Abstract
Channelrhodopsins act as photoreceptors for control of motility behavior in flagellates and are widely used as genetically targeted tools to optically manipulate the membrane potential of specific cell populations (“optogenetics”). The first two channelrhodopsins were obtained from the model organism Chlamydomonas reinhardtii (CrChR1 and CrChR2). By homology cloning we identified three new channelrhodopsin sequences from the same genus, CaChR1, CyChR1 and CraChR2, from C. augustae, C. yellowstonensis and C. raudensis, respectively. CaChR1 and CyChR1 were functionally expressed in HEK293 cells, where they acted as light-gated ion channels similar to CrChR1. However, both, which are similar to each other, differed from CrChR1 in current kinetics, inactivation, light intensity dependence, spectral sensitivity, and dependence on the external pH. These results show that extensive channelrhodopsin diversity exists even within the same genus, Chlamydomonas. The maximal spectral sensitivity of CaChR1 was at 520 nm at pH 7.4, about 40 nm red-shifted as compared to that of CrChR1 under the same conditions. CaChR1 was successfully expressed in Pichia pastoris and exhibited an absorption spectrum identical to the action spectrum of CaChR1-generated photocurrents. The red-shifted spectra and the lack of fast inactivation in CaChR1- and CyChR1-generated currents are features desirable for optogenetics applications.
INTRODUCTION
Light regulates the swimming behavior of most photosynthetic microorganisms, helping them find optimal living conditions. Shortly after the discovery of retinal-containing proteins (rhodopsins) mediating photomotility behavior in archaea (1–2), retinal was found to be also essential for light control of motility of the phylogenetically distant green flagellate alga Chlamydomonas reinhardtii (3). The algal rhodopsins remained unidentified until a C. reinhardtii expressed sequence tag (EST) project (4) revealed the presence of proteins homologous to the archaeal receptors. Knock-down experiments (5–7) demonstrated that photoexcitation of two rhodopsins in C. reinhardtii identified with EST information led to generation of the transmembrane currents known to drive photomotility behavior in native cells (8). When introduced into animal cells, these proteins act as light-activated ion channels, whereupon they were named “channelrhodopsins” (9–10).
The two channelrhodopsin homologs from C. reinhardtii were designated as ChR1 and ChR2 (9–10), and independently as CSRA and CSRB (for Chlamydomonas sensory rhodopsins A and B (5–6), and Acop-1 and Acop-2 (for archaeal type Chlamydomonas opsin 1 and 2 (11). Similar proteins were subsequently identified in related algae: VChR1 and VChR2 in Volvox carteri (12–13) and MChR1 in Mesostigma viride (14). In this work we found channelrhodopsins with different properties even in the same genus Chlamydomonas. To distinguish them, we precede the channelrhodopsin abbreviations with the initial letters of the genus and species of their native host, following the practice in earlier microbial rhodopsin studies. Today, channelrhodopsins are used in many fields of biomedical research, especially neuroscience, to manipulate the membrane potential by light, a technique referred to as “optogenetics” (15–16). This approach offers an unprecedented specificity and both temporal and spatial precision of stimulation in cultured cells and living animals. So far channelrhodopsins have been used to study normal brain and cardiac function, stem cell differentiation, and a wide range of mental and neurological disorders, such as Parkinson’s disease, autism, schizophrenia, drug abuse, and depression (17). Moreover, channelrhodopsins are under study for future gene therapy for vision restoration, with promising results already obtained in rodent models (18–19). Therefore, further research into channelrhodopsins is valuable for purposes that reach beyond understanding mechanisms of algal phototaxis.
CrChR1 and CrChR2 each possess characteristics desirable for optogenetics applications, but neither of them is ideal. The kinetics of the currents generated by CrChR1 is faster than those of the currents generated by CrChR2 (20–21), which allows for photostimulation at higher stimulus frequencies. On the other hand, CrChR1-generated currents show rapid inactivation, i.e., a decrease in the current amplitude upon sustained illumination, which is much slower for CrChR2-generated currents (20–21). The maximal spectral sensitivity of CrChR1-generated currents in native C. reinhardtii cells is at 505 nm, compared to 470 nm for CrChR2-generated currents (5). This is an advantage for optogenetics, as using long-wavelength excitation light minimizes its scattering by biological tissues and absorption by hemoglobin. However, in heterologous systems the long-wavelength spectral sensitivity of CrChR1 can only be observed at non-physiological acidic pH (7, 20).
The model organism C. reinhardtii is only one of at least several hundred different species in the genus Chlamydomonas (http://www.algaebase.org). Their habitats include soil, temporary pools, eutrophic lakes, and melting snow. Rhodopsin-mediated photocurrents similar to those in C. reinhardtii have been recorded in many other Chlamydomonas species besides C. reinhardtii ((22) and our unpublished observations). Their photocurrents strongly suggested that they too harbor channelrhodopsin homologs, but it was unclear whether characteristics of CrChR1 and CrChR2 were universal for the whole genus. We hypothesized that channelrhodopsin variants from Chlamydomonas species inhabiting ecological niches other than those of C. reinhardtii may differ from their C. reinhardtii counterparts, and may offer alternative, possibly advantageous, properties for optogenetic applications. C. augustae and C. yellowstonensis are “snow algae”, i.e., species that contribute to microbial communities of regions permanently or seasonally covered with snow and are often responsible for the appearance of colored snow (for review see (23)). An extreme psychrophilic strain of C. raudensis was isolated from a permanently ice-covered lake in Antarctica (for review see (24)).
Using homology cloning, we identified three new channelrhodopsin variants in these Chlamydomonas species. The sequences from C. augustae and C. yellowstonensis contain molecular determinants characteristic of CrChR1, as compared to CrChR2. Yet, photocurrents measured upon their expression in HEK293 (human embryonic kidney) cells differ from those generated by the “classical” CrChR1 in several important aspects. We partially purified channelrhodopsin from C. augustae by expression in the methylotrophic yeast Pichia pastoris and showed that its absorption spectrum closely matched the action spectrum of its channel activity. Our findings revealed that remarkable channelrhodopsin diversity exists even within the same genus Chlamydomonas. The slower inactivation of photocurrents and red-shifted spectral sensitivity at physiological pH make the new channelrhodopsins from C. augustae and C. yellowstonensis promising candidates for future optogenetic applications, although their expression in target cells needs to be optimized.
MATERIALS AND METHODS
Strains and culture conditions
Algal strains were obtained from the Culture Collection of Algae at the University of Texas (UTEX), Austin, TX and the National Center for Culture of Marine Phytoplankton (CCMP), West Boothbay Harbor, ME. Chlamydomonas augustae (UTEX SNO134) and Chlamydomonas yellowstonensis (UTEX B SNO155) were grown in Bold 1NV medium (25) under illumination of 14 μmol photon × m−2 × s−1 at 4°C or 16° (C. augustae), or 4°C (C. yellowstonensis). Chlamydomonas raudensis (CCMP 1619) was grown in modified Bold’s basal medium (26) containing vitamins and three times the amount of nitrogen and vitamins as described at the website of the Culture Collection of Algae and Protozoa (UK) under illumination of 5 μmol photon × m−2 × s−1 at 4°C. Illumination was set to a 16 h light, 8 h dark cycle and was provided by cool-white fluorescent lamps.
Homology cloning of new channelopsin sequences
Algae were inoculated from plates into 25 mL liquid medium in 250 mL flasks and grown for 18 hr at 16°C (C. yellowstonensis) or 4°C (C. raudensis). Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA). First strand cDNA was synthesized with the Transcriptor first strand cDNA synthesis kit (Roche Diagnostics, Mannheim, Germany) using oligo-d(T) primer. The degenerate primers were designed according to the conserved regions of the four earlier known channelopsins from C. reinhardtii and V. carteri, and their degeneracy was reduced by including only the most frequently used codons. The forward primer was SYHB-F (5′-TGC GGN TGG GAG GAG RTN TA-3′), and the reverse primers were SYOG-R (5′ AGR ATR TGC TCR TGR ATC -3′) for C. augustae and C. yellowstonensis, and SYLPEF-R (5′-RCC CTT SGG NAC SGT RTG-3′) for C. raudensis. The PCR (polymerase chain reaction) program was as follows: denaturation at 98°C for 2 min, followed by 30 cycles of 98°C for 30 s, 49°C for 45 s, 72°C for 55 s, and final extension at 72°C for 5 min. PCR fragments were cloned into the pGEM-T Easy vector (Promega, Madison, WI) and sequenced. For the fragments that showed homology with channelopsins, 3′ and 5′ RACE (rapid amplification of cDNA ends) was performed using the SMARTer RACE cDNA amplification kit (Clontech Laboratories, Mountain View, CA). Overlapping RACE fragments were joined by fusion PCR to obtain full-length cDNA, which was cloned into the pGEM-T Easy vector and sequenced. Fragments encoding for the 7TM domains (residues 1-352 for the C. augustae and C. raudensis sequences, and 1-354 for C. yellowstonensis) were inserted between BamHI and NotI sites to replace the VcChR1 sequence in the pcDNA3.1/VcChR1-EYFP mammalian expression vector provided by K. Deisseroth (Stanford University). The vector map and sequence are available at the Optogenetics Resource Center website. The presence of a fluorescent tag is not expected to affect channelrhodopsin properties, as has been shown by quantitative comparison of photocurrents generated by YFP-, mCherry- and myc-tagged CrChR2 (27).
Whole-cell patch clamp recording in HEK293 cells
HEK293 cells were transfected using the TransPass COS/293 transfection reagent (New England Biolabs, Ipswich, MA). All-trans-retinal (Sigma) was added as a stock solution in ethanol at the final concentration of 2.5 μM, unless otherwise indicated. Measurements were performed 48-72 h after transfection with an Axopatch 200B amplifier (Molecular Devices, Union City, CA). The signals were digitized with a Digidata 1440A using pClamp 10 software (both from Molecular Devices). Patch pipettes with resistances 2–5 MΩ were fabricated from borosilicate glass and filled with the following solution (in mM): KCl 126, MgSO4 2, CaCl2 0.5, EGTA 5, HEPES 25, pH 7.2. The bath solution contained (in mM): NaCl 150, CaCl2 1.8, KCl 4, MgCl2 1, glucose 5, HEPES 10, pH 7.4, unless otherwise indicated. For experiments at pH 9, Tris was used in the bath solution instead of HEPES. Unless otherwise indicated, the holding potential was −60 mV. Light excitation was provided by a Polychrome IV light source (T.I.L.L. Photonics GMBH, Grafelfing, Germany) pulsed with a mechanical shutter (Uniblitz Model LS6, Vincent Associates, Rochester, NY; half-opening time 0.5 ms). The light intensity was attenuated with the built-in Polychrome system or with neutral density filters. Maximal quantum density at the focal plane of the 40x objective lens was ~2 × 1022 photons × m−2.
Channelopsin expression and purification from Pichia.
The 7TM domain of CaChR1 (1-352 residues) was cloned into the pPIC9K vector (Invitrogen, Carlsbad, CA) between EcoRI and NotI sites in frame with two TEV protease sites at the N-terminus and before a C-terminal 6His-tag. The sequence encoding EYFP was not included in this construct. The resultant plasmid was linearized with BspEI and transformed into the P. pastoris SMD1168 (his4, pep4) strain by electroporation according to the manufacturer's instructions. First, transformants were screened for plasmid integration by their ability to grow on hystidine-deficient plates, and second, for multiple inserts by their geneticin resistance. A single colony that grew on 4 mg/mL geneticin was selected. A starter culture was inoculated in 500 mL BMMY (buffered minimal methanol yeast) medium. Expression was induced by the addition of 0.5% methanol every 24 h in the presence of 30 μM all-trans-retinal. Cells were grown for two days, harvested by low-speed centrifugation and disrupted by a bead beater. Membrane fragments were collected by centrifugation for 1 h at 48,000 rpm and solubilized by incubation with 2 % dodecyl maltoside for 1 h. The protein was partially purified on a Ni-NTA agarose column (Qiagen, Hilden, Germany). The protein yield was 6.4 mg/L of culture, as estimated from absorbance at 520 nm.
Absorption spectroscopy
Absorption spectrum of partially purified CaChR1 in the UV visible range was recorded on a Cary 4000 spectrophotometer (Varian, Palo Alto, CA).
RESULTS
Channelopsin sequences from C. augustae, C. yellowstonensis and C. raudensis
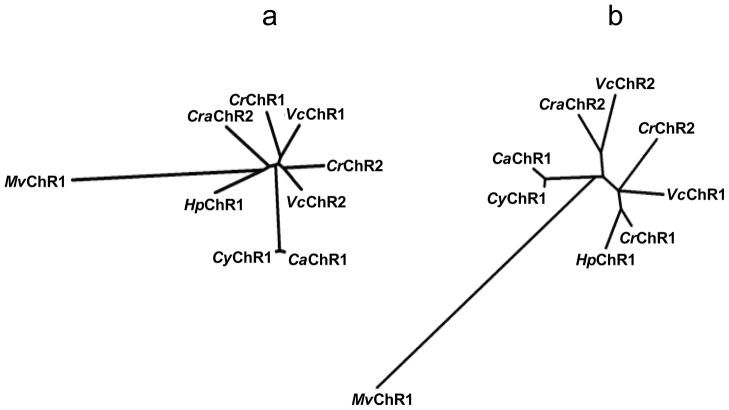
We cloned a channelopsin homolog from each of C. augustae (715 amino acid residues; nucleotide Acc. No. JN596951), C. yellowstonensis (717 residues, Acc. No. JN596948) and C. raudensis (635 residues, Acc. No. JN596949). As in all so far known channelopsins, the new proteins consist of a predicted 7TM (rhodopsin) domain responsible for light-gated channel activity and a C-terminal domain, the function of which is yet unknown. CrChR1/VcChR1 and CrChR2/VcChR2 form two distinct branches on the phylogenetic tree of their 7TM domains (Fig. 1a). The 7TM domains of the new Chlamydomonas sequences do not show closer homology with either the CrChR1/VcChR1 branch or CrChR2/VcChR2 branch, when their overall sequence homology is concerned (Fig. 1a). The 7TM domains from C. augustae and C. yellowstonensis are however very close to each other.
Figure 1.
Phylogenetic trees of the 7TM domains (a) and the C-terminal domains (b) of the so far known channelopsins constructed by the neighbor-joining method. CrChR1, channelrhodopsin 1 from C. reinhardtii; CrChR2, channelrhodopsin 2 from C. reinhardtii; VcChR1, channelrhodopsin 1 from V. carteri; VcChR2, channelrhodopsin 2 from V. carteri; MvChR1, channelrhodopsin 1 from M. viride; CaChR1, channelrhodopsin 1 from C. augustae; CyChR1, channelrhodopsin 1 from C. yellowstonensis; CraChR2, channelrhodopsin 2 from C. raudensis; HpChR1 channelrhodopsin 1 from Haematococcus pluvialis.
Two molecular determinants are conserved in CrChR1/VcChR1 and CrChR2/VcChR2 sequences, respectively, and shown to determine their different properties: 1) Glu87 (CrChR1 numbering) in the predicted first helix is responsible for pH-dependent color tuning and fast channel inactivation of CrChR1, as compared to CrChR2 (20); 2) Tyr226 (CrChR1)/Asn187 (CrChR2) in the predicted fifth helix confers differences in spectral sensitivity, inactivation and kinetics between CrChR1 and CrChR2 (21). According to these criteria, the sequences identified in C. augustae and C. yellowstonensis (CaChR1 and CyChR1, respectively) belong to the CrChR1/VcChR1 class (Fig. 2). This placement is confirmed by their red-shifted spectra (see below), characteristic of the CrChR1/VcChR1 class, as compared to the CrChR2/VcChR2 class (13, 20). In contrast, the sequence from C. raudensis (CraChR2) belongs to the CrChR2/VcChR2 class according to the two above-mentioned molecular determinants. However, since this sequence failed to generate photocurrents in HEK cells (see below), the lack of functional data makes this placement only tentative.
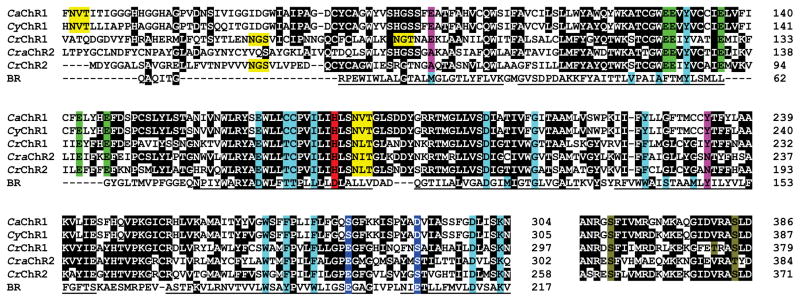
Figure 2.
Partial alignment of Chlamydomonas channelopsin and BR sequences. Black background indicates conserved identical residues. Turquoise background, positions of the residues that form the retinal-binding pocket in BR. Green background, conserved Glu residues in the predicted second helix. Magenta background, molecular determinants that differentiate CrChR1/VcChR1 from CrChR2/VcChR2. Red background, residues in the position of the proton donor in BR. Blue background, residues in the positions of Glu194 and Glu204 in BR. Yellow background, predicted glycosylation sites. Olive background, conserved residues known to be phosphorylated in CrChR1 or CrChR2. Underlined characters show the regions that form transmembrane helices in BR.
In the 7TM domain, residues at the critical sites characteristic of other known channelrhodopsin apoproteins (channelopsins) are conserved in all three new homologs. These include: 1) Glu in the position of the Schiff base proton acceptor (Asp85 according to bacteriorhodopsin (BR) numbering); 2) His in the position of the Schiff base proton donor (Asp96 in BR); 3) five Glu residues in or near the predicted second helix; 4) Cys128 and Asp156 (CrChR2 numbering) that form a predicted hydrogen bond between the third and fourth helices (28–30). Out of other residues known to form the retinal-binding pocket in BR, Tyr57, Gly122, Trp182, Asp212 and Lys216 (BR numbering) are conserved in new channelopsins, as they are in previously known ones. Positions of Tyr185 and Trp189 (BR numbering) are occupied by Phe residues, as in previously known channelopsins.
Phylogeny of the C-terminal domain does not match that of the 7TM domain, although CaChR1 and CyChR1 again show close similarity (Fig. 1b). No helices are predicted in the C-terminal domains of either of the new channelopsin sequences, in contrast to CrChR1 (5). As in previously known channelrhodopsins, the C-terminal domains of the new channelopsins contain several highly conserved regions with no homology to any other so far known protein interspersed with repeats that vary in length and amino acid composition among different channelopsin variants. Long stretches of Gly-Met repeats and Met-rich regions are found in CaChR1 and CyChR1, and Gln repeats are found in all three new channelopsins. Such repeats, known as homopolymeric tracts (31), occur in many eukaryotic proteins and have been associated with protein-protein (32) or protein-membrane (33) interactions. A highly conserved region of about 40 residues at the very end of the C-terminal domains present in all so far known channelrhodopsins but MvChR1 shows homology to domains in fibrinogen and ABC transporters that are responsible for protein multimerization and protein-protein interaction. In algal cells channelrhodopsins are confined to the membrane area above the eyespot (7, 11) and are associated with acetylated microtubules of the daughter four-membered flagellar rootlet (34). It is plausible that the function of the C-terminal domains is to control appropriate subcellular positioning of channelrhodopsins.
The extracellular N-terminal regions of CaChR1 and CyChR1 contain a predicted conserved N-glycosylation site. Such sites, although at different positions, are also predicted in the N-termini of CrChR1 and several other channelopsins, but not in CraChR2 (Fig. 2). Another such site conserved in all so far known channelopsins is located at the cytoplasmic end of the third predicted transmembrane helix. The existence of predicted N-glycosylation sites in the cytoplasmic domains of proteins has been mostly ignored, although it was confirmed biochemically in at least one example, the α-subunit of mammalian Na+,K+-ATPase (35). Occurrence of glycosylation in heterologously expressed CrChR2 was demonstrated by treatment with peptidyl N-glycosidase F, but its site(s) were not determined (36). A requirement for glycosylation for correct folding and targeting of channelrhodopsins may explain why no functional channelrhodopsin has been produced by expression in E. coli, despite many attempts. All three new channelopsin sequences lack an additional α-helix predicted in the N-terminus of CrChR1 as a signal peptide, as do CrChR2 and both VcChR1 and VcChR2. However, such a helix is predicted in the sequence from Haematococcus pluvialis (nucleotide Acc. No. JN596950). The putative role of this helix in protein targeting needs further investigation.
In CrChR1 and CrChR2, three and one phosphorylated residues, respectively, were identified by phosphoproteomics of the eyespot fractions (37). These residues are found in the cytoplasmic loop next to the 7TM domain that is highly conserved in all so far known channelopsin sequences, with the exception of MvChR1. Out of the three phosphorylated residues of CrChR1, Ser359 is conserved in all five Chlamydomonas channelopsins (Fig. 2), and the corresponding residue (Ser321) is the only phosphorylated site detected in CrChR2 (37). Thr374 is unique for CrChR1 (with Val found at this site in other sequences), and Ser377 is conserved in four sequences with Thr conservative substitution in CraChR2 (Fig. 2). Possibly channelrhodopsin phosphorylation plays a role in the adaptation processes which enable the large range of light intensities over which Chlamydomonas algae exhibit phototaxis (38).
Proteins from psychrophilic organisms show characteristic biases in amino acid composition, compared to their meso- and thermophilic homologs, that are believed to increase flexibility at low temperatures. Among these are decreased percentages of Pro, Arg and Ala residues, and an increased percentage of Ile residues (39–40). The same trends are observed in Chlamydomonas channelopsins: the combined percentages of Pro, Arg and Ala residues in the sequences of CrChR1, CaChR1 and CyChR1 are 20%, 15.7% and 16%, respectively, whereas the percentages of Ile are 4.9%, 5.9% and 5.2%, respectively. Other trends, such as an increased percentage of Gly, were not detected in channelopsins.
Functional characterization of new channelrhodopsins
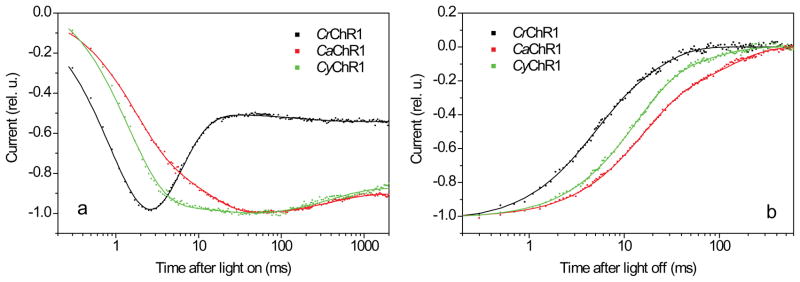
The 7TM domains of all three new channelopsins showed robust expression in the plasma membrane of HEK293 cells, as indicated by fluorescence of their EYFP-tags. CaChR1 and CyChR1 exhibited light-gated channel activity in this system, but no currents could be recorded upon expression of CraChR2. The kinetics of the currents generated by CaChR1 and CyChR1 were however quite different from that generated by CrChR1. Figure 3 shows typical signals recorded at the maximal light intensity under our standard conditions, i.e., bath pH 7.4, and holding potential (Vhold) -60 mV (for more details see Methods). Upon switching on the light, currents generated by CrChR1 underwent a rapid initial rise with a time constant (τ) ~ 1 ms, reached a peak and rapidly (τ ~ 5 ms) decreased under sustained illumination, i.e., inactivated to a lower level (Fig. 3a, black line). In many cells the currents showed a subsequent slight increase with τ ~ 200 ms. This behavior closely resembled the results reported for CrChR1 earlier by others (41). In contrast, the rise of CaChR1- and CyChR1-generated currents was biphasic. The first rapid phase was similar to that of CrChR1-generated currents, but it was followed by a slower rising phase with τ ~ 20 ms (Fig. 3a, red and green lines). The relative contributions of these two components varied from cell to cell. After reaching a peak, CaChR1- and CyChR1-generated currents exhibited very slow inactivation (τ ~ 500 ms).
Figure 3.
(a) Typical kinetics of light-induced currents generated in HEK293 cells by CrChR1 (black dots), CaChR1 (red dots) and CyChR1 (green dots). (b) Decay of the same currents after 2-s illumination. The currents in (a) were normalized to the peak amplitude, and the currents in (b), to the plateau level, and fitted with three (a) or two (b) exponential functions (solid lines). The excitation wavelength was 520 nm for CaChR1 and CyChR1, and 480 nm for CrChR1, which corresponded to their spectral maxima (see below). The traces are average signals measured in response to a series of light pulses delivered with 30-s time intervals. Cells expressing ChRs were selected for EYFP fluorescence before measurements. Bath pH was 7.4, Vhold was -60 mV. For complete ionic conditions, see Methods section.
For all three tested channelrhodopsin variants the response to the first flash showed a larger peak relative to the plateau level (measured at the end of the light pulse), which was not fully recovered even after 30 min dark interval, suggesting a contribution of a very slow adaptation process, or irreversible bleaching of an unstable fraction of the pigment. However, no difference was observed between responses to the second and all subsequent flashes recorded with 30 s dark intervals. Under these conditions, the peak to plateau ratio at the maximal light intensity was 1.7 ± 0.2 (mean ± SEM, n = 8) for CrChR1, close to the earlier reported results (42). For both CaChR1 and CyChR1 this ratio was significantly smaller: 1.2 ± 0.1 (mean ± SEM, n = 12 and n = 6, respectively). The absolute plateau amplitude was 101 ± 25 pA (mean ± SEM, n = 8) for CrChR1, 64 ± 9 pA (mean ± SEM, n = 12) for CaChR1, and 49 ± 13 pA (mean ± SEM, n = 6) for CyChR1. After switching off the light, the currents decayed biexponentially with τ ~ 15 and ~ 120 ms for CaChR1, ~ 13 and ~ 150 ms for CyChR1, which was slower than ~ 4 and ~ 18 ms measured for CrChR1 (Fig. 3b), but close to that for CrChR2 (20).
The most widely used channelrhodopsin variant, CrChR2, generates large enough currents in HEK cells even without the addition of exogenous retinal ((21) and our own unpublished observations), indicating that its trace amount present in these cells is sufficient for reconstitution of functional protein. However, currents in cells transfected with the new channelrhodopsins or CrChR1 were considerably smaller if no exogenous retinal was added: their plateau levels were only ~ 8% for CaChR1 and ~ 26% for CrChR1 (data for CyChR1 are not shown), relative to the results obtained with the respective channelrhodopsins in the presence of 2.5 μM exogenous retinal (our standard conditions).
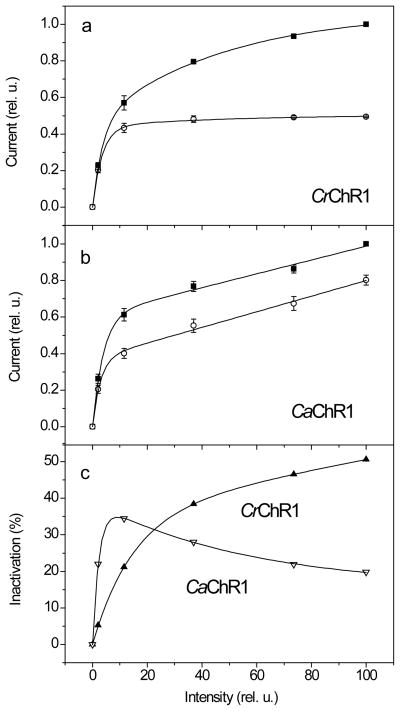
For CrChR1-generated currents, the dependence of the plateau amplitude on the stimulus intensity saturated earlier than that of the peak (Fig. 4a), as was shown previously (41). In fact, the curve for the peak amplitude was biphasic and showed two levels of saturation, the first of which corresponded to that of the plateau level, whereas the second was at more than 10-fold higher light intensity. Therefore, the magnitude of light inactivation, calculated as the difference between the peak and plateau amplitudes relative to the peak amplitude, increased with light intensity and did not saturate even at the highest available intensities (Fig. 4c, black squares). Similar results were reported earlier also for CrChR2 (43) and VcChR1/VcChR2 hybrid (44). In contrast, the curves for both peak and plateau amplitudes of CaChR1-generated currents consisted of two phases (Fig. 4b), so that the magnitude of light inactivation reached the maximum at 10% maximal light intensity and then declined (Fig. 4c, red circles). Similar behavior was observed for CyChR1-generated currents (data not shown).
Figure 4.
(a, b) The dependence of peak (solid squares) and plateau (open circles) amplitudes on the stimulus intensity for currents generated by CrChR1 (a) or CaChR1 (b). Data points are the mean normalized values ± SEM (n = 3 (a) and 5 (b)). For complete ionic conditions, see Methods section. (c) The dependence of light inactivation (calculated as the difference between the peak and plateau amplitudes shown in panels (a) and (b), relative to the peak amplitude) on the stimulus intensity for currents generated by CrChR1 (solid triangles) and CaChR1 (open triangles).
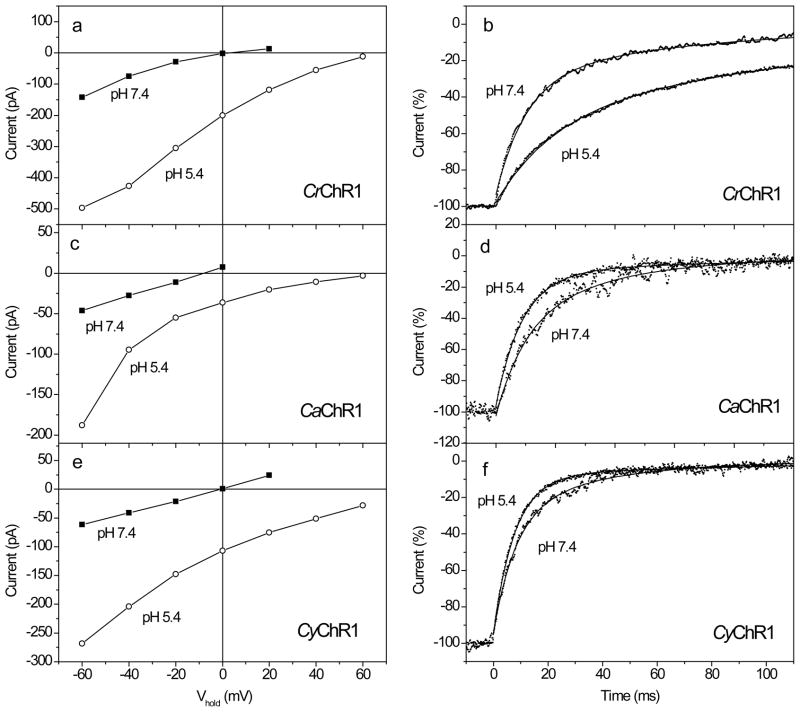
CaChR1- and CyChR1-generated currents showed a typical dependence on the holding potential (Vhold), as described earlier for CrChR1 and other channelrhodopsins (Fig. 5a, c, e). The reversal potentials (Vr) were similar to that for CrChR1 and close to zero under our experimental conditions. Since CrChR1 is known to be a highly proton-selective channel (9), we tested CaChR1 and CyChR1 for proton permeability by measuring current-voltage relationships (I-V curves) under variable external pH. Acidification of the external medium caused an increase in the current amplitude at a given voltage and a shift of the reversal potential to more positive values. The magnitude of this shift for CaChR1 and CyChR1 was similar to that for CrChR1 (Fig. 5a, c, e). Therefore, it can be concluded that both CaChR1 and CyChR1 are highly selective for protons, as is CrChR1 (7, 9). It has been reported for CrChR1 that the rate of current decay after switching off the light decreases at acidic bath pH (20), which we also observed in our experiments (Fig. 5b). In contrast, for both CaChR1 and CyChR1 the decay rate slightly accelerated upon a change of the bath pH from 7.4 to 5.4 (Fig. 5d, f).
Figure 5.
(a, c, e) Typical current-voltage relationships (I-V curves) for the plateau level measured at the end of a 2-s excitation light pulse upon an increase of Vhold in 20 mV steps from −60 mV at the bath pH 7.4 (solid squares) and 5.4 (open circles) in HEK293 cells transfected with CrChR1, CaChR1, or CyChR1. The wavelength was 520 nm for CaChR1 and CyChR1, and 480 nm for CrChR1, which corresponded to their spectral maxima (see below). (b, d, e) Normalized current decay traces recorded from cells transfected with CrChR1, CaChR1, or CyChR1 at holding potential (Vhold) −60 mV. Traces at the bath pH 7.4 or 5.4 (indicated in the panels) were recorded from the same cell. Note the opposite effects of pH changes on the decay kinetics in CrChR1 and the new channelrhodopsins. Zero time corresponds to the end of a 2-s excitation light pulse. Excitation light was as in a, c, e. Experimental data (dots) were fitted with two exponential functions (solid lines).
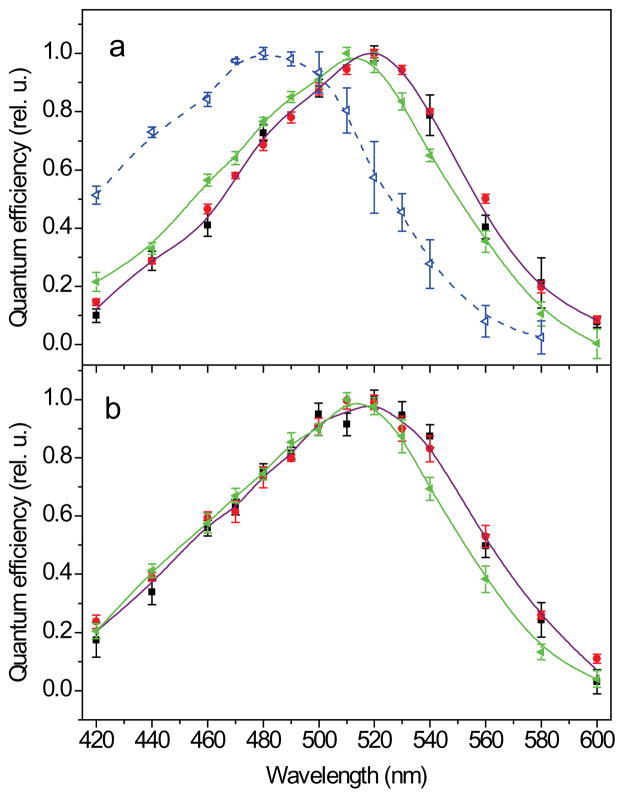
The spectral sensitivity of photocurrents was analyzed under low intensity light, as described earlier for MvChR1-generated currents (14). The spectral maxima for the new Chlamydomonas channelrhodopsins, CaChR1 and CyChR1, were at 520 nm at pH 7.4 (Fig. 6a and b, black squares), which is 40 nm longer than that of the action spectrum of CrChR1-generated currents in oocytes, and the absorption spectrum of purified CrChR1 at neutral pH (7, 20). To rule out a possible influence of a different expression system and/or a different algorithm of construction of the spectra, we also measured the action spectrum of currents generated by CrChR1 in HEK cells. It showed a maximum at ~480 nm at pH 7.4 (Fig. 6a, blue open triangles), which confirmed the earlier published results obtained in oocytes (7, 20). The spectrum had a broad shape with significant absorption above 500 nm, indicating that the red-shifted protonated form of the pigment dominating the spectrum at low pH (7, 20) also contributes to the spectrum at pH 7.4. For both CaChR1 and CyChR1, the spectra measured at neutral (7.4) and acidic (5.4) pH were identical (Fig. 6a and b, black and red symbols), in contrast to the spectrum of CrChR1, which showed a significant red shift upon acidification of the medium (7, 20). However, a small ~10 nm blue shift was observed upon the pH change from 7.4 to 9 for both CaChR1 and CyChR1 (Fig. 6a and b, green solid triangles).
Figure 6.
(a, b) The action spectra of photoelectric currents generated in HEK293 cells by CaChR1 (a) or CyChR1 (b) at the bath pH 7.4 (black squares), 5.4 (red circles) or 9.0 (green solid triangles). For comparison, the action spectrum of ChR1 from C. reinhardtii measured at pH 7.4 is shown in panel A (blue open triangles, dashed line).
The maximum of the action spectrum of photocurrents generated by CrChR1 in native C. reinhardtii cells is 505 nm (5), whereas that of currents generated by CrChR1 in heterologous systems at neutral pH is blue-shifted by at least 25 nm (Fig. 6a; (7, 20)). Since most studies on heterologously expressed CrChR1 were carried out with its 7TM domain, one possible explanation for this difference is that in native cells the spectral properties of CrChR1 are altered by the C-terminal domain. However, the spectrum of the currents generated by the full-length CrChR1 in HEK cells was also blue-shifted (data not shown), which rules out that the C-terminal domain presence is sufficient for the red-shift at neutral pH.
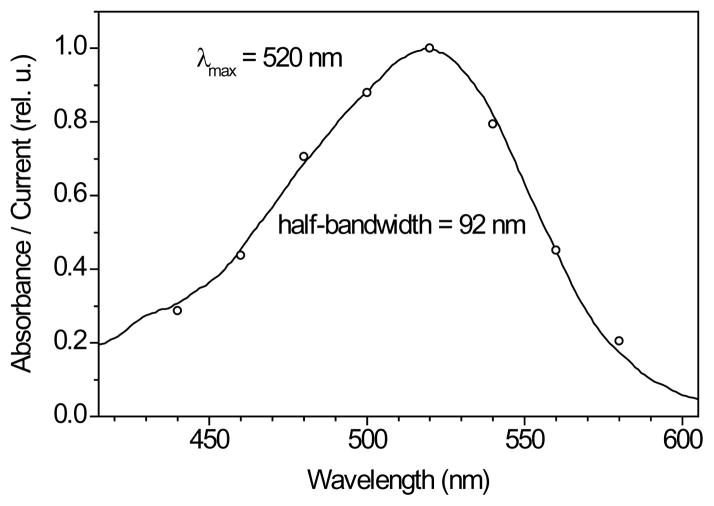
Expression of CaChR1 in P. pastoris in the presence of all-trans-retinal yielded photoactive pigment, in contrast to the previously reported unsuccessful attempts to produce functional CrChR1 in this system (7). The absorption spectrum of CaChR1 partially purified from Pichia exactly matched the action spectrum of photocurrents generated by this pigment in HEK cells, which indicated that its native state was essentially preserved in detergent (Fig. 7). Spectroscopic characterization of purified CaChR1 will be reported separately.
Figure 7.
Comparison of the absorption spectrum of partially purified CaChR1 in detergent (black line) with the action spectrum of photocurrents (open circles), pH 7.4.
DISCUSSION
In recent years, a growing demand for more efficient and diversified optogenetic tools has caused proliferation of studies aimed at molecular engineering of the known channelrhodopsins, mostly CrChR2 (e.g., (45-47)). However, this approach is limited by the current minimal understanding of the structure-function relationships of channelrhodopsin molecules. One possibility to advance this knowledge, and to expand the toolbox of channelrhodopsin probes for optogenetic applications, is to look for natural channelrhodopsin variants in other algal species, in which the occurrence of such proteins can be inferred from physiological observations. Analysis of existing genomic and EST databases is an efficient means to search for new channelopsins. Sequences from C. reinhardtii, V. carteri and M. viride have all been discovered in this way. MvChR1 from the latter organism turned out to be the most divergent variant to date to act as a light-gated cation channel (14).
However, DNA sequencing projects have been initiated only for a handful of phototactic algal species, therefore homology cloning of new channelopsins is required. Previously homology cloning has been successfully used by us (48) and others (49) to clone microbial opsin genes from the cryptophyte Cryptomonas and the glaucophyte Cyanophora, respectively. These genes, however, are not closely related to known channelopsins from green algae and do not confer ion channel activity when expressed in HEK cells (our unpublished observations).
Currently, we applied homology cloning to identify new channelopsin sequences in three Chlamydomonas species, in which a reasonably high homology with the earlier known genes from C. reinhardtii was expected. Each species yielded only one channelopsin gene, whereas the completely sequenced genomes of C. reinhardtii and V. carteri each contain two. In C. reinhardtii, CrChR1 predominantly activates a fast, high-saturating current component, whereas CrChR2 is mainly responsible for a slow (delayed), low-saturating current component (5). The same two components were detected in native cells of all presently tested flagellate species that generate rhodopsin-mediated photocurrents, with a possible exception of M. viride (22). Therefore it is likely that most algae, including C. augustae, C. yellowstonensis and C. raudensis, also harbor at least two channelrhodopsin homologs. In C. reinhardtii, CrChR1 is more abundant than CrChR2 at both mRNA (Dr. S. Merchant, UCLA, personal communication) and protein (6) levels. Our cloning efforts yielded mostly CrChR1-like sequences ostensibly reflecting their higher abundance also in other algae. Although no direct tests have been performed to determine the function of the channelopsin sequences cloned in this work in the native algae, there is little doubt that they serve as photomotility receptors, as do their homologs in C. reinhardtii. Cells of all three Chlamydomonas species generate photocurrents similar to those of C. reinhardtii (our unpublished observations), and phototactic behavior has been reported in two of them (24, 50).
Our experiments with CraChR2 in HEK cells showed a lack of channel activity that is currently unclear. One possibility is that this sequence, cloned from an extreme psychrophilic alga, is not correctly folded at 37°C. It has been noted however that C. raudensis exhibits strong positive phototaxis at the non-permissive growth temperature 29°C (24). Furthermore, channelrhodopsin from the mesophilic alga H. pluvialis also did not produce currents in HEK cells in our experiments, despite its being examined near the organism's optimal growth temperature.
Photocurrents generated by CaChR1 and CyChR1 differ from that of CrChR1 in their kinetics, inactivation and light dependence, suggesting their photochemical reaction cycles are at least quantitatively different from that of CrChR1. The dependence of the spectral sensitivity of CaChR1- and CyChR1-generated currents on the external pH confirmed the role of the residue in the position of Glu87 (CrChR1 numbering) in color tuning, as both these proteins also contain a Glu residue in this position (Glu94 and Glu95, respectively). In contrast, MvChR1, which contains a non-carboxylated residue in this position, does not show the spectral shift over the entire tested pH range from 5.3 to 9 (14). This residue was suggested to contribute to the formation of a trimodal counterion of the protonated Schiff base characteristic of CrChR1-like channelrhodopsins in contrast to CrChR2-like ones (20). Therefore, protonation/deprotonation of this residue would alter the chromophore polarity and change the absorption spectrum. Interestingly, in both CaChR1 and CyChR1 the spectral transition occurs at a pH several units higher than that in CrChR1.
Moreover, CaChR1 and CyChR1 differ from the earlier known CrChR1 in that the spectral maxima of their protonated forms is at 520 nm, compared to 497 nm of CrChR1 (7). The red-shifted spectrum of VcChR1, peaking at 530 nm, has been attributed to the residues in the positions of Ser141 and Ala215 (BR numbering), which differentiates this protein from all other known channelrhodopsins (12). However, both CaChR1 and CyChR1 contain Gly and Ser, respectively, in these positions, as do all other known channelopsins except VcChR1, i.e. their red-shifted spectral sensitivity must be due to other structural reasons. This result points to the involvement of additional structural factors defining the pH dependence of the spectral sensitivity.
Comparative sequence analysis reveals that both CaChR1 and CyChR1 differ from CrChR1 and, in fact, from all other reported channelrhodopsins, in the residues found in the positions of Glu194 and Glu204 (BR numbering). Glu194 is conserved in all reported channelopsins, except CaChR1 and CyChR1, in which it is replaced by Ser. Conversely, the position of Glu204 is occupied by Ser in all previously described channelopsins, but it is Asp in CaChR1 and CyChR1. In BR, Glu194 and Glu204 are part of the extracellular hydrogen-bonded network that forms the proton release pathway (reviewed by (51)) and are known to contribute to the pH dependence of spectral transitions (52). It is plausible that these residues also play a role in regulation of the pH dependence and/or color tuning in channelrhodopsins, although further studies are needed to test this hypothesis. Replacement of Ser with Glu at the site corresponding to Glu204 in BR led to the total abolishment of channel activity in MvChR1 (our unpublished observations), alluding to the importance of this site for channelrhodopsin function.
The slower inactivation of photocurrents and red-shifted spectral sensitivity of CaChR1 and CyChR1 are features desirable for optogenetic use. However, the currents generated by these proteins in HEK cells were smaller than those generated by CrChR1, which are smaller than those produced by CrChR2. Nevertheless, if small currents result from inefficient expression/targeting, these can potentially be improved by the addition of signal peptides and/or ER export sequences, as has been recently done for halorhodopsin use as an optogenetic tool (53).
Acknowledgments
We thank Dr. K. Deisseroth (Stanford University) for the pcDNA3.1/VcChR1-EYFP mammalian expression vector, Dr. V. Jayaraman (UT Houston Health Science Center) for the HEK293 cell line and advice on whole-cell patch clamp recording, and Dr. H. Buelow (Albert Einstein College of Medicine) for the information about the possibility of cytoplasmic protein N-glycosylation. We also thank the late Dr. K.D. Ridge for his help with HEK293 cell culture. The work was supported by Award Number RC1AG035779 from the National Institute on Aging, R37GM027750 from the National Institute of General Medical Sciences, the Hermann Eye Fund, and endowed chair AU-0009 from the Robert A. Welch Foundation. Sing-Yi Hou was supported by a Taiwan Government Scholarship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes on Aging, General Medical Sciences, or the National Institutes of Health.
References
- 1.Bogomolni R, Spudich JL. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc Natl Acad Sci USA. 1982;79:6250–6254. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spudich JL, Bogomolni RA. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984;312:509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster KW, Saranak J, Patel N, Zarrilli G, Okabe M, Kline T, Nakanishi K. A rhodopsin is the functional photoreceptor for phototaxis in the unicelullar eukaryote Chlamydomonas. Nature. 1984;311:756–759. doi: 10.1038/311756a0. [DOI] [PubMed] [Google Scholar]
- 4.Asamizu E, Miura K, Kucho K, Inoue Y, Fukuzawa H, Ohyama K, Nakamura Y, Tabata S. Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Research. 2000;7:305–307. doi: 10.1093/dnares/7.5.305. [DOI] [PubMed] [Google Scholar]
- 5.Sineshchekov OA, Jung KH, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Govorunova EG, Jung KW, Sineshchekov OA, Spudich JL. Chlamydomonas sensory rhodopsins A and B: Cellular content and role in photophobic responses. Biophys J. 2004;86:2342–2349. doi: 10.1016/S0006-3495(04)74291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berthold P, Tsunoda SP, Ernst OP, Mages W, Gradmann D, Hegemann P. Channelrhodopsin-1 initiates phototaxis and photophobic responses in Chlamydomonas by immediate light-induced depolarization. Plant Cell. 2008;20:1665–77. doi: 10.1105/tpc.108.057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litvin FF, Sineshchekov OA, Sineshchekov VA. Photoreceptor electric potential in the phototaxis of the alga Haematococcus pluvialis. Nature. 1978;271:476–478. doi: 10.1038/271476a0. [DOI] [PubMed] [Google Scholar]
- 9.Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–8. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 10.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–5. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T, Yamasaki K, Fujita S, Oda K, Iseki M, Yoshida K, Watanabe M, Daiyasu H, Toh H, Asamizu E, Tabata S, Miura K, Fukuzawa H, Nakamura S, Takahashi T. Archaeal-type rhodopsins in Chlamydomonas: model structure and intracellular localization. Biochem Biophys Res Commun. 2003;301:711–717. doi: 10.1016/s0006-291x(02)03079-6. [DOI] [PubMed] [Google Scholar]
- 12.Zhang F, Prigge M, Beyriere F, Tsunoda SP, Mattis J, Yizhar O, Hegemann P, Deisseroth K. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat Neurosci. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kianianmomeni A, Stehfest K, Nematollahi G, Hegemann P, Hallmann A. Channelrhodopsins of Volvox carteri are photochromic proteins that are specifically expressed in somatic cells under control of light, temperature, and the sex inducer. Plant Physiol. 2009;151:347–366. doi: 10.1104/pp.109.143297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govorunova EG, Spudich EN, Lane CE, Sineshchekov OA, Spudich JL. New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. mBio. 2011;2:e00115–11. doi: 10.1128/mBio.00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow BY, Chuong AS, Klapoetke NC, Boyden ES. Synthetic physiology strategies for adapting tools from nature for genetically targeted control of fast biological processes. Methods Enzymol. 2011;497:425–443. doi: 10.1016/B978-0-12-385075-1.00018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kokaia M, Sorensen AT. The treatment of neurological diseases under a new light: the importance of optogenetics. Drugs Today (Barc) 2011;47:53–62. doi: 10.1358/dot.2011.47.1.1543306. [DOI] [PubMed] [Google Scholar]
- 18.Tomita H, Sugano E, Isago H, Tamai M. Channelrhodopsins provide a breakthrough insight into strategies for curing blindness. J Genet. 2009;88:409–415. doi: 10.1007/s12041-009-0062-6. [DOI] [PubMed] [Google Scholar]
- 19.Doroudchi MM, Greenberg KP, Liu J, Silka KA, Boyden ES, Lockridge JA, Arman AC, Janani R, Boye SE, Boye SL, Gordon GM, Matteo BC, Sampath AP, Hauswirth WW, Horsager A. Virally delivered channelrhodopsin-2 safely and effectively restores visual function in multiple mouse models of blindness. Mol Ther. 2011;19:1220–1229. doi: 10.1038/mt.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsunoda SP, Hegemann P. Glu 87 of channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. Photochem Photobiol. 2009;85:564–569. doi: 10.1111/j.1751-1097.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Sugiyama Y, Hikima T, Sugano E, Tomita H, Takahashi T, Ishizuka T, Yawo H. Molecular determinants differentiating photocurrent properties of two channelrhodopsins from Chlamydomonas. J Biol Chem. 2009;284:5685–5696. doi: 10.1074/jbc.M807632200. [DOI] [PubMed] [Google Scholar]
- 22.Sineshchekov OA, Spudich JL. Sensory rhodopsin signaling in green flagellate algae. In: Briggs WR, Spudich JL, editors. Handbook of Photosensory Receptors. Wiley-VCH; Weinheim: 2005. pp. 25–42. [Google Scholar]
- 23.Hoham RW, Duval B. Microbial ecology of snow and freshwater ice with emphasis on snow algae. In: Jones HG, Pomeroy JW, Walker DA, Hoham RW, editors. Snow Ecology: an Interdisciplinary Examination of Snow-covered Ecosystems. Cambridge University Press; Cambridge: 2001. pp. 168–228. [Google Scholar]
- 24.Morgan-Kiss RM, Priscu JC, Pocock T, Gudynaite-Savitch L, Huner NP. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol Mol Biol Rev. 2006;70:222–252. doi: 10.1128/MMBR.70.1.222-252.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starr RC, Zeikus JA. UTEX - the culture collection of algae at the University of Texas at Austin. J Phycol. 1993;29(suppl):1–106. [Google Scholar]
- 26.Bischoff HW, Bold HC. Phycological studies IV. Some soil algae from Enchanted Rock and related algal species. University of Texas Publication. 1963;6318:1–95. [Google Scholar]
- 27.Nikolic K, Grossman N, Grubb MS, Burrone J, Toumazou C, Degenaar P. Photocycles of channelrhodopsin-2. Photochem Photobiol. 2009;85:400–411. doi: 10.1111/j.1751-1097.2008.00460.x. [DOI] [PubMed] [Google Scholar]
- 28.Radu I, Bamann C, Nack M, Nagel G, Bamberg E, Heberle J. Conformational changes of channelrhodopsin-2. J Am Chem Soc. 2009;131:7313–7319. doi: 10.1021/ja8084274. [DOI] [PubMed] [Google Scholar]
- 29.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 30.Nack M, Radu I, Gossing M, Bamann C, Bamberg E, von Mollard GF, Heberle J. The DC gate in channelrhodopsin-2: crucial hydrogen bonding interaction between C128 and D156. Photochem Photobiol Sci. 2010;9:194–198. doi: 10.1039/b9pp00157c. [DOI] [PubMed] [Google Scholar]
- 31.Hancock JM, Simon M. Simple sequence repeats in proteins and their significance for network evolution. Gene. 2005;345:113–118. doi: 10.1016/j.gene.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Xiao H, Jeang KT. Glutamine-rich domains activate transcription in yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:22873–22876. doi: 10.1074/jbc.273.36.22873. [DOI] [PubMed] [Google Scholar]
- 33.Weiss JL, Evans NA, Ahmed T, Wrigley JD, Khan S, Wright C, Keen JN, Holzenburg A, Findlay JB. Methionine-rich repeat proteins: a family of membrane-associated proteins which contain unusual repeat regions. Biochim Biophys Acta. 2005;1668:164–174. doi: 10.1016/j.bbamem.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Mittelmeier TM, Boyd JS, Lamb MR, Dieckmann CL. Asymmetric properties of the Chlamydomonas reinhardtii cytoskeleton direct rhodopsin photoreceptor localization. J Cell Biol. 2011;193:741–753. doi: 10.1083/jcb.201009131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart GW. Nuclear and cytoplasmic glycosylation. In: Varki A, Cummings R, Esko J, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2009. pp. 171–182. [PubMed] [Google Scholar]
- 36.Kirsch T. Ph D Thesis. J.W. Goethe University; Frankfurt: 2007. Functional expression of channelrhodopsin 2 (ChR2) in the methylotrophic yeast Pichia pastoris and its biophysical characterization. [Google Scholar]
- 37.Wagner V, Ullmann K, Mollwo A, Kaminski M, Mittag M, Kreimer G. The phosphoproteome of a Chlamydomonas reinhardtii eyespot fraction includes key proteins of the light signaling pathway. Plant Physiol. 2008;146:772–788. doi: 10.1104/pp.107.109645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zacks DN, Spudich JL. Gain setting in Chlamydomonas reinhardtii: Mechanism of phototaxis and the role of the photophobic response. Cell Mot Cytoskeleton. 1994;29:225–230. doi: 10.1002/cm.970290305. [DOI] [PubMed] [Google Scholar]
- 39.Violot S, Aghajari N, Czjzek M, Feller G, Sonan GK, Gouet P, Gerday C, Haser R, Receveur-Brechot V. Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by X-ray diffraction and small angle X-ray scattering. J Mol Biol. 2005;348:1211–24. doi: 10.1016/j.jmb.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 40.Thorvaldsen S, Hjerde E, Fenton C, Willassen NP. Molecular characterization of cold adaptation based on ortholog protein sequences from Vibrionaceae species. Extremophiles. 2007;11:719–732. doi: 10.1007/s00792-007-0093-y. [DOI] [PubMed] [Google Scholar]
- 41.Hegemann P, Ehlenbeck S, Gradmann D. Multiple photocycles of channelrhodopsin. Biophys J. 2005;89:3911–8. doi: 10.1529/biophysj.105.069716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–14. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishizuka T, Kakuda M, Araki R, Yawo H. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci Res. 2006;54:85–94. doi: 10.1016/j.neures.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Ernst OP, Sanchez Murcia PA, Daldrop P, Tsunoda SP, Kateriya S, Hegemann P. Photoactivation of channelrhodopsin. J Biol Chem. 2008;283:1637–43. doi: 10.1074/jbc.M708039200. [DOI] [PubMed] [Google Scholar]
- 45.Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–92. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 46.Kleinlogel S, Feldbauer K, Dempski RE, Fotis H, Wood PG, Bamann C, Bamberg E. Ultra light-sensitive and fast neuronal activation with the Ca2+-permeable channelrhodopsin CatCh. Nat Neurosci. 2011;14:513–518. doi: 10.1038/nn.2776. [DOI] [PubMed] [Google Scholar]
- 47.Berndt A, Schoenenberger P, Mattis J, Tye KM, Deisseroth K, Hegemann P, Oertner TG. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc Natl Acad Sci USA. 2011;108:7595–7600. doi: 10.1073/pnas.1017210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sineshchekov OA, Govorunova EG, Jung KH, Zauner S, Maier UG, Spudich JL. Rhodopsin-mediated photoreception in cryptophyte flagellates. Biophys J. 2005;89:4310–4319. doi: 10.1529/biophysj.105.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frassanito AM, Barsanti L, Passarelli V, Evangelista V, Gualtieri P. A rhodopsin-like protein in Cyanophora paradoxa: gene sequence and protein immunolocalization. Cell Mol Life Sci. 2010;67:965–971. doi: 10.1007/s00018-009-0225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams CR, Bees MA. A tale of three taxes: photo-gyro-gravitactic bioconvection. J Exp Biol. 2011;214:2398–2408. doi: 10.1242/jeb.051094. [DOI] [PubMed] [Google Scholar]
- 51.Lanyi JK. Proton transfers in the bacteriorhodopsin photocycle. Biochim Biophys Acta. 2006;1757:1012–1018. doi: 10.1016/j.bbabio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 52.Balashov SP, Imasheva ES, Ebrey TG, Chen N, Menick DR, Crouch RK. Glutamate-194 to cysteine mutation inhibits fast light-induced proton release in bacteriorhodopsin. Biochemistry. 1997;36:8671–8676. doi: 10.1021/bi970744y. [DOI] [PubMed] [Google Scholar]
- 53.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]