Abstract
Aldehyde- and NHS-activated magnetic microspheres were used to immobilize trypsin (CHO-trypsin and NHS-trypsin), and their performance for protein digestion was evaluated by reversed phase liquid chromatography-electrospray ionization-tandem mass spectrometry using an LTQ Orbitrap Velos instrument. NHS-trypsin provided greater sequence coverage and identified more peptides for the digestion of bovine serum albumin. A one-minute digestion at room temperature using the immobilized trypsin also identified more peptides (96 ± 6 vs. 48 ± 1) and produced higher sequence coverage (90 ± 2% vs. 75 ± 2%) than traditional free trypsin digestion for 12 hours at 37 °C. Analysis of 15 nM (0.001 mg/mL) BSA digested by NHS-trypsin in 1 min. at room temperature consistently yielded one detected peptide; 150 nM BSA generated 22 peptides. Peptide intensity and protein spectral count were used to evaluate the run-to-run digestion reproducibility of NHS-trypsin with a three-protein-mixture. Three high intensity peptides for each protein generated intensity ratios from 0.70 to 1.09 and spectral count ratios from 0.78 to 1.18. Finally, RAW 264.7 cell lysates were digested by NHS-trypsin for 10 min. and 30 min. at room temperature; 604 and 697 protein groups, respectively, were identified by RPLC-ESI-MS/MS, with a peptide false discovery rate of less than 1%. Digestion by solution phase trypsin for 12 hours at 37 °C resulted in identification of 878 protein groups.
Keywords: protein digestion, trypsin-immobilized magnetic microspheres, reproducibility, RPLC-ESI-MS/MS
1. Introduction
Bottom-up sequencing is widely used for protein identification and posttranslational modification analysis [1]. In this approach, proteins are first digested with an enzyme into peptides, and then analyzed by mass spectrometry [2, 3].
Trypsin is a widely used enzyme for protein digestion due to its high cleavage specificity. Immobilized trypsin provides advantages in protein digestion. Auto-digestion is reduced and the effective trypsin concentration is increased over typical solution digestion conditions, which can result in significantly shorter digestion times [4]. Several solid supports have been used for proteolytic enzyme immobilization, including monolithic materials [5–15], polymer particles [16], and magnetic particles [17–21]. Magnetic particles are attractive due to the simplicity of their capture and replacement. Zhang’s group synthesized aldehyde-functionalized magnetic particles for trypsin immobilization and achieved complete protein digestion within 5 min. in an Eppendorf tube [17]. They further packed the trypsin-immobilized magnetic particles into a microchip channel using a magnet, and applied the microreactor for digestion of standard proteins [18]. Complete protein digestion was achieved in 10 s with the microreactor. Our group also prepared NHS-activated magnetic particles for trypsin immobilization [21]. The beads were packed into a capillary to form a microreactor for on-line protein digestion. Two model proteins, insulin chain b oxidized and α-casein, were digested in 1 min. in a two-dimensional capillary electrophoresis system. Commercial immobilized trypsin beads or column have also been used for protein quantitation [22–25].
Several issues remain in the use of these reagents. First, different functional groups have been used for immobilization, including aldehyde [6,7,10,17], epoxy [19], and succinimide [8,9,12,21]. However, relatively few publications have appeared that compare these immobilization chemistries. Veuthey compared epoxy, carbonyldiimidazole (CDI), and ethylenediamine (EDA) Convective Interaction Media® (CIM) monolithic disks for trypsin immobilization; immobilization on CIM® EDA disk previously derivatized with glutaraldehyde was the most efficient for digestion of standard proteins [26]. Hugon-Chapuis used CNBr-activated Sepharose, NHS-activated Sepharose, and glutaraldehyde-activated silica to immobilize trypsin for digestion of standard peptide and protein digestion; CNBr-activated Sepharose produced the best performance [27]. However, the surface functional group density of the materials is not mentioned, which affects the immobilized amount of trypsin.
Second, for analysis of standard proteins, higher sequence coverage and more peptides are sometimes obtained with immobilized trypsin than with free trypsin [4,10,12,17]. This phenomenon is not well understood.
Third, two parameters, sequence coverage and number of peptides, are commonly used for evaluating reproducibility of protein digestion with immobilized trypsin. Unfortunately, these parameters are not particularly useful for determining the precision of protein quantitation. Several groups have applied commercial immobilized trypsin beads or column to 18O-labeling based protein quantitation, and their results showed high peptide labeling efficiency and good reproducibility [22–24].
2. Materials and methods
2.1 Materials and Chemicals
Bovine pancreas TPCK-treated trypsin, bovine serum albumin, bovine heart cytochrome c, equine myoglobin, urea, ammonium bicarbonate, dithiothreitol (DTT), iodoacetamide (IAA), N-hydroxysulfosuccinimide sodium salt (NHS), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), 2-morpholinoethanesulfonic acid monohydrate (MES monohydrate), sodium hydroxide, glutaraldehyde (25% in water), sodium cyanoborohydride (NaCNBH3), potassium phosphate dibasic trihydrate, potassium phosphate monobasic, benzamidine, and ethanolamine were purchased from Sigma–Aldrich (St. Louis, MO, USA). EnzChek® peptidase/protease assay kit (E33758) was purchased from Invitrogen (Grand Island, NY, USA). Acetonitrile (ACN) and formic acid (FA) were purchased from Fisher Scientific (Pittsburgh, PA, USA). Water was deionized by a Nano Pure system from Thermo scientific (Marietta, OH, USA).
Carboxyl functionalized magnetic microspheres (BioMag® Plus carboxyl, mean diameter ~1.5 μm, surface titration, ~240 μmol/g) were purchased from Bangs Laboratories, Inc. (Fishers, IN, USA). Amine functionalized magnetic microspheres (diameter ~1 μm, surface functional group density ~250 μmol/g) were purchased from Bioclone Inc. (San Diego, CA, USA). A magnet was purchased on-line from http://www.supermagnetman.net. ZipTip C18 (ZTC18S096) was purchased from Millipore (Bedford, MA, USA).
Dulbecco’s Modified Eagle’s Medium (DMEM) with L-glutamine and fetal bovine serum (FBS) were purchased from ATCC (Manassas, VA, USA). Mammalian Cell-PE LB™ Buffer used for cell lysis was purchased from G-Biosciences (St. Louis, MO, USA). Complete, mini protease inhibitor cocktail (provided in EASYpacks) was purchased from Roche (Indianapolis, IN, USA).
2.2 Preparation of magnetic microspheres based immobilized trypsin
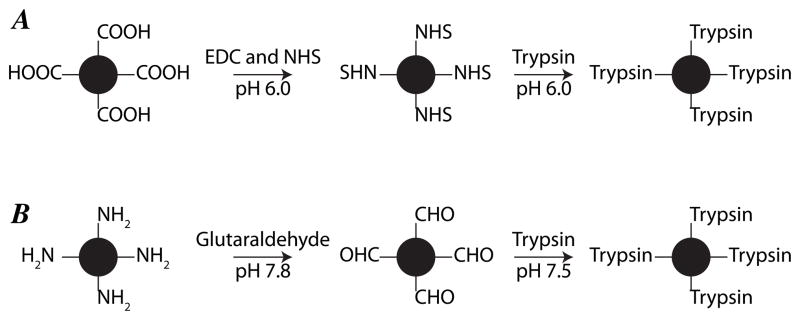
The procedure for activation of carboxyl functionalized magnetic microspheres and trypsin immobilization was similar to our previous work, Figure 1A [21]. Carboxyl functionalized magnetic microspheres (3 mg) were first washed with MES buffer (pH 6.0). Then, the carboxyl groups were activated with 200 μL EDC (50 mg/mL in 50 mM MES buffer, pH 6.0) and 200 μL NHS (50 mg/mL in 50 mM MES buffer, pH 6.0) for 30 min at room temperature with gentle rotation. The reaction solution was removed while the beads were held with a magnet, and the NHS-activated magnetic microspheres were washed four times with 1 mL MES buffer (pH 6.0). Subsequently, 360 μL trypsin solution (2.5 mg/mL, dissolved in 50 mM MES buffer, pH 6.0) containing 50 mM benzamidine was mixed with the activated and washed microspheres, and the reaction was incubated at 4°C for 24 h with gentle rotation. The immobilized trypsin microspheres were washed with MES buffer (pH 6.0) and 20% acetonitrile to remove the adsorbed trypsin. The unreacted active groups on the microspheres were blocked with 400 μL ethanolamine solution (0.4 M, pH 8.4) at 4 °C for 3 h with gentle rotation. After washing three times with 1 mL of 20 mM ammonium bicarbonate (pH 8.0) buffer, the NHS-activated immobilized trypsin (NHS-trypsin) magnetic microspheres were stored in 600 μL 20 mM ammonium bicarbonate (pH 8.0) at 4 °C.
Figure 1.
Procedure for preparation of NHS-activated magnetic microsphere immobilized trypsin (NHS-trypsin) (A) and aldehyde-activated magnetic microsphere immobilized trypsin (CHO-trypsin) (B).
The procedure for activation of amine functionalized magnetic microspheres and trypsin immobilization was similar to that reported by the Zhang group, Figure 1B [17]. First, amine functionalized magnetic microspheres (3 mg) were washed with phosphate buffer (pH 7.8). Second, the amine groups were activated with 400 μL 5% glutaraldehyde solution in 50 mM phosphate buffer (pH 7.8) at room temperature for 3 h with gentle rotation. The aldehyde-activated magnetic microspheres were washed four times with 1 mL 50 mM phosphate buffer (pH 7.8). Third, 360 μL trypsin solution (2.5 mg/mL, dissolved in 50 mM phosphate buffer, pH 7.5) containing 50 mM benzamidine and 7 mg/mL NaCNBH3 was mixed with the activated and washed microspheres, and the reaction was left at 4°C for 24 h with gentle rotation. Fourth, phosphate buffer and 20% acetonitrile solution were used to wash the immobilized trypsin microspheres to remove the adsorbed trypsin. Fifth, 400 μL ethanolamine solution (0.4 M, pH 8.4) was used to block the unreacted active group on the microspheres at 4 °C for 3 h with gentle rotation. After washing three times with 1 mL 20 mM ammonium bicarbonate (pH 8.0), the prepared aldehyde-activated immobilized trypsin (CHO-trypsin) magnetic microspheres were stored in 600 μL 20 mM ammonium bicarbonate (pH 8.0) at 4 °C.
After immobilization, the bicinchoninic acid (BCA) method was used to estimate the amount of trypsin bound to the magnetic microspheres [28].
2.3 Sample preparation
Bovine serum albumin (BSA, 1 mg/mL) dissolved in 100 mM ammonium bicarbonate (pH 8.0) was denatured at 90 °C for 15 min, followed by standard reduction and alkylation with DTT and IAA. The resulting protein solution (~1 mg/mL) was stored at −20 °C before use.
Cytochrome c (5 mg/mL) and myoglobin (5 mg/mL) dissolved in 8 M urea and 100 mM ammonium bicarbonate (pH 8.0) were denatured at 52 °C for 1 h, followed by standard reduction and alkylation with DTT and IAA. The resulting protein solutions were diluted with 100 mM ammonium bicarbonate to 0.8 mg/mL with urea concentration at ~1.3 M, and stored at −20 °C.
RAW 264.7 cells were cultured in a T25 flask at 37 °C and 5% CO2 in DMEM with L-glutamine and 10% FBS. The flask was washed with cold PBS buffer twice. Then, 1 mL mammalian cell-PE LB™ buffer (pH 7.5) supplemented with complete protease inhibitor was added to the flask, and the flask was shaken gently for 10 min on ice. The cell lysate was transfer to a 1.5 mL centrifuging tube and incubated on ice for 15 min. Subsequently, the cell lysate was centrifuged at 18,000 g for 15 min, and the supernatant was collected for measurement of protein concentration with the BCA method. After that, 300 μL cell lysate (~54 μg proteins) was denatured at 90 °C for 20 min, followed by reduction with DTT (3.3 mM) at 65 °C for 1 h and alkylation with IAA (8.3 mM) at room temperature for 30 min in dark. Then, 1.2 mL cold acetone was added to the protein solution and incubated at −20 °C for 12 h, followed by centrifugation at 18,000 g for 15 min. The protein pellet was washed with cold acetone again, and dried at room temperature. Finally, the protein pellet was redissolved in 100 μL 1 M urea and 100 mM ammonium bicarbonate buffer (pH 8.0), and stored at −20 °C.
2.4 Protein digestion
The BSA solution (1 mg/mL) was diluted to 0.5, 0.1, 0.01, and 0.001 mg/mL with 100 mM ammonium bicarbonate buffer (pH 8.0). For digestion using immobilized trypsin, 1 mg/mL (20 μL), 0.5 mg/mL (20 μL), 0.1 mg/mL (50 μL), 0.01 mg/mL (100 μL) and 0.001 mg/mL (100 μL) BSA solution was added to 200 μg immobilized trypsin magnetic microspheres. Digestion was performed at room temperature for 1 min. under rotation. For 0.5 mg/mL (20 μL) BSA solution, the digestion was also performed for 5 and 10 min. After digestion, the digests from 1 and 0.5 mg/mL BSA were diluted to 0.1 mg/mL with 2% ACN and 0.1% FA solution, followed by RPLC-ESI-MS/MS analysis. The digests from 0.1 mg/mL BSA were directly analyzed by RPLC-ESI-MS/MS. The digests from 0.01 mg/mL and 0.001 mg/mL BSA were first dried with a concentrator (Eppendorf, Hamburg, Germany), redissolved in 15 μL 2% ACN and 0.1% FA solution, desalted with ZipTip C18, dried again, and redissolved in 10 μL 2% ACN and 0.1% FA solution for RPLC-ESI-MS/MS analysis. For traditional free trypsin (in-solution) digestion, 0.5 mg/mL, 0.01 mg/mL (100 μL) and 0.001 mg/mL (100 μL) BSA solutions were digested at 37 °C for 12 h with trypsin and protein ratio of 1:40 (w/w). After digestion, the digests were acidified with FA (final concentration 0.5% (v/v)). Then, the digests were treated with the same procedures as those for immobilized trypsin digestion.
The BSA, cytochrome c, and myoglobin solutions were mixed to form a three-protein-mixture (BSA, 0.25 mg/mL; cytochrome c, 0.1 mg/mL; myoglobin, 0.05 mg/mL). A 20-μL aliquot of the three-protein-mixture was added to 300 μg immobilized trypsin magnetic microspheres (NHS-trypsin). The digestion was performed at room temperature for 1 min. under rotation. The digests were then diluted to 0.1 mg/mL with 2% ACN and 0.1% FA solution, followed by RPLC-ESI-MS/MS analysis.
Forty microliters of the protein solution from the RAW 264.7 cells was added to 400 μg immobilized trypsin magnetic microspheres (NHS-trypsin), and digestion was performed at room temperature for 10 min. under rotation. Another 20 μL of the protein solution from the RAW 264.7 cells was also digested with 400 μg NHS-trypsin microspheres at room temperature for 30 min. under rotation. The two digests were first desalted with ZipTip C18, then dried and redissolved in 40 μL and 20 μL 2% ACN, respectively, and 0.1% FA solution for RPLC-ESI-MS/MS analysis. For traditional free trypsin digestion, 40 μL of protein solution from RAW 264.7 cells was digested at 37 °C for 12 h with trypsin at a trypsin protein ratio of 1:30 (w/w). The digests were acidified with FA at a final concentration of 0.5% (v/v). The digests were then desalted, dried, and redissolved in 40 μL 2% ACN and 0.1% FA for RPLC-ESI-MS/MS analysis.
2.5 RPLC-ESI-MS/MS analysis
A nanoACQUITY UltraPerformance LC® (UPLC®) system (Waters) was use for separation of the protein digests. Buffer A (0.1% FA in water) and buffer B (0.1% FA in ACN) were used as mobile phases for gradient separation. Protein digests were automatically loaded onto a commercial C18 reversed phase column (100 μm×100 mm, 1.7 μm particle, BEH130C18, column temperature 40 °C) with 1% buffer B for 5 min. at a flow rate of 1.2 μL/min, followed by 3-step gradient separation. For standard protein digests analysis, the gradient was 2 min. from 1% B to 10% B, 33 min. to 40% B, 2 min. to 85% B, and maintained for 8 min. For RAW 264.7 cell lysate digests analysis, the gradient was 2 min. from 1% B to 10% B, 60 min. to 40% B, 1 min. to 85% B, and maintained for 7 min. The column was equilibrated for 14 min. with 1% buffer B prior to the next sample analysis. The eluted peptides from the C18 column were pumped through a manually pulled capillary tip for electrospray, and analyzed by LTQ-Orbitrap Velos instrument (Thermo Fisher Scientific). The electrospray voltage was 1.75 kV, and the ion transfer tube temperature was 300 °C. The mass spectrometer was programmed in a data dependent mode. Full MS scans were acquired in the Orbitrap mass analyzer over 395–1900 m/z range with resolution 60,000. Ten most intense peaks (for standard protein digests analysis) and 20 most intense peaks with charge state ≥ 2 (for cell lysate digests analysis) were selected for sequencing and fragmented in the ion trap with normalized collision energy of 35%, activation q = 0.25, activation time of 10 ms, and one microscan. For all sequencing events except the spectral count experiment, dynamic exclusion was enabled. For standard protein digest analysis, peaks selected for fragmentation more than once within 30 s were excluded from selection for 180 s. For cell lysate digests analysis, peaks selected for fragmentation more than once within 45 s were excluded from selection for 45 s. For standard proteins analysis, 1 μL digests were injected for each run, and each sample was analyzed in triplicate runs. For cell lysate analysis, 2 μL digests were used for each run, and each sample was analyzed in quadruplicate.
2.6 Data processing and analysis
Database searching of raw files was performed in Proteome Discoverer 1.2 with the SEQUEST search engine against ipi.bovin.v3.68.fasta (for BSA and cytochrome c), equine.fasta (for myoglobin), and ipi.mouse.v3.85.fasta (for RAW 264.7 cell lysate). Database searching against the corresponding decoy database was also performed to evaluate the false discovery rate (FDR) of peptide identification. The database searching parameters included up to two missed cleavages allowed for full tryptic digestion, precursor mass tolerance 50 ppm, fragment mass tolerance 1.0 Da, cysteine carbamidomethylation as a fixed modification, and oxidation of methionine as variable modification. For cytochrome c and myoglobin, acetylation of lysine was also set as variable modification.
For standard proteins analysis, peptides identified with confidence value of high were considered as a positive identification. For cell lysate analysis, on peptide level, peptide confidence value was set as high and the FDR was less than 1%; on protein level, peptides per protein was set as 1, count only rank 1 peptides, and count peptide only in top scored proteins. Protein grouping was enabled for standard proteins and cell lysate analysis.
The intensity of peptides used in this work was extracted manually from the raw files with Xcalibur software. The mass tolerance was 20 ppm. Spectral count of proteins used in this work was obtained directly from database searching results in Proteome Discoverer 1.2.
3 Results and discussion
3.1 Comparison of NHS-trypsin and CHO-trypsin
NHS-trypsin was prepared from carboxyl-functionalized magnetic microspheres using EDC and NHS activation, Figure 1A, and CHO-trypsin was prepared from amine-terminated magnetic microspheres using glutaraldehyde activation, Figure 1B. The carboxyl and amine-functionalized magnetic microspheres had similar functional group density (~240 vs. ~250 μmol/g), and the amount of immobilized trypsin was also similar (86 μg/mg and 88 μg/mg microsphere, respectively).
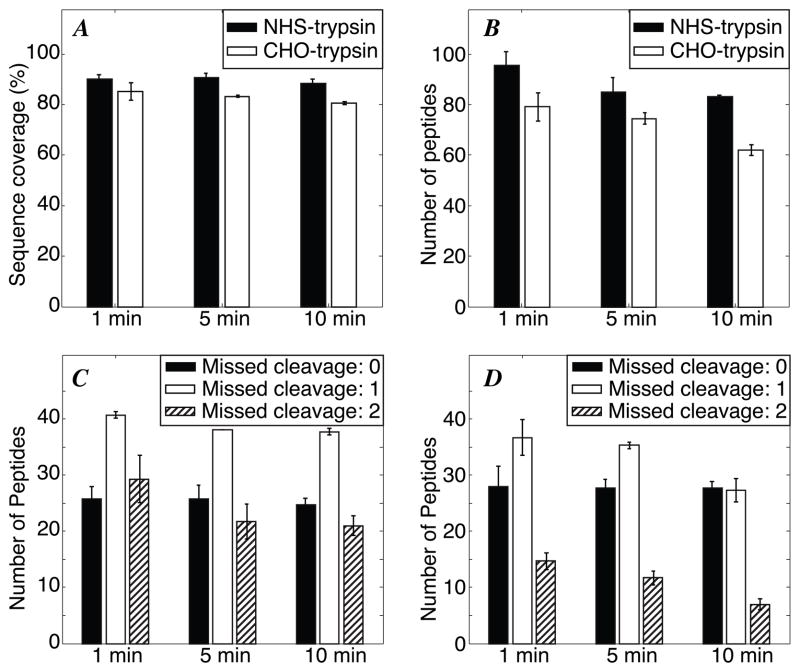
Both types of microspheres were used to digest 0.5 mg/mL BSA for 1, 5 and 10 min at room temperature. The NHS-trypsin beads generated ~8% greater sequence coverage and ~25% more peptide IDs than the CHO-trypsin beads, Figure 2A–B.
Figure 2.
Kinetic analysis of 0.5 mg/mL BSA digest (in triplicate). A - Sequence coverage. B - number of peptides. C - missed cleavage distribution with NHS-trypsin. D – missed cleavage distribution with CHO-trypsin digestion. Error bars are ± one standard deviation of the measurement.
Interestingly, both sequence coverage and the number of peptide IDs decreased slightly with longer digestion time. This decrease in coverage was associated with more complete digestion, which generated fewer peptides with one or two missed cleavages, Figure 2C–D. The resulting peptides are, on average, shorter with longer digestion time, leading to the modest decrease in coverage at longer digestion times. For the same digestion time, CHO-trypsin generates fewer peptides with one or two missed cleavages and more peptides with no missed cleavage. In order to understand the phenomenon more clearly, an EnzChek® peptidase/protease assay kit was used to compare the activity of NHS-trypsin and CHO-trypsin. Equal amounts of NHS-trypsin and CHO-trypsin microspheres (40 μL) were mixed with 40 μL of EnzChek® peptidase/protease substrate. After digestion for ~1 min at room temperature, the fluorescent signal from the digests was measured. The fluorescent signal intensity from digests with NHS-trypsin and CHO-trypsin was 2200 and 4800 RFU, respectively. The results indicate that CHO-trypsin is more efficient in cleaving the substrate, which is consistent with the missed cleavages results mentioned above. Improved digestion may be due to the longer spacer (glutaraldehyde) between trypsin and the microspheres, which should provide easier access to the trypsin active site.
Non-specific adsorption of peptides on the NHS-trypsin and CHO-trypsin microspheres was also evaluated, S-Figure 1 in supporting material. The details of the experiment are also presented in the supporting material. Many peptide peaks were obtained from 0.1 mg/mL BSA digests in the m/z range 800–4000 with NHS-trypsin and CHO-trypsin digestion, and the signal intensity was about 1.0×104, S-Figure 1 A and C. Several peptide peaks were obtained from the eluate of NHS-trypsin microspheres with 20%ACN and 0.1%FA, and the signal intensity was only about 300, S-Figure 1B, which demonstrates that the amount of peptides adsorbed on the NHS-trypsin microspheres is very small, about 1% of total BSA digests (calculation by ratio of peptide intensity). More peptide peaks and higher signal intensity were obtained from the eluate of CHO-trypsin microspheres with 20%ACN and 0.1%FA, S-Figure 1 D, which indicates that the non-specific adsorption of peptides on the CHO-trypsin microspheres is stronger than that on NHS-trypsin microspheres. The higher nonspecific binding from the CHO-trypsin microspheres may be due to the longer spacer (glutaraldehyde) on the CHO-trypsin microspheres, which makes the peptides easier to adsorb on the microspheres by hydrophobic interaction.
The toxicity of reagents used in the preparation of NHS-trypsin and CHO-trypsin should also be considered. Glutaraldehyde and NaCNBH3 were used in the preparation of CHO-trypsin. EDC and NHS were used in NHS-trypsin preparation. The toxicity of these reagents were obtained from their materials safety data sheet (MSDS) provided by Sigma–Aldrich. Glutaraldehyde is respiratory sensitizer, skin sensitizer, is toxic by inhalation, and corrosive. NaCNBH3 is a flammable solid, is corrosive, and is highly toxic by inhalation and skin absorption. Signal word for these two reagents is “Danger”. For NHS, there are no known OSHA hazards. EDC is an irritant, and its signal word is “Danger”. Therefore, the reagents used in the preparation of NHS-trypsin are of lower toxicity than those used for CHO-trypsin. Based on these safety issues, higher non-specific adsorption of peptides, and its lower sequence coverage, we eliminated the CHO-trypsin from further study.
3.2 Comparison between NHS-trypsin and free trypsin digestion
NHS-trypsin digestion (1 min. room temperature) was also compared with free trypsin digestion (12 h, 37 °C) of BSA for samples ranging from 0.001 to 0.5 mg/mL BSA, Table 1. We consistently detected one peptide (2% coverage) at a BSA concentration of 0.001 mg/mL (15 nM) for the NHS-trypsin digestion. The coverage increased dramatically at higher concentrations, reaching 90% coverage for BSA concentration of 0.5 mg/mL. In contrast, the free trypsin digestion produced significantly poorer coverage at all concentrations studied. Digestion with immobilized trypsin for 1 min at room temperature was superior to digestion with solution-phase trypsin at all substrate concentrations.
Table 1.
calibration curve data
| [BSA] | 0.001 mg/mL | 0.01 mg/mL | 0.1 mg/mL | 0.5 mg/mL | 1 mg/mL |
|---|---|---|---|---|---|
| NHS-trypsin (1 min @ 20°C) | 2% coverage, 1 peptide | 42 ± 5% coverage, 22 ± 4 peptides | 92 ± 0.4% coverage, 98 ± 2 peptides | 90 ± 2% coverage, 96 ± 6 peptides | 91 ± 2% coverage, 88 ± 6 peptides |
| solution trypsin (12 hr @ 37 °C) | 0% coverage, 0 peptide | 26 ± 3% coverage, 14 ± 2 peptides | n/d | 75 ± 2% coverage, 48 ± 1 peptides | n/d |
n/d = not determined.
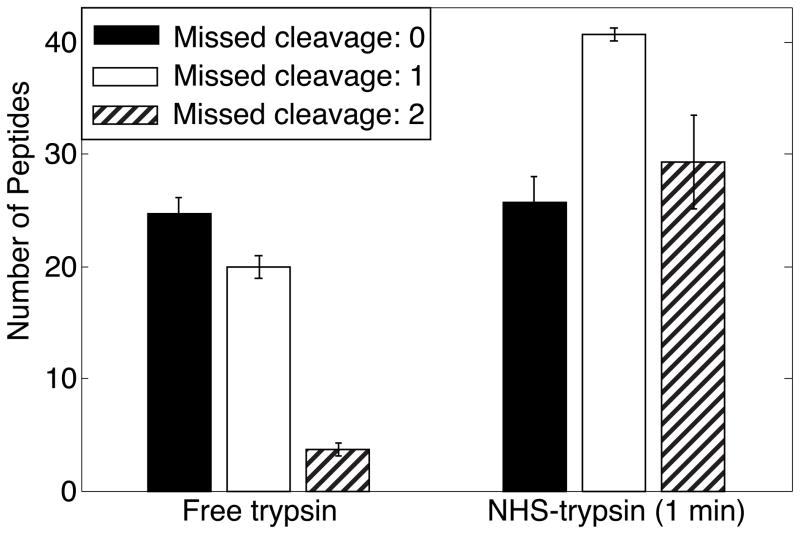
NHS-trypsin digestion generated more peptides with 1 and 2 missed cleavages than that free trypsin digestion, and the number of peptides with 0 missed cleavages was comparable, Figure 3. The fewer missed cleavages of free trypsin digestion leads to fewer peptides in the digests, resulting in fewer peptides and lower sequence coverage.
Figure 3.
Missed cleavage distribution of 0.5 mg/mL BSA digested with free trypsin (37 °C for 12 hr) and NHS-trypsin (room temperature for 1 min). Analysis by RPLC-ESI-MS/MS (in triplicate). Error bars are ± one standard deviation of the measurement.
3.3 Reproducibility of NHS-trypsin preparation and digestion
Three batches of NHS-trypsin beads were prepared and the amount of immobilized trypsin was determined to be 86 ± 7 μg/mg microspheres. The reproducibility of NHS-trypsin digestion was evaluated by digesting a mixture of three proteins (BSA, 0.25 mg/mL; cytochrome C, 0.1 mg/mL; myoglobin, 0.05 mg/mL). The run-to-run reproducibility was determined by digesting the protein mixture using three aliquots of one batch of NHS-trypsin; three aliquots from each digestion were analyzed by RPLC-ESI-MS/MS.
Batch-to-batch reproducibility was determined by using aliquots from three batches for digestion; each aliquot was analyzed by three times RPLC-ESI-MS/MS. The number of peptides and sequence coverage of the three proteins are presented in S-Table 1 and S-Table 2 in supporting material. For run-to-run digestion (S-Table 1), the RSD of the number of peptides was better than 10% and the RSD for the sequence coverage was better than 4%. For batch-to-batch digestion (S-Table 2), the RSD of number of peptides and the sequence coverage of the three proteins were better than 6%.
Peptide intensity was used for the quantitative evaluation of run-to-run and batch-to-batch digestion reproducibility. Three peptides with relatively high intensities were chosen for each protein, and their intensities were extracted from the raw files with Xcalibur software manually with mass tolerance 20 ppm. The extracted spectra of these peptides are shown in S-Figure 2 in supporting material. The peptide intensity from triplicates analysis was averaged and the mean was used for calculation of ratio between runs or batches. The ratios of peptide intensity between runs and batches are shown in Table 2. The peptide intensity ratios between runs range from 0.70 to 1.09, and the ratios between batches range from 0.57 to 1.24. Considering the difference of peptide intensity from RPLC-ESI-MS/MS analysis itself, the results indicate that the run-to-run digestion with NHS-trypsin is reasonably reproducible.
Table 2.
Peptide intensity ratios of cytochrome c, bovine serum albumin and myoglobin obtained with NHS-trypsin run-to-run and batch-to-batch digestion after RPLC-ESI-MS/MS analysis in triplicates.
| Sequence | Charge | m/z | Ratio of intensity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Run2/Run1 | Run3/Run2 | Run3/Run1 | Batch2/Batch1 | Batch3/Batch2 | Batch3/Batch1 | ||||
| Cytochrome c | GEREDLIAYLKK | 3 | 478.9374 | 0.92 | 0.91 | 0.84 | 0.94 | 0.81 | 0.76 |
| TGQAPGFSYTDANK | 2 | 728.8392 | 1.04 | 0.86 | 0.90 | 1.17 | 0.91 | 1.06 | |
| TGPNLHGLFGR | 2 | 584.8159 | 1.09 | 0.86 | 0.94 | 1.24 | 0.84 | 1.04 | |
| Bovine serum albumin | SLHTLFGDELcK | 3 | 473.9035 | 0.92 | 0.83 | 0.77 | 0.91 | 0.68 | 0.62 |
| KVPQVSTPTLVEVSR | 3 | 547.3178 | 0.90 | 0.83 | 0.75 | 0.95 | 0.70 | 0.67 | |
| YIcDNQDTISSK | 2 | 722.3263 | 0.99 | 0.78 | 0.77 | 1.16 | 0.57 | 0.66 | |
| Myoglobin | ALELFRNDIAAK | 2 | 680.8832 | 0.71 | 0.99 | 0.70 | 1.06 | 0.84 | 0.90 |
| YKELGFQG | 2 | 471.2407 | 0.81 | 0.93 | 0.75 | 1.02 | 0.98 | 1.00 | |
| HLKTEAEMK | 2 | 543.7848 | 0.98 | 0.71 | 0.70 | 1.18 | 0.80 | 0.94 | |
To confirm the quantitative reproducibility of run-to-run digestion with NHS-trypsin, the spectra count [29] was also determined, Table 3. Spectra count is defined as the total number of spectra identified from a protein. The three-protein-mixture (BSA, 0.25 mg/mL; cytochrome c, 0.1mg/mL; myoglobin, 0.05 mg/mL) was used as the sample. The RPLC-ESI-MS/MS analysis of the run-to-run digests was performed in triplicate with dynamic exclusion turned off. The ratios of spectra count between runs range from 0.78 to 1.18, which also indicates that the run-to-run digestion with NHS-trypsin is quantitatively reproducible.
Table 3.
Spectra count of bovine serum albumin, cytochrome c and myoglobin obtained with NHS-trypsin run-to-run digestion after RPLC-ESI-MS/MS analysis in triplicates
| Spectral count | ||||||
|---|---|---|---|---|---|---|
| Bovine serum albumin | Average | Cytochrome c | Average | Myoglobin | Average | |
| Run1 | 672 | 695.00 | 188 | 193.33 | 58 | 59.33 |
| 727 | 194 | 61 | ||||
| 686 | 198 | 59 | ||||
| Run2 | 682 | 679.67 | 182 | 180.00 | 76 | 70.00 |
| 679 | 181 | 68 | ||||
| 678 | 177 | 66 | ||||
| Run3 | 558 | 543.00 | 161 | 155.67 | 52 | 56.00 |
| 570 | 161 | 55 | ||||
| 501 | 145 | 61 | ||||
| Run2/Run1 | 0.98 | 0.93 | 1.18 | |||
| Run3/Run2 | 0.80 | 0.86 | 0.80 | |||
| Run3/Run1 | 0.78 | 0.81 | 0.94 | |||
3.4 Storage of NHS-trypsin
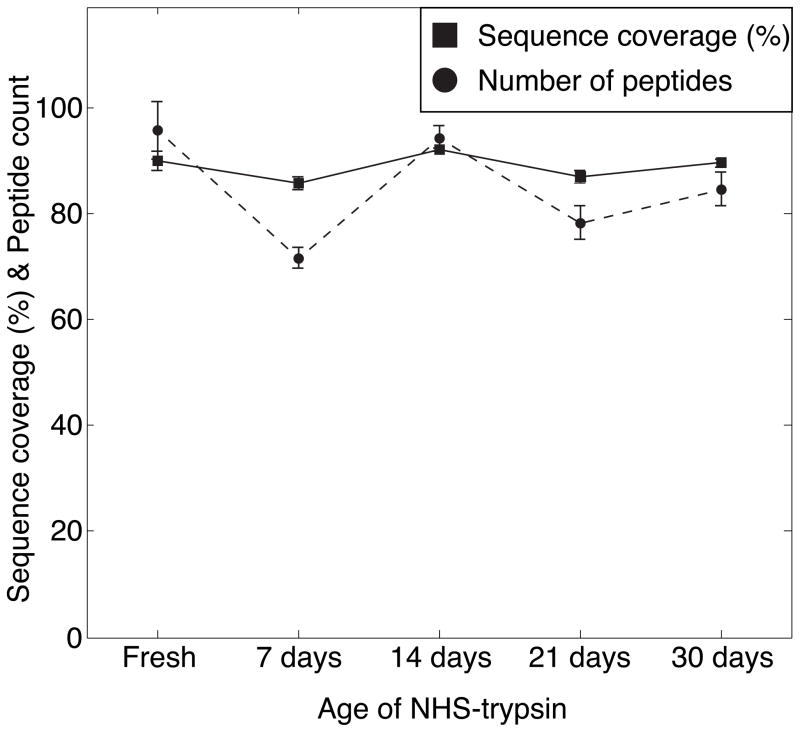
The effect of storage on the activity of NHS-trypsin was also investigated for digestion of 0.5 mg/mL BSA. The digests were diluted to 0.1 mg/mL with 2% ACN and 0.1% FA solution, followed by RPLC-ESI-MS/MS analysis in triplicate. After SEQUEST database searching the data were filtered with peptide confidence value as high in Proteome Discoverer 1.2. The sequence coverage and number of peptides of BSA were analyzed, Figure 4. The sequence coverage and number of peptides are comparable, which demonstrates that the NHS-trypsin can be stored at 4 °C for at least 1 month without loss of activity.
Figure 4.
Sequence coverage and number of peptides of 0.5 mg/mL BSA digested by fresh NHS-trypsin and that stored at 4 °C for 7 days, 14 days, 21 days and 30 days. Error bars are ± one standard deviation of the triplicate measurement.
3.5 Application of NHS-trypsin for proteome analysis of RAW 264.7 cells
To evaluate the potential of NHS-trypsin for high-throughput and large-scale proteomics analysis, NHS-trypsin was used for digestion of a cell lysate prepared from RAW 264.7 cells. Digestion using NHS-trypsin was performed at room temperature for 10 min. and 30 min. The same sample was also treated with free trypsin (in-solution) digestion (37 °C, 12 h). The digests were analyzed by RPLC-ESI-MS/MS in quadruplicate, followed by SEQUEST database searching and results control in Proteome Discoverer 1.2. Combination of the results from the quadruplicate runs 604, 697 and 878 protein groups were identified by NHS-trypsin (10 min.), NHS-trypsin (30 min.), and free trypsin digestion, respectively, with FDR of peptide identification less than 1%.
The number of protein groups identified with NHS-trypsin digestion is about 30% (10 min.) and 20% (30 min.) lower than with free trypsin digestion. Interestingly, with on-column immobilized trypsin digestion, the number of identified proteins is usually comparable to or higher than that with free trypsin digestion [10,13, 30]. In our work, NHS-trypsin digestion was performed in an Eppendorf tube, which may result in lower enzyme-to-substrate ratio than for on-column digestion [30], so the number of identified proteins is lower than that with free trypsin digestion.
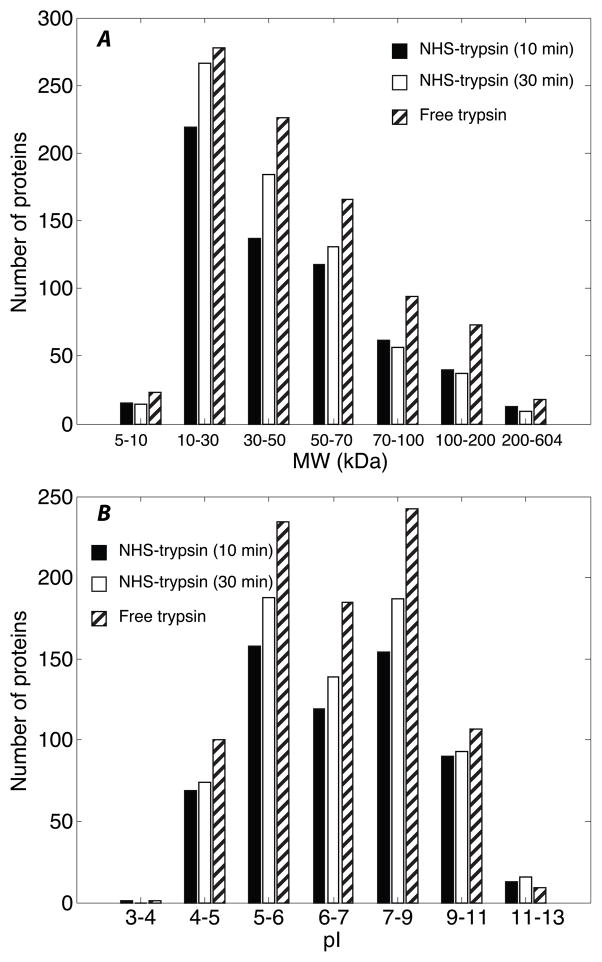
The molecular weight (MW) and pI distributions of identified proteins with NHS-trypsin and free trypsin digestion were determined, Figure 5. The MW of the proteins ranges from 5 kDa to 604 kDa, and the distribution from NHS-trypsin and free trypsin digestion is similar, which indicates that proteins with low and very high MW can both be digested efficiently with NHS-trypsin. The pI distributions of identified proteins with NHS-trypsin and free trypsin digestion from 3 to 13 are also similar, which demonstrates that acidic and basic proteins can both be digested with NHS-trypsin.
Figure 5.
Distribution of identified proteins from RAW 264.7 cells with NHS-trypsin (10 min. and 30 min.) and free trypsin digestion after RPLC-ESI-MS/MS analysis (in quadruplicate). A - Molecular weight. B – pI.
In addition, the overlap of identified proteins with NHS-trypsin (30 min.) and free trypsin digestion was also determined, S-Figure 3 in supporting material. Combination of the results from NHS-trypsin (30 min.) and free trypsin digestion, 1104 proteins were identified. Only about 43% (471 proteins) of these proteins were identified with both methods, which indicates that some extent complementarity exists for NHS-trypsin and free trypsin digestion.
4 Conclusions
Aldehyde- and NHS-activated magnetic microspheres based immobilized trypsin (CHO-trypsin and NHS-trypsin) were prepared and compared. NHS-trypsin provided greater sequence coverage and identified more peptides for the digestion of bovine serum albumin. A one-minute digestion at room temperature using the immobilized trypsin also identified more peptides and produced higher sequence coverage than traditional free trypsin digestion for 12 hours at 37 °C. Analysis of 15 nM (0.001 mg/mL) BSA digested by NHS-trypsin in 1 min. at room temperature consistently yielded one detected peptide. The run-to-run digestion of a three-protein-mixture with NHS-trypsin was quantitatively reproducible. For proteome analysis of RAW 264.7 cells, 604 and 697 protein groups were identified by RPLC-ESI-MS/MS with NHS-trypsin digestion for 10 min. and 30 min. at room temperature, with a peptide false discovery rate of less than 1%.
Supplementary Material
Highlights.
Comprehensive comparison of chemistry for immobilization of trypsin to magnetic microbeads
Evaluation of precision of digestion in terms of peak intensity and spectral counts.
One minute digestion with immobilized beads generated more peptides than 12 digestion using conventional solution-phase digestion
Obtained peptides from digestion of 15 nM protein
Acknowledgments
We acknowledge the assistance of Dr. Bill Boggess of mass spectrometry & proteomics facility at the University of Notre Dame in the establishment of RPLC-ESI-MS/MS system. This work was supported by a grant from the National Institutes of Health (R01GM096767).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aebersold R, Mann M. Nature. 2003;422:198. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat Biotechnol. 1999;17:994. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 3.Chait BT. Science. 2006;314:65. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 4.Ma JF, Zhang LH, Liang Z, Shan YC, Zhang YK. Trends Anal Chem. 2011;30:691. [Google Scholar]
- 5.Peterson DS, Rohr T, Svec F, Fréchet JMJ. J Proteome Res. 2002;1:563. doi: 10.1021/pr0255452. [DOI] [PubMed] [Google Scholar]
- 6.Dulay MT, Baca QJ, Zare RN. Anal Chem. 2005;77:4604. doi: 10.1021/ac0504767. [DOI] [PubMed] [Google Scholar]
- 7.Ye ML, Hu S, Schoenherr RM, Dovichi NJ. Electrophoresis. 2004;25:1319. doi: 10.1002/elps.200305841. [DOI] [PubMed] [Google Scholar]
- 8.Palm AK, Novotny MV. Rapid Commun Mass Spectrom. 2004;18:1374. doi: 10.1002/rcm.1500. [DOI] [PubMed] [Google Scholar]
- 9.Duan JC, Sun LL, Liang Z, Zhang J, Wang H, Zhang LH, Zhang WB, Zhang YK. J Chromatogr, A. 2006;1106:165. doi: 10.1016/j.chroma.2005.11.102. [DOI] [PubMed] [Google Scholar]
- 10.Ma JF, Liang Z, Qiao XQ, Deng QL, Tao DY, Zhang LH, Zhang YK. Anal Chem. 2008;80:2949. doi: 10.1021/ac702343a. [DOI] [PubMed] [Google Scholar]
- 11.Sun LL, Ma JF, Qiao XQ, Liang Y, Zhu GJ, Shan YC, Liang Z, Zhang LH, Zhang YK. Anal Chem. 2010;82:2574. doi: 10.1021/ac902835p. [DOI] [PubMed] [Google Scholar]
- 12.Liang Y, Tao DY, Ma JF, Sun LL, Liang Z, Zhang LH, Zhang YK. J Chromatogr A. 2011;1218:2898. doi: 10.1016/j.chroma.2011.02.073. [DOI] [PubMed] [Google Scholar]
- 13.Feng S, Ye ML, Jiang XG, Jin WH, Zou HF. J Proteome Res. 2006;5:422. doi: 10.1021/pr0502727. [DOI] [PubMed] [Google Scholar]
- 14.Schoenherr RM, Ye M, Vannatta M, Dovichi NJ. Anal Chem. 2007;79:2230. doi: 10.1021/ac061638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicoli R, Rudaz S, Stella C, Veuthey JL. J Chromatogr, A. 2009;1216:2695. doi: 10.1016/j.chroma.2008.10.046. [DOI] [PubMed] [Google Scholar]
- 16.Yuan HM, Zhang LH, Hou CY, Zhu GJ, Tao DY, Liang Z, Zhang YK. Anal Chem. 2009;81:8708. doi: 10.1021/ac900310y. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Xu XQ, Deng CH, Yang PY, Zhang XM. J Proteome Res. 2007;6:3849. doi: 10.1021/pr070132s. [DOI] [PubMed] [Google Scholar]
- 18.Liu JY, Lin S, Qi DW, Deng CH, Yang PY, Zhang XM. J Chromatogr, A. 2007;1176:169. doi: 10.1016/j.chroma.2007.10.094. [DOI] [PubMed] [Google Scholar]
- 19.Lin S, Yao GP, Qi DW, Li Y, Deng CH, Yang PY, Zhang XM. Anal Chem. 2008;80:3655. doi: 10.1021/ac800023r. [DOI] [PubMed] [Google Scholar]
- 20.Chen WY, Chen YC. Anal Chem. 2007;79:2394. doi: 10.1021/ac0614893. [DOI] [PubMed] [Google Scholar]
- 21.Li YH, Wojcik R, Dovichi NJ. J Chromatogr, A. 2011;1218:2007. doi: 10.1016/j.chroma.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirza SP, Greene AS, Olivier M. J Proteome Res. 2008;7:3042. doi: 10.1021/pr800018g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Ferrer D, Hixson KK, Smallwood H, Squier TC, Petritis K, Smith RD. Anal Chem. 2009;81:6272. doi: 10.1021/ac802540s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sevinsky JR, Brown KJ, Cargile BJ, Bundy JL, Stephenson JL., Jr Anal Chem. 2007;79:2158. doi: 10.1021/ac0620819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda T, Saito N, Tomita M, Ishihama Y. Mol Cell Proteomics. 2009;8:2770. doi: 10.1074/mcp.M900240-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicoli R, Gaud N, Stella C, Rudaz S, Veuthey JL. J Pharm Biomed Anal. 2008;48:398. doi: 10.1016/j.jpba.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 27.Cingöz A, Hugon-Chapuis F, Pichon V. J Chromatogr, A. 2008;1209:95. doi: 10.1016/j.chroma.2008.08.120. [DOI] [PubMed] [Google Scholar]
- 28.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 29.Liu HB, Sadygov RG, Yates JR., III Anal Chem. 2004;76:4193. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 30.Klammer AA, MacCoss MJ. J Proteome Res. 2006;5:695. doi: 10.1021/pr050315j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.