Abstract
Regulation of protein function through oxidative modification has emerged as an important molecular mechanism modulating various biological processes. Here, we report a proteomic study of redox-sensitive proteins in Arabidopsis cells subjected to H2O2 treatment. Four gel-based approaches were employed, leading to the identification of four partially overlapping sets of proteins whose thiols underwent oxidative modification in the H2O2-treated cells. Using a method based on differential labeling of thiols followed by immunoprecipitation and Western blotting, five of the six selected putative redox-sensitive proteins were confirmed to undergo oxidative modification following the oxidant treatment in Arabidopsis leaves. Another method, which is based on differential labeling of thiols coupled with protein electrophoretic mobility shift assay, was adopted to reveal that one of the H2O2-sensitive proteins, a homologue of cytokine-induced apoptosis inhibitor 1 (AtCIAPIN1), also underwent oxidative modification in Arabidopsis leaves after treatments with salicylic acid or the peptide elicitor flg22, two inducers of defense signaling. The redox-sensitive proteins identified from the proteomic study are involved in various biological processes such as metabolism, the antioxidant system, protein biosynthesis and processing, and cytoskeleton organization. The identification of novel redox-sensitive proteins will be helpful toward understanding of cellular components or pathways previously unknown to be redox-regulated.
Keywords: redox proteomics, oxidative stress, hydrogen peroxide, AtCIAPIN1, salicylic acid, flg22, Arabidopsis
Short abstract
Through four redox proteomic methods, we identified a number of Arabidopsis proteins that underwent rapid oxidative modifications in Arabidopsis cells upon H2O2 treatment. We also established two methods for detailed analysis of individual putative redox-sensitive proteins. The identification of the oxidant-sensitive proteins would be greatly helpful toward in-depth characterization of other signaling pathways previously unknown to be redox-regulated.
Introduction
Rapid generation of reactive oxygen species (ROS) is one of the cellular physiological events that occur in plants during exposure to various environmental stresses.1−4 Excessive accumulation of ROS can cause damages to cellular molecules such as proteins, nucleic acids and lipids.4 To counteract oxidative stress, plants evolved complex antioxidant systems.5 On the other hand, ROS also functions as key signaling molecules in physiological responses and development.1,3,5
Mitochondria, chloroplasts, and microbodies are considered as major sources of ROS production, and cellular ROS levels are determined by their production and scavenging systems.3,5 During the plant hypersensitive response against avirulent pathogens, host cells actively produce high levels of ROS.1 This oxidative burst is contributed mainly by plasma membrane-bound NADPH oxidases.1,6 Pathogen infection also triggers production of nitric oxide (NO) which coordinates with ROS to regulate hypersensitive cell death and defense gene induction.2,7,8
Redox signals are largely sensed by redox-sensitive proteins which can undergo different forms of oxidative modifications.4,9,10 Some modifications are irreversible such as oxidation of cysteine thiols to sulfonic acids (P-SO3H) and nitration of tyrosine and tryptophan. Others are reversible such as oxidation of thiols to sulfenic acid (P-SOH), S-nitrosylation, and intra- or intermolecular disulfide bond formation. Irreversible protein oxidation often leads to permanent loss of function, whereas reversible oxidation can protect the protein from irreversible oxidation and/or act as a molecular switch for regulation of protein function.
Cysteine thiols in many proteins are highly redox-sensitive and their redox states often determine protein activities and functions. Several proteomic approaches have been developed for detection and identification of oxidant-sensitive thiol proteins.9−11 Generally, these methods are based on differential labeling of reduced versus oxidized thiols with specific thiol-reactive reagents. Following thiol labeling, proteins are separated by two-dimensional gel electrophoresis (2-DE), and differentially labeled proteins are identified by mass spectrometry. In addition to the gel-based methods, a modified isotope coded affinity tag technology (OxICAT) has been developed to identify redox-sensitive proteins in E. coli(12) and also in yeast.13 Fu et al. compared the merits and limitations of ICAT and DIGE in identifying redox-sensitive proteins in heart tissues and found that the two methods are complementary.14 Recently, a method termed isotopic tandem orthogonal proteolysis activity based protein profiling (isoTOP-ABPP) was developed and used to quantitatively profile the intrinsic reactivity of cysteine residues in human cells.15
Proteomic approaches have been used to detect and identify oxidant-sensitive proteins in plants (reviewed in refs (10,16)). These reports include detection and identification of glutathionylated Arabidopsis proteins under in vivo or in vitro experimental conditions.17,18 A method based on thiol affinity chromatography was used to identify 65 Arabidopsis proteins that contain disulfide bonds under normal conditions,19 although the study did not reveal whether these disulfides are redox-regulated under an environmental stress. Hancock et al. identified 5 Arabidopsis proteins that became oxidized following the treatment of Arabidopsis suspension cells with 10 mM H2O2.20 Alvarez et al. identified 22 proteins in Arabidopsis with altered redox states and/or abundance in response to methyl jasmonate treatment.21 Proteomic approaches have also been employed for identification of a large number of proteins from several plant species that are potential targets of thioredoxins (Trxs).22−31 In addition, using a biotin switch labeling method, a number of S-nitrosylated Arabidopsis proteins have been identified following treatments with the NO donor S-nitrosoglutathione or gaseous NO32 and during the hypersensitive response.33
Here, we report the identification of proteins whose thiols underwent oxidative modifications in Arabidopsis cells following a 10 min treatment with a sublethal dose of H2O2. A majority of them have not been previously reported to undergo oxidative modifications under redox-perturbing conditions. We also established two methods for confirmation and detailed analysis of in vivo redox states of individual proteins in Arabidopsis plants challenged with H2O2 or two chemical inducers of the plant defense response. These results indicate that the Arabidopsis cells undergo rapid cellular reprogramming through oxidative modification of proteins involved in multiple cellular processes.
Materials and Methods
Arabidopsis T87 Cell Culture and Treatments
Arabidopsis thaliana (Columbia ecotype) suspension cell line T87 was kindly provided by Dr. Kevin Lutke (Donald Danforth Plant Science Center, St. Louis, MO). The T87 cell culture was maintained in 50 mL liquid medium (1/2 MS salts, 3% sucrose, 0.2 g/L KH2PO4, 0.2 mg/L 2,4-D, and B5 vitamins) in 250 mL flasks by gentle agitation (50 rpm) in dark at 25 °C. Cells were subcultured weekly by transferring 3 mL culture to a new flask with 50 mL culture medium.
For the oxidant treatment of T87 cells, a 0.5 M H2O2 stock solution was added to the cell culture (3 days after subculturing) to a final concentration of 5 mM. For control samples, the same amount of H2O was added to the culture cells. The cells were harvested 10 min after H2O2 treatment by filtering through a glass vacuum filter (VWR, West Chester, PA) and immediately frozen in liquid nitrogen for further analysis.
Protein Extraction and Labeling
All steps of protein sample preparation were carried out under reduced light. All buffers were degassed before use.
For the direct 5-(iodoacetamido)fluorescein (IAF) labeling method and the blocking-IAF labeling method, 500 mg cells were ground in the homogenization buffer [20 mM Tris-HCl (pH 8.0), 5 mM EDTA, 0.5% Triton X-100, 100 mM NaCl and 1% protease inhibitor cocktail (Sigma, St. Louis, MO)] supplemented either with 200 μM IAF (Molecular Probe Inc., Eugene, OR) (the direct IAF labeling method), or 10 mM N-ethylmaleimide (NEM) (the blocking-IAF labeling method). The homogenate was incubated on ice for 30 min and centrifuged for 45 min at 12000× g at 4 °C. The supernatant was mixed with an equal volume of ice-cold phenol (Tris-buffered, pH 6.4–6.8) and centrifuged at 12000× g for 15 min at 4 °C to separate phenol and aqueous phases. The upper aqueous phase was removed leaving the interface intact, and the phenol phase was extracted twice with 50 mM Tris-HCl, pH 8.0 and then mixed with 5 volumes of cold 0.1 M ammonium acetate in methanol and left at −20 °C overnight to precipitate proteins. After centrifugation at 12000× g for 15 min, the protein pellet was washed five times with 1 mL methanol and air-dried for 10 min in a fume hood. For the direct IAF labeling method, the pellet was resuspended in the rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 20 mM DTT and 0.5% IPG buffer) (GE Healthcare). After centrifugation at 12000× g for 3 min, the supernatant was transferred to a new tube, and the protein concentration was determined with the CB-X Protein Assay kit (G-Biosciences, St. Louis, MO). For the blocking-IAF labeling method, the pellet was resuspended in the reduction buffer (50 mM Tris-HCl, pH 8.0, 7 M urea, 2 M thiourea, 1% CHAPS and 1 mM DTT). After sitting at room temperature for 30 min, IAF was added to a final concentration of 200 μM and the mixture was incubated at room temperature in dark for 30 min. The reaction was stopped by adding 5 volumes of methanol and free IAF was removed by precipitation with centrifugation. The pellet was resuspended in the rehydration buffer and protein concentration was determined as above.
For the direct N-(biotinoyl)-N′-(iodoacetyl)-ethylenediamine (BIAM) tagging method, cells were broken in the homogenization buffer (described above) supplemented with 100 μM BIAM (Invitrogen, Carlsbad, California). The protein homogenate was incubated at room temperature for 10 min and centrifuged at 12000× g for 10 min. Supernatant (2.5 mL) was taken and passed through a PD-10 desalting column (GE Healthcare) equilibrated with the homogenization buffer to remove free BIAM. BIAM-tagged proteins in the sample were then immunoprecipitated with 300 μL NeutrAvidin agarose resin (Thermo Scientific, Rockford, IL). Proteins were eluted from the resin by 6 M guanidine-HCl, pH 1.5 and purified with Perfect-FOCUS (G-Biosciences). The protein pellet was resuspended in the rehydration buffer and protein concentration was determined as above.
For the blocking-BIAM tagging method, cells were broken in the homogenization buffer supplemented with 10 mM NEM. The protein homogenate was incubated at room temperature for 15 min and then centrifuged at 12000× g for 10 min. The supernatant was taken and cysteine was added to a final concentration of 10 mM to eliminate remaining NEM. After incubation at room temperature for 10 min, DTT was added to a final concentration of 1 mM and the sample was kept at room temperature for another 10 min. The samples were cleaned by passing through PD-10 desalting columns (GE Healthcare) equilibrated with the homogenization buffer. After that, 100 μM BIAM was added and the samples were further processed as described in the direct BIAM tagging method.
Two-dimensional Gel Electrophoresis (2-DE)
Protein samples (200 μg) were mixed with the rehydration buffer to bring the volume to 180 μL, and the samples were then applied to 11 cm IPG strips (pH 4–7, Bio-Rad, Hercules, CA). The IPG strips were rehydrated overnight at room temperature. Isoelectric focusing (IEF) was carried out on a Bio-Rad PROTEAN IEF Cell at 20 °C with a maximum current of 50 μA/strip and the following settings: 250 V for 30 min, 500 V for 1 h, a gradient increase to 8000 V in 2.5 h, and remaining at 8000 V until reaching 35000 Vh. The strips were then transferred to 8–16% Criterion Precast Gels (Bio-Rad) for the second dimension electrophoresis using Criterion Cell system (Bio-Rad). SDS-PAGE was run at 60 V for 15 min and then 200 V until the bromphenol blue dye reached the gel end.
Protein Image Scanning
The gels were scanned using a Typhoon 9410 scanner (GE Healthcare). Gels with IAF-labeled samples were scanned using 488 nm laser and a 520 nm band-pass emission filter. Postelectrophoretic staining with the SYPRO Ruby (Bio-Rad) fluorescent dye was performed according to the manufacturer’s instructions and subsequent scanning was performing using 488 nm laser and 610 nm band-pass emission filter. All gels were scanned at 100 μm pixel size, and the photomultiplier tube was set to ensure maximum pixel intensity between 40000 and 80000 to avoid saturation.
Two-dimensional Gel Image Analysis
The 2-D gel images were analyzed using the Progenesis SameSpots software version 2.0 as described in the user’s instruction (Nonlinear Dynamics, Durham, NC). Briefly, a control sample was chosen as a reference image, and the software will automatically generate alignment vectors between the reference image and the other samples. After alignment, the images of three replicate samples for the same treatment were grouped together and the aligned images were analyzed for spot volume quantification and volume ratio normalization of different samples in the same treatment group. Statistical, quantitative, and qualitative analysis sets were created between the control group and each treated group. Protein spots with more than 2 fold increase or decrease in the IAF labeling intensities (for the IAF labeling methods) or SYPRO Ruby labeling intensities (for the BIAM tagging methods) with a p value smaller than 0.05 (Student’s t test) between the H2O2-treated samples and control samples were picked for identification.
Spot Picking and Mass Spectrometry
A spot picking list generated from Phoretix 2D Evolution gel analysis software (Nonlinear Dynamics Ltd.) was exported to Gelpix (Genetix Inc., Boston, MA). The excised spots were then digested with trypsin as previously described.21 Protein digests were subjected to nano-LC–ESI–MS/MS analysis. Nano-LC was performed with a nanoLC-2D (Eksigent, Dublin, Ireland) equipped with a capillary trap LC Packings PepMap (DIONEX, Sunnyvale, CA) and LC Packings C18 Pep Map 100 (75 μm, 15 cm) connected to the MS. Peptides (5 μL injections) were desalted for 10 min with a flow rate 5 μL/min of 90.5% solvent A (0.1% formic acid in Milli-Q water). Peptides were then resolved on a gradient from 9.5 to 35% solvent B (0.1% formic acid in ACN) for 4 min, from 35 to 45% B for 31 min, and from 45 to 90.5% B over the final 6 min at 200 nL/min flow rate.
The MS analysis was performed on an ABI QSTAR XL (AB Sciex, Foster City, CA) hybrid QTOF MS/MS mass spectrometer equipped with a nanoelectrospray source (Protana XYZ manipulator). Positive mode nanoelectrospray was generated from fused-silica PicoTip emitters with a 10 μm aperture (New Objective, Woburn, MA) at 2.5 kV. The m/z response of the instrument was calibrated daily with manufacturer standards. TOF mass and product ion spectra were acquired using information dependent data acquisition (IDA) in Analyst QS v1.1 with the following parameters: mass ranges for TOF MS and MS/MS were m/z 300–2000 and 70–2000, respectively. Every second, a TOF MS precursor ion spectrum was accumulated, followed by three product ion spectra, each for 3 s. The switching from TOF MS to MS/MS is triggered by the mass range of peptides (m/z 300–2000), precursor charge state (2–4) and ion intensity (>50 counts). The DP, DP2, and FP settings were 60, 10, and 230, respectively, and rolling collision energy was used.
Protein Database Search
The peptide tandem mass spectra were processed using Analyst QS software v1.1 (AB Sciex) and searched against the NCBInr_20081206 database (7463447 entries) using an in-house version of MASCOT v2.20 (Matrix Science Inc., Boston, MA). The following parameters were selected: tryptic peptides with ≤1 missed cleavage site; precursor and MS/MS fragment ion mass tolerance of 0.8 and 0.8 Da, respectively; variable carbamidomethylation and fluorescein modification of cysteines for direct IAF labeling method; variable carbamidomethylation, N-ethylmaleimide and fluorescein modification of cysteines for blocking-IAF labeling method; variable carbamidomethylation and IED-biotin modification of cysteines for direct BIAM tagging method; variable carbamidomethylation, N-ethylmaleimide and IED-biotin modification of cysteines for blocking-BIAM tagging method; and variable oxidation of methionine for all methods. The Mascot results were imported into Scaffold 3.0 (ProteomeSoftware, Portland, OR) and protein identification based on the following criteria were reported: protein probability ≥99% with at least 2 peptide with peptide probability ≥95%
Investigation of in vivo Redox State of Individual Proteins by Differential Labeling of Thiols Coupled with Immunoprecipitation and Western Blotting
For each gene under investigation, the genomic sequence from ∼2 kb upstream of its start codon to the codon before stop codon was cloned into pCR-Blunt II-TOPO (Invitrogen) and then subcloned into the binary vector pBAR-FLAG to fuse in frame with the FLAG tag at its C-terminus. Agrobacteria carrying the resulting construct was used to transform Arabidopsis. Stable transgenic Arabidopsis lines were generated that express each putative redox-sensitive protein fused with the FLAG tag driven by its native promoter. Three week-old plants of the transgenic lines (grown at 22 °C with 50% humidity under 9 h light/15 h dark photoperiod and at a light intensity of 125 mol m–2 s–1) were used for analysis.
For AtCIAPIN1, eEF1α and AtPTP1, transgenic plants were vacuum-infiltrated with 5 mM H2O2 or water. Ten minutes later, 0.1 g leaves were collected and total proteins were extracted by homogenization in the extraction buffer (20 mM Tris-HCl, pH 8.0, 5 mM EDTA, 0.5% Triton X-100, 100 mM NaCl, 1% protease inhibitor cocktail) supplemented with 100 μM BIAM. After incubation at room temperature for 10 min, the samples were centrifuged at 12000× g for 10 min. Supernatant was taken and excessive BIAM was removed by Zeba Spin Desalting Columns (Thermo Scientific) pre-equilibrated with the extraction buffer. FLAG-tagged protein was then affinity-purified by anti-FLAG M2 Affinity Gel (Sigma) according to the manufacturer’s instructions. Immunoprecipitated protein was separated by SDS-PAGE and immunoblotted with either anti-FLAG M2-Peroxidase (HRP) antibody (Sigma) or Horseradish Peroxidase-Conjugated Streptavidin (Thermo Scientific).
For AtNAP1;1, AtPDIL1-1 and 14-3-3 λ, after the oxidant treatment of leaves as described earlier, total protein was extracted in the extraction buffer supplemented with 5 mM iodoacetamide (IAM). After incubation at room temperature for 10 min and centrifugation at 12000× g for 10 min, the supernatant was taken and DTT was added to a final concentration of 7.5 mM. After incubation at room temperature for 15 min, the samples were cleaned by passing through Zeba Spin Desalting Columns. The samples were further processed as described for AtCIAPIN1 and eEF1α.
Protein Electrophoretic Mobility Shift Assay (PEMSA) for Analysis of in vivo Redox States of AtCIAPIN1
Stable transgenic plants expressing AtCIAPIN1-FLAG driven by its native promoter were vacuum-infiltrated with either water, 5 mM H2O2, 10 μM flg22, or 250 μM salicylic acid (SA). Leaves were harvested 10 min after H2O2 treatment or 30 min after all other three treatments. PEMSA was performed as described previously34 with modifications. Briefly, leaf samples were homogenized in the urea buffer [8 M urea, 100 mM Tris (pH 8.2), 1 mM EDTA] supplemented with 30 mM iodoacetic acid (IAA). After 15 min at 37 °C, the extracts were centrifuged at 12000× g for 10 min. Supernatant was taken and free IAA was removed by acetone precipitation of proteins. The protein pellet was resuspended in the urea buffer containing 3.5 mM DTT and incubated at 37 °C for 30 min. After that, IAM was added to a final concentration of 10 mM and samples were incubated at 37 °C for 15 min. Protein samples were then separated by urea-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membrane, and immunoblotted with the anti-FLAG M2 HRP-conjugated antibody (Sigma). Mobility standards were prepared by treating total leaf proteins extracted in the urea buffer with 3.5 mM DTT at 37 °C for 30 min followed by labeling all free thiols with either 30 mM IAA or 10 mM IAM at 37 °C for 15 min, respectively.
Results and Discussion
Methods for Differential Labeling of Reduced versus Oxidized Protein Thiols
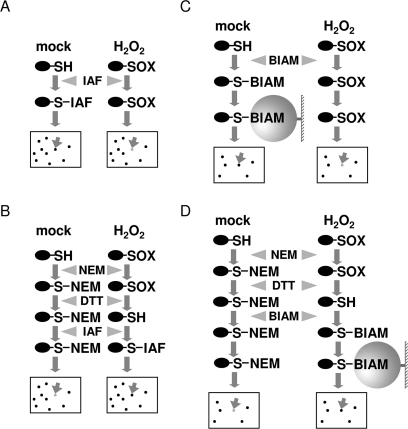
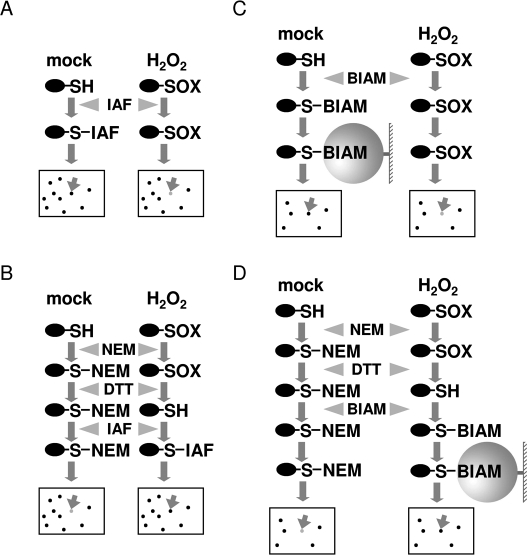
We used two distinct thiol labeling approaches (IAF labeling and BIAM tagging) for detection of redox-sensitive proteins. For each approach, two methods (the direct labeling method and the blocking-labeling method) were employed. The procedures are schematically represented in Figure 1. For the direct IAF labeling method (Figure 1A), proteins are extracted from oxidant-treated and control cells and free thiols are directly labeled with the fluorescent probe 5-(iodoacetamido)fluorescein (IAF). After that, proteins are separated by 2-DE and the IAF labeling patterns are imaged using Typhoon 9410. If a particular protein becomes oxidized in the cells following oxidant treatment, the protein spot will show a reduced labeling intensity compared with the corresponding spot in the control sample. The direct labeling method is relatively straightforward. However, thiol/disulfide exchange may occur during protein extraction.9 An alternative method (Figure 1B) is to freeze the thiol/disulfide states of proteins by first extracting proteins in the presence of a thiol alkylation reagent such as N-ethylmaleimide (NEM) which reacts and blocks free thiols. Oxidized thiols present in the samples are then reduced with DTT and the newly generated free thiols are labeled with IAF. In this blocking-IAF labeling method, if a protein becomes oxidized in the oxidant-treated cells, it is expected to display an increased labeling intensity in the treated sample compared with that in the control sample. The blocking-IAF labeling method could only detect reversibly oxidized (DTT-reducible) proteins.
Figure 1.
Schematic drawing of the proteomics methods used in this study. (A) Direct IAF labeling method. (B) Blocking-IAF labeling method. (C) Direct BIAM tagging method. (D) Blocking-BIAM tagging method. −SH, a reduced (free) thiol; −SOX, an oxidized thiol.
One drawback of the IAF labeling methods is the limited resolution and sensitivity of 2-DE for resolving complex total proteins from plant tissues. Hence, we developed another labeling approach, BIAM tagging, in an attempt to at least partially overcome this drawback (Figure 1C,D). In this approach, IAF is replaced by N-(biotinoyl)-N′-(iodoacetyl)-ethylenediamine (BIAM) which is also thiol-reactive and contains a biotin moiety. Following direct BIAM tagging or blocking-BIAM tagging, tagged proteins are affinity-purified using NeutrAvidin agarose resin, separated by 2-DE, and stained with SYPRO Ruby. In this approach, only proteins that contain reduced thiols (in the direct BIAM tagging method) or reversibly oxidized thiols (in the blocking-BIAM tagging method) in original tissue samples can be affinity-purified. As a result, the complexity of protein samples subjected to 2-DE is reduced.
Establishment of Experimental Conditions for Induction of Oxidative Stress in Arabidopsis Cell Cultures
We aimed to identify proteins that undergo oxidative modification in Arabidopsis suspension cells following treatment with hydrogen peroxide (H2O2). Arabidopsis suspension cells offer an advantage over whole plant tissues for achieving more uniform exposure of all cells to the oxidant, which increases the sensitivity for detecting oxidatively modified proteins. It has been reported that no substantial cell death was caused by treatment with H2O2 at a concentration as high as 10 mM for 6 h35 or even at 88 mM for 16 h.36
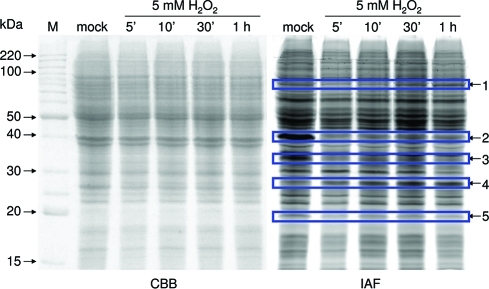
In this study, Arabidopsis suspension cells were treated with 5 mM H2O2 to induce oxidative stress. The direct IAF labeling method was used to evaluate whether the oxidant treatment leads to detectable changes in protein redox state in the suspension cells. At different time points following the 5 mM H2O2 treatment, cells were harvested and free thiols were labeled with IAF during protein extraction. Proteins were then separated by one-dimensional SDS-PAGE (1-DE). As shown in Figure 2, multiple bands showed altered IAF labeling intensities within 5 min of the treatment (marked by arrows), indicating that some proteins in the suspension cells were oxidized (with decreased IAF labeling intensities) or reduced (indicated by Arrow 4 with an elevated IAF labeling intensity). The most striking change came from a 40 kDa band (indicated by Arrow 2), whose IAF labeling intensity significantly diminished during the 5 min-1 h treatment. Since we are interested in early responsive redox-sensitive proteins, in our subsequent redox proteomics experiments, cells were harvested after being treated with 5 mM H2O2 for 10 min. The short oxidant treatment was also intended to minimize the chance of false positives resulting from changes in abundance of a thiol-containing protein, which is more likely to occur after a longer period of the oxidant treatment.
Figure 2.
Effect of H2O2 treatment on protein oxidation. Arabidopsis suspension cells were treated with 5 mM H2O2 for different time periods (as indicated). For each sample, 150 μg protein was labeled with IAF and separated by discontinuous SDS-PAGE with a 12% separating gel and a 4% stacking gel. The IAF labeling pattern (right panel) was scanned by Typhoon 9410. Total proteins were visualized by Commassie Brilliant Blue (CBB) staining (left panel). The arrows point to proteins that displayed altered IAF labeling intensities after H2O2 treatment. Mock: cells were treated with H2O for 10 min.
Detection and Identification of H2O2–Sensitive Thiol-Containing Proteins in Arabidopsis Cell Cultures
Three biological replicates were included for each proteomic method for detection and identification of proteins that underwent oxidative modifications in Arabidopsis suspension cells following the oxidant treatment. After IAF labeling of thiol-containing proteins or affinity-purification of BIAM-tagged proteins, protein samples were separated by 2-DE. For the IAF-labeled samples, the IAF labeling patterns of gels were imaged. Then the gels were stained with SYPRO Ruby which helps to locate IAF-labeled protein spots for spot picking. For the BIAM tagging methods, the gels were stained with SYPRO Ruby to detect differentially tagged proteins.
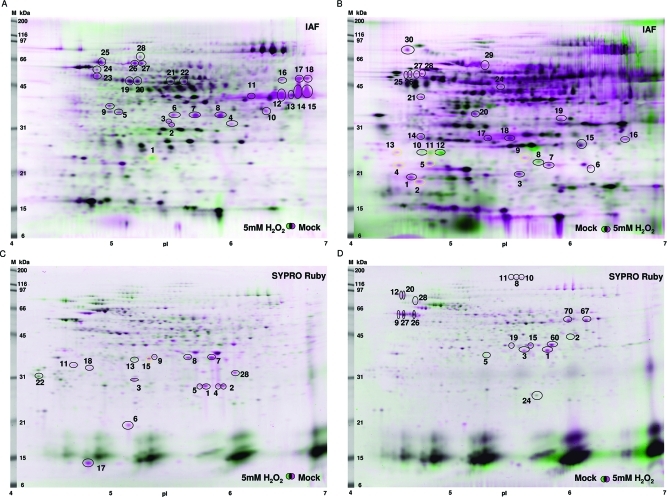
Protein spots with an IAF labeling intensity (for the IAF labeling methods) or SYPRO Ruby staining intensity (for the BIAM tagging methods) differing by more than 2-fold (with a p value of <0.05) as determined by the Progenesis Samespots software were marked for identification by mass spectrometry (MS) (shown as circled spots in Figure 3). In direct labeling methods, gel images from control and oxidant-treated samples were pseudocolored as red and green, respectively (Figure 3A,C). In blocking-labeling methods, gel images from control and oxidant-treated samples were pseudocolored as green and red, respectively (Figure 3B,D). Gel images from control and oxidant-treated samples were then superimposed. The spots in pink represent proteins that were in more oxidized states in the H2O2-treated samples than in the control samples whereas the spots in green were in more reduced states in the treated samples. The spots that appear black were proteins with relatively similar labeling intensities between control and treated samples.
Figure 3.
Detection of redox-sensitive proteins upon H2O2 treatment. IAF labeling images (for the IAF labeling methods) or SYPRO Ruby stained images (for the BIAM tagging methods) from oxidant-treated samples and control samples were pseudocolored and superimposed. Differentially labeled spots (marked by circles in black or yellow) were numbered and excised for MS identification. The MW markers and pI markers are indicated. The yellow circled spots were not identified by MS analysis while the black circled spots yielded positive identification. The images are based on the results from (A) direct IAF labeling, (B) blocking-IAF labeling, (C) direct BIAM tagging, and (D) blocking-BIAM tagging. For each method, three biological replicates were conducted and a representative superimposed image is presented.
Some IAF-labeled spots could not be detected by SYPRO Ruby staining, suggesting that they were at very low abundance and generally could not be identified due to the sensitivity limitation of the mass spectrometer utilized for identification. Such spots were marked by yellow circles in Figure 3. In total, 27 spots from direct IAF labeling, 22 from blocking-IAF labeling, 15 from direct BIAM tagging, and 19 from blocking-BIAM tagging yielded positive identification. In total, 84 proteins were identified as putative oxidant-sensitive proteins (Table 1, Table S1, S2, Supporting Information). Among them, 13 proteins were identified by more than one method.
Table 1. List of the Redox-Sensitive Proteins Identified in This Studya.
| gene locus | protein function | number of peptides matchedb | coverage (%)b | methodc |
|---|---|---|---|---|
| Primary metabolism | ||||
| AT1G64190 | 6-phosphogluconate dehydrogenase | 9 | 23.0 | D/I |
| AT1G77120 | alcohol dehydrogenase 1 (ADH1) | 2 | 8.2 | B/B |
| AT3G48000 | aldehyde dehydrogenase 2B4 (ALDH2B4) | 7 | 15.0 | B/B |
| AT3G17940 | aldose 1-epimerase, putative | 5 | 16.0 | D/I |
| AT1G14810 | aspartate semialdehyde dehydrogenase | 4 | 15.0 | D/B |
| AT3G55440 | cytosolic triose phosphate isomerase | 10 | 42.0 | B/I |
| AT1G74030 | enolase 1 (ENO1) | 4 | 8.6 | D/I |
| AT2G36530 | enolase 2 (ENO2) | 3 | 9.7 | D/I |
| AT3G52930 | fructose-bisphosphate aldolase, putative | 11 | 49.0 | D/I |
| AT3G04120 | GAPDH C subunit 1 (GAPC-1) | 8 | 24.0 | D/I |
| AT1G13440 | GAPDH C subunit 2 (GAPC-2) | 4 | 13.0 | D/I |
| AT1G53240 | malate dehydrogenase (NAD), mitochondrial | 3 | 12.0 | D/I |
| AT5G43330 | malate dehydrogenase, cytosolic, putative | 2 | 5.4 | D/I |
| AT1G79550 | phosphoglycerate kinase (PGK) | 6 | 20.0 | B/I |
| AT5G50850 | pyruvate dehydrogenase | 2 | 6.1 | D/B |
| AT2G19940 | semialdehyde dehydrogenase family protein | 4 | 12.0 | D/I |
| AT1G01800 | short-chain dehydrogenase/reductase family protein | 7 | 35.0 | D/B |
| Antioxidant system | ||||
| AT3G11630 | 2-Cys peroxiredoxin (2-Cys PrxA) | 4 | 22.0 | B/I |
| AT1G07890 | ascorbate peroxidase (APX1) | 2 | 9.6 | B/I D/B |
| AT1G75270 | dehydroascorbate reductase 2 (DHAR2) | 2 | 9.9 | B/B |
| AT5G42980 | encodes a cytosolic thioredoxin | 4 | 40.0 | D/B |
| AT3G10920 | manganese superoxide dismutase (MSD1) | 2 | 9.5 | B/I |
| AT1G65980 | thioredoxin-dependent peroxidase 1 (TPX1) | 2 | 6.2 | D/B |
| Translational and post-translational control | ||||
| translation | ||||
| AT3G09200 | 60S acidic ribosomal protein P0 | 2 | 10.0 | D/I |
| AT1G07920 | elongation factor 1-alpha | 6 | 19.0 | D/I |
| AT5G60390 | elongation factor 1-alpha | 6 | 19.0 | D/I |
| AT1G07940 | elongation factor 1-alpha | 6 | 19.0 | D/I |
| AT1G07930 | elongation factor 1-alpha | 6 | 19.0 | D/I |
| AT5G19510 | elongation factor 1B alpha-subunit 2 (eEF1Balpha2) | 4 | 26.0 | D/B |
| AT4G11120 | translation elongation factor Ts (EF-Ts) | 3 | 7.8 | D/B |
| protein folding | ||||
| AT2G04030 | a chloroplast-targeted 90-kDa heat shock protein | 3 | 4.7 | B/B |
| AT1G21750 | protein disulfide isomerase-like protein (AtPDIL1-1) | 14 | 29.0 | B/B |
| AT1G77510 | protein disulfide isomerase-like protein (AtPDIL1-2) | 10 | 30.0 | B/B |
| AT3G54960 | protein disulfide isomerase-like protein (AtPDIL1-3) | 10 | 21.0 | B/B |
| AT5G60640 | protein disulfide isomerase-like protein (AtPDIL1-4) | 10 | 17.0 | B/I B/B |
| AT3G16110 | protein disulfide isomerase-like protein (AtPDIL1-6) | 5 | 9.7 | D/I B/B |
| AT2G47470 | protein disulfide isomerase-like protein (AtPDIL2-1) | 5 | 16.0 | D/B B/B |
| AT1G56340 | calreticulin1 (CRT1) | 2 | 4.5 | B/I |
| AT2G28000 | chaperonin-60 alpha (CPN60A) | 15 | 30.0 | D/I B/I B/B |
| AT3G56070 | cytosolic cyclophilin (CYP2) | 4 | 24.0 | B/I |
| AT1G60420 | DC1 domain-containing protein/PDI-like protein | 10 | 19.0 | D/I |
| AT3G12580 | heat shock protein 70(HSP70) | 15 | 29.0 | D/I |
| AT5G28540 | luminal-binding protein 1(BIP1) | 18 | 32.0 | D/I |
| AT4G37910 | mitochondrial heat shock protein 70-1 (MTHSC70-1) | 24 | 44.0 | D/I B/I |
| AT4G26110 | nucleosome assembly protein 1;1 (NAP1;1) | 3 | 8.6 | B/I |
| AT3G62030 | rotamase/CYP 4 | 2 | 8.1 | B/I |
| protein degradation | ||||
| AT5G35590 | 20S proteasome alpha subunit A1 (PAA1) | 2 | 9.8 | D/B |
| AT1G16470 | 20S proteasome alpha subunit B1 (PAB1) | 5 | 20.0 | B/I |
| AT2G27020 | 20S proteasome alpha subunit G1 (PAG1) | 2 | 8.4 | D/B |
| AT3G26340 | 20S proteasome beta subunit E, putative | 2 | 9.5 | B/I |
| AT5G42790 | 26S proteasome alpha subunit F1 (PAF1) | 6 | 25.0 | D/I B/I |
| AT4G12060 | double Clp-N motif protein | 2 | 9.1 | B/I |
| AT4G20850 | Tripeptidyl Peptidase II | 10 | 7.6 | B/B |
| AT2G17190 | ubiquitin family protein | 4 | 9.3 | B/I |
| post-translational modifications | ||||
| AT1G50370 | phosphoprotein phosphatase | 2 | 6.6 | D/I |
| AT5G53140 | protein phosphatase 2C, putative (PP2C) | 4 | 9.0 | B/I |
| AT1G71860 | protein tyrosine phosphatase 1(PTP1) | 4 | 13.0 | D/I |
| Cytoskeleton | ||||
| AT5G09810 | actin 7 (ACT7) | 10 | 31.0 | D/I B/I |
| AT4G14960 | tubulin alpha-6 chain (TUA6) | 11 | 36.0 | D/I |
| AT5G62690 | tubulin beta-2 (TUB2) | 5 | 11.0 | D/I |
| Others | ||||
| AT5G10450 | 14-3-3 lambda | 4 | 21.0 | B/I |
| AT4G37000 | accelerated cell death 2 (ACD2) | 4 | 14.0 | D/I |
| AT1G12910 | anthocyanin1 (AtAN11) | 3 | 8.4 | B/I |
| AT5G10920 | argininosuccinate lyase (AtArgH) | 5 | 8.5 | D/I |
| AT4G24830 | arginosuccinate synthase family protein | 11 | 25.0 | D/I |
| AT5G18400 | AtCIAPIN1 | 3 | 11.0 | D/B |
| AT1G78900 | catalytic subunit A of the vacuolar ATP synthase | 19 | 35.0 | D/I B/I |
| AT1G13870 | deformed roots and leaves 1 (DRL1) | 2 | 7.9 | D/B |
| AT3G25530 | gamma-hydroxybutyrate dehydrogenase | 2 | 7.6 | D/I |
| AT4G34540 | isoflavone reductase family protein | 2 | 6.9 | D/I |
| AT3G07720 | kelch repeat-containing protein | 2 | 6.7 | D/I |
| AT5G48480 | lactoylglutathione lyase | 4 | 27.0 | B/I |
| AT3G44300 | nitrilase 2 (NIT2) | 5 | 15.0 | D/B B/B |
| AT4G08790 | nitrilase, putative | 2 | 6.8 | D/I |
| AT1G63000 | nucleotide-rhamnose synthase/epimerase-reductase | 4 | 14.0 | D/I D/B |
| AT3G62530 | PBS lyase, HEAT-like repeat-containing protein | 3 | 13.0 | B/I |
| AT3G07090 | PPPDE putative thiol peptidase family protein | 3 | 13.0 | D/I |
| AT4G14000 | Putative methyltransferase family protein | 4 | 19.0 | B/I |
| AT2G38230 | pyridoxine biosynthesis 1.1 (ATPDX1.1) | 5 | 18.0 | D/I |
| AT5G01410 | pyridoxine biosynthesis 1.3 (ATPDX1.3) | 3 | 11.0 | B/I |
| AT1G07750 | RmlC-like cupins superfamily protein | 4 | 14.0 | D/B B/B |
| AT2G41530 | S-formylglutathione hydrolase | 2 | 7.4 | D/I |
| AT1G09760 | U2 small nuclear ribonucleoprotein A | 4 | 20.0 | D/I B/I |
| AT4G21580 | zinc-binding dehydrogenase family protein | 6 | 20.0 | D/I |
More detailed information on protein identification and properties are listed in Tables S1 and S2 (Supporting Information).
For proteins identified from multiple spots, derived from the spot yielding most numbers of uniquely matched peptides.
The method by which the protein was identified: D/I, direct IAF labeling. B/I, blocking-IAF labeling. D/B, direct BIAM tagging. B/B, blocking-BIAM tagging.
One common problem with gel-based proteomics methods is that the limited resolution of 2-DE might lead to false positives. Multiple protein IDs were obtained from some of the protein spots particularly in the gel areas where proteins were not well separated. These spots might contain protein(s) that were not differentially labeled but have similar molecular weights and isoelectric points to redox-sensitive proteins. Among proteins identified in this study, two nonthiol-containing proteins were found. One is AT1G16470, identified from Spot 18 in the blocking-IAF labeling method. This spot was also identified as a cytosolic triose phosphate isomerase which was reported to be S-nitrosylated in the hypersensitive response.33 The other is AT1G09760, identified from both Spot 8 in the direct IAF labeling method and Spot 19 in the blocking-IAF labeling method. These two spots also contained peptides from AtPDX1.1 and AtPDX1.3, respectively. Such nonthiol-containing false positive proteins were not found with the BIAM tagging methods.
Some spots were assigned with multiple proteins that share very high sequence similarities due to the fact that a peptide might match to more than one protein. For instance, Spot 17 and 18 in Figure 3A were identified as translation elongation factor 1α (eEF1α), which is encoded by 4 distinct genes (At1g07920, At1g07930, At1g07940 and At5g60390). However, these 4 genes encode proteins with identical amino acid sequences.
Oxidative Modification of Identified Proteins in Arabidopsis Plants upon H2O2 Treatment
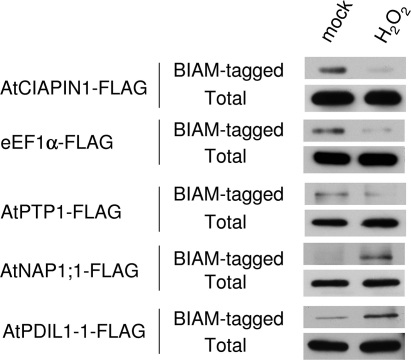
To make a rough assessment of the reliability of the proteomics methods, six putative redox-sensitive proteins were chosen and their in vivo redox state under oxidant treatment were individually investigated in Arabidopsis plants. These proteins include eEF1α and protein tyrosine phosphatase 1 (AtPTP1) identified from the direct IAF labeling method, AtCIAPIN1 from the direct BIAM tagging method, nucleosome assembly protein 1;1 (AtNAP1;1) and 14-3-3 λ from the blocking-IAF labeling method, and protein disulfide isomerase like 1-1 (AtPDIL1-1) from the blocking-BIAM tagging method.
Stable transgenic Arabidopsis lines expressing each of the six proteins fused with the FLAG tag driven by its native promoter were generated. Soil-grown plant leaves were vacuum-infiltrated with either 5 mM H2O2 or water (as a control) and harvested after 10 min of the treatment. To determine the redox state of the proteins previously identified from the direct labeling methods, total protein was extracted in the BIAM-containing buffer to tag free thiols. The FLAG-tagged protein was affinity-purified with anti-FLAG M2 affinity gel and detected by immunoblotting with horseradish peroxidase-conjugated streptavidin to determine the amount of BIAM attached to the recombinant protein. The total amount of the recombinant proteins was determined by immunoblotting with the anti-FLAG antibody. If a protein was oxidatively modified by the treatment, we would expect reduced BIAM tagging of this protein. Indeed, as shown in Figure 4, the amount of BIAM-tagged recombinant AtCIAPIN1, eEF1α and AtPTP1 was greatly reduced in the H2O2-treated samples. For the proteins identified from the blocking methods, free thiols were first alkylated by IAM and samples were then treated with DTT followed by BIAM labeling to tag thiols that were originally in a reversibly oxidized state. Therefore, enhanced BIAM tagging of a protein would be expected if it underwent oxidation during the treatment, which was indeed the case for AtNAP1;1 and AtPDIL1-1 (Figure 4). However, we failed to detect BIAM tagging of the recombinant 14-3-3 λ in either control or treated samples in our experimental conditions, suggesting that 14-3-3 λ may be unsusceptible to BIAM labeling. It is also possible that 14-3-3 λ constitutively exists in its reduced state during the H2O2 treatment and thus represents a false positive from the redox proteomics experiment. Another possibility is that 14-3-3 λ might indeed undergo redox changes in the oxidant-treated suspension cells, but such a change did not occur in the oxidant-treated leaves.
Figure 4.
Oxidative modification of identified proteins in planta upon H2O2 treatment. Transgenic plants expressing the protein of interest fused with the FLAG tag were vacuum infiltrated with either water (mock) or 5 mM H2O2. For analysis of AtCIAPIN1, eEF1α, and AtPTP1, free thiols in the total protein were labeled with BIAM during protein extraction. For analysis of AtNAP1;1 and AtPDIL1-1, free thiols in the samples were first alkylated by IAM. Samples were then treated with DTT and newly generated free thiols were labeled by BIAM. After that, FLAG-tagged protein from each sample was affinity purified, separated by SDS-PAGE, and detected by HRP-Conjugated Streptavidin (to determine the amount of BIAM attached to the FLAG-tagged protein) or by the anti-FLAG M2 antibody (to determine the amount of the total recombinant protein).
Although it is impractical to individually verify the redox states of all 84 putative redox-sensitive proteins identified from the redox proteomics approaches, the result that 5 of the 6 selected proteins were confirmed to undergo oxidative modification in the oxidant-treated leaves indicates that at least a majority of the putative redox-sensitive proteins identified in this study are likely real positives.
AtCIAPIN1 Alters its Redox State in Response to the Treatment with Flg22 or SA
Rapid production of ROS is usually associated with the plant defense response. It is interesting to investigate whether the H2O2-sensitive proteins identified are also sensitive to redox-perturbing chemicals such as flg22 and SA. Flg22 is a peptide derived from the bacterial flagellin which triggers the innate immune response.37 Flg22 has been found to induce ROS production catalyzed by the NADPH oxidase AtrbohD.38 SA is a key regulator of the plant defense response which causes perturbation of cellular redox homeostasis.39
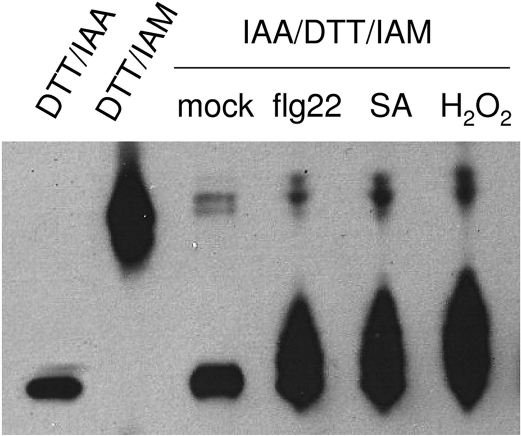
A modified Protein Electrophoretic Mobility Shift Assay (PEMSA)34 method was used to analyze the redox profiles of AtCIAPIN1 which contains 11 cysteine residues. Soil-grown transgenic plants expressing AtCIAPIN1-FLAG were vacuum infiltrated with either water, flg22, SA or H2O2. Total protein was extracted and free thiols were carboxymethylated by iodoacetic acid (IAA). Thiols that were in a reversibly oxidized state in the samples were reduced by DTT and amidomethylated by iodoacetamide (IAM). The IAM adducts are neutral while the ionized IAA adducts of free thiols lead to faster protein migration toward the anode during urea-PAGE. Therefore, AtCIAPIN1 proteins with more oxidized thiols in the original leaf samples tend to migrate more slowly than those with more free thiols. As shown in Figure 5, in the control sample (lane 3), the large majority of the AtCIAPIN1-FLAG protein had the similar mobility to the AtCIAPIN1-FLAG whose thiols were all labeled by IAA (lane 1), suggesting that they were in a reduced state. However, the samples from the leaves treated with flg22, SA, or H2O2 all resulted in multiple bands with slower mobility than that in lane 1. These bands represent AtCIAPIN1-FLAG with different numbers of oxidized thiols. This result indicates that flg22 and SA, like H2O2, lead to the oxidative modification of AtCIAPIN1 presumably through alteration of cellular redox homeostasis.
Figure 5.
AtCIAPIN1-FLAG underwent oxidative modification in leaves upon flg22, SA and H2O2 treatment. Lane 1 and lane 2 are mobility standards corresponding to fully reduced and fully oxidized AtCIAPIN1-FLAG protein, respectively. They were prepared by treating total leaf protein extracts with DTT followed by labeling all free thiols with either IAA (lane 1) or IAM (lane 2). For the other samples, total proteins were extracted in the buffer containing IAA to carboxymethylate free thiols. The samples were then treated with DTT followed by amidomethylating newly generated thiols with IAM. The protein samples were then separated by urea-PAGE and AtCIAPIN1-FLAG protein was detected by immunoblotting with the anti-FLAG M2 antibody.
Categorization of Proteins Identified as Redox-sensitive Proteins
Our study led to the identification of 84 putative redox-sensitive proteins (Table 1 and Table S1, Supporting Information). All peptides identified from the MS analysis are listed in Table S2 (Supporting Information). These proteins can be classified into multiple functional groups, as discussed below.
Metabolism
Seventeen of the redox-sensitive proteins are enzymes involved in primary metabolic processes. Strikingly, at least 8 of them are involved in glycolysis, including 2 cytosolic GAPDHs (GapC1 and GapC2), triose phosphate isomerase, phosphoglycerate kinase (PGK), aldose 1-epimerase, aldolase, and 2 enolases. Several of them or their paralogs have been identified as S-nitrosylated proteins in Arabidopsis.32 It has been reported that in an in vitro assay, GAPDH activity can be inhibited by oxidation or nitrosylation and restored by reductant treatment.20 Inactivation of glycolysis enzymes likely acts to redirect the metabolic flux from glycolysis to the pentose phosphate pathway (PPP) to generate NADPH, the reducing power for cellular antioxidant systems. Such a mechanism was reported during the oxidative stress in yeast.40,41
It has been found that in response to H2O2 stress, yeast GAPDH becomes oxidized and mediates phosphorelay from the Mak2/3 sensor kinase to the Mcs response regulator to activate a MAPK cascade.42 More recently, S-nitrosylated GAPDH was found to bind to an E3-ubiquitin-ligase and then translocates to the nucleus to initiate a cell death cascade through activation of p53.43 It is intriguing to speculate that in addition to modulating the glycolytic pathway, oxidative modifications of GAPDH may play a role in other signaling pathways in plants.
Our results also suggest the presence of redox control on the metabolic processes downstream of glycolysis. There are two pathways for the conversion of pyruvate to acetyl-CoA. This reaction can be catalyzed by pyruvate dehydrogenase (PDH) under normal conditions. During ethanolic fermentation, pyruvate is converted by pyruvate decarboxylase (PDC) to acetaldehyde. The detoxification of acetaldehyde by aldehyde dehydrogenases (ALDH) produces acetate, which can then be used by acetyl-CoA synthetase (ACS) to synthesize acetyl-CoA. This PDC-ALDH-ACS pathway, also termed the PDH bypass, has been reported in yeast44 and recently also in Arabidopsis.45 In our results, the ALDH was found to be oxidized upon H2O2 treatment while the PDH became more reduced. Although further studies are needed to evaluate potential redox regulation of ALDH and PDH activity, it is possible that the two pathways are synchronously and probably differentially regulated under oxidative stress.
Antioxidant System
Six well-known proteins of the antioxidant system were identified, including dehydroascorbate reductase 2 (DHAR2) which serves to maintain a reduced ascorbate pool through oxidation of reduced GSH to glutathione disulfide, thioredoxin-dependent peroxidase 1 (TPX1), a cytosolic thioredoxin, a mitochondrial manganese superoxide dismutase MSD1, the cytosolic l-ascorbate peroxidase APX1, and the chloroplast 2-cysteine peroxiredoxin PrxA.
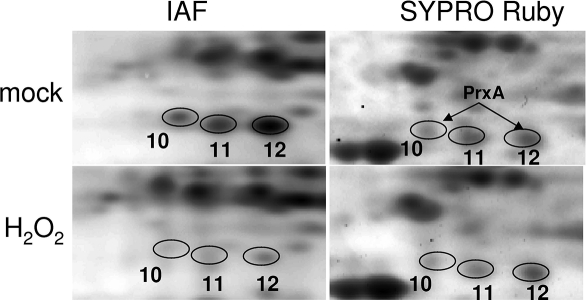
A cluster of three protein spots (Spot 10, 11, and 12 in Figure 3B) detected from the blocking-IAF labeling method showed a reduction in IAF labeling in oxidant-treated samples compared to the control samples, suggesting that these proteins might became more reduced following the H2O2 treatment. We were not able to identify Spot 11. However, the two other spots were found to be the same protein, a 2-Cys peroxiredoxin (PrxA), which is localized in chloroplasts and protects photosynthetic machinery from oxidative damage.46,47 Peroxiredoxins reduce hydrogen peroxide and alkyl hydroperoxides and therefore modulates peroxide levels. During the reaction, its active site cysteine (the peroxidatic cysteine) is oxidized to a sulfenic acid intermediate which forms an intermolecular disulfide with a second cysteine on the other identical substrate. The disulfide can then be reduced to thiol by glutathione or thioredoxin. There are several possibilities which lead to its reduced IAF labeling in the H2O2-treated cells. One possibility is that the oxidant treatment might cause a rapid drop in the PrxA protein level, which can be ruled out since PrxA protein levels were found to be similar in the control samples and the H2O2-treated samples based on SYPRO Ruby staining (Figure 6). The second possibility is that thiols of some PrxA proteins might be in an oxidized state under normal conditions. The oxidant treatment might activate the antioxidant system, which leads to temporary reduction of the oxidized thiol by such as thioredoxin or glutathione in a manner similar to reduction of NPR1.48
Figure 6.
Comparison of the IAF-labeled protein spots and SYPRO Ruby-stained protein spots from a portion of 2-D gels from the blocking-IAF labeling method. The numbered protein spots correspond to those in Figure 3B. Note the reduced IAF labeling of Spots 10, 11, and 12 in the H2O2-treated sample compared to that in the mock-treated sample. Spot 10 and Spot 12 were identified as PrxA. Staining of the gels with SYPRO Ruby revealed that the abundance of these proteins was not changed by the oxidant treatment.
Protein Synthesis, Folding, Modification, and Degradation
Regulation of protein synthesis and post-translational modifications allows cells to obtain a rapid response to an environmental signal. Not surprisingly, a large group of the oxidant sensitive proteins listed in Table 1 are involved in protein synthesis, folding, modification, and degradation.
eEF1α was identified from the direct IAF labeling method and was confirmed to undergo oxidation in the oxidant-treated leaves (Figure 4). Inhibition of protein synthesis in response to oxidative stress has been reported in mammalian cells and in yeast, mediated by phosphorylation of the elongation initiation factor eIF2α.49,50 Our result suggests another level of redox regulation of protein synthesis through regulation of translational elongation by oxidative modification of eEF1α. However, the consequence of eEF1α oxidation under oxidative stress remains to be defined.
Among proteins in this category, 6 protein disulfide isomerase-like proteins (AtPDILs) were found. All of the 6 PDILs were identified by the blocking methods, indicating that their oxidative modification is reversible. One of them, AtPDIL1-1, was confirmed to be oxidized in the H2O2-treated leaves (Figure 4). Oxidative stress is expected to increase mis-folded proteins in the ER and thus causes ER stress, which is also called unfolded protein response (UPR). Protein disulfide isomerases (PDIs), which catalyze the formation and breakage of disulfide bonds within substrate proteins and thus facilitate their correct folding, are key protein folding catalysts activated during UPR in animals and yeast. It has been reported that the redox state of PDILs is regulated by a thiol-disulfide oxidoreductase OsEro1 in rice,51 but little is known about the influence of oxidative stress on their functions in vivo.
The nucleosome assembly protein 1 (AtNAP1;1) was identified from the blocking-IAF labeling method. AtNAP1;1 is a histone chaperone involved in nucleosome assembly/disassembly and regulates the expression of some genes in the nucleotide excision repair pathway.52 The oxidation of AtNAP1;1 may act to regulate nucleosome formation and downstream gene expression.
Besides protein folding, several proteins involved in protein degradation were identified as targets of H2O2, indicating the involvement of protein degradation systems in coping with the accumulation of unfolded and mis-folded proteins under oxidative stress. Among them, the 20S proteasome alpha subunit A1, B1, G1 and F1 were also found to be S-glutathionylated upon tert-butylhydroperoxide treatment.18
Three protein phosphatases were found to be oxidant-sensitive, including two serine/threonine phosphatases and a tyrosine protein phosphatase known as AtPTP1.53 It has been reported that AtPTP1 dephosphorylates MAPKs54,55 and is inactivated by H2O2 treatment in vitro.55 Our result further reveals that AtPTP1 was oxidized in vivo in the oxidant-treated plants (Figure 4) which likely inactivates AtPTP1. Protein phosphatases are well-known targets of H2O2 which oxidizes their conserved catalytic cysteine residues to render them inactive.56,57 Inactivation of AtPTP1 as well as other protein phosphatases might serve to enhance the oxidative stress-induced MAPK cascade.55,58
Cytoskeleton Proteins
Another group of the oxidant sensitive proteins are components of the cytoskeleton, including actin 7 (ACT7), tubulin alpha-6 chain (TUA6), and tubulin beta-2 (TUB2). Actin and tubulin have previously been found to be sensitive to oxidation in animals59,60 and Arabidopsis.18 In yeast, actin is considered as an oxidative stress sensor whose oxidation regulates stress-triggered cell death.61
Other Proteins
In this group of proteins, special attention was paid to AtCIAPIN1, which was identified from the direct BIAM tagging method. AtCIAPIN1 encodes a putative iron–sulfur protein homologous to human cytokine-induced apoptosis inhibitor 1 (Ciapin1),62 and the yeast Dre2 protein, a negative regulator of H2O2-induced cell death.63 In yeast, the Tah18-Dre2 complex is part of an electron transfer chain involved in an early step of cytosolic Fe–S protein biogenesis and this complex can be functionally replaced by human Ndor1-Ciapin1 complex, with Ndor1 being a human homologue of Tah18.64 Recently, AtCIAPIN1 was identified in a yeast 2-hybrid screen to interact with ATR3, an Arabidopsis homologue of Tah18.65 These results suggest that this protein complex is involved in a pathway conserved among eukaryotes. AtCIAPIN1 was confirmed to be oxidized in leaves upon H2O2 treatment (Figure 4). In addition, AtCIAPIN1 also underwent oxidative modification when plants were treated with flg22 and SA (Figure 5), two inducers of plant defense response. It has been reported that the levels of H2O2 induced by flg22 and SA generally do not exceed several μM.66,67 Our results indicate that AtCIAPIN1 could be oxidatively modified by low levels of endogenously produced ROS. Alternatively, AtCIAPIN1 in the SA- or flg22-treated samples might not be directly oxidized by endogenous ROS, but by another protein through thiol-disulfide exchange reaction. The exact function of AtCIAPIN1 and its homologues is still unknown, but the sensitivity of AtCIAPIN1 to even low levels of ROS and the role of its yeast homologue plays in H2O2-stimulated cell death suggest that AtCIAPIN1 might be an ancient redox sensor which integrates redox signals to the process of cell death.
Four Proteomic Methods are Complementary
The direct IAF or BIAM labeling methods are less tedious than the blocking-labeling methods, but they present a higher risk of false positives due to possible thiol-disulfide exchanges occurring during protein extraction. Additionally, direct labeling can potentially identify both reversibly and irreversibly oxidized proteins whereas the blocking-labeling methods only identify reversibly oxidized proteins. The BIAM-tagging methods reduced the complexity of protein samples loaded onto 2-DE, thus reduced the chance of false positives. However, since affinity purification of BIAM tagged proteins by NeutrAvidin agrose resin requires a nondenaturing condition, proteins need to be labeled by BIAM in their native configurations. Therefore, it is possible that some redox sensitive cysteines may be inaccessible to bulky BIAM probe for steric reasons, as reported before.68
We initially expected that many redox-sensitive proteins would be identified by more than one method. However, our results showed that each method identified a very different set of proteins. The fact that only a small portion of them were identified by more than one method might be partly explained by the following examples. Assuming that protein X contains three thiols that are present in the reduced form in the control sample and the oxidant treatment causes two of the thiols to be oxidized. Such a change could be detected by the direct IAF labeling method because the protein will show a 3 fold (3:1) IAF labeling intensity in the control sample compared to that in the treated sample. In the direct BIAM tagging method, however, this protein in both the control and treated samples will be purified by the NeutrAvidin agarose resin regardless of the number of thiols tagged by BIAM. Therefore, the change in the redox state of such a protein will not be detected by the direct BIAM tagging method. On the other hand, assuming that protein Y also contains three free thiols but only one of them becomes oxidized in the oxidant-treated sample. Such a protein will not be identified by the direct IAF labeling method in our experiment as we set 2-fold difference in IAF labeling intensity as a threshold for identifying differentially labeled proteins. However, it will possibly be identified by the blocking-IAF labeling method because one thiol in protein Y from the oxidant-treated sample will be labeled with IAF but none of the thiols in protein Y from the control sample will be labeled with IAF. The above examples illustrate that the four methods we used are complementary in identifying redox-sensitive proteins.
Concluding Remarks
In summary, through the proteomics approaches, we have identified a number of Arabidopsis proteins that undergo rapid oxidative modifications in Arabidopsis suspension-cultured cells in response to a sublethal dose of H2O2 treatment. Each of the four proteomics approaches has its distinct limitations and advantages, leading to the identification of four sets of proteins with small overlaps. To evaluate the validity and physiological relevance of the proteomics approaches used, we also established two methods for detailed analysis of individual putative redox-sensitive proteins in Arabidopsis plants. One such protein, AtCIAPIN1, was also shown to be oxidized upon treatment with SA and Flg22, two inducers of defense response. The identification of oxidant-sensitive proteins will be helpful toward in-depth characterization of other signaling pathways previously unknown to be redox-regulated.
Acknowledgments
We thank Joseph Jez (Department of Biology, Washington University, St. Louis) for advice and Anita Snyder for editing the manuscript. This work was supported by Research Grants Council of Hong Kong (grant no. HKBU261910 and HKBU1/CRF/10 to Y. X.) and by the National Institutes of Health (grant no. GM076420 to Y.X.).
Supporting Information Available
Table S1. Summary of redox-sensitive proteins identified by the four proteomic methods.
Table S2. The list of identified peptides from the differentially labeled proteins.
This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Lamb C.; Dixon R. A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Delledonne M.; Zeier J.; Marocco A.; Lamb C. Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. U.S.A. 2001, 98 (23), 13454–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R.; Vanderauwera S.; Gollery M.; Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9 (10), 490–8. [DOI] [PubMed] [Google Scholar]
- Moller I. M.; Jensen P. E.; Hansson A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–81. [DOI] [PubMed] [Google Scholar]
- Apel K.; Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biol. 2004, 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Torres M. A.; Dangl J. L.; Jones J. D. G. Arabidopsis gp91(phox) homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 2002, 99 (1), 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M.; Xia Y. J.; Dixon R. A.; Lamb C. Nitric oxide functions as a signal in plant disease resistance. Nature 1998, 394 (6693), 585–588. [DOI] [PubMed] [Google Scholar]
- Durner J.; Wendehenne D.; Klessig D. F. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. U.S.A. 1998, 95 (17), 10328–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi P.; Bonetto V. Redox proteomics: Identification of oxidatively, modified proteins. Proteomics 2003, 3 (7), 1145–1153. [DOI] [PubMed] [Google Scholar]
- Maeda K.; Hägglund P.; Finnie C.; Svensson B.. Proteomics of disulfide and cysteine oxidoreduction. In Plant Proteomics; Finnie C., Ed.; Blackwell Publishing: Oxford, U.K., 2006; pp 71–97. [Google Scholar]
- Hurd T. R.; Prime T. A.; Harbour M. E.; Lilley K. S.; Murphy M. P. Detection of reactive oxygen species-sensitive thiol proteins by redox difference gel electrophoresis: implications for mitochondrial redox signaling. J. Biol. Chem. 2007, 282 (30), 22040–22051. [DOI] [PubMed] [Google Scholar]
- Leichert L. I.; Gehrke F.; Gudiseva H. V.; Blackwell T.; Ilbert M.; Walker A. K.; Strahler J. R.; Andrews P. C.; Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. U.S.A. 2008, 105 (24), 8197–8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes N.; Reichmann D.; Tienson H.; Leichert L. I.; Jakob U. Using quantitative redox proteomics to dissect the yeast redoxome. J. Biol. Chem. 2011, 10.1074/jbc.M111.296236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C.; Hu J.; Liu T.; Ago T.; Sadoshima J.; Li H. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J. Proteome Res. 2008, 7 (9), 3789–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E.; Wang C.; Simon G. M.; Richter F.; Khare S.; Dillon M. B.; Bachovchin D. A.; Mowen K.; Baker D.; Cravatt B. F. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468 (7325), 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinalducci S.; Murgiano L.; Zolla L. Redox proteomics: basic principles and future perspectives for the detection of protein oxidation in plants. J. Exp. Bot. 2008, 59 (14), 3781–3801. [DOI] [PubMed] [Google Scholar]
- Ito H.; Iwabuchi M.; Ogawa K. The sugar-metabolic enzymes aldolase and triose-phosphate isomerase are targets of glutathionylation in Arabidopsis thaliana: detection using biotinylated glutathione. Plant Cell Physiol. 2003, 44 (7), 655–660. [DOI] [PubMed] [Google Scholar]
- Dixon D. P.; Skipsey M.; Grundy N. M.; Edwards R. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 2005, 138 (4), 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.; Lee J.; Kim Y.; Bae D.; Kang K. Y.; Yoon S. C.; Lim D. Defining the plant disulfide proteome. Electrophoresis 2004, 25 (3), 532–541. [DOI] [PubMed] [Google Scholar]
- Hancock J. T.; Henson D.; Nyirenda M.; Desikan R.; Harrison J.; Lewis M.; Hughes J.; Neill S. J. Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxidez in Arabidopsis. Plant Physiol. Biochem. 2005, 43 (9), 828–835. [DOI] [PubMed] [Google Scholar]
- Alvarez S.; Zhu M.; Chen S. Proteomics of Arabidopsis redox proteins in response to methyl jasmonate. J. Proteomics 2009, 73 (1), 30–40. [DOI] [PubMed] [Google Scholar]
- Motohashi K.; Kondoh A.; Stumpp M. T.; Hisabori T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 2001, 98 (20), 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H.; Wong J. H.; Lee Y. M.; Cho M. J.; Buchanan B. B. A strategy for the identification of proteins targeted by thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 2001, 98 (8), 4794–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y.; Koller A.; del Val G.; Manieri W.; Schurmann P.; Buchanan B. B. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. U.S.A. 2003, 100 (1), 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. H.; Balmer Y.; Cai N.; Tanaka C. K.; Vensel W. H.; Hurkman W. J.; Buchanan B. B. Unraveling thioredoxin-linked metabolic processes of cereal starchy endosperm using proteomics. FEBS Lett. 2003, 547 (1–3), 151–156. [DOI] [PubMed] [Google Scholar]
- Balmer Y.; Vensel W. H.; Tanaka C. K.; Hurkman W. J.; Gelhaye E.; Rouhier N.; Jacquot J. P.; Manieri W.; Schuurmann P.; Droux M.; Buchanan B. B. Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2004, 101 (8), 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C.; Le Marechal P.; Meyer Y.; Miginiac-Maslow M.; Issakidis-Bourguet E.; Decottignies P. New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics 2004, 4 (9), 2696–2706. [DOI] [PubMed] [Google Scholar]
- Wong J. H.; Cal N.; Balmer Y.; Tanaka C. K.; Vensel W. H.; Hurkman W. J.; Buchanan B. B. Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry 2004, 65 (11), 1629–1640. [DOI] [PubMed] [Google Scholar]
- Maeda K.; Finnie C.; Svensson B. Identification of thioredoxin h-reducible disulphides in proteornes by differential labelling of cysteines: Insight into recognition and regulation of proteins in barley seeds by thioredoxin h. Proteomics 2005, 5 (6), 1634–1644. [DOI] [PubMed] [Google Scholar]
- Balmer Y.; Vensel W. H.; Cai N.; Manieri W.; Schurmann P.; Hurkman W. J.; Buchanan B. B. A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc. Natl. Acad. Sci. U.S.A. 2006, 103 (8), 2988–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhalfioui F.; Renard M.; Vensel W. H.; Wong J.; Tanaka C. K.; Hurkman W. J.; Buchanan B. B.; Montrichard F. Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiol. 2007, 144 (3), 1559–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C.; Saalbach G.; Durner J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005, 137 (3), 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas M. C.; Campostrini N.; Matte A.; Righetti P. G.; Perazzolli M.; Zolla L.; Roepstorff P.; Delledonne M. Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 2008, 8 (7), 1459–1469. [DOI] [PubMed] [Google Scholar]
- Bersani N. A.; Merwin J. R.; Lopez N. I.; Pearson G. D.; Merrill G. F. Protein electrophoretic mobility shift assay to monitor redox state of thioredoxin in cells. Methods Enzymol. 2002, 347, 317–326. [DOI] [PubMed] [Google Scholar]
- Desikan R.; Reynolds A.; Hancock J. T.; Neill S. J. Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochem. J. 1998, 330, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L. J.; Heazlewood J. L.; Herald V.; Holtzapffel R.; Day D. A.; Leaver C. J.; Millar A. H. The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 2002, 32 (6), 891–904. [DOI] [PubMed] [Google Scholar]
- Chinchilla D.; Bauer Z.; Regenass M.; Boller T.; Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 2006, 18 (2), 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Shao F.; Li Y.; Cui H.; Chen L.; Li H.; Zou Y.; Long C.; Lan L.; Chai J.; Chen S.; Tang X.; Zhou J. M.; Pseudomonas A syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 2007, 1 (3), 175–185. [DOI] [PubMed] [Google Scholar]
- Foyer C. H.; Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17 (7), 1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D.; Grant C. M. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 2003, 374, 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser M.; Wamelink M. M.; Kowald A.; Gerisch B.; Heeren G.; Struys E. A.; Klipp E.; Jakobs C.; Breitenbach M.; Lehrach H.; Krobitsch S. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 2007, 6 (4), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morigasaki S.; Shimada K.; Ikner A.; Yanagida M.; Shiozaki K. Glycolytic enzyme GAPDH promotes peroxide stress signaling through multistep phosphorelay to a MAPK cascade. Mol. Cell 2008, 30 (1), 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N.; Hara M. R.; Kornberg M. D.; Cascio M. B.; Bae B. I.; Shahani N.; Thomas B.; Dawson T. M.; Dawson V. L.; Snyder S. H.; Sawa A. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat. Cell Biol. 2008, 10 (7), 866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubekeur S.; Camougrand N.; Bunoust O.; Rigoulet M.; Guerin B. Participation of acetaldehyde dehydrogenases in ethanol and pyruvate metabolism of the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 2001, 268 (19), 5057–5065. [DOI] [PubMed] [Google Scholar]
- Wei Y.; Lin M.; Oliver D. J.; Schnable P. S. The roles of aldehyde dehydrogenases (ALDHs) in the PDH bypass of Arabidopsis. BMC Biochem. 2009, 10, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M.; Dietz K. J. The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 1997, 12 (1), 179–190. [DOI] [PubMed] [Google Scholar]
- Baier M.; Dietz K. J. Protective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis. Evidence from transgenic Arabidopsis. Plant Physiol. 1999, 119 (4), 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y.; Spoel S. H.; Pajerowska-Mukhtar K.; Mou Z.; Song J.; Wang C.; Zuo J.; Dong X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321 (5891), 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H. P.; Zhang Y.; Zeng H.; Novoa I.; Lu P. D.; Calfon M.; Sadri N.; Yun C.; Popko B.; Paules R.; Stojdl D. F.; Bell J. C.; Hettmann T.; Leiden J. M.; Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 2003, 11 (3), 619–633. [DOI] [PubMed] [Google Scholar]
- Dunand-Sauthier I.; Walker C. A.; Narasimhan J.; Pearce A. K.; Wek R. C.; Humphrey T. C. Stress-activated protein kinase pathway functions to support protein synthesis and translational adaptation in response to environmental stress in fission yeast. Eukaryot. Cell 2005, 4 (11), 1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda Y.; Kumamaru T.; Kawagoe Y. ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc. Natl. Acad. Sci. U.S.A. 2009, 106 (33), 14156–14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Zhu Y.; Gao J.; Yu F.; Dong A.; Shen W. H. Molecular and reverse genetic characterization of NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) genes unravels their function in transcription and nucleotide excision repair in Arabidopsis thaliana. Plant J. 2009, 59 (1), 27–38. [DOI] [PubMed] [Google Scholar]
- Xu Q.; Fu H. H.; Gupta R.; Luan S. Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell 1998, 10 (5), 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. F.; Li H.; Gupta R.; Morris P. C.; Luan S.; Kieber J. J. ATMPK4, an arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol. 2000, 122 (4), 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.; Luan S. Redox control of protein tyrosine phosphatases and mitogen-activated protein kinases in plants. Plant Physiol. 2003, 132 (3), 1149–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeen A.; Andersen J. N.; Myers M. P.; Meng T. C.; Hinks J. A.; Tonks N. K.; Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 2003, 423 (6941), 769–773. [DOI] [PubMed] [Google Scholar]
- van Montfort R. L. M.; Congreve M.; Tisi D.; Carr R.; Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature 2003, 423 (6941), 773–777. [DOI] [PubMed] [Google Scholar]
- Kovtun Y.; Chiu W. L.; Tena G.; Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. U.S.A. 2000, 97 (6), 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I.; Rossi R.; Milzani A.; Di Simplicio P.; Colombo R. The actin cytoskeleton response to oxidants: From small heat shock protein phosphorylation to changes in the redox state of actin itself. Free Radic. Biol. Med. 2001, 31 (12), 1624–1632. [DOI] [PubMed] [Google Scholar]
- Lassing I.; Schmitzberger F.; Bjornstedt M.; Holmgren A.; Nordlund P.; Schutt C. E.; Lindberg U. Molecular and structural basis for redox regulation of beta-actin. J. Mol. Biol. 2007, 370 (2), 331–348. [DOI] [PubMed] [Google Scholar]
- Farah M. E.; Amberg D. C. Conserved actin cysteine residues are oxidative stress sensors that can regulate cell death in yeast. Mol. Biol. Cell 2007, 18 (4), 1359–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibayama H.; Takai E.; Matsumura I.; Kouno M.; Morii E.; Kitamura Y.; Takeda J.; Kanakura Y. Identification of a cytokine-induced antiapoptotic molecule anamorsin essential for definitive hematopoiesis. J. Exp. Med. 2004, 199 (4), 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernis L.; Facca C.; Delagoutte E.; Soler N.; Chanet R.; Guiard B.; Faye G.; Baldacci G. A newly identified essential complex, Dre2-Tah18, controls mitochondria integrity and cell death after oxidative stress in yeast. PLoS One 2009, 4 (2), e4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz D. J.; Stumpfig M.; Dore C.; Muhlenhoff U.; Pierik A. J.; Lill R. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 2010, 6 (10), 758–765. [DOI] [PubMed] [Google Scholar]
- Varadarajan J.; Guilleminot J.; Saint-Jore-Dupas C.; Piegu B.; Chaboute M. E.; Gomord V.; Coolbaugh R. C.; Devic M.; Delorme V. ATR3 encodes a diflavin reductase essential for Arabidopsis embryo development. New Phytol. 2010, 187 (1), 67–82. [DOI] [PubMed] [Google Scholar]
- Rao M. V.; Paliyath G.; Ormrod D. P.; Murr D. P.; Watkins C. B. Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes. Salicylic acid-mediated oxidative damage requires H2O2. Plant Physiol. 1997, 115 (1), 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R.; Denoux C.; Gambetta S.; Dewdney J.; Ausubel F. M.; De Lorenzo G.; Ferrari S. The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 2008, 148 (3), 1695–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. R.; Yoon H. W.; Kwon K. S.; Lee S. R.; Rhee S. G. Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal. Biochem. 2000, 283 (2), 214–221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.