Abstract
Multidrug resistance protein Pdr5p is a yeast ATP-binding cassette (ABC) transporter in the plasma membrane. It confers multidrug resistance by active efflux of intracellular drugs. However, the highly polymorphic Pdr5p from clinical strain YJM789 loses its ability to expel azole and cyclohexmide. To investigate the role of amino acid changes in this functional change, PDR5 chimeras were constructed by segmental replacement of homologous BY4741 PDR5 fragments. Functions of PDR5 chimeras were evaluated by fluconazole and cycloheximide resistance assays. Their expression, ATPase activity, and efflux efficiency for other substrates were also analyzed. Using multiple lines of evidence, we show that an alanine-to-methionine mutation at position 1352 located in the predicted short intracellular loop 4 significantly contributes to the observed transport deficiency. The degree of impairment is likely correlated to the size of the mutant residue.
Introduction
ATP-binding cassette (ABC) transporters are integral membrane proteins that carry diverse substrates across plasma membrane [1], [2]. Active extrusion of xenobiotics mediated by ABC transporters is one of the most common resistance mechanisms developed by microorganisms. [3], [4], [5]. As the most abundant ABC transporter in Saccharomyces cerevisiae, Pdr5p plays a major role in cellular detoxification via efflux of a wide variety of drugs and substrates in an ATP-dependent manner [6], [7], [8], [9]. Predicted topology of Pdr5p shows that it carries two highly hydrophobic transmembrane domains (TMDs) and two nucleotide binding domains (NBDs) [10], [11]. In a functional Pdr5p transporter, several amino acid residues of the TMDs determine substrate specificity [12], whereas some other amino acid residues of the NBDs are responsible for nucleotide binding and/or hydrolysis, then couple the liberated energy to conformational changes at the TMDs, which allow the translocation of drug substrates across plasma membrane barrier [13], [14]. Residues in NBDs may also selectively affect substrate selection [15]. The mechanism for how Pdr5p recognizes and transports a large number of structurally distinct substrates still remains unclear [16].
Previous reports have described many site-directed mutations in NBDs or TMDs of Pdr5p [11], [12], [13], [15], [17], [18], [19], leading to elucidation of the roles of single amino acid in the overall function of the protein. These studies emphasized on alterations affecting ATP hydrolysis and/or binding, as well as transport of substrates through the membrane. Pathogenic strain YJM789 was derived from a clinical S. cerevisiae isolate (YJM128) collected from the lung of an AIDS patient [20]. PDR5 in YJM789 strain contains 274 nucleotide differences (5.3% divergence) resulting in 80 amino acid changes from PDR5 in the lab strain BY4741 (hereafter referred to as BPDR5), whereas at the whole genome level the nucleotide difference between these two strains is only 0.43% [21]. Interestingly, most of the amino acid differences occur in two TMDs [22]. Our earlier work presented evidence that the drug-efflux function of Pdr5p in YJM789 strain is impaired by its dramatic sequence divergence although YJM789 Pdr5p localized to plasma membrane similarly to BY4741 Pdr5p. Further study illustrated that the sequence divergence in the TMD2 segment leads to fluconazole and cycloheximide hypersensitivity of YJM789, which contains a part of the predicted C-terminal NBD and the whole C- terminal TMD coding regions of PDR5 in YJM789 strain (hereafter referred to as YPDR5) [22].
In this study, the mutated sites in YPDR5 TMD2 segment that impair Pdr5p drug-efflux function were traced by the segment replacement method. The results show an A1352M mutation in the predicted intracellular loop 4 (ICL4) [11], [23], [24], [25] can cause severe decrease of drug export ability.
Results
Mutation sites in block S2A and S2B are critical for Pdr5p-mediated drug resistance
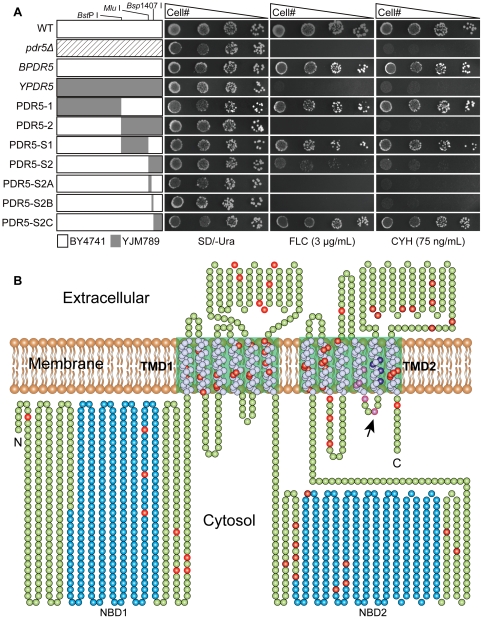
Previous studies found that Pdr5p in YJM789 strain cannot export its drug substrates such as cyclohexmide and fluconazole efficiently [22], [26]. In this study, YPDR5 and BPDR5 were overexpressed from high-copy plasmids. As shown in Fig. 1A, cells expressing BPDR5 were able to grow as well as wild type BY4741 on media containing 3 µg/mL fluconazole or 75 ng/mL cyclohexmide, while cells expressing YPDR5 were phenotypically similar to the pdr5 null mutant G1. It shows that the dramatic sequence divergence in YPDR5 disrupts its drug-efflux function under overexpression background. By exchanging of NBD-TMD coding regions in BPDR5 with their counterparts in YPDR5, PDR5 chimeras PDR5-1 and PDR5-2 were constructed. Drug susceptibility analysis revealed that cells expressing PDR5-1 had similar drug resistance patterns as cells expressing BPDR5, while cells expressing PDR5-2 was as same as pdr5 null mutant. This result suggests that sequence divergence in the C-terminal NBD-TMD region of YPDR5 can abrogate the Pdr5p-mediated drug resistance.
Figure 1. Identification of residues important for Pdr5p function and distribution of mutations in YJM789 Pdr5p.
(A) Segmental replacement of BY4741 PDR5 and drug resistance profiles of pdr5Δ S. cerevisiae cells expressing wild type and chimeric PDR5s by spot assay. Various regions of BY4741 PDR5 are segmentally replaced by corresponding segments from YPDR5. Segments from BY4741 and YJM789 PDR5 are shown as white and grey rectangles, respectively. The unique Bsp I, Mlu I, Bsp1407 I restriction sites, which were used to construct PDR5 chimeras are marked. Five-fold serial dilutions of pdr5Δ mutant carrying different PDR5 constructs were spotted on SD/-Ura media in the absence and the presence of 3 µg/mL fluconazole or 75 ng/mL cycloheximide. Plates were incubated for 72 hours at 30°C. (B) The topological model is based on the molecular modeling study of Pdr5p [11]. Two-dimensional schematic cartoon of Pdr5p shows the 12 predicted membrane spanning helices (white spheres) and two NBDs (blue spheres). The green boxed areas indicate the TMDs. Divergent residues between BY4741 and YJM789 are shown as red, purple and dark blue spheres, respectively. Ala 1352 is indicated by black arrow.
To identify which amino acid residues are important for the efflux function of Pdr5p, additional PDR5 chimeras PDR5-S1 and PDR5-S2 were created by dividing the C-terminal half divergent sequence of PDR5-2 into two segments S1 and S2. After transforming PDR5-S1 or PDR5-S2 overexpression plasmid into the fluconazole-sensitive pdr5 null mutant G1, their drug sensitivity to fluconazole was tested. As shown in Fig. 1A, cells expressing PDR5-S1 presented the similar drug resistant phenotypes as cells expressing BPDR5, whereas cells expressing PDR5-S2 were highly susceptible to fluconazole. Hence, sequence divergence in the segment S2 from YPDR5 can cause reduction in Pdr5p-mediated fluconazole resistance.
In order to narrow down the critical regions for Pdr5p function, segment S2 from YPDR5 was subdivided into 3 blocks (S2A, S2B and S2C) to replace their homologous regions in BPDR5 and the resulted PDR5 chimeras were designated as PDR5-S2A, PDR5-S2B and PDR5-S2C, respectively. Comparison of drug resistance phenotypes shows that the replacement of block S2C did not affect the Pdr5p-mediated fluconazole efflux, while PDR5-S2A and PDR5-S2B resulted in hypersusceptibility to fluconazole and cyclohexmide as compared to BPRD5 (Fig. 1A). Sequence analysis shows that PDR5-S2A and PDR5-S2B possess 5 (Fig. 1B, purple spheres) and 4 amino acid mutations (Fig. 1B, dark blue spheres), respectively. These changed residues could disturb Pdr5p–mediated drug efflux.
A1352M mutant strain is sensitive to fluconazole and cyclohexmide
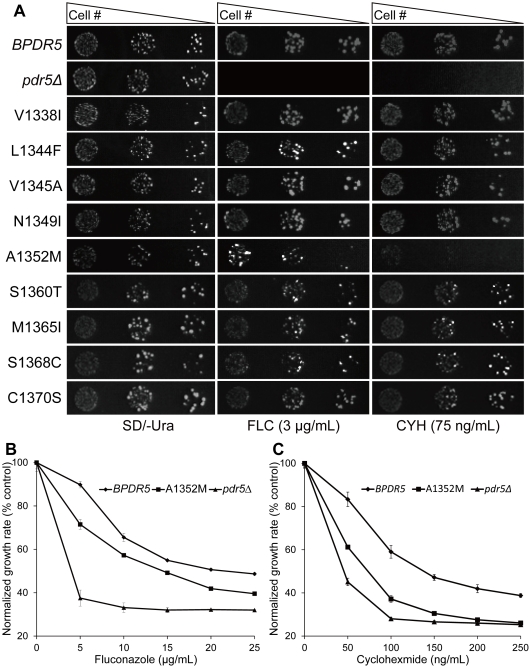
In order to investigate which mutations in the S2A and S2B regions can alter the capacity of Pdr5p to confer drug resistance, as well as whether there are any additive or synergistic effects in mutation combinations, nine mutants expressing variants of PDR5 including V1338I, L1344F, V1345A, N1349I, A1352M, S1360T, M1365I, S1368C and C1370S were constructed (See Table 1). The drug resistance patterns of all above Pdr5p mutants were indistinguishable from WT except A1352M mutant (Fig. 2A), which was hypersensitive to fluconazole and cyclohexmide. A similar phenotype was observed when relative fluconazole (Fig. 2B) and cyclohexmide (Fig. 2 C) resistance of A1352M mutant, BPDR5 and pdr5Δ mutant was determined in liquid media. Taken together, these results demonstrate that one single amino acid substitution of alanine 1352 with methionine is sufficient to impair Pdr5p function. In addition, the fact that all four single mutations separated from PDR5-S2B (S1360T, M1365I, S1368C and C1370S) could functionally complement the pdr5Δ mutant suggests the existence of synergistic effects among these mutations (Fig. 2A).
Table 1. Strains and plasmids.
| Strains or plasmids | Relevant characteristics | Reference |
| Strains | ||
| S. cerevisiae BY4741 | MATa his3 leu2 met15 ura3 | Yeast Knock-out (YKO) deletion collection |
| S. cerevisiae YJM789 | MATα hoΔ::hisG gal2 lys2 | [35] |
| S. cerevisiae G1 | MATa his3 leu2 met15 ura3 pdr5::loxP | This study |
| E. coli DH5a | SupE44, ΔlacU169 (φ80 lacZΔM15), hsdR17, recA1, endA1, gyrA96, thi-1, relA1, Nalr | |
| Plasmids | ||
| pUG6 | loxP-kanMX6-loxP | EUROSCARF |
| pSH47 | URA3, Cre | EUROSCARF |
| YEplac195 | URA3 | |
| pTA2 | TOYOBO | |
| YEplac195-BPDR5 | URA3, expressing BY4741 PDR5 | This study |
| YEplac195-YPDR5 | URA3, expressing YJM789 PDR5 | This study |
| PDR5-1 | This study | |
| PDR5-2 | This study | |
| PDR5-S1 | This study | |
| PDR5-S2 | This study | |
| PDR5-S2A | This study | |
| PDR5-S2B | This study | |
| PDR5-S2C | This study | |
| YEplac195-PDR5-V1338I | This study | |
| YEplac195-PDR5-L1344F | This study | |
| YEplac195-PDR5-V1345A | This study | |
| YEplac195-PDR5-N1349I | This study | |
| YEplac195-PDR5-A1352M | This study | |
| YEplac195-PDR5-S1360T | This study | |
| YEplac195-PDR5-M1365I | This study | |
| YEplac195-PDR5-S1368C | This study | |
| YEplac195-PDR5-C1370S | This study | |
| YEplac195-PDR5-A1352G | This study | |
| YEplac195-PDR5-A1352V | This study | |
| YEplac195-PDR5-A1352P | This study | |
| YEplac195-PDR5-A1352S | This study | |
| YEplac195-PDR5-A1352L | This study | |
Figure 2. Drug resistance profiles of cells expressing PDR5 variants.
(A) Nine mutated sites distributed throughout blocks S2A and S2B were separated by site-directed mutagenesis and fluconazole sensitivities of mutant variants were determined by spot assay. Cells were grown overnight and reinoculated to OD600 = 0.1, then 4 µL of five-fold serial dilutions were spotted onto drug free or drug containing (3 µg/mL fluconazole or 75 ng/mL cycloheximide) SD/-Ura plates. The plates were incubated at 30°C for 72 hours. (B) Resistance of BPDR5 (⧫), pdr5Δ (▴) and A1352 (▪) mutants to increasing concentrations of fluconazole. Cells from overnight cultures were inoculated to OD600 = 0.1. Growth was estimated on the basis of OD600 and plotted as percentage growth, where the growth in drug free media was taken to be 100%. The mean values of three independent experiments are plotted and the error bars represent the S.D. (C) Growth of BPDR5 (⧫), pdr5Δ (▴) and A1352 (▪) mutants was compared as described in B, except that the experiment was performed in the presence of increasing concentrations of cycloheximide.
A1352M mutant strain is transport-deficient
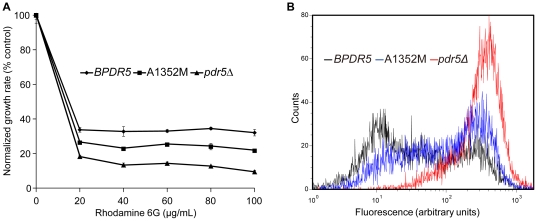
Furthermore, we analyzed the capability of A1352M mutant to transport rhodamine 6G (R6G) in vivo. Null mutants of pdr5 were known to cause increased accumulation of R6G. As shown in Fig. 3A, A1352M mutant was more sensitive to R6G that BPDR5, the flow cytometry results (Fig. 3B) show that intracellular accumulation of R6G was much higher in A1352M mutant than that in cells expressing BPDR5. It demonstrates that transport capabilities of A1352M mutant are compromised. As a result, the observation that A1352M mutant conferred lower fluconazole and cycloheximide resistance than BPDR5 resulted from transport deficiency of Pdr5p.
Figure 3. Pdr5p-mediated efflux of rhodamine 6G in BPDR5 and mutant cells.
(A) Rhodamine 6 G resistance of BPDR5 (□), pdr5Δ (▴) and A1352 (▪) mutants were compared in liquid culture as described in materials and methods. Growth in the absence of R6G was served as control. Growth at each concentration of R6G is shown as (% control). The mean values of three independent experiments are plotted and the error bars represent the S.D. (B) The transport capabilities of BPDR5, pdr5Δ and A1352M were compared with flow cytometry. The histogram derived from the CellQuest software depicts fluorescence intensities for BPDR5 (black), pdr5Δ (red) and A1352M (blue).
Mutational analysis of A1352 in Pdr5p
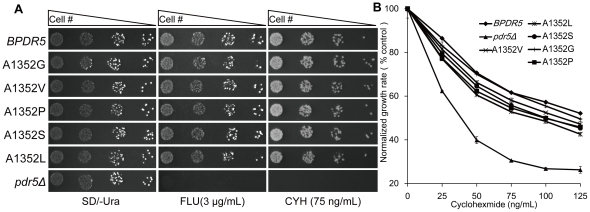
We further substituted the alanine residue in position 1352 with glycine (A1352G), serine (A1352S), proline (A1352P), valine (A1352V), and leucine (A1352L). The consequence of single residue changes in position 1352 to modulate Pdr5p mediated cycloheximide resistance was examined by spot assay after transformation of G1 with the corresponding overexpression plasmids. In order to compare the growth patterns of the mutants in different concentrations of cycloheximide, drug sensitivity analysis based on quantitative measurements of optical densities of liquid cell cultures was performed simultaneously as described in the method. As shown in Fig. 4, the capacities of A1352 mutants in conferring cycloheximide resistance were all slightly lower than that of BPDR5. The discrepancies of drug resistance for distinct mutations at position 1352 demonstrate that the size of residue at this position can affect the Pdr5p-mediated drug export.
Figure 4. Drug resistance profiles of pdr5Δ S. cerevisiae cells expressing wild type and A1352 mutant variants of PDR5 on agar plates and in liquid culture.
(A) Drug sensitivity of A1352 mutant variants determined by spot assay. Serial dilutions of BPDR5 cells, pdr5Δ and A1352 mutant variants were spotted on SD/-Ura agar plates in the absence (left panel) and the presence (right panel) of 75 ng/mL cycloheximide, respectively. The plates were incubated at 30°C for 72 hours. (B) Quantitative analysis of drug resistance mediated by Pdr5p mutants. Cells carrying empty plasmid or overexpressing BPDR5, A1352 mutant variants as indicated, were subjected to analysis in liquid culture as described in materials and methods. OD600 values at 12 h were analyzed. Growth in drug free cultures of each strain was served as control. Normalized growth rates are shown as the means ± standard deviations (error bars) for three independent experiments at each concentration of cycloheximide.
Mutations in A1352 elicit alteration in Pdr5p function
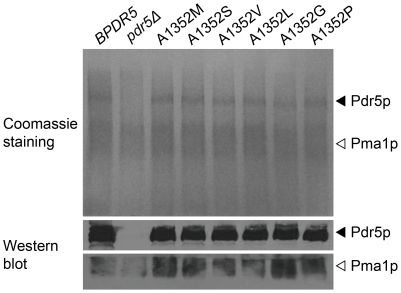
To ascertain that the observed differences in cycloheximide resistance are due to alterations in Pdr5p function rather than poor Pdr5p expression level or mislocalization of Pdr5p variants, plasma membranes from A1352 mutants were prepared and analyzed by SDS-PAGE. The presence of a specific band corresponded to Pdr5p in membranes from mutant variants of PDR5 but was absent in membranes from pdr5Δ indicates that BY4741 Pdr5p and its A1352 mutant proteins are properly expressed and localized to plasma membrane with comparable levels (Fig. 5). This was further confirmed by immunoblotting with antibodies against Pdr5p (Fig. 5).
Figure 5. Membrane localization of Pdr5p and its mutant variants.
Isolated plasma membranes (15 µg/lane) were electrophoresed on an SDS–8% polyacrylamide gel and visualized with Coomassie blue (top panel). Pdr5p (middle panel) and Pma1p (bottom panel) were immunodetected with specific antibodies as described in the materials and methods section. Filled arrowheads indicate Pdr5p and open arrowheads indicate Pma1p. Coomassie blue stained SDS-PAGE gel (upper panel) and western blot (lower panel) of the same strains are shown.
Pdr5p-specific ATPase activity is unaffected by mutations in A1352
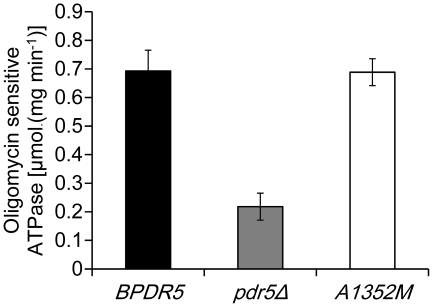
Energy released from ATP hydrolysis drives the substrate transport by Pdr5p. In order to examine whether the observed changes in cycloheximide resistance was accompanied by alterations in Pdr5p-specific ATPase activities, plasma membranes isolated from mutants carrying Pdr5p variants were used to determine their ATPase activities. It is apparent from Fig. 6 that, compared to plasma membranes from the cells expressing BPDR5, there was no significant difference in plasma membrane ATPase activity of cells expressing mutant PDR5, the plasma membranes from pdr5 null mutant exhibit considerably reduced ATPase activity. Therefore, mutations at A1352 do not seem to interfere with Pdr5p-specific ATPase activity.
Figure 6. The effect of A1352M mutation on the ATPase activity of Pdr5p.
Plasma membrane preparations (0.25 µg/well) from BPDR5 (dark), pdr5Δ (gray) and A1352M (white) were incubated in 25 µL reaction buffer containing 300 mM Mes-Tris (pH 7.4), 4 mM ATP, 5 mM MgCl2, 0.2 mMammonium molybdate, 50 mM KNO3, and 10 mM NaN3 at 30°C for 20 min. The oligomycin-sensitive activity was determined as the difference in the ATPase activity in the presence or absence of 20 µg/mL oligomycin. The figure shows the means of three independent experiments, and the error bars represent standard error.
Discussion
S. cerevisiae Pdr5p belongs to the ABC super-family of primary transporters which directly couples ATP hydrolysis to substrate transport. It not only expels azoles and its derivatives but also extrudes a variety of structurally unrelated compounds [1], [26]. To understand the structure-function relationships of Pdr5p, we took advantage of the highly polymorphic Pdr5p from clinical strain YJM789 that have lost its ability to export azoles. By exchanging segments of BY4741 PDR5 with their counterparts from YJM789 PDR5, we found that A1352M substitution could cause considerable reduction in Pdr5p mediated drug resistance.
As shown in Fig. 1B, Pdr5p shares a common architecture with other ABC transporters, which contains two TMDs and each of the two is comprised of six membrane-spanning helices connected by three extracellular loops (ECLs) and two intracellular loops (ICLs). Recent molecular modeling study on Pdr5p [11] shows that A1352 (The purple sphere indicated by arrow in Fig. 1B) is located in the ICL4 which connects TMH 10 and TMH 11 in TMD2. It is widely accepted that interactions between the membrane-bound domains and cytosolic domains of ABC transporters are critical for their transport capabilities. The high-resolution structure of Staphylococcus aureus Sav1866 suggests that the NBDs interact with the TMDs via ICLs [27]. Analysis of P-glycoprotein by chemical cross-linking suggests that ICL4 within TMD2 forms an interface with NBD1 [28]. Studies of CFTR (Cystic Fibrosis Transmembrane Regulator) mutants demonstrate a tertiary interaction between the surface of NBD1 and ICL4 in TMD2 [29]. ICL4-NBD1 interaction in Yor1p (another ABC transporter in S. cerevisea) has been defined by chemical cross-linking experiments. [30]. All above studies supported an interface with a trans arrangement in which ICL4 within TMD2 interacts with the NBD of the opposite half of the molecule.
Despite their distinct molecular architecture, it could be inferred from the predicted topological models that ICL4 in Pdr5p corresponds to ICL4 in above mentioned ABC transporters. It is possible that the ICL4 of Pdr5p is involved in interdomain cross-talk. Reduction of drug resistance in A1352M mutant may attributable to the alterations of ICL4 that uncouple interdomain cross-talk. However, substitution of A1352 with more rigid amino acid residue proline does not show significant reduction in the drug resistance and the mutant possess the same level of ATPase activity as BPDR5 (data not shown). Genetic and biochemical studies from Pdr5p strongly suggest that the Pdr5p signaling interface is in the cis rather than the trans configuration, interactions between the TMH2 and NBD1 occur via the ICL1 [13], [14].Therefore, it is unlikely that the reduced resistance to drug substrates in the A1352M mutant was a consequence of uncoupling of interdomain cross-talk.
The reported crystal structures of Sav1866 [27] and mouse P-glycoprotein [31] suggest that the transmembrane helices in TMDs form a substrate-binding pocket. Modeling of Pdr5p shows that Ala1352 is pointing toward the predicted substrate-binding pocket [11]. It is more likely that the transport-deficiency of A1352M mutant is caused by the increased length of residue which results in affecting accessibility of binding pocket to intracellular substrates. We also increased the residue size at position 1352 by substituting the Ala with Gly (A1352G), Ser (A1352S), Val (A1352V) and Leu (A1352L). Western blot result showed that the expression levels of all above A1352 mutants were similar to that of BPDR5. The drug resistance assays also support that substitutions of Ala residue with larger residues tend to have more severe impact on Pdr5p, resulting in increased sensitivity to cycloheximide.
Previous work shows that A1352 and the negatively charged E1353 in ICL4 are very conserved by aligning PDR5 with other fungal ABC transporters, and E1353 is predicted to located at the edge of substrate-binding pocket [11].When positively charged Lys (K) and uncharged Gln (Q) were used to substitute the similar-size E1353, only E1353K mutant shows significant decrease in drug efflux. (data not shown). This implies that the E1353 may play a role in neutralizing positively charged substrates such as rhodamine 6G. It requires further investigation to understand whether the importance comes from its role in substrate recognition.
Interestingly, the A1352 was identified to be under positive selection by branch-site model (P ω>1, posterior probability of NEB was 0.98, and BEB was 0.93). Although this significance is not extremely high, the fast evolution of A1352 indicates that this amino acid is responsible for the changing of Pdr5p function in drug transport.
In summary, mutation of A1352 in ICL4 could disturb Pdr5p mediated drug resistance pattern without affecting its ATPase activity, expression level or protein localization. It hints that the ICL4 of Pdr5p may play an important role in substrates recognition and export. While, there is no report about ICL4 function in Pdr5p, our observation of the functional importance of A1352 might encourage further studies on this region.
Materials and Methods
Strains, media and growth conditions
S. cerevisiae BY4741 (MAT a, his3Δ, leu2Δ, met15Δ, ura3Δ) and YJM789 (MAT α, hoΔ::hisG, gal2Δ, lys2Δ) were used for PDR5 cloning. pdr5 null mutant G1 (MAT a, his3Δ, leu2Δ, met15Δ, ura3Δ, pdr5Δ::loxP) was constructed as previously described [32]. Escherichia coli strain DH5α was used for plasmid manipulation.
E. coli cells were grown in LB medium. Yeast strains were grown on standard rich YPD medium. SD uracil-deficient medium was used to select ura3 auxotrophic derivatives of transformants. Solid media were supplemented with 2% agar.
Chemicals
Stock solutions of fluconazole (FLC) and Rhodamine 6G (R6G) were prepared in 20% dimethyl sulfoxide at final concentrations of 5 mg/mL and 10 mM. Cycloheximide (CYH) solution (1 mg/mL) was purchased from Sigma-Aldrich.
Plasmid construction and transformation
Construction of plasmids YEplac195-BPDR5, YEplac195-YPDR5, YEplac195-PDR5-1 and YEplac195-PDR5-2 has been described previously [22]. An 1851 bp segment was amplified by PCR from YEplac195-YPDR5 with forward primer #01 and reverse primer #02. The PCR product was digested with BstP I and Mlu I and ligated to YEplac195-BPDR5 cut accordingly producing PDR5-S1. PDR5-S2 was similarly generated using forward primer #03, reverse primer #04 and Mlu I – Sph I as restriction enzymes. The PCR product of primer pair #03 and #04 was also ligated into the pTA2 vector to obtain pTA-S2 for further modification.
For substituting blocks S2C in YEplac195-BPDR5 by its counterpart from YEplac195-YPDR5, YEplac195-BPDR5 was cut with restriction enzymes Pst I - Bsp1407 I, the resulted restriction fragment of 209 bp was recovered and used to replace its counterpart sequence in pTA-S2, yielding intermediate plasmids pTA-S2C; pTA-S2C was then digested with single restriction site enzymes Mlu I and Sph I and ligated to YEplac195-BPDR5 cut accordingly, creating PDR5-S2C.
PDR5-S2A and PDR5-S2B were constructed as follows. Inverse PCR amplification was performed using PAGE-grade PCR primers #05 and #06 which containing the desired non-synonymous base pair substitutions indicated by uppercase letters and pTA-S2C plasmid DNA isolated from E. coli DH5α as template. After Dpn I digestion of the methylated template plasmid, self-ligation of PCR product was conducted in a reaction mixture which includes both T4 Polynucleotide Kinase (TOYOBO) and T4 DNA ligase (Fermentas). The resulted plasmid was labeled pTA-S2A. The same procedure was applied to construct pTA-S2B using primer pair #07 and #08. Substitutions of nucleotides introduced by primers were confirmed by DNA sequencing. Mlu I – Sph I fragments from pTA-S2A and pTA-S2B were used to replace the homologous fragments in YEplac195-BPDR5 generating PDR5-S2B and PDR5-S2A, respectively. The sequences of primers used for plasmid construction are listed in Table 2.
Table 2. Primers used in construction of PDR5 chimeras.
| Primer | Sequence(5′-3′) |
| #01 | CATTAATCCATTGGCTTACTT |
| #02 | aactacgcgtTAGAGTAAAACCCG |
| #03 | atatacgcgtCTGCAGCTGGACAG |
| #04 | acatgcatgcACCGATGAGATAACCTAGAAAT |
| #05 | tcttttaACcaagttGCAgaaagtgctgcaaacttag |
| #06 | aatGAcaaGtagacccattgaaccaaCgtaaacgtag |
| #07 | tctttatCtttctGtggtgttatgaccacacccag |
| #08 | CatggtgaacaataaagAtgctaagtttgcagcac |
Bold: modified nucleotides.
Underlined: modified or newly generated endonuclease restriction sites.
Yeast transformations were performed by LiAc/SS carrier DNA/PEG high efficiency transformation protocol [33].
Site-directed mutagenesis of PDR5
The Mlu I – Sph I DNA fragment from YEplac195-BPDR5 was used to replace the homologous fragment in pTA-S2 creating pTA-mu. pTA-mu was then served as the template for site-directed mutagenesis to create PDR5 mutants. Site directed mutagenesis was performed using the Quick Change kit (Stratagene). Mutations were introduced into YEplac195-BPDR5 from pTA-mu derivatives bearing desired nucleotide substitutions by exchanging the Mlu I – Sph I fragments in plasmids.
All PCR amplifications were performed with the KOD-Plus system (TOYOBO). After every cloning, the entire chimeric construct was sequenced by using the Big Dye Terminator Cycle sequencing kit and an ABI 3730xl DNA analyzer to ensure that the construct remained in-frame and no unwanted mutations had been introduced.
Drug sensitivity analysis
Each transformed yeast strain to be tested was grown in SD/-Ura medium to late exponential phase. Then cells were washed and subsequently resuspended in sterile water to an OD600 of 0.1. For spot assay, aliquots (4 µL) of 5-fold serial dilutions were spotted onto SD/-Ura plates lacking or containing drugs to be tested. Plates were incubated at 30°C for 72 hours. For quantitative measurements of optical densities of liquid cell cultures, cultures were treated with different concentrations of cycloheximide. 150 µL normalized cultures were seeded onto NUNC 96 well optical bottom plates (in triplicate) and covered with adhesive plate sealers in Power Wave XS microplate spectrophotometer (BioTek) with constant shaking at 30°C. OD600s for each well were recorded using Gen5 software every 15 minutes for 24 hours.
Preparation of purified membrane vesicles and immunoblots
Plasma membranes were prepared as described previously [15]. The protein concentration was determined by Bradford assay. Equivalent amounts of protein were separated by SDS-PAGE with 8% gel. Immunoblotting was done using standard methods. Pdr5p specific goat polyclonal antibody yN-18 and Pma1 specific antibody yN-20 were from Santa Cruz Biotechnology. HRP labeled donkey anti-goat polyclonal antibody was from Beyotime Co. Proteins were visualized using the ECL chemiluminescence detection system (BeyoECL Plus).
ATPase activity assays
Pdr5p-associated OM-sensitive ATPase activity of the isolated plasma membrane fractions was measured as described previously [15]. Absorbance data were collected using a Power Wave XS microplate spectrophotometer equipped with Gen5 software (BioTek).
Rhodamine 6G transport assay
Transport of rhodamine 6G was determined as described previously [13]. The data were analyzed with a CellQuest program. Fluorescence was expressed in arbitrary units.
Detecting positive selection
10 PDR5 gene sequences of different S. cerevisiae strains were selected from website (http://www.sanger.ac.uk/research/projects/genomeinformatics/sgrp.html). Pdr5p sequences were aligned using ClustalW with the default parameter settings, and were then reversely translated into nucleotide sequences using MEGA4 [34]. The evolutionary rate of amino acid sites was detected by using the branch-site model of positive selection, as implemented in codeml program in PAML package [27].
Acknowledgments
We thank Dr. Di Xia and Dr. Robert M Rutledge for sharing the atomic model of Pdr5p, Dr. Lutz Schmitt for technical support with ATPase Activity Assays. We also thank Lin Xu, Brandon Barker and Ryan Alexander Coots for reading and helping us to improve the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by National Natural Science Foundation of China (No.31170032) and Zhejiang Provincial Natural Science Foundation of China (No.Y2100685). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 2.Higgins CF. ABC transporters: physiology, structure and mechanism–an overview. Res Microbiol. 2001;152:205–210. doi: 10.1016/s0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- 3.Glavinas H, Krajcsi P, Cserepes J, Sarkadi B. The role of ABC transporters in drug resistance, metabolism and toxicity. Curr Drug Deliv. 2004;1:27–42. doi: 10.2174/1567201043480036. [DOI] [PubMed] [Google Scholar]
- 4.Ejendal KF, Hrycyna CA. Multidrug resistance and cancer: the role of the human ABC transporter ABCG2. Curr Protein Pept Sci. 2002;3:503–511. doi: 10.2174/1389203023380521. [DOI] [PubMed] [Google Scholar]
- 5.Leonessa F, Clarke R. ATP binding cassette transporters and drug resistance in breast cancer. Endocr Relat Cancer. 2003;10:43–73. doi: 10.1677/erc.0.0100043. [DOI] [PubMed] [Google Scholar]
- 6.Kolaczkowski M, van der Rest M, Cybularz-Kolaczkowska A, Soumillion JP, Konings WN, et al. Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J Biol Chem. 1996;271:31543–31548. doi: 10.1074/jbc.271.49.31543. [DOI] [PubMed] [Google Scholar]
- 7.Rogers B, Decottignies A, Kolaczkowski M, Carvajal E, Balzi E, et al. The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J Mol Microbiol Biotechnol. 2001;3:207–214. [PubMed] [Google Scholar]
- 8.Ernst R, Klemm R, Schmitt L, Kuchler K. Yeast ATP-binding cassette transporters: cellular cleaning pumps. Methods Enzymol. 2005;400:460–484. doi: 10.1016/S0076-6879(05)00026-1. [DOI] [PubMed] [Google Scholar]
- 9.Bauer BE, Wolfger H, Kuchler K. Inventory and function of yeast ABC proteins: about sex, stress, pleiotropic drug and heavy metal resistance. Biochim Biophys Acta. 1999;1461:217–236. doi: 10.1016/s0005-2736(99)00160-1. [DOI] [PubMed] [Google Scholar]
- 10.Dassa E, Bouige P. The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol. 2001;152:211–229. doi: 10.1016/s0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 11.Rutledge RM, Esser L, Ma J, Xia D. Toward understanding the mechanism of action of the yeast multidrug resistance transporter Pdr5p: a molecular modeling study. Journal of structural biology. 2011;173:333–344. doi: 10.1016/j.jsb.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tutulan-Cunita AC, Mikoshi M, Mizunuma M, Hirata D, Miyakawa T. Mutational analysis of the yeast multidrug resistance ABC transporter Pdr5p with altered drug specificity. Genes to Cells. 2005;10:409–420. doi: 10.1111/j.1365-2443.2005.00847.x. [DOI] [PubMed] [Google Scholar]
- 13.Sauna ZE, Bohn SS, Rutledge R, Dougherty MP, Cronin S, et al. Mutations Define Cross-talk between the N-terminal Nucleotide-binding Domain and Transmembrane Helix-2 of the Yeast Multidrug Transporter Pdr5 Possible Conservation of A Signaling Interface for Coupling ATP Hydrolysis to Drug Transport. Journal of Biological Chemistry. 2008;283:35010–35022. doi: 10.1074/jbc.M806446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ananthaswamy N, Rutledge R, Sauna ZE, Ambudkar SV, Dine E, et al. The Signaling Interface of the Yeast Multidrug Transporter Pdr5 Adopts a Cis Conformation, and There Are Functional Overlap and Equivalence of the Deviant and Canonical Q-Loop Residues. Biochemistry. 2010;49:4440–4449. doi: 10.1021/bi100394j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst R, Kueppers P, Klein CM, Schwarzmueller T, Kuchler K, et al. A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5069–5074. doi: 10.1073/pnas.0800191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golin J, Ambudkar SV, May L. The yeast Pdr5p multidrug transporter: how does it recognize so many substrates? Biochem Biophys Res Commun. 2007;356:1–5. doi: 10.1016/j.bbrc.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Egner R, Rosenthal FE, Kralli A, Sanglard D, Kuchler K. Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Molecular Biology of the Cell. 1998;9:523–543. doi: 10.1091/mbc.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Thozee CP, Cronin S, Goj A, Golin J, Ghislain M. Subcellular trafficking of the yeast plasma membrane ABC transporter, Pdr5, is impaired by a mutation in the N-terminal nucleotide-binding fold. Mol Microbiol. 2007;63:811–825. doi: 10.1111/j.1365-2958.2006.05562.x. [DOI] [PubMed] [Google Scholar]
- 19.Egner R, Bauer BE, Kuchler K. The transmembrane domain 10 of the yeast Pdr5p ABC antifungal efflux pump determines both substrate specificity and inhibitor susceptibility. Mol Microbiol. 2000;35:1255–1263. doi: 10.1046/j.1365-2958.2000.01798.x. [DOI] [PubMed] [Google Scholar]
- 20.McCusker JH, Clemons KV, Stevens DA, Davis RW. Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics. 1994;136:1261–1269. doi: 10.1093/genetics/136.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, et al. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12825–12830. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan WJ, Jiang HF, Guo XX, Mancera E, Xu L, et al. Antagonistic Changes in Sensitivity to Antifungal Drugs by Mutations of an Important ABC Transporter Gene in a Fungal Pathogen. PLoS ONE. 2010;5:e11309. doi: 10.1371/journal.pone.0011309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 25.Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 26.Winzeler EA, Richards DR, Conway AR, Goldstein AL, Kalman S, et al. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- 27.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 28.Zolnerciks JK, Wooding C, Linton KJ. Evidence for a Sav1866-like architecture for the human multidrug transporter P-glycoprotein. The FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21:3937–3948. doi: 10.1096/fj.07-8610com. [DOI] [PubMed] [Google Scholar]
- 29.Serohijos AWR, Hegedűs T, Aleksandrov AA, He L, Cui L, et al. Phenylalanine-508 mediates a cytoplasmic–membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proceedings of the National Academy of Sciences. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pagant S, Brovman EY, Halliday JJ, Miller EA. Mapping of interdomain interfaces required for the functional architecture of Yor1p, a eukaryotic ATP-binding cassette (ABC) transporter. The Journal of biological chemistry. 2008;283:26444–26451. doi: 10.1074/jbc.M803912200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, et al. Structure of P-Glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature protocols. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 34.He L, Aleksandrov AA, Serohijos AW, Hegedus T, Aleksandrov LA, et al. Multiple membrane-cytoplasmic domain contacts in the cystic fibrosis transmembrane conductance regulator (CFTR) mediate regulation of channel gating. The Journal of biological chemistry. 2008;283:26383–26390. doi: 10.1074/jbc.M803894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geladopoulos TP, Sotiroudis TG, Evangelopoulos AE. A malachite green colorimetric assay for protein phosphatase activity. Analytical biochemistry. 1991;192:112–116. doi: 10.1016/0003-2697(91)90194-x. [DOI] [PubMed] [Google Scholar]