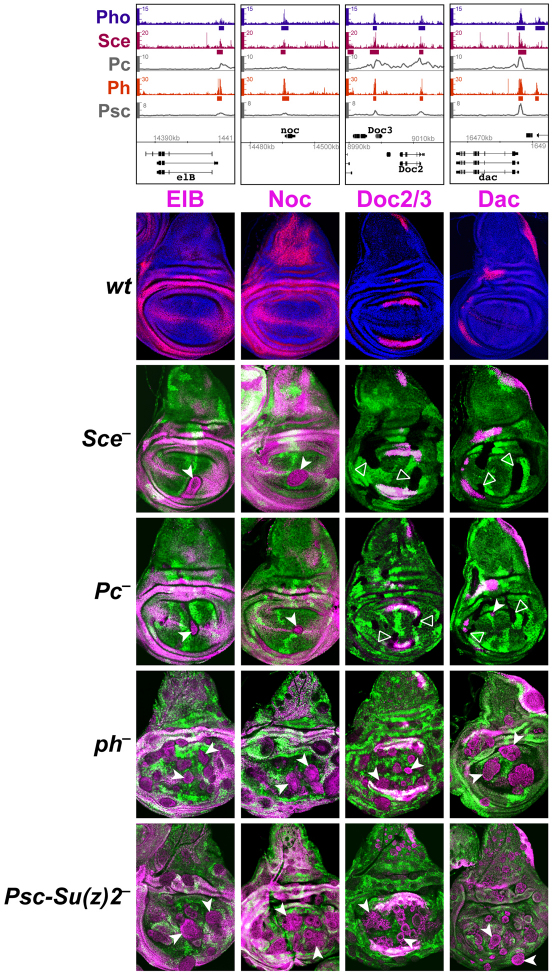
Fig. 4.
PRC1 subunits are differentially required for repression of elB, noc, Doc2/3 and dac. (Top) ChIP profiles at the PRC1-bound genes elB, noc, Doc2/3 and dac, represented as in Fig. 3. (Below) Regulation of elB, noc, Doc2/3 and dac in wing imaginal discs with cell clones that lack individual PRC1 subunits. Row 1 shows wing discs from wild-type third instar larvae stained with antibodies against ElB, Noc, Doc2/3 and Dac proteins (magenta in each case) and co-stained with Hoechst (blue) to visualize nuclei. Rows 2 to 5 show wing discs with clones of cells lacking individual PRC1 subunits as indicated. Clones are marked by the absence of GFP and discs were analyzed 72 to 96 hours (rows 2 and 3) or 48 to 72 hours (rows 4 and 5) after clone induction. ElB, Noc, Doc2/3 and Dac proteins are all widely misexpressed in Ph or Psc-Su(z)2 mutant clones (white arrowheads). ElB and Noc are also misexpressed in Sce and Pc mutant clones (white arrowheads) but the Doc2/3 and dac genes remain stably repressed (open arrowheads).
In a first set of experiments, expression of the eve, dac and pros genes was analyzed in wild-type, Sce, ph and Psc-Su(z)2 mutant embryos. In the case of Sce, the SceKO homozygous embryos were derived from females with an SceKO mutant germ line and therefore completely lacked Sce protein (Scem–z–). In the case of ph0 and Psc-Su(z)2, the mutant embryos lacked zygotic ph and Psc-Su(z)2 function but contained the maternally deposited wild-type products of these genes [phm+z– and Psc-Su(z)2m+z–]. In wild-type embryos, eve, dac and pros are each expressed in a specific set of cells in the CNS of late stage embryos (Fig. 3). eve becomes misexpressed in most cells of the nervous system of ph0 or Psc-Su(z)2 mutant embryos (Fig. 3) (see Dura and Ingham, 1988; Oktaba et al., 2008). By contrast, eve expression remains confined in its normal pattern in embryos that lack Sce protein (Fig. 3). Similarly, we found that dac and pros are also widely misexpressed in the nervous system of ph0 or Psc-Su(z)2 mutant embryos, but their expression patterns in Sce mutant embryos are indistinguishable from those in wild-type embryos (Fig. 3). PcG regulation of eve, dac and pros genes thus differs from that of HOX genes in that their repression in embryos requires Ph, Psc and Su(z)2 proteins but not Sce.
We next analyzed expression of the PRC1-bound genes elB, noc, Doc2/3, dac, en, eve and cad in imaginal discs with clones of SceKO, Pc, ph0 or Psc-Su(z)2 homozygous mutant cells. In wild-type animals, elB, noc, Doc2/3, dac and en are all expressed in characteristic patterns in the wing imaginal disc, whereas eve and cad are inactive in the wing disc (Fig. 4; supplementary material Fig. S3). Each of these genes becomes widely misexpressed in ph0 or Psc-Su(z)2 mutant clones in regions of the disc where these genes are normally not expressed (Fig. 4; supplementary material Fig. S3). By contrast, Doc2/3, dac, eve and cad all remained stably repressed in Sce or Pc mutant clones (Fig. 4; supplementary material Fig. S3). en also remained repressed in Sce or Pc mutant clones in most areas of the wing disc, but the gene was misexpressed in such clones in the presumptive dorsal wing hinge (supplementary material Fig. S3). Thus, among all the PRC1-bound genes analyzed here, only elB and noc were widely misexpressed in Sce or Pc mutant clones (Fig. 4).
One of the 129 PRC1-bound genes is wingless (wg) (Oktaba et al., 2008; Schwartz et al., 2006) (supplementary material Table S1). wg encodes a signaling molecule that acts as a positive regulator of elB, noc, dac and other genes involved in imaginal disc patterning (Lecuit and Cohen, 1997; Weihe et al., 2004). We have not been able to detect strong upregulation of Wg protein expression in ph0, Psc-Su(z)2, Sce or Pc mutant clones even though the Wg protein signal is often slightly increased in an erratic pattern in large clones of ph0 or Psc-Su(z)2 mutant cells (supplementary material Fig. S3). Misexpression of elB and noc in clones of PcG mutant cells (Fig. 4) is therefore unlikely to be simply an indirect consequence of the overexpression of Wg protein in the mutant clones. Finally, we note that we have not been able to detect misexpression of nubbin, homothorax or vestigial in either ph0 or Psc-Su(z)2 mutant clones in wing discs (data not shown). These three genes are all bound by PRC1 and are positively or negatively regulated by Wg in the wing imaginal disc. It is possible that they are repressed by PRC1 in other tissues or at other stages of development.
Taken together, these analyses suggest that several genes identified as PRC1 targets by genome-wide profiling are indeed directly repressed by PRC1 but with important differences with respect to the requirement for individual PRC1 subunits. PRC1 target genes can be grouped into two classes. Class I genes, such as elB, noc, en and the HOX genes, are derepressed in ph, Psc-Su(z)2, Sce and Pc mutants, whereas class II genes, such as Doc2/3, dac, pros, eve and cad, are only derepressed in ph and Psc-Su(z)2 mutants but remain stably repressed in Sce and Pc mutants. Similar observations were made in Sce RNAi-treated imaginal discs (supplementary material Fig. S4). The global reduction of H2A-K118ub1 levels in these discs (Fig. 1D) thus correlates with loss of repression of class I genes, whereas class II genes remain repressed.
Psc-Su(z)2 and ph act as tumor suppressor genes but Sce does not
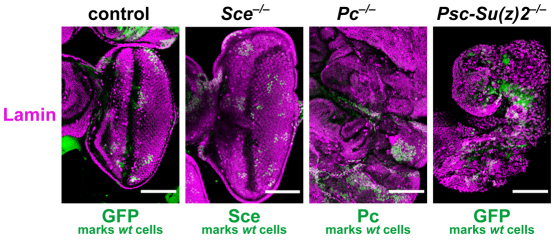
We previously reported that clones of Psc-Su(z)2 or ph mutant cells in imaginal wing discs show a tumor phenotype caused by hyperproliferation of the mutant cells, but that Sce or Pc mutant clones show normal cell proliferation (Beuchle et al., 2001; Oktaba et al., 2008). Recent studies reported that under conditions in which cell competition is removed, not only Psc-Su(z)2 and ph but also Pc and Sce mutant cell clones show hyperproliferation and tumor formation (Classen et al., 2009). Interestingly, this hyperproliferation of Pc and Sce mutant cells was only observed in eye imaginal discs and not in wing discs (Classen et al., 2009). Because Classen et al. (Classen et al., 2009) had used the Sce1 allele for their analyses, we re-examined the role of PRC1 components under the same assay conditions but with the SceKO allele. Specifically, we used the eyeless-FLP/cell-lethal system to generate eye imaginal discs that consisted mainly of Sce, Pc or Psc-Su(z)2 mutant cells (Fig. 5). Psc-Su(z)2 and Pc mutant discs showed the tumor phenotype as previously reported (Classen et al., 2009) but SceKO mutant discs were morphologically indistinguishable from wild-type discs (Fig. 5). Taken together, these analyses show that Psc-Su(z)2, ph and Pc have tumor suppressor activity to restrict cell proliferation in imaginal discs but that Sce does not show this activity. Tumor suppression by PRC1-type complexes therefore does not seem to require H2A monoubiquitylation.
Fig. 5.
Sce lacks the tumor suppressor activity shown by other PRC1 subunits. Eye imaginal discs consisting mainly of Sce–/–, Pc–/– or Psc-Su(z)2–/– mutant cells were generated with the eyeless-FLP/cell-lethal system. Discs were stained with antibody against Lamin to visualize all nuclei; wild-type cells were visualized by staining with antibodies against Sce or Pc or by the GFP marker protein. The SceKO, PcXT109 and Psc-Su(z)2P3C alleles were used to permit direct comparison with the results reported by Classen et al. (Classen et al., 2009). Control shows clones of wild-type tissue (non-green) generated in the background of wild-type cells marked by GFP.
Sce and PR-DUB synergize to repress HOX genes
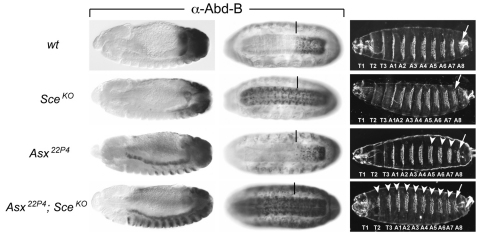
Finally, we investigated the interplay between H2A monoubiquitylation and deubiquitylation in the control of HOX gene repression. Recent studies identified PR-DUB as a major deubiquitinase for H2A-K118ub1 in Drosophila (Scheuermann et al., 2010). PR-DUB comprises the catalytic subunit Calypso (Bap1) and its essential co-factor Asx. Mutants lacking Calypso or Asx show misexpression of HOX genes and corresponding homeotic transformations (Gaytán de Ayala Alonso et al., 2007; Scheuermann et al., 2010), but they show no detectable misexpression of any of the other PcG target genes analyzed here (data not shown). We previously reported that Asx22P4 Sce1 double-homozygous embryos that were derived from heterozygous mothers show severe homeotic transformations that are comparable to the transformations observed in Sce1 mutant embryos derived from females with an Sce1 mutant germ line (Scheuermann et al., 2010). The same is true for Asx22P4 SceKO double-homozygous embryos; the homeotic transformations in such animals are more severe than those in Asx22P4 or SceKO single homozygotes and resemble the transformations observed in SceKO homozygous embryos that lack maternally deposited Sce protein (compare the Asx22P4 SceKO embryo in Fig. 6 with the SceKOm–z– embryo in supplementary material Fig. S2). As illustrated in Fig. 6, this is explained by the observation that repression of HOX genes in the epidermis of Asx22P4 SceKO double-mutant embryos is lost more rapidly than in Asx22P4 or SceKO single mutants and that the double-mutant embryos show much more widespread HOX gene misexpression in the epidermis of late-stage embryos compared with the single mutants. This suggests that the partial rescue of HOX gene repression by maternally deposited Sce protein in SceKO homozygotes is abolished in the absence of PR-DUB. At present, we can only speculate how H2A deubiquitylation contributes to transcriptional repression (see Discussion), but the strong genetic synergy between PR-DUB and Sce in HOX gene silencing supports the idea that H2A ubiquitylation by Sce is important for this process.
Fig. 6.
Sce and PR-DUB synergize to repress HOX genes. Lateral views of stage 13 (left column) and ventral views of stage 16 (middle column) Drosophila embryos, stained with antibody against the HOX protein Abd-B. The right-hand column shows ventral views of embryonic cuticles. The genotypes are wild type (first row), SceKO homozygous (second row), Asx22P4 homozygous (third row), SceKO and Asx22P4 double homozygous (fourth row); in all cases the mutant embryos were derived from heterozygous females. In stage 16 embryos, a vertical bar marks the anterior margin of the normal Abd-B expression domain in parasegment (ps) 10 in the CNS. In embryonic cuticles, an arrow marks the denticle belt of the eighth abdominal segment. (Row 1) In wild-type embryos, Abd-B is expressed from ps 10-14, with lowest levels present in ps 10 and highest levels in ps 14. In the embryonic cuticle, every thoracic (T) and abdominal (A) segment contains a characteristic belt of denticles, specified by the level of the HOX protein expressed in that particular segment. (Row 2) In stage 13 SceKO embryos, Abd-B is still repressed in most cells anterior to ps 10 and only a few Abd-B-positive cells are detected in more anterior regions. In stage 16 SceKO embryos, Abd-B is expressed throughout the CNS but misexpression in the epidermis is low. The embryonic cuticle appears indistinguishable from that of wild type. (Row 3) In stage 13 Asx22P4 embryos, Abd-B is misexpressed in the anterior visceral mesoderm and also in a subset of cells in the epidermis of abdominal segments. In stage 16 Asx22P4 embryos, Abd-B is misexpressed in a subset of cells in the epidermis of every segment and in a few rare cells in the CNS. Within ps 10-14, there is a partial loss of Abd-B expression in the CNS but an increase in Abd-B levels in the epidermis (not visible here) and consequently A5-A7 in the cuticle are partially transformed into copies of A8 (white arrowheads). (Row 4) In stage 13 Asx22P4 SceKO double-mutant embryos, Abd-B misexpression in the epidermis is more widespread than in the single mutants and also extends throughout the thoracic and anterior abdominal segments. In stage 16 Asx22P4 SceKO embryos, Abd-B is strongly misexpressed throughout the epidermis and CNS and in more cells in every segment compared with the single mutants. Segments T2-A7 in the cuticle are extensively, but not completely, transformed into copies of A8 (white arrowheads).
DISCUSSION
In this study, we analyzed how PRC1 regulates target genes in Drosophila to investigate how the distinct chromatin-modifying activities of this complex repress transcription in vivo. Because H2A monoubiquitylation is thought to be central to the repression mechanism of PRC1-type complexes, we focused on the role of Sce. The following main conclusions can be drawn from the work reported here. First, in the absence of Sce, bulk levels of H2A-K118ub1 are drastically reduced but the levels of the PRC1 subunits Psc and Ph are undiminished. Sce is therefore the major E3 ligase for H2A monoubiquitylation in developing Drosophila but is not required for the stability of other PRC1 subunits. Second, PRC1-bound genes fall into two classes. Class I target genes are misexpressed if any of the PRC1 subunits is removed. Class II target genes are misexpressed in the absence of Ph or Psc-Su(z)2 but remain stably repressed in the absence of Sce or Pc. At class II target genes, Ph and the Psc-Su(z)2 proteins work together to repress transcription by a mechanism that does not require Sce and Pc and is therefore independent of H2A monoubiquitylation. Third, removal of the Ph, Psc-Su(z)2 or Pc proteins results in imaginal disc tumors that are characterized by unrestricted cell proliferation. However, removal of Sce does not cause this phenotype, suggesting that this tumor suppressor activity by the PcG system does not require H2A monoubiquitylation. Finally, our analyses reveal that PRC1 subunits are essential for repressing the elB, noc, dac and pros genes outside of their normal expression domains in developing Drosophila. This expands the inventory of developmental regulator genes in Drosophila for which PcG repression has been demonstrated in a functional assay. Below we discuss the implications of the findings reported in this study.
Molecular role of Sce in PRC1 and dRAF
In the Sce33M2 allele Arg65 is mutated to Cys, but this mutant Sce protein is undetectable and therefore does not appear to be stable in vivo (Fig. 1A,B). The crystal structure of the Ring1B-Bmi1 complex provides a molecular explanation for this observation: the Arg70 residue in Ring1B that corresponds to Arg65 in Sce is thought to be critical for interaction with Bmi1 (Buchwald et al., 2006; Li et al., 2006). A likely scenario therefore is that the SceArg65Cys protein in Drosophila is unstable and is degraded because it is unable to associate with Psc or its paralog Su(z)2. Interestingly, removal of Sce protein has no detectable effect on the levels of the Psc and Ph proteins (Fig. 1C). Psc is therefore stable in the absence of its binding partner Sce. This is in contrast to the situation in mice in which Ring1B mutant ES cells show a drastic reduction in the levels of the Ring1B partner protein Bmi1 and its paralog Mel18 (Pcgf62) and also a reduction in the levels of Mph2 (Phc2) and Mpc2 (Cbx4) (Leeb and Wutz, 2007). The interdependence between PRC1 subunits for protein stability is therefore different in mammals and Drosophila.
Reconstitution of the Drosophila PRC1 core complex in a baculovirus expression system suggests that Sce is important for complex stability (Francis et al., 2001). At present, we do not know whether the Psc, Ph and Pc proteins still form a complex in vivo in the absence of Sce. It is currently unknown whether Psc, Ph and Pc are still bound to all PRC1 target genes in the absence of Sce. However, the finding that class II genes remain repressed in the absence of Sce, even though their repression depends on Psc-Su(z)2 and Ph, argues against a crucial role of Sce in the targeting of these other PRC1 subunits to these genes. Interestingly, the repression of all class II target genes analyzed here always requires both the Ph and the Psc-Su(z)2 proteins. A possible explanation for this observation is that Ph and Psc-Su(2) still form a PRC1 subcomplex in the absence of Sce and that this complex is fully functional to repress class II target genes. Alternatively, it is possible that Ph and Psc-Su(z)2 repress class II target genes as components of as yet uncharacterized complexes that are distinct from PRC1 and dRAF.
Transcriptional repression by Psc-Su(z)2 and Ph
In vitro, Psc and Su(z)2 proteins compact nucleosome templates, inhibit nucleosome remodeling by SWI/SNF complexes and repress transcription on chromatin templates (Francis et al., 2004; Francis et al., 2001; Lo et al., 2009). The observation that repression of class II target genes requires Psc-Su(z)2 and Ph but not Pc and Sce supports the idea that the chromatin-modifying activities of Psc-Su(z)2 identified in vitro are the main mechanism by which PRC1 represses these genes. Previous structure/function analyses in Drosophila showed that the same domains of the Psc protein responsible for chromatin compaction and remodeling inhibition in vitro are crucial for HOX gene repression in vivo (King et al., 2005). Chromatin modification by Psc and Su(z)2 is therefore also crucial for repression of class I target genes. Regulation of the class I target gene en further illustrates this point. In some tissues (e.g. in the dorsal hinge region of the wing imaginal disc; supplementary material Fig. S3, white arrowhead) repression of en requires all PRC1 core subunits, but in other tissues (e.g. in the pouch of the wing imaginal disc; supplementary material Fig. S3, open arrowhead) en remains repressed in the absence of Sce and Pc, and only Psc-Su(z)2 and Ph seem to be crucial to keep the gene inactive. At present, the molecular mechanism of Ph is not well understood. In vitro, Ph protein has the capacity to inhibit chromatin remodeling and transcription but it does so less effectively than Psc (Francis et al., 2001; King et al., 2002). At the target genes analyzed here, Ph is required for transcriptional repression wherever Psc-Su(z)2 is required, suggesting that Ph and Psc-Su(z)2 act in concert in this repression. Nevertheless, it is possible that repression of other PRC1 target genes requires a different subset of PRC1 subunits, or that, as in the case of en, the subunit requirement for repression changes depending on the cell type.
H2A ubiquitylation and deubiquitylation in transcriptional repression
In mammals, Ring1B and Ring1A are responsible for the bulk of H2A-K119 monoubiquitylation (de Napoles et al., 2004; Wang, H. et al., 2004). Similarly, Sce generates the bulk of H2A-K118 monoubiquitylation in Drosophila, both in tissue culture cells (Lagarou et al., 2008) and in the developing organism (this study). The requirement for Sce at class I target genes is consistent with the idea that H2A monoubiquitylation of their chromatin is part of the repression mechanism. Repression of a subset of class I genes, namely the HOX genes, also requires the H2A deubiquitinase PR-DUB (Gaytán de Ayala Alonso et al., 2007; Scheuermann et al., 2010). Moreover, PR-DUB and Sce strongly synergize to repress HOX genes. Specifically, the phenotype of Sce PR-DUB double mutants (Fig. 6) suggests that H2A monoubiquitylation becomes ineffective for HOX gene repression if PR-DUB is absent. However, embryos that lack PR-DUB alone show a 10-fold increase in the bulk levels of H2A-K118ub1 and we estimate that ∼10% of all H2A molecules become monoubiquitylated in these animals [see figure 4A in Scheuermann et al. (Scheuermann et al., 2010)]. How could this conundrum be explained? One possibility is that H2A monoubiquitylation and deubiquitylation at HOX gene chromatin need to be regulated in a precisely balanced manner. However, an alternative explanation considers H2A-K118ub1 levels in the context of ubiquitin homeostasis. In particular, the high H2A-K118ub1 levels in PR-DUB mutants suggest that Sce generates widespread H2A monoubiquitylation at most Sce-bound genes and possibly also elsewhere in the genome, but that in wild-type animals PR-DUB continuously deubiquitylates H2A-K118ub1 at these locations and thereby recycles ubiquitin. The observation that PR-DUB is widely co-bound with Sce, not only at HOX but also at many other class I and class II target genes, is consistent with this idea. It is tempting to speculate that the widespread H2A monoubiquitylation in PR-DUB mutants sequesters a substantial fraction of the pool of free ubiquitin. It is therefore possible that removal of PR-DUB effectively depletes the pool of free ubiquitin in the nucleus to an extent that H2A monoubiquitylation at HOX target genes becomes inefficient and, consequently, their repression can no longer be maintained. According to this model, the crucial function of PR-DUB would not be the deubiquitylation of H2A-K118ub1 at HOX genes but rather at class II target genes and elsewhere in the genome where Sce ‘wastefully’ monoubiquitylates H2A.
Supplementary Material
Acknowledgments
We thank S. Bickel, S. Cohen, M. Frasch, G. Struhl, P. Verrijzer and M. Vidal for the generous gift of antibodies; J. de Graaf and V. Benes (EMBL Genomics Core Facility) for help with hybridization of microarrays; and J. Gagneur for discussions.
Footnotes
Funding
This work was supported by funds from the Deutscher Akademischer Austausch Dienst (DAAD); European Molecular Biology Laboratory (EMBL); and the Max-Planck Institute (MPI) of Biochemistry. Deposited in PMC for immediate release.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.074450/-/DC1
References
- Balicky E. M., Young L., Orr-Weaver T. L., Bickel S. E. (2004). A proposed role for the Polycomb group protein dRING in meiotic sister-chromatid cohesion. Chromosoma 112, 231–239 [DOI] [PubMed] [Google Scholar]
- Beuchle D., Struhl G., Müller J. (2001). Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128, 993–1004 [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- Breen T. R., Duncan I. M. (1986). Maternal expression of genes that regulate the bithorax complex of Drosophila melanogaster. Dev. Biol. 118, 442–456 [DOI] [PubMed] [Google Scholar]
- Brown J. L., Kassis J. A. (2010). Spps, a Drosophila Sp1/KLF family member, binds to PREs and is required for PRE activity late in development. Development 137, 2597–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. (2006). Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busturia A., Morata G. (1988). Ectopic expression of homeotic genes caused by the elimination of the Polycomb gene in Drosophila imaginal epidermis. Development 104, 713–720 [DOI] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- Cao R., Wang H., He J., Erdjument-Bromage H., Tempst P., Zhang Y. (2008). Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol. Cell. Biol. 28, 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Sharma S., Keelan D. J., Lewis E. B. (1990). The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 9, 4277–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen A.-K., Bunker B. D., Harvey K. F., Vaccari T., Bilder D. (2009). A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nat. Genet. 41, 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. (2002). Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196 [DOI] [PubMed] [Google Scholar]
- de Napoles M., Mermoud J. E., Wakao R., Tang Y. A., Endoh M., Appanah R., Nesterova T. B., Silva J., Otte A. P., Vidal M., et al. (2004). Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663–676 [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea F. J., Cohen S. M. (1995). Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development 121, 4215–4225 [DOI] [PubMed] [Google Scholar]
- Duncan I. M. (1982). Polycomblike: a gene that appears to be required for the normal expression of the bithorax and antennapedia gene complexes of Drosophila melanogaster. Genetics 102, 49–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dura J. M., Ingham P. (1988). Tissue- and stage-specific control of homeotic and segmentation gene expression in Drosophila embryos by the polyhomeotic gene. Development 103, 733–741 [DOI] [PubMed] [Google Scholar]
- Dura J. M., Randsholt N. B., Deatrick J., Erk I., Santamaria P., Freeman J. D., Freeman S. J., Weddell D., Brock H. W. (1987). A complex genetic locus, polyhomeotic, is required for segmental specification and epidermal development in D. melanogaster. Cell 51, 829–839 [DOI] [PubMed] [Google Scholar]
- Eskeland R., Leeb M., Grimes G., Kress C., Boyle S., Sproul D., Gilbert N., Fan Y., Skoultchi A., Wutz A., et al. (2010). Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38, 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Huang J., Wang J. (2011). Loss of the Polycomb group gene polyhomeotic induces non-autonomous cell overproliferation. EMBO Rep. 12, 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis N. J., Saurin A. J., Shao Z., Kingston R. E. (2001). Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8, 545–556 [DOI] [PubMed] [Google Scholar]
- Francis N. J., Kingston R. E., Woodcock C. L. (2004). Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577 [DOI] [PubMed] [Google Scholar]
- Frasch M., Hoey T., Rushlow C., Doyle H., Levine M. (1987). Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 6, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch C., Beuchle D., Müller J. (2003). Molecular and genetic analysis of the Polycomb group gene Sex combs extra/Ring in Drosophila. Mech. Dev. 120, 949–954 [DOI] [PubMed] [Google Scholar]
- Gambetta M. C., Oktaba K., Müller J. (2009). Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96 [DOI] [PubMed] [Google Scholar]
- Gaytán de Ayala Alonso A., Gutiérrez L., Fritsch C., Papp B., Beuchle D., Müller J. (2007). A genetic screen identifies novel polycomb group genes in Drosophila. Genetics 176, 2099–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W. J., Golic K. G. (2003). Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100, 2556–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N., Fanti L., Melgar T., García E., Pimpinelli S., Guerrero I., Vidal M. (2004). The Drosophila Polycomb group gene Sex combs extra encodes the ortholog of mammalian Ring1 proteins. Mech. Dev. 121, 449–462 [DOI] [PubMed] [Google Scholar]
- Ji H., Wong W. H. (2005). TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics 21, 3629–3636 [DOI] [PubMed] [Google Scholar]
- Jürgens G. (1985). A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316, 153–155 [Google Scholar]
- Kahn T. G., Schwartz Y. B., Dellino G. I., Pirrotta V. (2006). Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J. Biol. Chem. 281, 29064–29075 [DOI] [PubMed] [Google Scholar]
- Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Minoda A., Riddle N. C., Ernst J., Sabo P. J., Larschan E., Gorchakov A. A., Gu T., et al. (2011). Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471, 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I. F. G., Francis N. J., Kingston R. E. (2002). Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol. Cell. Biol, 22, 7919–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I. F. G., Emmons R. B., Francis N. J., Wild B., Müller J., Kingston R. E., Wu C.-T. (2005). Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol. Cell. Biol, 25, 6578–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T., Papp B., Fischle W., Köcher T., Schelder M., Fritsch C., Wild B., Wilm M., Müller J. (2006). A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 20, 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. (2002). Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong C., Adryan B., Bell I., Meadows L., Russell S., Manak J. R., White R. (2008). Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 4, e1000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarou A., Mohd-Sarip A., Moshkin Y. M., Chalkley G. E., Bezstarosti K., Demmers J. A. A., Verrijzer C. P. (2008). dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 22, 2799–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence P. A., Johnston P., Struhl G. (1983). Different requirements for homeotic genes in the soma and germ line of Drosophila. Cell 35, 27–34 [DOI] [PubMed] [Google Scholar]
- Lecuit T., Cohen S. (1997). Proximal-distal axis formation in the Drosophila leg. Nature 388, 139–145 [DOI] [PubMed] [Google Scholar]
- Leeb M., Wutz A. (2007). Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J. Cell Biol. 178, 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S. S., Weiss A., Erdjument-Bromage H., Shao Z., Tempst P., Kingston R. E. (2002). The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22, 6070–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 [DOI] [PubMed] [Google Scholar]
- Li Z., Cao R., Wang M., Myers M. P., Zhang Y., Xu R.-M. (2006). Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J. Biol. Chem. 281, 20643–20649 [DOI] [PubMed] [Google Scholar]
- Lo S. M., Francis N. J. (2010). Inhibition of chromatin remodeling by polycomb group protein posterior sex combs is mechanistically distinct from nucleosome binding. Biochemistry 49, 9438–9448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S. M., Ahuja N. K., Francis N. J. (2009). Polycomb group protein Suppressor 2 of zeste is a functional homolog of Posterior Sex Combs. Mol. Cell. Biol. 29, 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P. M., Struhl G. (1986). A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature 324, 537–545 [DOI] [PubMed] [Google Scholar]
- Mardon G., Solomon N. M., Rubin G. M. (1994). dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120, 3473–3486 [DOI] [PubMed] [Google Scholar]
- Martin E. C., Adler P. N. (1993). The Polycomb group gene Posterior Sex Combs encodes a chromosomal protein. Development 117, 641–655 [DOI] [PubMed] [Google Scholar]
- Misaghi S., Ottosen S., Izrael-Tomasevic A., Arnott D., Lamkanfi M., Lee J., Liu J., Oandapos Rourke K., Dixit V. M., Wilson A. C. (2009). Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol. Cell. Biol. 29, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A., Cléard F., Mishra R. K., Karch F., Verrijzer C. P. (2005). Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev. 19, 1755–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A., van der Knaap J. A., Wyman C., Kanaar R., Schedl P., Verrijzer C. P. (2006). Architecture of a polycomb nucleoprotein complex. Mol. Cell 24, 91–100 [DOI] [PubMed] [Google Scholar]
- Müller J., Kassis J. A. (2006). Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16, 476–484 [DOI] [PubMed] [Google Scholar]
- Müller J., Verrijzer P. (2009). Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19, 150–158 [DOI] [PubMed] [Google Scholar]
- Müller J., Hart C. M., Francis N. J., Vargas M. L., Sengupta A., Wild B., Miller E. L., Oandapos Connor M. B., Kingston R. E., Simon J. A. (2002). Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208 [DOI] [PubMed] [Google Scholar]
- Nègre N., Hennetin J., Sun L. V., Lavrov S., Bellis M., White K. P., Cavalli G. (2006). Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 4, e170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M., Klymenko T., Fraterman S., Papp B., Oktaba K., Köcher T., Cohen A., Stunnenberg H. G., Wilm M., Müller J. (2007). Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 26, 4078–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktaba K., Gutiérrez L., Gagneur J., Girardot C., Sengupta A. K., Furlong E. E. M., Müller J. (2008). Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell 15, 877–889 [DOI] [PubMed] [Google Scholar]
- Papp B., Müller J. (2006). Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 20, 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S., Melfi R., Pirrotta V. (2001). Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 15, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim I., Lee H.-H., Frasch M. (2003). The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development 130, 3187–3204 [DOI] [PubMed] [Google Scholar]
- Sarma K., Margueron R., Ivanov A., Pirrotta V., Reinberg D. (2008). Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol. Cell. Biol. 28, 2718–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann J. C., de Ayala Alonso A. G., Oktaba K., Ly-Hartig N., McGinty R. K., Fraterman S., Wilm M., Muir T. W., Müller J. (2010). Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465, 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoorlemmer J., Marcos-Gutiérrez C., Were F., Martínez R., García E., Satijn D. P., Otte A. P., Vidal M. (1997). Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 16, 5930–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Cavalli G. (2009). Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136, 3531–3542 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Kahn T. G., Nix D. A., Li X.-Y., Bourgon R., Biggin M., Pirrotta V. (2006). Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38, 700–705 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Kahn T. G., Stenberg P., Ohno K., Bourgon R., Pirrotta V. (2010). Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 6, e1000805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Raible F., Mollaaghababa R., Guyon J. R., Wu C. T., Bender W., Kingston R. E. (1999). Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98, 37–46 [DOI] [PubMed] [Google Scholar]
- Simon J. A., Kingston R. E. (2009). Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell. Biol. 10, 697–708 [DOI] [PubMed] [Google Scholar]
- Smouse D., Goodman C., Mahowald A., Perrimon N. (1988). polyhomeotic: a gene required for the embryonic development of axon pathways in the central nervous system of Drosophila. Genes Dev. 2, 830–842 [DOI] [PubMed] [Google Scholar]
- Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009). Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Peng C. Y., Nair S., Skeath J. B., Spana E. P., Doe C. Q. (1998). Biochemical analysis of ++Prospero protein during asymmetric cell division: cortical Prospero is highly phosphorylated relative to nuclear Prospero. Dev. Biol. 204, 478–487 [DOI] [PubMed] [Google Scholar]
- Strübbe G., Popp C., Schmidt A., Pauli A., Ringrose L., Beisel C., Paro R. (2011). Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc. Natl. Acad. Sci. USA 108, 5572–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. (1981). A gene product required for correct initiation of segmental determination in Drosophila. Nature 293, 36–41 [DOI] [PubMed] [Google Scholar]
- Tolhuis B., de Wit E., Muijrers I., Teunissen H., Talhout W., van Steensel B., van Lohuizen M. (2006). Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 38, 694–699 [DOI] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004). Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- Wang L., Brown J. L., Cao R., Zhang Y., Kassis J. A., Jones R. S. (2004). Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14, 637–646 [DOI] [PubMed] [Google Scholar]
- Wang R., Taylor A. B., Leal B. Z., Chadwell L. V., Ilangovan U., Robinson A. K., Schirf V., Hart P. J., Lafer E. M., Demeler B., et al. (2010). Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 18, 966–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe U., Dorfman R., Wernet M. F., Cohen S. M., Milán M. (2004). Proximodistal subdivision of Drosophila legs and wings: the elbow-no ocelli gene complex. Development 131, 767–774 [DOI] [PubMed] [Google Scholar]
- White R. A., Wilcox M. (1984). Protein products of the bithorax complex in Drosophila. Cell 39, 163–171 [DOI] [PubMed] [Google Scholar]
- Yu H., Mashtalir N., Daou S., Hammond-Martel I., Ross J., Sui G., Hart G. W., Rauscher F. J., Drobetsky E., Milot E., et al. (2010). The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol. Cell. Biol. 30, 5071–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.