Abstract
Cancer chemopreventive response to D,L-sulforaphane (SFN), a synthetic racemic analogue of broccoli constituent L-sulforaphane, is partly attributable to apoptosis induction, but the mechanism of cell death is not fully understood. The present study demonstrates a critical role for adapter protein p66Shc in SFN -induced apoptosis. Immortalized mouse embryonic fibroblasts (MEF) derived from p66shc knockout mice were significantly more resistant to SFN-induced apoptosis, collapse of mitochondrial membrane potential, and reactive oxygen species (ROS) production compared with MEF obtained from the wild-type mice. Notably, a spontaneously immortalized and non-tumorigenic human mammary epithelial cell line (MCF-10A) was resistant to SFN-induced ROS production and apoptosis. Stable overexpression of manganese superoxide dismutase in MCF-7 and MDA-MB-231 human breast cancer cells conferred near complete protection against SFN-induced apoptosis and mitochondrial membrane potential collapse. SFN treatment resulted in increased S36 phosphorylation and mitochondrial translocation of p66shc in MDA-MB-231 and MCF-7 cells, and SFN-induced apoptosis was significantly attenuated by RNA interference of p66shcin both cells. SFN-treated MDA-MB-231 and MCF-7 cells also exhibited a marked decrease in protein level of peptidyl prolyl isomerase (Pin1), which is implicated in mitochondrial translocation of p66shc. However, stable overexpression of Pin1 failed to alter proapoptotic response to SFN at least in MCF-7 cells. Finally, SFN-induced S36 phosphorylation of p66Shc was mediated by protein kinase Cβ (PKCβ), and pharmacological inhibition of PKCβ significantly inhibited apoptotic cell death resulting from SFN exposure. In conclusion, the present study provides new insight into the mechanism of SFN-induced apoptosis involving PKCβ-mediated S36 phosphorylation of p66shc.
Keywords: Sulforaphane, p66Shc, Apoptosis, Chemoprevention
INTRODUCTION
Cancer chemoprevention is a rapidly emerging sub-discipline in oncology focusing on development of novel agents to reduce disease-related cost, mortality, and morbidity associated with cancer [Lippman and Hawk, 2009; Shu et al., 2010]. Edible fruits and vegetables have attracted attention for the discovery of cancer chemopreventive agents [Surh, 2003]. Cruciferous vegetables (e.g., broccoli and watercress) constitute one such example of edible plants from which cancer chemopreventive compounds have been identified [Hecht, 2000; Fahey et al., 2001]. Cancer chemopreventive effect of cruciferous vegetables is partly attributed to chemicals with an isothiocyanate functional group [Hecht, 2000]. D,L-Sulforaphane(SFN), a synthetic racemic analogue of broccoli-derived L-isomer, is a promising cancer chemopreventive agent with efficacy against chemically-induced neoplasia as well as oncogene-driven spontaneous cancer development in rodents [Zhang et al., 1994; Chung et al., 2000; Singh et al., 2009]. SFN-mediated growth retardation of human cancer cells transplanted in athymic mice has also been reported [Singh et al., 2004b].
Elucidation of the mechanism underlying cancer chemopreventive response to SFN has been the topic of intense research over the past decade. Seminal contributions from colleagues around the world concerning mechanisms of cancer chemoprevention by SFN include: inhibition of CYP2E1 [Barcelo et al., 1996], cell cycle arrest [Gamet-Payrastre et al., 2000; Singh et al., 2004a; Kim et al., 2010], apoptosis induction [Gamet-Payrastre et al., 2000; Singh et al., 2004b; Singh et al., 2005], inhibition of angiogenesis [Bertl et al., 2006] induction of autophagy as a protective mechanism against apoptotic cell death [Herman-Antosiewicz et al., 2006], inhibition of histone deacetylase [Myzak et al., 2004], protein binding [Mi et al., 2007], induction of phase 2 enzymes [Li et al., 2006; Kensler and Wakabayashi, 2010], epigenetic repression of hTERT [Meeran et al., 2010], and inhibition of breast cancer stem cells [Li et al., 2010].
Mechanism by which SFN causes cell cycle arrest and apoptotic cell death continues to expand. For example, SFN-induced G2/M phase cell cycle arrest in human prostate cancer cells was mediated by checkpoint kinase 2-mediated phosphorylation of cell division cycle 25C [Singh et al., 2004a]. In KB and YD-10B human oral squamous carcinoma cells, G2/M phase cell cycle arrest resulting from SFN exposure was associated with a significant increase in the p21 protein level [Kim et al., 2010]. Furthermore, SFN treatment increased the p21 promoter activity and resulted in induction of p21 expression in tumor xenograft in vivo [Kim et al., 2010]. SFN-mediated suppression of many cellular pathways implicated in apoptosis control have also been described, including nuclear factor-κB, Akt, anti-apoptotic proteins (Bcl-2, Bcl-xL), and signal transducer and activator of transcription 3 [Xu et al., 2005; Singh et al., 2005; Hahm and Singh, 2010]. We have also shown recently that while activation of signal transducer and activator of transcription 3 confers modest protection against SFN-induced apoptosis, mitochondria-derived reactive oxygen species (ROS) provide initial signal for apoptosis commitment [Singh et al., 2005; Xiao et al., 2009]. SFN-induced ROS production and apoptosis in human prostate cancer cells were significantly attenuated by boosting of cellular anti-oxidative capacity as well as depletion of mitochondrial DNA to disrupt mitochondrial electron transport chain [Singh et al., 2005; Xiao et al., 2009]. Nevertheless, both intrinsic (mitochondria-mediated) and extrinsic caspase cascades appear important for execution of SFN-induced apoptosis [Singh et al., 2005; Kim et al., 2006].
The present study extends these observations and determines the role of adapter protein p66Shcin proapoptotic response to SFN. This was a worthy research objective because electron transfer between cytochrome c and p66Shc, which is a splice variant of the cytoplasmic adapter proteins p52/p46 that are involved in signal transduction from activated tyrosine kinases to Ras [Pelicci et al., 1992], was shown to cause ROS-dependent and mitochondria-mediated apoptosis [Giorgio et al., 2005].
MATERIALS AND METHODS
REAGENTS
SFN (purity >99 %) was purchased from LKT Laboratories (St. Paul, MN), whereas 4′,6-diamidino-2-phenylindole (DAPI) was purchased from Sigma-Aldrich (St. Louis, MO). Stock solution of SFN was prepared in dimethyl sulfoxide (DMSO), and diluted with complete media immediately before use. An equal volume of DMSO was added to controls. MitoSOX Red, MitoTracker Green, 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1), and Alexa Fluor 568 goat anti-mouse antibody were purchased from Invitrogen-Life Technologies (Carlsbad, CA). Annexin V-FITC Apoptosis Detection Kit was purchased from BD Biosciences (San Diego, CA). Anti-actin antibody was from Sigma-Aldrich; an antibody against total p66Shc was from Santa Cruz Biotechnology (Santa Cruz, CA), antibody against S36 phosphorylated p66Shcwas from Santa Cruz Biotechnology or Abcam (Cambridge, MA), antibodies against cleaved caspase-3 and peptidyl prolyl isomerase (Pin1)were from Cell Signaling Technology (Danvers, MA). The p66Shc-targeted small interfering RNA (siRNA) was obtained from Santa Cruz Biotechnology, whereas a nonspecific control siRNA was from Qiagen (Valencia, CA). Protein kinase Cβ (PKCβ) inhibitor [3-(1-(3-imidazol-1-ylpropyl)-1H-indol-3-yl)-4- anilino-1H-pyrrole-2, 5-dione; hereafter abbreviated as PKCβ-I] was purchased from EMD Biosciences (Gibbstown, NJ).
CELL LINES
Immortalized mouse embryonic fibroblasts (MEF) derived from wild-type mice [hereafter abbreviated as p66Shc(+/+)MEF] and p66Shc knockout mice [abbreviated as p66Shc(−/−)MEF] were generously provided by Dr. T. Finkel (National Institutes of Health, Bethesda, MD) and cultured in Dulbecco’s modified essential medium supplemented with 10% fetal bovine serum, 0.1 mM non-essential amino acids, 0.1 μM 2-mercaptoethanol, and penicillin-streptomycin antibiotic mixture. MDA-MB-231, MCF-7, and MCF-10A cell lines were purchased from American Type Culture Collection (Manassas, VA) and maintained as described by us previously [Xiao et al., 2006]. Authentication of MDA-MB-231, MCF-7, and MCF-10Acell lines was done by Research Animal Diagnostic Laboratory (University of Missouri, Columbia, MO). The MDA-MB-231 and MCF-7 cells were last tested in February 2011, and found to be of human origin without any inter-species contamination. Moreover, the genetic profiles for MDA-MB-231 and MCF-7 cells were consistent with the corresponding genetic profiles in the American Type Culture Collection database. Cells were stably transfected with empty pcDNA3.1 vector or pcDNA3.1 vector encoding for myc-Pin1 or manganese-superoxide dismutase (Mn-SOD) and selected by culture in medium supplemented with 800 μg/mL G-418 over a period of 8 weeks.
WESTERN BLOTTING
Cells were treated with DMSO (control) or SFN (10 or 20 μM) for specified time periods, and both floating and attached cells were collected. Cells were lysed as described by us previously [Powolny et al., 2011]. Lysate proteins were resolved by sodium dodecyl-sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membrane. After blocking with a solution consisting of Tris-buffered saline supplemented with 0.05% Tween 20 and 5% (w/v) non-fat dry milk, the membrane was exposed to desired primary antibody for 1 hour at room temperature or overnight at 4°C. Following treatment with an appropriate secondary antibody, immunoreactive bands were visualized using Chemiluminescence method. The blots were stripped and re-probed with anti-actin antibody to correct for differences in protein loading. Change in protein level was determined by densitometric scanning of the immunoreactive band and corrected for actin loading control.
DETERMINATION OF APOPTOSIS AND MITOCHONDRIAL MEMBRANE POTENTIAL
Apoptosis induction was assessed by quantitation of histone-associated DNA fragment release into the cytosol using an ELISA kit from Roche Applied Sciences (Indianapolis, IN) or flow cytometry using Annexin V/Propidium Iodide Apoptosis Detection kit. Quantitation of histone-associated DNA fragment release into the cytosol was performed according to the manufacturer’s instructions. For quantitation of apoptosis by flow cytometry using Annexin V kit, cells were treated with DMSO or SFN for 24 hours. Cells were harvested and washed with phosphate buffered saline (PBS). Cells (1×105) were suspended in 100 μL binding buffer, and stained with 4 μL Annexin V and 2 μL propidium iodide solution for 15 minutes at room temperature in the dark. Samples were then diluted with 200 μL binding buffer. Stained cells were analyzed using a Coulter Epics XL Flow Cytometer. Mitochondrial membrane potential in DMSO-treated control and SFN-treated cells was determined by flow cytometry using JC-1 essentially as described by us previously [Xiao et al., 2009].
DETERMINATION OF ROS PRODUCTION
ROS production was measured by fluorescence microscopy or flow cytometry with the use of a chemical probe (MitoSOX Red). For fluorescence microscopy, cells were plated on coverslips and allowed to attach by overnight incubation. Cells were then treated with DMSO (control) or desired concentrations of SFN for 4 hours followed by incubation with 2.5 μM MitoSOX Red for 30 minutes at 37°C. Cells were then treated for 15 minutes with 200 nM MitoTracker green to stain mitochondria. After washing with PBS, cells were fixed with 2% paraformaldehyde for 1 hour at room temperature and examined under a Leica fluorescence microscope at 100× objective magnification. For flow cytometric analysis, cells were treated with DMSO (control) or desired concentrations of SFN for 4 hours and then incubated with 5 μM MitoSOX Red for 30 minutes. Cells were collected, washed with PBS, and fluorescence was detected using a Coulter Epics XL Flow Cytometer.
IMMUNOCYTOCHEMICAL ANALYSIS OF p66Shc LOCALIZATION
MDA-MB-231 (1.2 ×105) or MCF-7 cells (1×105) stably transfected with Mito-GFP were plated on coverslips in 12-well plates, allowed to attach, and then exposed to SFN or DMSO (control) for 8 hours. After washing with PBS, cells were fixed with 2% paraformaldehyde overnight at 4°C and permeabilized using 0.1% Triton X-100 in PBS for 10 minutes. Cells were washed with PBS, blocked with 0.5% bovine serum albumin and 0.15% glycine in PBS for 1 hour, and incubated with anti-p66Shc antibody overnight at 4°C. Cells were then washed with PBS and incubated with AlexaFluor 568 -conjugated secondary antibody for 1 hour at room temperature. Subsequently, cells were washed with PBS and treated with DAPI (10 ng/mL) for 5 minutes at room temperature to stain nucleus. Cells were washed twice with PBS and examined under a Leica fluorescence microscope at 100× objective magnification.
RNA INTERFERENCE OF p66Shc AND CELL VIABILITY ASSAY
MDA-MB-231 (1.8×105) or MCF-7 cells (1.5×105) were seeded in 6-well plates and allowed to attach by overnight incubation. Cells were then transfected with 200 nM of a control nonspecific siRNA or a p66Shc-targeted siRNA using OligoFECTAMINE (Invitrogen-Life Technologies). Twenty-four hours after transfection, cells were treated with DMSO (control) or specified concentrations of SFN for desired time period. Subsequently, cells were collected, washed with PBS, and processed for immunoblotting and analysis of histone-associated DNA fragment release into the cytosol. Effect of SFN treatment on cell viability was determined by trypan blue dye exclusion assay as previously described [Xiao et al., 2004].
RESULTS
p66Shc DEFICIENCY CONFERRED PROTECTION AGAINST SFN-INDUCED APOPTOSIS
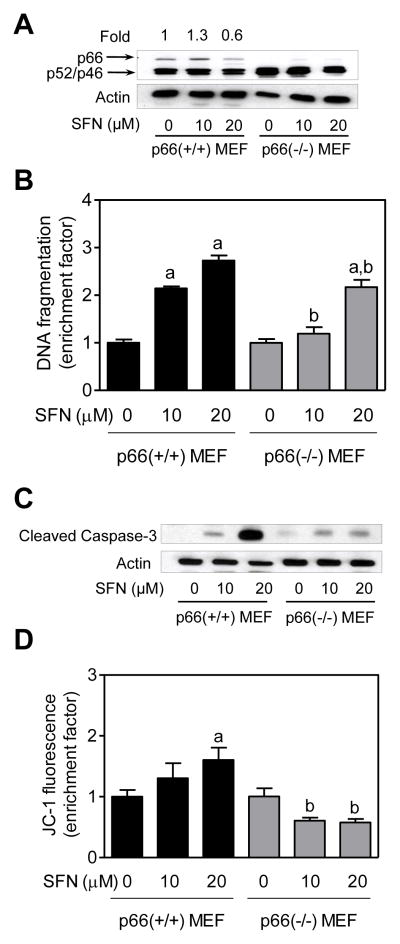
Initially we used p66 (+/+)MEF and p66(−/−)MEF to study the role of p66Shcin regulation of SFN-induced apoptosis. Immunoblotting confirmed absence of p66Shc protein (but not p52 or p46) in p66(−/−) MEF (Fig. 1A). SFN treatment resulted in a dose-dependent and statistically significant increase in histone-associated DNA fragment release into the cytosol (a measure of apoptosis)over DMSO -treated control in p66(+/+) MEF (Fig. 1B). On the other hand, p66(−/−) MEF were significantly more resistant to SFN-mediated release of histone-associated DNA fragments into the cytosol in comparison with p66(+/+) MEF (Fig. 1B). Consistent with these results, SFN-mediated cleavage of procaspase-3 was markedly reduced in p66(−/−) MEF in comparison with p66(+/+) MEF(Fig. 1C). Moreover, the p66(−/−) MEF resisted SFN-induced collapse of mitochondrial membrane potential as judged by flow cytometry for monomeric (green) JC-1 fluorescence (Fig. 1D). Collectively, these results indicated that (a) p66Shc expression was critical for SFN-induced apoptosis, and (b) p66Shc acted upstream of mitochondrial membrane potential collapse in execution of SFN-induced apoptosis.
Fig. 1.
Immortalized mouse embryonic fibroblasts (MEF) derived from p66Shc knockout mice were resistant to D,L-sulforaphane (SFN)-induced apoptosis. A: Western blotting for total p66Shc protein using lysates from MEF derived from wild -type mice [p66(+/+) MEF] and p66Shc knockout mice [p66(−/−) MEF] and treated for 24 hours with dimethyl sulfoxide (DMSO; control) or the indicated concentrations of SFN. Numbers above the bands represent densitometric quantitation of change in protein level relative to DMSO-treated control. B: Quantitation of histone-associated DNA fragment release into the cytosol(a measure of apoptosis), C: Western blotting for cleaved caspase-3, and D: Flow cytometric quantitation of monomeric (green) JC-1 fluorescence (a measure of mitochondrial membrane potential) in p66(+/+) MEF and p66(−/−) MEF after 24-hour treatment with DMSO (control) or the indicated concentrations of SFN. Each experiment was done at least twice and representative data from one such experiment are shown. For quantitative measurements (panels B and D) results are expressed as enrichment factor relative to DMSO-treated p66(+/+) MEF (mean ± SD, n = 3). Significantly different (P<0.05) compared with arespective DMSO-treated control for each cell line, and bbetween SFN-treated p66(+/+) MEF and SFN-treated p66(−/−) MEF by one-way ANOVA followed by Bonferroni’s test.
SFN CAUSED ROS PRODUCTION IN p66(+/+) MEF BUT NOT IN p66(−/−) MEF
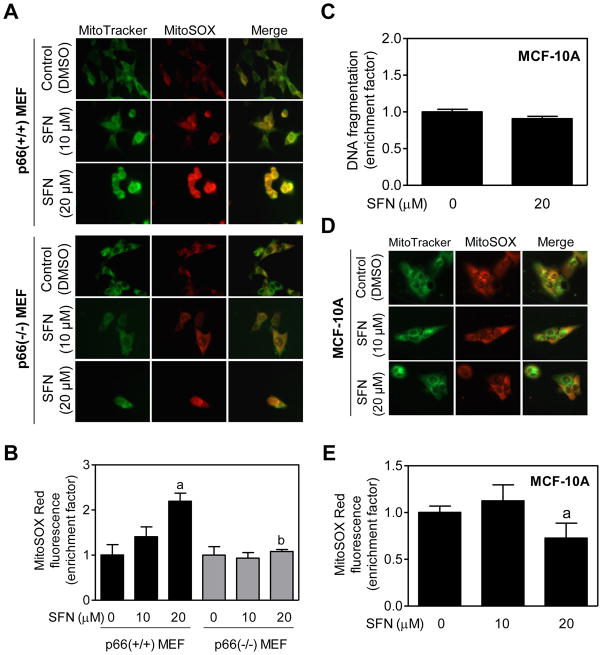
Previous studies have shown that electron transfer between cytochrome c and p66Shc leads to ROS production [Giorgio et al., 2005]. Because SFN-induced apoptosis is intimately linked to ROS production [Singh et al., 2005; Kim et al., 2006; Xiao et al., 2009], it was of interest to determine the role of p66Shcin ROS production by SFN. We addressed this question using MEF and a chemical probe MitoSOX Red, which is a derivative of hydroethidine containing a hexyl triphenylphosphonium cation for targeting to the mitochondria. The MitoSOX Red detects mitochondria generated superoxide anion [Robinson et al., 2006]. As can be seen in Fig. 2A, endogenous MitoSOX Red fluorescence was very weak and diffuse in DMSO-treated p66(+/+) MEF as well as DMSO -treated p66(−/−) MEF. On the other hand, SFN-treated p66(+/+) MEF were brightly stained with MitoSOX Red with appearance of yellow-orange color due to the merging of the MitoTracker green fluorescence and MitoSOX Red fluorescence (Fig. 2A). Intensity of SFN-induced yellow-orange color was relatively higher in the p66(+/+) MEF in comparison with those derived from the p66Shc knockout mice (Fig. 2A). These results were confirmed by flow cytometric quantitation of MitoSOX Red fluorescence in control (DMSO-treated) and SFN-treated MEF (Fig. 2B). These results pointed towards involvement of p66Shcin SFN -mediated ROS production.
Fig. 2.
D,L-Sulforaphane (SFN)treatment caused ROS generation in immortalized mouse embryonic fibroblasts (MEF) derived from wild-type mice [p66(+/+) MEF] but not in p66(−/−) MEF or MCF-10A cells. A: Fluorescence microscopy for MitoSOX Red fluorescence (a measure of reactive oxygen species generation) in p66(+/+) MEF and p66(−/−) MEF after 4-hour treatment with dimethyl sulfoxide (DMSO)or the indicated concentrations of SFN. Staining for mitochondria (MitoTracker green) and MitoSOX Red is indicated by green and red colors, respectively. B: Flow cytometric quantitation of MitoSOX Red fluorescence in p66(+/+) MEF and p66(−/−) MEF after 4-hour treatment with DMSO or the indicated concentrations of SFN. Results are expressed as enrichment of MitoSOX Red fluorescence relative to DMSO-treated p66(+/+) MEF (mean ± SD, n = 3). Significantly different (P<0.05) compared with arespective DMSO-treated control for each cell line, and bbetween SFN-treated p66(+/+) MEF and SFN-treated p66(−/−) MEF by one-way ANOVA followed by Bonferroni’s test. C: Quantitation of histone-associated DNA fragment release into the cytosol in MCF-10A cells after 24-hour treatment with DMSO or 20 μM SFN. Results shown are mean ± SD (n = 3). D: Fluorescence microscopy for MitoSOX Red fluorescence in MCF-10A cells after 4-hour treatment with DMSO or the indicated concentrations of SFN. Note that SFN treatment did not increase MitoSOX Red fluorescence over DMSO-treated control. E: Flow cytometric analysis of MitoSOX Red fluorescence in DMSO-treated control and SFN-treated MCF-10A cells (4-hour treatment). Results are shown as enrichment factor relative to DMSO-treated control (mean ± SD, n = 6). aSignificantly different(P<0.05)compared with DMSO-treated control by one-way ANOVA with Dunnett’s adjustment. All experiments were repeated at least twice.
MCF-10A CELLS WERE RESISTANT TO SFN-INDUCED ROS PRODUCTION AND APOPTOSIS
We have shown previously that a normal human prostate epithelial cell line (PrEC) is significantly more resistant to SFN-induced apoptosis compared with LNCaP human prostate cancer cells [Choi and Singh, 2005]. A similar differential was also observed for a normal bronchial epithelial cell line and a human lung cancer cell line [Choi and Singh, 2005]. We questioned whether this selectivity was translated to mammary cells. In the present study, we tested this possibility using MCF-10A cell line, which is a spontaneously immortalized and non-tumorigenic cell line originally derived from a fibrocystic breast disease. As shown in Fig. 2C, SFN treatment (20 μM, 24 hours) failed to increase release of histone-associated DNA fragments into the cytosol over DMSO-treated control in MCF-10A cells. MCF-10A cells were also resistant to SFN-induced ROS production as judged by fluorescence microscopy (Fig. 2D) or flow cytometry (Fig. 2E) using MitoSOX Red. Interestingly, basal MitoSOX Red-associated fluorescence was relatively more intense in MCF-10A cells (Fig. 2D) than in MEF (Fig. 2A). Relative resistance of MCF-10A cells to SFN-mediated growth inhibition compared with breast cancer cells has been demonstrated previously [Meeran et al., 2010].
SFN-INDUCED APOPTOSIS IN MCF-7 AND MDA-MB-231 BREAST CANCER CELLS WAS INHIBITED BY Mn-SOD OVEREXPRESSION
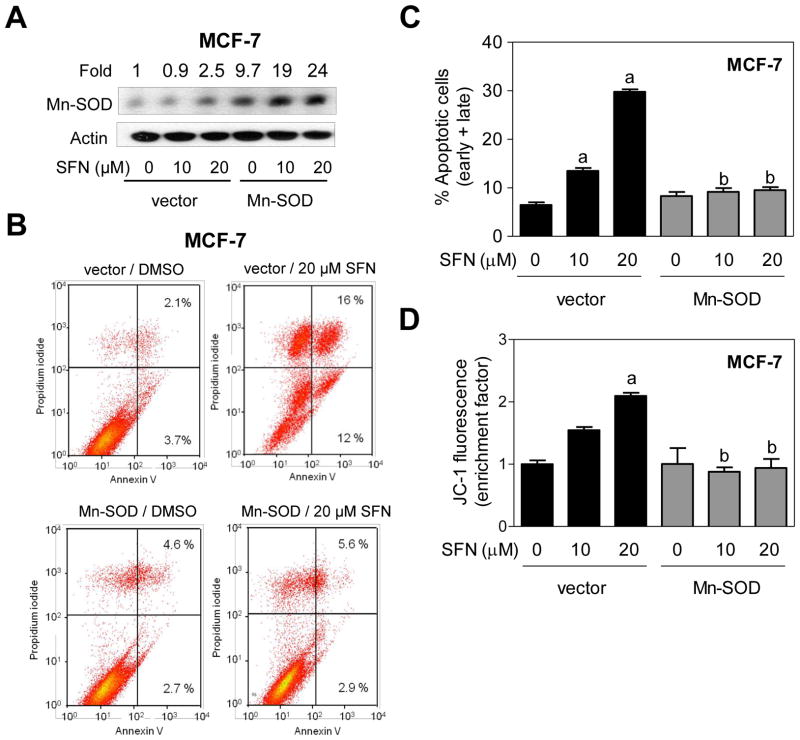
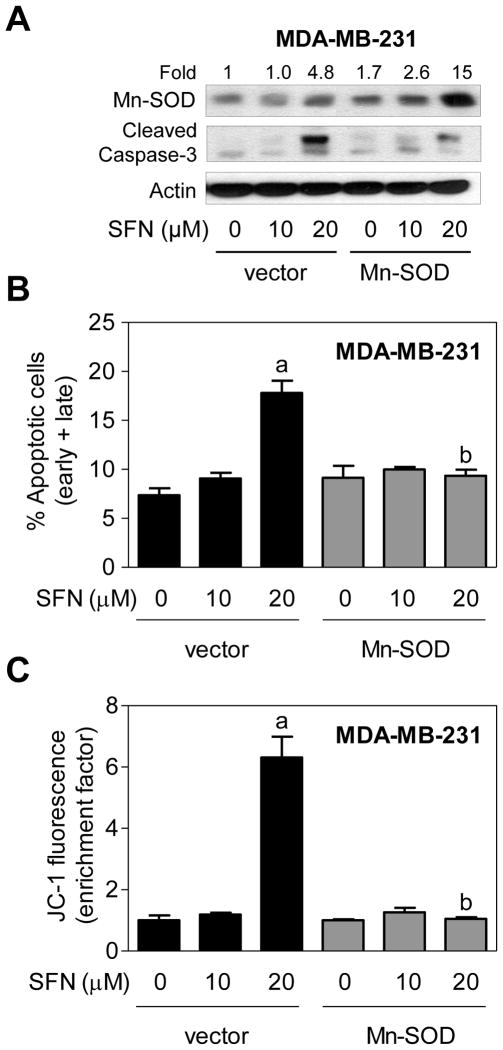
Next, we designed experiments to determine whether SFN-induced apoptosis was dependent on ROS generation using MCF-7 and MDA-MB-231 human breast cancer cells stably transfected with Mn-SOD. Level of Mn-SOD protein was 9.7-fold higher in MCF-7 cells stably transfected with a plasmid encoding for Mn-SOD compared with empty vector transfected cells (Fig. 3A). Interestingly, SFN treatment resulted in up-regulation of Mn-SOD protein in both empty vector transfected cells and Mn-SOD overexpressing MCF-7 cells (Fig. 3A). SFN-induced ROS production was markedly suppressed by Mn-SOD overexpression as judged by fluorescence microscopy using MitoSOX Red (results not shown). Moreover, Mn-SOD overexpression conferred near complete protection against SFN-induced apoptosis, as judged by Annexin Vassay (Fig. 3B, C), as well collapse of mitochondrial membrane potential (Fig. 3D). Similarly, stable overexpression of Mn-SOD (Fig. 4A) nearly fully blocked SFN-induced apoptosis (Fig. 4B) as well as mitochondrial membrane potential collapse (Fig. 4C) in MDA-MB-231 cells. Collectively, these results indicated that the (a) SFN-induced apoptosis in breast cancer cells was intimately linked to ROS generation, and (b) ROS acted upstream of mitochondrial membrane potential collapse during SFN-induced apoptosis.
Fig. 3.
Manganese-superoxide dismutase (Mn-SOD) overexpression attenuated D,L-sulforaphane (SFN)-induced apoptosis in MCF-7 cells. A: Immunoblotting for Mn-SOD using lysates from MCF-7 cells stably transfected with empty vector or vector encoding for Mn-SOD and treated for 24 hours with dimethyl sulfoxide (DMSO; control) or the indicated concentrations of SFN. Numbers above the bands represent densitometric quantitation of change in protein level relative to empty vector transfected cells treated with DMSO. B: Representative flow histograms depicting apoptotic fraction in MCF-7 cells stably transfected with empty vector or vector encoding for Mn-SOD and treated for 24 hours with DMSO (control) or 20 μM SFN. C: Quantitation of % apoptotic fraction (early + late apoptotic cells) in MCF-7 cells stably transfected with empty vector or vector encoding for Mn-SOD and treated for 24 hours with DMSO (control) or the indicated concentrations of SFN. D: Flow cytometric quantitation of monomeric (green) JC-1 fluorescence in MCF-7 cells stably transfected with empty vector or vector encoding for Mn-SOD and treated for 24 hours with DMSO (control) or the indicated concentrations of SFN. For quantitative measurements (panels C and D) results are expressed relative to DMSO-treated empty vector cells (mean ± SD, n = 3). Significantly different (P<0.05) compared with arespective DMSO-treated control for each cell line, and bbetween SFN-treated empty vector transfected cells and SFN-treated Mn-SOD overexpressing MCF-7 cells by one-way ANOVA followed by Bonferroni’s test. Each experiment was done at least twice and representative from one such experiment are shown.
Fig. 4.
Manganese-superoxide dismutase (Mn-SOD) overexpression inhibited D,L-sulforaphane (SFN)-induced apoptosis in MDA-MB-231 cells. A: Immunoblotting for Mn-SOD and cleaved caspase-3 using lysates from MDA-MB-231 cells stably transfected with empty vector or vector encoding for Mn-SOD and treated for 24 hours with dimethyl sulfoxide (DMSO; control) or the indicated concentrations of SFN. Numbers above the bands represent densitometric quantitation of change in protein level relative to empty vector transfected cells treated with DMSO. B: Quantitation of % apoptotic fraction (early + late apoptotic cells) in MDA-MB-231 cells stably transfected with empty vector or vector encoding for Mn-SOD and treated for 24 hours with DMSO (control) or the indicated concentrations of SFN. C: Flow cytometric quantitation of monomeric (green) JC-1 fluorescence in MDA-MB-231 cells stably transfected with empty vector or vector encoding for Mn-SOD and treated for 24 hours with DMSO (control) or the indicated concentrations of SFN. For quantitative measurements (panels B and C) results are expressed relative to DMSO-treated empty vector cells (mean ± SD, n = 3). Significantly different (P<0.05) compared with arespective DMSO-treated control for each cell line, and bbetween SFN-treated empty vector transfected cells and SFN-treated Mn-SOD overexpressing MDA-MB-231 cells by one-way ANOVA followed by Bonferroni’s test. Each experiment was done at least twice and representative from one such experiment are shown.
EFFECT OF p66Shc KNOCKDOWN ON SFN-INDUCED APOPTOSIS
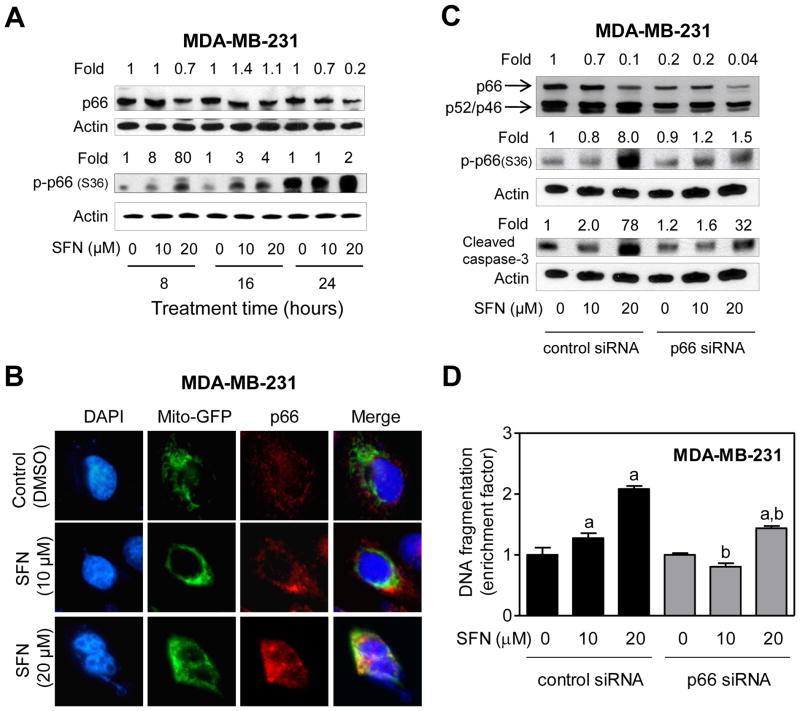
We used MDA-MB-231 and MCF-7 cells to further investigate the role of p66Shcin SFN-mediated apoptotic cell death. With the exception of 24-hour time point at 20 μM dose, the level of total p66Shc protein was not altered by more than 30–40% by SFN treatment in MDA-MB-231 cells (Fig. 5A). In contrast, SFN-treated MDA-MB-231 cells exhibited a marked increase in levels of S36 phosphorylated p66 Shc(2 -80-fold increase over DMSO-treated control), especially at the 8 hour time point with 20 μM dose (Fig. 5A). Previous studies have shown that S36 phosphorylation of p66Shc is important for its translocation to the mitochondria after recognition by Pin1 [Pinton et al., 2007]. We therefore tested the effect of SFN treatment on mitochondrial translocation of p66Shc using MDA-MB-231 cells stably transfected with Mito-GFP (Fig. 5B). SFN-treated MDA-MB-231 cells exhibited mitochondrial translocation of p66Shc as evidenced by merging of the p66Shc-associated red fluorescence and Mito-GFP-associated green fluorescence leading to appearance of yellow-orange color, which was rare in DMSO-treated control cells (Fig. 5B). As can be seen in Fig. 5C, the level of p66Shc protein was decreased by 80% upon transient transfection of MDA-MB-231 cells with a p66Shc-targeted siRNA in comparison with cells transfected with a nonspecific control siRNA. Moreover, SFN-mediated increase in S36 phosphorylation of p66Shc as well as cleavage of procaspase -3 was markedly suppressed by knockdown of p66Shc protein especially at the 20 μM dose (Fig. 5C). Consistent with these results and data shown in Fig. 1, SFN-mediated increase in histone-associated DNA fragment release into the cytosol was significantly higher in the control siRNA transfected cells than in p66Shc silenced MDA-MB-231 cells (Fig. 5D).
Fig. 5.
Knockdown of p66Shc conferred protection against D,L-sulforaphane (SFN)-induced apoptosis in MDA-MB-231 cells. A: Immunoblotting for total and S36 phosphorylated p66Shc using lysates from MDA-MB-231 cells treated with dimethyl sulfoxide (DMSO; control) or SFN (10 or 20 μM) for the indicated time periods. Numbers above the bands represent densitometric quantitation of change in protein level relative to corresponding DMSO-treated control at each time point. B: Immunofluorescence microscopic analysis of p66Shc in Mito-GFP transfected MDA-MB-231 cells after 8-hour treatment with DMSO or the indicated concentrations of SFN. C: Immunoblotting for total and S36 phosphorylated p66Shc and cleaved caspase-3 using lysates from MDA-MB-231 cells transiently transfected with a control (nonspecific) siRNA or a p66Shc-targeted siRNA and treated for 24 hours with DMSO or the indicated concentrations of SFN. Numbers on top of the immunoreactive bands represent densitometric quantitation of changes in protein levels relative to control siRNA transfected cells treated with DMSO. D: Quantitation of histone-associated DNA fragment release into the cytosol in MDA-MB-231 cells transiently transfected with a control siRNA or a p66Shc-targeted siRNA and treated for 24 hours with DMSO or the indicated concentrations of SFN. The results are expressed as enrichment factor relative to DMSO-treated MDA-MB-231 cells transfected with the control siRNA (mean ± SD, n = 3). Significantly different (P<0.05) compared with arespective DMSO-treated control, and bbetween SFN-treated control siRNA transfected cells and SFN-treated p66Shc siRNA transfected cells by one-way ANOVA followed by Bonferroni’s test. Each experiment was done at least twice, and representative data from a single experiment are shown.
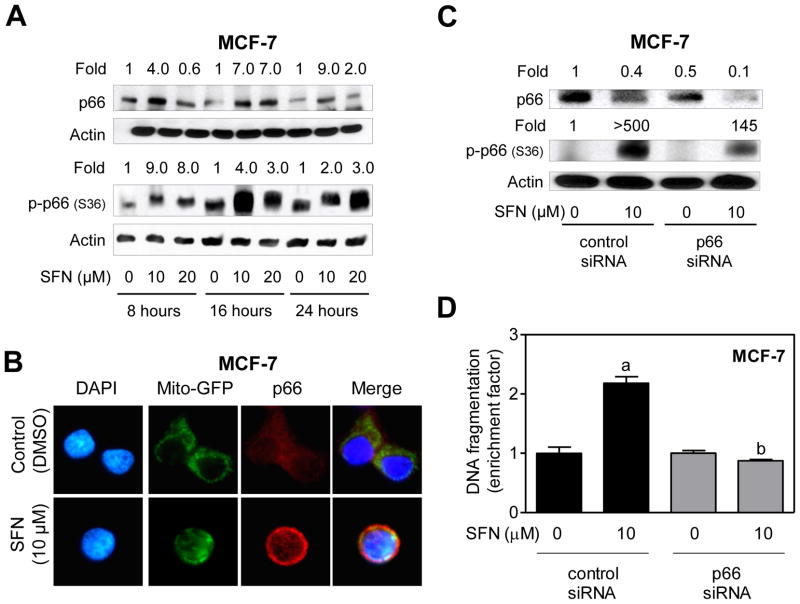
We questioned if the association between p66Shc expression status and SFN-induced apoptosis was unique to the MDA-MB-231 cell line, which lacks estrogen-receptor and expresses mutant p53. Because p53 has been shown to cooperate with p66Shc to control intracellular redox status, levels of oxidation-damaged DNA, and oxidative stress-induced apoptosis [Trinei et al., 2002], it was also of interest to determine whether p66Shc dependence of SFN-induced apoptosis was influenced by the p53 status. We addressed these question’s using MCF-7 cell line, which is estrogen-receptor positive and expresses wild -type p53. In sharp contrast to MDA-MB-231 cells (Fig. 5A), SFN treatment resulted in induction of total p66Shc protein in MCF -7 cells, which was clearly visible at 16-and 24 -hour time points with both 10 and 20 μM doses (Fig. 6A). However, similar to MDA-MB-231 cells, SFN-treated MCF-7 cells exhibited not only an increase in S36 phosphorylation of p66Shc(Fig. 6A) but also its mitochondrial translocation (Fig. 6B). For reasons not yet clear, collapse of mitochondria around DAPI-stained nuclei was also discernible in SFN-treated MCF-7 cells (Fig. 6B), which was not as pronounced in either DMSO-treated control MCF-7 cells (Fig. 6B) or in MDA-MB-231 cells (Fig. 5B). Nevertheless, a 50% decrease in the level of p66Shc protein by its RNA interference conferred significant protection against SFN-induced S36 phosphorylation of p66Shc(Fig. 6C) as well as apoptosis (Fig. 6D). These results indicated that while p66Shc dependence of SFN -induced apoptosis was neither a cell line-specific phenomenon nor influenced by the p53 status, the SFN-mediated induction of total p66Shc protein in MCF-7 cells may be p53-dependent.
Fig. 6.
RNA interference of p66Shc attenuated SFN-induced apoptosis in MCF-7 cells. A: Immunoblotting for total and S36 phosphorylated p66Shc using lysates from MCF -7 cells treated with dimethyl sulfoxide (DMSO; control) or SFN (10 or 20 μM) for the indicated time periods. Numbers above the bands represent densitometric quantitation of change in protein level relative to corresponding DMSO-treated control at each time point. B: Immunofluorescence microscopic analysis for p66Shc in Mito -GFP transfected MCF-7 cells after 8-hour treatment with DMSO or 10 μM SFN. C: Immunoblotting for total and S36 phosphorylated p66Shc using lysates from MCF -7 cells transiently transfected with a control (nonspecific) siRNA or a p66Shc- targeted siRNA and treated for 24 hours with DMSO or 10 μM SFN. Numbers on top of the immunoreactive bands represent changes in protein levels relative to control siRNA transfected cells treated with DMSO. D: Quantitation of histone-associated DNA fragment release into the cytosol in MCF-7 cells transiently transfected with a control siRNA or a p66Shc-targeted siRNA and treated for 24 hours with DMSO or 10 μM SFN. The results are expressed as enrichment factor relative to DMSO-treated cells transiently transfected with the control siRNA (mean ± SD, n = 3). Significantly different (P<0.05)compared with arespective DMSO-treated control, and bbetween SFN-treated control siRNA transfected cells and SFN-treated p66Shc siRNA transfected cells by one-way ANOVA followed by Bonferroni’s test. Each experiment was performed at least twice, and representative data from a single experiment are shown.
EFFECT OF PIN1 OVEREXPRESSION ON SFN-INDUCED APOPTOSIS
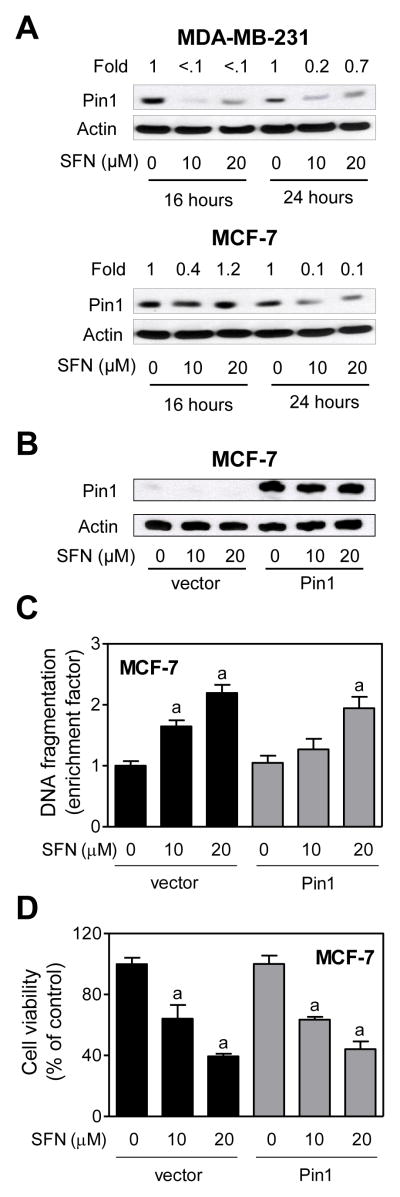
Because Pin1 is involved in regulation of oxidative stress-induced mitochondrial translocation of p66Shc as well as apoptosis [Pinton et al., 2007], we determined the effect of SFN treatment on its protein level using MDA-MB-231 and MCF-7 cells. Interestingly, SFN treatment resulted in a decrease in level of Pin1 protein in both cell lines at 16 and 24 hour time points (Fig 7A). However, stable overexpression of Pin1 (Fig. 7B) failed to confer any protection against SFN-induced apoptosis (Fig. 7C) or cell growth inhibition (Fig. 7D). These results suggested that Pin1 protein level per se did not have any meaningful impact on SFN-induced apoptosis at least in the MCF-7 cell line.
Fig. 7.
Peptidyl prolyl isomerase (Pin1)was dispensable for D,L-sulforaphane (SFN)-induced apoptosis. A: Immunoblotting for Pin1 using lysates from MDA-MB-231 and MCF-7 cells treated for 16-or 24 -hour with dimethyl sulfoxide (DMSO)or the indicated concentrations of SFN. Numbers above the bands represent densitometric quantitation of change in protein level relative to corresponding DMSO-treated control at each time point. B: Immunoblotting for Pin1 using lysates for MCF-7 cells stably transfected with empty vector or vector encoding for myc-Pin1 and treated for 24 hours with DMSO or the indicated concentrations of SFN. C: Histone-associated DNA fragment release into the cytosol, and D: cell viability in MCF-7 cells stably transfected with empty vector or vector encoding for myc-Pin1 and treated for 24 hours with DMSO or the indicated concentrations of SFN. Results are expressed relative to DMSO-treated cells transfected with empty vector (mean ± SD, n = 3). Significantly different (P<0.05) compared with arespective DMSO-treated control, and bbetween SFN-treated empty vector transfected cells and SFN-treated myc-Pin1 overexpressing cells by one-way ANOVA followed by Bonferroni’s test. Each experiment was done at least twice, and representative data from a single experiment are shown.
ROLE OF PKCβ IN SFN -INDUCED APOPTOSIS
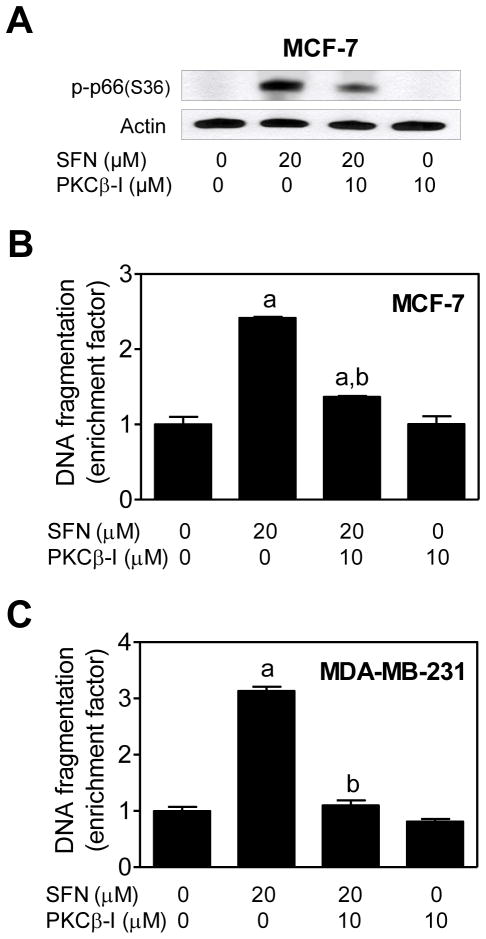
PKCβ has been implicated in S36 phosphorylation of p66Shc [Pinton et al., 2007]. We used a cell-permeable pharmacological inhibitor of PKCβ (hereafter abbreviated as PKCβ-I) to study its contribution in SFN-mediated apoptosis. SFN-mediated S36 phosphorylation of p66Shcwas inhibited markedly in the presence of PKCβ-I in both MCF-7 (Fig. 8A) and MDA-MB-231 cells (results not shown). SFN -induced apoptosis in MCF-7 (Fig. 8B) and MDA-MB-231 cells (Fig. 8C) was also significantly attenuated in the presence of PKCβ-I. These results indicated that the SFN-mediated S36 phosphorylation of p66Shcin MCF -7 and MDA-MB-231 cells was mediated by PKCβ.
Fig. 8.
Phosphorylation (S36) of p66Shc was mediated by protein kinase Cβ (PKCβ). A: Immunoblotting for S36 phosphorylated p66Shc using lysates from MCF -7 cells treated for 24 hours with 20 μM D,L-sulforaphane (SFN)and/or 10 μM PKCβ inhibitor (PKCβ-I). B: Histone-associated DNA fragment release into the cytosol in MCF-7 cells, and C: Histone-associated DNA fragment release into the cytosol in MDA-MB-231 cells after 24-hour treatment with 20 μM SFN and/or 10 μM PKCβ inhibitor (PKCβ-I). Results are expressed as enrichment factor relative to DMSO-treated control cells (mean ± SD, n = 3). Significantly different (P<0.05) compared with aDMSO-treated control, and bSFN treatment groups in the absence or presence of the PKCβ-I by one-way ANOVA followed by Bonferroni’s test.
DISCUSSION
Expression of total and tyrosine phosphorylated p66Shc have been shown to be strong and independent predictors of treatment failure in breast cancer patients [Frackelton et al., 2006]. The p66Shc protein is upregulated by steroid hormones in hormone-sensitive cancer cells [Lee et al., 2004]. Cells lacking p66Shc protein exhibit resistance towards oxidation of ROS-sensitive chemical probes and reduced accumulation of endogenous oxidative stress marker 8-oxo-guanosine [Trinei et al., 2002; Orsini et al., 2004]. The p66Shc knockout mice exhibit diminished levels of systemic (isoprostane) as well as intracellular (8-oxo-guanosine) markers of oxidative stress [Trinei et al., 2002; Nemoto and Finkel, 2002; Napoli et al., 2003]. Genetic deletion of p66 Shc in cells also confers protection against apoptosis induced by ultraviolet radiation, staurosporin, and growth factor deprivation [Migliaccio et al., 1999; Orsini et al., 2004; Pacini et al., 2004]. Results of the present study indicate that p66Shc protein plays an important role in SFN-induced apoptosis. This conclusion is based on the following observations: (a) immortalized MEF derived from p66Shc knockout mice are significantly more resistant to SFN-induced apoptosis and collapse of mitochondrial membrane potential compared with those derived from wild -type mice, and (b) siRNA-mediated knockdown of p66Shc protein confers significant protection against SFN-induced apoptosis in human breast cancer cells. Moreover, the p66Shc dependence of SFN-induced apoptosis is neither a cell line-specific phenomenon nor influenced by the p53 or estrogen-receptor status because MDA -MB-231 and MCF-7 cells behave similarly. Another important conclusion from the present study is that p66Shc protein functions upstream of ROS production and collapse of mitochondrial membrane potential in execution of SFN-induced apoptosis. However, it remains to be determined whether SFN treatment increases p66Shc-mediated oxidation of reduced cytochrome c leading to ROS production.
We have shown previously that normal prostate and bronchial epithelial cells are significantly more resistant to SFN-induced apoptosis compared with prostate cancer and lung cancer cells [Choi and Singh, 2005]. A similar selectivity is discernible in breast cancer cells as MCF-10A cell line is resistant to SFN-induced apoptosis (Fig. 2C). Previous studies from our laboratory have also indicated that SFN-induced apoptosis in prostate cancer cells is initiated by ROS [Singh et al., 2005]. The results of the present study in breast cancer cells are consistent with these previous observations because MCF-7 and MDA-MB-231 cells with stable overexpression of Mn-SOD are nearly fully resistant to SFN-induced apoptosis.
Despite lack of a mitochondria-targeting sequence in p66 Shc, the PKCβ-mediated S36 phosphorylation of p66Shc has been shown to cause its mitochondrial translocation in an oxidative environment [Pinton et al., 2007]. Oxidant-induced mitochondrial translocation of p66Shc is mediated by Pin1 [Pinton et al., 2007]. Interestingly, Pin1 has been shown to facilitate cytokine-induced survival of eosinophils by suppressing Bax activation [Shen et al., 2009]. We found that SFN treatment robustly increases S36 phosphorylation of p66Shc. Moreover, SFN-mediated mitochondrial translocation of p66Shc is discernible in both MDA-MB-231 and MCF-7 cells. However, stable overexpression of Pin1 fails to confer any protection against SFN-induced apoptosis at least in MCF-7 cells. These results suggest that p66Shc dependence of SFN-induced apoptosis is not influenced by Pin1 expression level.
It is interesting to note that SFN treatment markedly decreases Pin1 protein level in both MDA-MB-231 and MCF-7 cells. The Pin1 protein, which catalyzes cis/trans isomerization of phospho-Ser/Thr-Pro bonds, has entertained intense scrutiny in the past couple of years in the context of cancer. The Pin1 is highly expressed in HER-2 positive human breast cancers [Lam et al., 2008]. The Pin1 ablation is highly effective in preventing Neu-or R as-mediated induction of cyclin D1 and mammary carcinogenesis in mice [Wulf et al., 2004]. Khanal et al. (2010) demonstrated that Pin1 interacted with mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1 leading to enhanced HER-2 expression and cellular transformation. Moreover, Pin1 induction was shown to contribute to epithelial-mesenchymal transition in tamoxifen-resistant breast cancer cells [Kim et al., 2009]. The Pin1 overexpression is also associated with poor differentiation and survival in oral squamous cell carcinoma [Leung et al., 2009]. Finally, Pin1 has been shown to be a target of Notch1 in human breast cancers [Rustighi et al., 2009]. Further studies are needed to determine if suppression of Pin1 protein level by SFN(Fig. 7A) contributes to its cancer chemopreventive activity. We have shown previously that oral SFN administration significantly inhibits pulmonary metastasis in a transgenic mouse model of prostate cancer [Singh et al., 2009]. Because Pin1 is implicated in epithelial-mesenchymal transition, a process by which polarized epithelial cells assume a phenotype to become highly motile, it is possible that anti-metastatic effect of SFN is partly mediated by Pin1 suppression. Future determination of the effect of SFN treatment on epithelial -mesenchymal transition would partly validate this hypothesis.
In conclusion, the present study indicates that SFN treatment causes PKCβ-mediated S36 phosphorylation of p66Shc and that this protein functions upstream of ROS production and collapse of mitochondrial membrane potential in execution of SFN-induced apoptosis.
Acknowledgments
This investigation was supported by United States Public Health Service grant RO1 CA115498-06 awarded by the National Cancer Institute. Immortalized mouse embryonic fibroblast derived from wild-type and p66Shc knockout mice were generously provided by Dr. T. Finkel and S. Nemoto. This research project used the UPCI Flow Cytometry Facility supported in part by United States Public Health Service grant P30CA047904.
References
- Barcelo S, Gardiner JM, Gescher A, Chipman JK. CYP2E1-mediated mechanism of anti-genotoxicity of the broccoli constituent sulforaphane. Carcinogenesis. 1996;17:277–282. doi: 10.1093/carcin/17.2.277. [DOI] [PubMed] [Google Scholar]
- Bertl E, Bartsch H, Gerhauser C. Inhibition of angiogenesis and endothelial cell functions are novel sulforaphane-mediated mechanisms in chemoprevention. Mol Cancer Ther. 2006;5:575–585. doi: 10.1158/1535-7163.MCT-05-0324. [DOI] [PubMed] [Google Scholar]
- Choi S, Singh SV. Bax and Bak are required for apoptosis induction by sulforaphane, a cruciferous vegetable-derived cancer chemopreventive agent. Cancer Res. 2005;65:2035–2043. doi: 10.1158/0008-5472.CAN-04-3616. [DOI] [PubMed] [Google Scholar]
- Chung FL, Conaway CC, Rao CV, Reddy BS. Chemoprevention of colonic aberrant crypt foci in Fischer rats by sulforaphane and phenethyl isothiocyanate. Carcinogenesis. 2000;21:2287–2291. doi: 10.1093/carcin/21.12.2287. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry. 2001;56:5–51. doi: 10.1016/s0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- Frackelton AR, Jr, Lu L, Davol PA, Bagdasaryan R, Hafer LJ, Sgroi DC. p66 Shc and tyrosine-phosphorylated Shc in primary breast tumors identify patients likely to relapse despite tamoxifen therapy. Breast Cancer Res. 2006;8:R73. doi: 10.1186/bcr1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamet-Payrastre L, Li P, Lumeau S, Cassar G, Dupont MA, Chevolleau S, Gasc N, Tulliez J, Terce F. Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res. 2000;60:1426–1433. [PubMed] [Google Scholar]
- Giorgio M, Migliaccio E, Orsini F, Paolucci D, Moroni M, Contursi C, Pelliccia G, Luzi L, Minucci S, Marcaccio M, Pinton P, Rizzuto R, Bernardi P, Paolucci F, Pelicci PG. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Hahm ER, Singh SV. Sulforaphane inhibits constitutive and interleukin-6-induced activation of signal transducer and activator of transcription 3 in prostate cancer cells. Cancer Prev Res. 2010;3:484–494. doi: 10.1158/1940-6207.CAPR-09-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht SS. Inhibition of carcinogenesis by isothiocyanates. Drug Metab Rev. 2000;32:395–411. doi: 10.1081/dmr-100102342. [DOI] [PubMed] [Google Scholar]
- Herman-Antosiewicz A, Johnson DE, Singh SV. Sulforaphane causes autophagy to inhibit release of cytochrome C and apoptosis in human prostate cancer cells. Cancer Res. 2006;66:5828–5835. doi: 10.1158/0008-5472.CAN-06-0139. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal P, Namgoong GM, Kang BS, Woo ER, Choi HS. The prolyl isomerase Pin1 enhances HER-2 expression and cellular transformation via its interaction with mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1. Mol Cancer Ther. 2010;9:606–616. doi: 10.1158/1535-7163.MCT-09-0560. [DOI] [PubMed] [Google Scholar]
- Kim MR, Choi HK, Cho KB, Kim HS, Kang KW. Involvement of Pin1 induction in epithelial-mesenchymal transition of tamoxifen-resistant breast cancer cells. Cancer Sci. 2009;100:1834–1841. doi: 10.1111/j.1349-7006.2009.01260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim EH, Eom YW, Kim WH, Kwon TK, Lee SJ, Choi KS. Sulforaphane sensitizes tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-resistant hepatoma cells to TRAIL-induced apoptosis through reactive oxygen species-mediated up-regulation of DR5. Cancer Res. 2006;66:1740–1750. doi: 10.1158/0008-5472.CAN-05-1568. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kwon KH, Jung JY, Han HS, Shim JH, Oh S, Choi KH, Choi ES, Shin JA, Leem DH, Soh Y, Cho NP, Cho SD. Sulforaphane increases cyclin-dependent kinase inhibitor, p21 protein in human oral carcinoma cells and nude mouse animal model to induce G2/M cell cycle arrest. J Clin Biochem Nutr. 2010;46:60–67. doi: 10.3164/jcbn.09-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PB, Burga LN, Wu BP, Hofstatter EW, Lu KP, Wulf GM. Prolyl isomerase Pin1 is highly expressed in Her2-positive breast cancer and regulates erbB2 protein stability. Mol Cancer. 2008;7:91. doi: 10.1186/1476-4598-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Igawa T, Chen SJ, Van Bemmel D, Lin JS, Lin FF, Johanasson SL, Christman JK, Lin MF. p66Shc protein is upregulated by steroid hormones in hormone-sensitive cancer cells and in primary prostate carcinomas. Int J Cancer. 2004;108:672–678. doi: 10.1002/ijc.11621. [DOI] [PubMed] [Google Scholar]
- Leung KW, Tsai CH, Hsiao M, Tseng CJ, Ger LP, Lee KH, Lu PJ. Pin1 overexpression is associated with poor differentiation and survival in oral squamous cell carcinoma. Oncol Rep. 2009;21:1097–1104. doi: 10.3892/or_00000329. [DOI] [PubMed] [Google Scholar]
- Li W, Yu SW, Kong AN. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. J Biol Chem. 2006;281:27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang T, Korkaya H, Liu S, Lee HF, Newman B, Yu Y, Clouthier SG, Schwartz SJ, Wicha MS, Sun D. Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin Cancer Res. 2010;16:2580–2590. doi: 10.1158/1078-0432.CCR-09-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman SM, Hawk ET. Cancer prevention: from 1727 to milestones of the past 100 years. Cancer Res. 2009;69:5269–5284. doi: 10.1158/0008-5472.CAN-09-1750. [DOI] [PubMed] [Google Scholar]
- Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5:e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi L, Wang X, Govind S, Hood BL, Veenstra TD, Conrads TP, Saha DT, Goldman R, Chung FL. The role of protein binding in induction of apoptosis by phenethyl isothiocyanate and sulforaphane in human non-small lung cancer cells. Cancer Res. 2007;67:6409–6416. doi: 10.1158/0008-5472.CAN-07-0340. [DOI] [PubMed] [Google Scholar]
- Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- Napoli C, Martin-Padura I, de Nigris F, Giorgio M, Mansueto G, Somma P, Condorelli M, Sica G, De Rosa G, Pelicci P. Deletion of the p66Shc longevity gene reduces systemic and tissue oxidative stress, vascular cell apoptosis, and early atherogenesis in mice fed a high-fat diet. Proc Natl Acad Sci USA. 2003;100:2112–2116. doi: 10.1073/pnas.0336359100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295:2450–2452. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- Orsini F, Migliaccio E, Moroni M, Contursi C, Raker VA, Piccini D, Martin-Padura I, Pelliccia G, Trinei M, Bono M, Puri C, Tacchetti C, Ferrini M, Mannucci R, Nicoletti I, Lanfrancone L, Giorgio M, Pelicci PG. The life span determinant p66Shc localizes to mitochondria where it associates with mitochondrial heat shock protein 70 and regulates trans-membrane potential. J Biol Chem. 2004;279:25689–25695. doi: 10.1074/jbc.M401844200. [DOI] [PubMed] [Google Scholar]
- Pacini S, Pellegrini M, Migliaccio E, Patrussi L, Ulivieri C, Ventura A, Carraro F, Naldini A, Lanfrancone L, Pelicci P, Baldari CT. p66SHC promotes apoptosis and antagonizes mitogenic signaling in T cells. Mol Cell Biol. 2004;24:1747–1757. doi: 10.1128/MCB.24.4.1747-1757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Grignani F, Pawson T, Pelicci PG. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- Pinton P, Rimessi A, Marchi S, Orsini F, Migliaccio E, Giorgio M, Contursi C, Minucci S, Mantovani F, Wieckowski MR, Del Sal G, Pelicci PG, Rizzuto R. Protein kinase C beta and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science. 2007;315:659–663. doi: 10.1126/science.1135380. [DOI] [PubMed] [Google Scholar]
- Powolny AA, Bommareddy A, Hahm ER, Normolle DP, Beumer JH, Nelson JB, Singh SV. Chemopreventative potential of the cruciferous vegetable constituent phenethyl isothiocyanate in a mouse model of prostate cancer. J Natl Cancer Inst. 2011;103:571–584. doi: 10.1093/jnci/djr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci USA. 2006;103:15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, Kaplan F, Capobianco A, Pece S, Di Fiore PP, Del Sal G. The prolyl-isomerase Pin 1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol. 2009;11:133–142. doi: 10.1038/ncb1822. [DOI] [PubMed] [Google Scholar]
- Shen ZJ, Esnault S, Schnizel A, Borner C, Malter JS. The peptidyl-prolyl isomerase Pin1 facilitates cytokine-induced survival of eosinophils by suppressing Bax activation. Nat Immunol. 2009;10:257–265. doi: 10.1038/ni.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu L, Cheung KL, Khor TO, Chen C, Kong AN. Phytochemicals: cancer chemoprevention and suppression of tumor onset and metastasis. Cancer Metastasis Rev. 2010;29:483–502. doi: 10.1007/s10555-010-9239-y. [DOI] [PubMed] [Google Scholar]
- Singh SV, Herman-Antosiewicz A, Singh AV, Lew KL, Srivastava SK, Kamath R, Brown KD, Zhang L, Baskaran R. Sulforaphane-induced G2/M phase cell cycle arrest involves checkpoint kinase 2-mediated phosphorylation of cell division cycle 25C. J Biol Chem. 2004a;279:25813–25822. doi: 10.1074/jbc.M313538200. [DOI] [PubMed] [Google Scholar]
- Singh AV, Xiao D, Lew KL, Dhir R, Singh SV. Sulforaphane induces caspase-mediated apoptosis in cultured PC-3 human prostate cancer cells and retards growth of PC-3 xenografts in vivo. Carcinogenesis. 2004b;25:83–90. doi: 10.1093/carcin/bgg178. [DOI] [PubMed] [Google Scholar]
- Singh SV, Srivastava SK, Choi S, Lew KL, Antosiewicz J, Xiao D, Zeng Y, Watkins SC, Johnson CS, Trump DL, Lee YJ, Xiao H, Herman-Antosiewicz A. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. J Biol Chem. 2005;280:19911–19924. doi: 10.1074/jbc.M412443200. [DOI] [PubMed] [Google Scholar]
- Singh SV, Warin R, Xiao D, Powolny AA, Stan SD, Arlotti JA, Zeng Y, Hahm ER, Marynowski SW, Bommareddy A, Desai D, Amin S, Parise RA, Beumer JH, Chambers WH. Sulforaphane inhibits prostate carcinogenesis and pulmonary metastasis in TRAMP mice in association with increased cytotoxicity of natural killer cells. Cancer Res. 2009;69:2117–2125. doi: 10.1158/0008-5472.CAN-08-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surh YJ. Cancer chemoprevention with dietary phytochemicals. Nature Rev Cancer. 2003;3:768–780. doi: 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- Trinei M, Giorgio M, Cicalese A, Barozzi S, Ventura A, Migliaccio E, Milia E, Padura IM, Raker VA, Maccarana M, Petronilli V, Minucci S, Bernardi P, Lanfrancone L, Pelicci PG. A p53-p66Shc signaling pathway controls intracellular redox status, levels of oxidation-damaged DNA and oxidative stress-induced apoptosis. Oncogene. 2002;21:3872–3878. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 2004;23:3397–3407. doi: 10.1038/sj.emboj.7600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D, Choi S, Johnson DE, Vogel VG, Johnson CS, Trump DL, Lee YJ, Singh SV. Diallyl trisulfide-induced apoptosis in human prostatecancer cells involves c -Jun N-terminal kinase and extracellular-signal regulated kinase-mediated phosphorylation of Bc1–2. Oncogene. 2004;23:5594–5606. doi: 10.1038/sj.onc.1207747. [DOI] [PubMed] [Google Scholar]
- Xiao D, Powolny AA, Antosiewicz J, Hahm ER, Bommareddy A, Zeng Y, Desai D, Amin S, Herman-Antosiewicz A, Singh SV. Cellular responses to cancer chemopreventive agent D, L-sulforaphane in human prostate cancer cells are initiated by mitochondrial reactive oxygen species. Pharm Res. 2009;26:1729–1738. doi: 10.1007/s11095-009-9883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D, Vogel V, Singh SV. Benzyl isothiocyanate-induced apoptosis in human breast cancer cells is initiated by reactive oxygen species and regulated by Bax and Bak. Mol Cancer Ther. 2006;5:2931–2945. doi: 10.1158/1535-7163.MCT-06-0396. [DOI] [PubMed] [Google Scholar]
- Xu C, Shen G, Chen C, Gelinas C, Kong AN. Suppression of NF-kappa B and NF-kappa B-regulated gene expression by sulforaphane and PEITC through IkappaBalpha, IKK pathway in human prostate cancer PC-3 cells. Oncogene. 2005;24:4486–4495. doi: 10.1038/sj.onc.1208656. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kensler TW, Cho CG, Posner GH, Talalay P. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc Natl Acad Sci USA. 1994;91:3147–3150. doi: 10.1073/pnas.91.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]